Abstract
Fungal conidia contain chemicals that inhibit germination and appressorium formation until they are well dispersed in a favorable environment. Recently, such self-inhibitors were found to be present on the conidia of Magnaporthe grisea, and plant surface waxes were found to relieve this self-inhibition. To determine whether the self-inhibitors suppress the expression of early genes involved in the germination and differentiation of conidia, the calmodulin gene was chosen as a representative early gene, because it was found to be expressed early in Colletotrichum gloeosporioides and Colletotrichum trifolii differentiation. After calmodulin cDNA and genomic DNA from M. grisea were cloned, the promoter of the calmodulin gene was fused to a reporter gene, that for green fluorescent protein (GFP), and transformed into the M. grisea genome. Confocal microscopic examination and quantitation of expression of GFP green fluorescence showed (i) that the expression of the calmodulin gene decreased significantly when self-inhibition of M. grisea appressorium formation occurred because of high conidial density or addition of exogenous self-inhibitors and (ii) that the expression level of this gene was restored when self-inhibition was relieved by the addition of plant surface waxes. The increase in fluorescence correlated with the percentage of conidia that formed appressoria. The induction of calmodulin was also confirmed by RNA blotting. Concanavalin A inhibited surface attachment of conidia, GFP expression, and appressorium formation without affecting germination. The high correlation between GFP expression and appressorium formation strongly suggests that calmodulin gene expression and appressorium formation require surface attachment.
Conidia of many fungal species contain chemicals that prevent germination and appressorium formation until they are well dispersed in a favorable environment for colonization of plant hosts (16, 19). These chemicals, called self-inhibitors, are often lipophilic molecules. Recent evidence suggests that the self-inhibitors diffuse into the lipophilic plant cuticle upon contact of the conidia with the host and thus relieve self-inhibition (12). Contact with the host surface induces expression of a set of early genes that are required for the conidia to respond to further host signals. One of the early genes is the calmodulin gene, whose transcription in Colletotrichum gloeosporioides was found to be induced by hard-surface contact maximally at 2 h and then to decline (15). Seven unique genes were found to be induced early during hard-surface treatment of C. gloeosporioides conidia (18). Subsequently, the conidia responded to host signals that caused the transcriptional activation of another set of genes, leading to the induction of germination and appressorium formation (13). At which stage in this chain of events the self-inhibitors exert their effect is not known. How the self-inhibitors affect conidial differentiation is also not known.
Recently, self-inhibitors were found to be present on the conidia of Magnaporthe grisea, and plant surface waxes were found to relieve this self-inhibition (12). To determine whether the self-inhibitors suppress the expression of early genes involved in conidium differentiation, calmodulin was chosen as a representative of the early genes, because the calmodulin gene was found to be expressed early during the germination and differentiation of conidia of both C. gloeosporioides (15) and Colletotrichum trifolii (3).
Since the experimental investigation of the effects of self-inhibitors involves measurement of gene expression in a small number of conidia, quantitation of the expression of a readily measurable reporter gene would be a convenient approach. Therefore, to measure the expression of the calmodulin (cam) gene, the promoter of this gene from M. grisea was cloned and fused to a reporter gene, that for green fluorescent protein (GFP), and incorporated into the M. grisea genome. Confocal microscopic quantitation of expression of GFP showed that expression of the cam gene decreased significantly when self-inhibition of M. grisea appressorium formation occurred because of high conidial density or addition of exogenous self-inhibitors, and expression of this gene was restored when self-inhibition was relieved by lowering the conidial density or by the addition of plant surface waxes. The enhanced fluorescence correlated with the percentage of conidia that formed appressoria. The induction of calmodulin was also confirmed by RNA blotting. Furthermore, we tested whether conidial attachment to a surface was necessary for early gene expression and used concanavalin A (ConA) to block conidial attachment. The results showed that ConA inhibited surface attachment of conidia, GFP expression, and appressorium formation without affecting germination. The high correlation between GFP expression and appressorium formation strongly suggests that cam gene expression and appressorium formation require surface attachment.
MATERIALS AND METHODS
Fungal and bacterial cultures and reagents.
M. grisea was cultured on V8 plates and grown at 24°C for ∼10 days. The conidia, harvested by gently scraping cultures in petri dishes flooded with sterilized distilled water, were filtered through Miracloth (Calbiochem) and recovered by centrifugation. Cycloheximide and ConA were purchased from Sigma. Escherichia coli DH5α was used for propagating all plasmids. Restriction and modification enzymes and Taq DNA polymerase were from Life Technologies, Inc. (BRL).
Preparation of genomic DNA.
To prepare genomic DNA of M. grisea, a previously described method (7) was followed with some modifications. Mycelia (1 g) were ground in liquid nitrogen with a mortar and pestle. After the powder was resuspended in a solution containing 10 ml of 7 M urea, 2% sodium dodecyl sulfate (SDS), and 5 mM EDTA (pH 8.0), the suspension was extracted with a phenol-chloroform (1:1 [vol/vol] mixture) and chloroform. Then the DNA was precipitated with an equal volume of isopropanol and recovered by centrifugation at a low speed to pellet the high-molecular-weight DNA. The pellet was then resuspended in a solution containing 0.75 ml of 150 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl (pH 8.0) and treated with RNase A and proteinase K. Finally, after extraction with phenol-chloroform and chloroform, the genomic DNA was precipitated with 2.5 volumes of ethanol and resuspended in TE (10 mM Tris-HCl [pH 8.0] containing 1 mM EDTA).
Northern blots.
Total RNA, isolated from conidia by the LiCl method as described previously (18), was dissolved in a solution containing 50% formamide, 16% formaldehyde, 20 mM MOPS [3-(N-morpholino)propanesulfonic acid], 5 mM sodium acetate, and 1 mM EDTA (pH 7.0); incubated for 15 min at 65°C; and chilled on ice. These denatured RNA samples were subjected to electrophoresis on a 1% agarose gel containing 2.2 M formaldehyde and blotted onto Nytran membranes. The blots were prehybridized for ∼4 h at 65°C in 6× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate [pH 7.6])–2× Denhardt’s solution–0.1% SDS–100 μg of sheared salmon sperm DNA per ml and hybridized for ∼16 h in the same solution with 106 cpm of a 32P-labeled cam cDNA probe per ml. The cDNA probe had been prepared by randomly primed labeling. The membranes were washed twice for 10 min each time at room temperature in 2× SSC–0.1% SDS, briefly washed at 65°C with 0.2× SSC–0.1% SDS, and exposed to X-ray film at −80°C in the presence of an intensifying screen.
Southern and dot blots.
Genomic DNA was isolated as described above from mycelium grown in liquid culture. The genomic DNA was digested to completion with restriction enzymes, subjected to electrophoresis on a 1% agarose gel, and transferred to Nytran membranes. The conditions for prehybridization, hybridization, and washing were the same as those described above for RNA blots.
GFP fusion with the calmodulin gene (camMg) promoter.
The GFP expression vector pTEFEGFP (29) was kindly provided by John H. Andrews, University of Wisconsin, Madison. A segment containing an engineered form (EGFP) of the Aspergillus victoria GFP cDNA (4) and a 200-bp terminator region derived from the Aspergillus awamori glucoamylase gene (9, 22) was cut out of pTEFEGFP by HincII digestion and ligated to pKS-pcamMg [calmodulin gene promoter in the Bluescript pKS(+) vector] at the HincII site, yielding an in-frame fusion with 33 amino acids of N-terminal CAMMg and a 27-amino-acid linker. This pcamEGFP expression vector was then inserted at the SalI site with a hygromycin cassette containing the E. coli hph gene, which conferred resistance to the antibiotic hygromycin B, and with the promoter and the terminator of Aspergillus trpC from plasmid pCSN43 (Fungal Genetic Stock Center) (27).
M. grisea protoplast transformation.
M. grisea protoplast transformation was done as described previously (1, 21) with some modifications. Conidia from a 7-day-old M. grisea culture were grown in potato dextrose broth at room temperature overnight with vigorous shaking; mycelia were harvested with Miracloth (Calbiochem) and washed with 2 volumes of 0.6 M MgSO4. The mycelial mat (1 g) was then digested with 75 mg of Novozyme (InterSpex Products, Inc., Foster City, Calif.) in 20 ml of freshly prepared osmotic medium (1.2 M MgSO4, 10 mM NaH2PO4 [pH 5.8]). After 2 to 3 h of gentle shaking at room temperature, protoplasts were filtered through nylon mesh, overlaid with ST buffer (0.6 M sorbitol, 100 mM Tris [pH 7.0]) and collected at the interface of the osmotic medium after centrifugation. The protoplasts were washed in STC buffer (1.2 M sorbitol, 10 mM Tris [pH 7.5], 10 mM CaCl2) three times. Transformation was carried out by placing 3 × 106 protoplasts with 3 to 10 μg of DNA on ice for 10 min before 1 ml of PTC (60% polyethelene glycol 4000, 10 mM Tris [pH 7.5], 10 mM CaCl2) was added. After 20 min, 3 ml of TB3 (3 g of yeast extract, 3 g of Casamino Acids hydrolysate, 10 g of glucose, 200 g of sucrose per liter) was added and the mixture was incubated for 6 h at room temperature. Molten TB3 with agar (40 ml) containing 200 μg of hygromycin B per ml was then added and poured into two plates. After ∼7 days, hygromycin-resistant colonies were transferred to V8 plates containing 200 μg of hygromycin B per ml.
M. grisea conidial surface lipid extraction.
M. grisea conidial surface lipid extraction was done as described previously (12). Conidia from 10- to 15-day-old cultures were harvested in sterile water by filtering them through Miracloth. Spores (5 × 108) were then collected on Whatman 1 paper in a Buchner funnel, and 50 ml of a 2:1 (vol/vol) mixture of chloroform-methanol was added. The spores resting in the funnel were stirred for 10 s and quickly filtered by application of vacuum suction to collect the conidial surface lipid extract, and the lipids were recovered as described previously (12). The surface lipids were finally dispersed in water with a model 250 sonifier equipped with a microprobe (Branson Ultrasonic, Danbury, Conn.).
Appressorium formation.
M. grisea conidia were placed on a polystyrene petri dish surface in 100 μl of water or water containing various additions. The polystyrene petri dish lid contained wet filter paper, and high humidity was maintained by wrapping the petri dish with parafilm. After ∼18 h, the effects of conidial density, conidial surface lipids, plant surface wax (cabbage leaf surface wax isolated by dipping mature leaves in chloroform for 30 s), cycloheximide, and ConA on appressorium formation were determined by examining 40 to 50 conidia per sample for appressorium formation. The results from three experiments were averaged.
Confocal microscopy.
M. grisea conidia of GFP transformants were placed on a polystyrene petri dish surface in 100 μl of water or water containing various additions for 2 h or for the periods of time indicated in the figures for the time course experiments. Then the solution was removed, and the conidia attached to the surface were fixed with 7 μl of 3% paraformaldehyde in 50 mM phosphate buffer (pH 7.4) and covered with a glass coverslip for confocal microscopic analysis. Untreated conidia were collected by centrifugation, resuspended in fixer, and placed on a polystyrene petri dish surface for confocal microscopic analysis. The GFP fluorescence of the untreated conidia was regarded as the level at zero time in the time course experiments and as the basal level in other experiments.
GFP fluorescence images were collected with a Bio-Rad model MRC-600 confocal microscope equipped with a Nikon 20× lens objective (aperture, 0.75) and fluorescein isothiocyanate filters (excitation/emission, 488/510 nm). Quantitation of the fluorescence intensity of each cell was done by measuring the histogram of a rectangle surrounding each cell. Usually the fluorescence of 10 conidia was quantitated except as otherwise noted below, and statistical analysis (means and standard deviations) was performed with CA-Cricket graph III. Experiments were repeated at least twice, and typical results are shown. The digitized images were stored as red-green-blue tagged-image-format files. The final images were prepared with Adobe Photoshop 3.0 (Adobe Systems, Mountain View, Calif.).
The increase (Δ) in fluorescence per conidium arising from attachment in the ConA experiments was calculated as follows: ΔF = na × ΔFa + nu × ΔFu − nA × ΔFA, where na is the fraction of attached conidia, nu is the fraction of unattached conidia, ΔFa and ΔFu are the increase in fluorescence (after subtraction of the fluorescence of untreated conidia) per attached and unattached conidia, respectively, nA is the fraction of attached conidia even at high concentrations of ConA, and ΔFA is the increase in fluorescence for these attached conidia. Finally, percentages of enhanced fluorescence per conidium with different concentrations of ConA were calculated, with the percentage of enhanced fluorescence per conidium without ConA being considered 100.
RESULTS
Cloning and sequencing of calmodulin cDNA and genomic DNA from M. grisea.
To determine at what stage self-inhibitors exert their effect on sequential gene expression events that occur during conidial germination and appressorium formation, we chose to examine the expression of the calmodulin gene, one of the early genes expressed during fungal conidial differentiation. Since calmodulin cDNA and genomic DNA from M. grisea had not been cloned, we first cloned calmodulin cDNA and genomic DNA from M. grisea. As the calmodulin gene is very conserved, degenerate primers corresponding to 7 N-terminal amino acids and 6 C-terminal amino acids plus the stop codon were used to clone M. grisea calmodulin cDNA by reverse transcription-PCR as was previously done for cam from C. gloeosporioides (15). The 450-bp cDNA reverse transcription-PCR product along with an 856-bp genomic DNA amplified with the same two primers and with M. grisea genomic DNA as the template were cloned into the pCR2.1 vector (Invitrogen) and sequenced. At the amino acid level, M. grisea calmodulin is identical to calmodulins of Neurospora crassa (20), Aspergillus oryzae (30), C. gloeosporioides (15), and C. trifolii (GenBank accession no. U15993 [6]). The cloned 856-bp genomic DNA contains the entire open reading frame interrupted by four introns. The first intron, later found in the gene, is missing in this PCR product because the primer used for PCR amplification linked exons 1 and 2 together. To obtain the promoter region, the 450-bp cDNA was used to screen a Lambda Fix II (9- to 23-kb) genomic library. Lambda DNA from a genomic clone that contains the cam gene was subjected to restriction analysis and Southern blotting by hybridization with an ∼400-bp SstI fragment from the 5′ end of the genomic DNA obtained by PCR. A 1.9-kb SstI fragment that hybridized with the probe was subcloned into the Bluescript pKS(+) vector, yielding pKS-pcamMg, and sequenced. This sequence together with the sequence of the genomic DNA obtained by PCR revealed the total sequence of the cam gene of M. grisea and showed the presence of five introns, including the first one that was found immediately after the first codon of the open reading frame (GenBank accession no. AF 103729). Southern blots of the M. grisea genomic DNA showed two bands in the PstI digest and two bands in the HindIII digest (data not shown). The restriction map of camMg genomic DNA showed that there is one HindIII and one PstI site within the genomic DNA. Thus, the Southern blot analysis indicates that the genome of M. grisea contains one copy of the camMg gene.
Incorporation of a camMg promoter fusion with the GFP reporter into the M. grisea genome.
Calmodulin gene expression in a few conidia had to be measured in order to determine at what stage of sequential gene expression the self-inhibitors exert their effect. Direct measurement of transcription in the few conidia that would be encountered under conditions of low conidial density would be extremely difficult. Measurement of the expression of a marker gene that can readily be examined might be a practical way to approach the problem. For this purpose, we fused the promoter of camMg to EGFP, with a hygromycin cassette attached for selection, and incorporated the construct into the M. grisea genome. To confirm the integration of EGFP into the M. grisea genome in the transformant [cam(p)::EGFP::Hph] and determine the number of EGFP copies integrated, one hygromycin B-resistant transformant was selected for genomic DNA preparation. DNA dot blotting was carried out, and the levels of hybridization of EGFP to aliquots of the genomic DNA and to known amounts of EGFP DNA were compared (data not shown). Quantitative comparison, in which a genome size of 38 Mb (11) was assumed, indicated that one copy of the cam gene was present per genome. This result indicated that a single copy of EGFP was incorporated into the genome.
GFP fluorescence as a measure of cam gene expression.
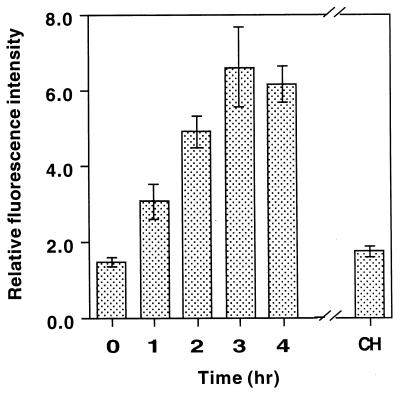
To determine whether GFP expression driven by the cam promoter reflects cam gene expression, which occurs early in fungal differentiation, we established a time course of the increase in GFP fluorescence that occurred during a 4-h period of surface attachment of conidia (Fig. 1). GFP fluorescence increased for up to 2 to 3 h of surface attachment and reached its maximum at a fourfold level. Therefore, for further study, a 2-h treatment was routinely used. In addition, in the presence of 0.3 mM cycloheximide (a protein synthesis inhibitor), GFP fluorescence at 2 h was close to the basal level observed at zero time (Fig. 2), indicating that cycloheximide blocked the enhancement of GFP fluorescence and that the enhanced green fluorescence observed under our experimental conditions was due to newly synthesized GFP. Statistical analysis of results obtained by measuring the fluorescence of 10 conidia gave an average value that represented the relative level of cam gene expression, with an average standard deviation of 11%, although the absolute value of GFP fluorescence may vary among different batches of conidia. These results suggested that GFP fluorescence was suitable for measuring cam gene expression.
FIG. 1.
Time course of development of GFP fluorescence by conidia at a low density. Conidia from the cam(p)::EGFP::Hph transformant were placed on a polystyrene surface at a low density (104/ml) for a 4-h period. Then the GFP fluorescence images were collected by confocal microscopy as described in Materials and Methods. The GFP fluorescence of six conidia observed at 2 h in the presence of 0.3 mM cycloheximide (CH) is shown at the right.
FIG. 2.
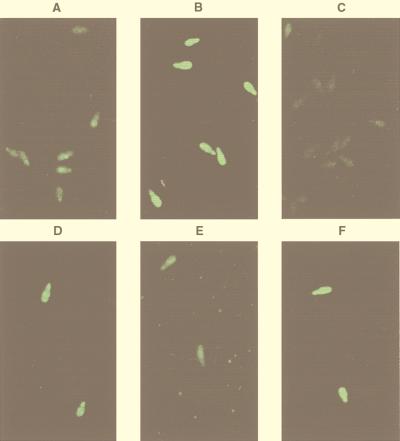
Levels of GFP fluorescence of M. grisea conidia affected by conidial density, conidial surface lipids as self-inhibitors, and plant surface wax. Conidia were placed on a polystyrene surface for 2 h at a high density (105/ml) (A), at a high density (105/ml) with plant surface wax (0.2 μg/μl) (B), without wax (C), at a low density (104/ml) (D), at a low density (104/ml) with conidial surface lipids (0.2 μg/μl) (E), and at a low density (104/ml) with conidial surface lipids (0.2 μg/μl) plus plant surface wax (0.3 μg/μl) (F). The fluorescence images were collected by confocal microscopy as described in Materials and Methods.
Inhibition of cam gene expression by self-inhibitors and restoration by plant surface wax.
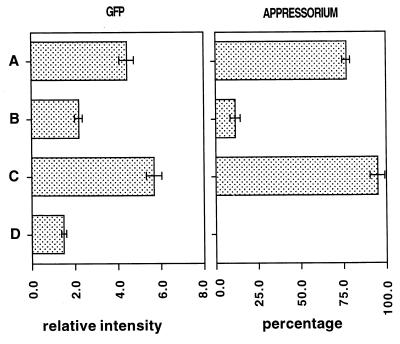
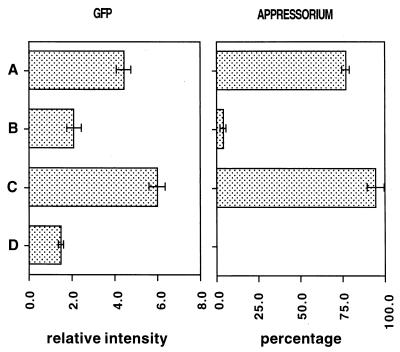
The effects of self-inhibitors are often manifested by inhibition of germination and appressorium formation with increasing conidial density. Therefore, to test whether self-inhibitors block the expression of the cam gene, the fluorescence of conidia attached to the surface at low (104/ml) and high (105/ml) densities was examined. Since inhibition caused by self-inhibitors is known to be reversed by plant surface wax, we also tested the effect of the addition of plant surface wax. Typical confocal images showing the levels of fluorescence are shown in Fig. 2A, B, and D. GFP fluorescence was much lower at the higher conidial density (Fig. 2A and D). Quantitation of the fluorescence data showed that at 105 conidia per ml, the increase in GFP fluorescence resulting from a 2-h hard-surface treatment was less than 25% of that observed for 104 conidia per ml. The level of appressorium formation was also much less at the higher conidial density (Fig. 3). The inhibition of the development of GFP fluorescence by a high conidial density could be prevented by plant surface wax (Fig. 2A and B). Quantitation of the data showed that the inhibition of development of GFP fluorescence by a high density of conidia was fully restored by plant surface wax. Inhibition of appressorium formation by a high density of conidia was also fully reversed by plant surface wax (Fig. 3).
FIG. 3.
Levels of GFP fluorescence of M. grisea conidia (left) and percentages of appressorium formation (right) at a low conidial density and at a high density with or without plant surface wax. Conidia were subjected to hard-surface treatment for 2 h at a low density (104/ml) (A), at a high density (105/ml) (B) and at a high density (105/ml) with cabbage leaf surface wax (0.2 μg/μl) (C) and to no treatment (D).
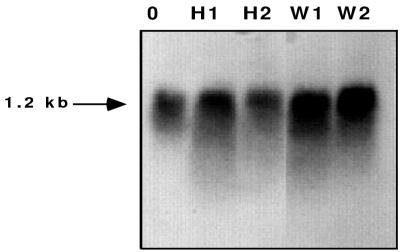
With conidia at high density, a direct measurement of the transcript level could also be done to test whether the change in GFP expression was reflected in changes in the transcript level. Total RNA was prepared from M. grisea conidia spread into polystyrene petri dishes at a high density with or without 0.25 μg of cabbage wax per μl for 1 and 2 h. RNA blots confirmed the enhancement of the level of calmodulin mRNA by plant surface wax, consistent with the notions that plant surface wax relieves self-inhibition and causes restoration of cam gene expression in M. grisea (Fig. 4).
FIG. 4.
Northern blot showing reversal of inhibition of cam gene expression caused by a high conidial density and plant surface wax. The number of hours on a polystyrene surface without (lanes with H prefix) or with (lanes with W prefix) cabbage leaf surface wax (0.25 μg/μl) are indicated (conidial density, ∼105/ml). Lane 0 contains nontreated conidia as a control. Total RNA (10 μg/lane) was loaded, and the ethidium bromide staining of 28S and 18S rRNAs was the same for all lanes. The probe was a 32P-labeled, 450-bp cDNA. Experimental details are noted in the text.
A direct test for the effects of self-inhibitors was made by adding self-inhibitors isolated from the surfaces of the conidia to fresh conidia attached to the surface at a low density (104/ml). Inhibition of the development of fluorescence by the added self-inhibitors was obvious in the confocal images (Fig. 2D and E). Quantitation of the fluorescence showed that the increase in GFP fluorescence with 104 conidia per ml with the added self-inhibitors was only near 20% of that observed without the self-inhibitors (Fig. 5). The addition of plant surface wax to the conidia with exogenous self-inhibitors caused the recovery of the development of fluorescence (Fig. 2E and F). Quantitation of the data showed that the inhibition of the development of GFP fluorescence by the addition of self-inhibitors was prevented by plant surface wax (Fig. 5). Inhibition of appressorium formation by the addition of self-inhibitors was also fully reversed by plant surface wax (Fig. 5). The self-inhibitors at a higher concentration blocked appressorium formation completely, inhibited more strongly the development of GFP fluorescence, and required a higher concentration of plant cuticular wax to reverse the effect (data not shown).
FIG. 5.
(Left) Inhibition of development of GFP fluorescence in M. grisea conidia at a low conidial density by conidial surface lipids and reversal of this inhibition by plant surface wax. (Right) Percentages of appressorium formation under the same conditions as in the left panel. Shown are results with a low conidial density (104/ml) (A), a low conidial density (104/ml) with conidial surface lipids (0.2 μg/μl) (B), a low conidial density (104/ml) with conidial surface lipids (0.2 μg/μl) plus plant surface wax (0.3 μg/μl) (C), and untreated conidia (D).
All of these results suggested that cam gene expression involved in the early stage of conidial appressorium formation was inhibited by self-inhibitors and that this inhibition was fully reversed by plant surface wax.
Attachment to the surface is necessary for the early expression of the calmodulin gene and appressorium formation.
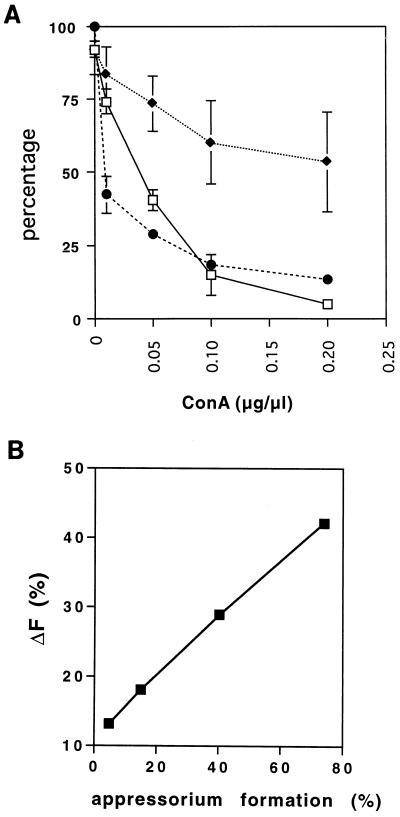
The mucilage from M. grisea conidia is thought to be used to attach the conidia to a surface (11). We tested whether such an attachment is necessary for early expression of the calmodulin gene and differentiation into appressoria. The lectin ConA is known to block the attachment of M. grisea conidia to the surface by binding to the mucilage of conidia. Experiments were carried out with a high conidial density (105/ml) and added cabbage wax (0.25 μg/μl) to relieve self-inhibition of expression of calmodulin and differentiation into appressoria. A high conidial density (105/ml) was used in these experiments because the fluorescence of a small fraction of the conidial population had to be determined and this would have been difficult to do with a low conidial density. An increase in the concentration of ConA from 0 to 0.2 μg/μl caused inhibition of conidial attachment to the surface and of appressorium formation and led to a virtually complete inhibition of appressorium formation at 0.2 μg/μl (Fig. 6A). In the presence of ConA, a fraction of the conidia were resting on but not attached to the hard surface. When the unattached conidia were recovered by pipetting, we found that the fluorescence of the unattached conidia was not as high as that of the conidia attached to the surface. As the ConA concentration increased from 0 to 0.2 μg/μl, the fluorescence of the attached conidia decreased. However, even at a very high concentration of ConA (1 μg/μl), a small fraction (∼20%) of the conidia attached to the surface and showed relatively high fluorescence. This fraction of conidia was unable to form appressoria because the high concentration of ConA inhibited appressorium formation completely but not germination. Since the fluorescence from the fraction of conidia that were unable to form appressoria still contributed to the total fluorescence increase, this portion of the increase was subtracted from the total increase before we tested for the correlation of the increase in fluorescence with the percentage of appressorium formation. This subtracted fluorescence value for conidia (attached and unattached), which can be affected by ConA at 2 h (ΔF), is shown in Fig. 6A, and the correlation between ΔF and the percentage of appressorium formation is shown in Fig. 6B. The result showed a strong correlation between the fluorescence increase indicative of cam gene expression and appressorium formation, suggesting that calmodulin gene expression may be needed for appressorium formation.
FIG. 6.
(A) Effects of ConA on conidial attachment (⧫), fluorescence increase (●), and appressorium formation (□). (B) Correlation between fluorescence increase and percentages of appressorium formation. Conidia at 105/ml were placed on a polystyrene surface in 100 μl of phosphate buffer with plant surface wax (0.25 μg/μl) in the presence of different concentrations of ConA. Two sets of duplicate samples were prepared for each concentration of ConA. Two samples were left overnight for observation of appressorium formation. After 2 h, the unattached conidia of the other two samples were removed by pipetting and the volume was adjusted to 200 μl. Conidial density was determined under a microscope with a hemacytometer. The removed conidia in 100 μl were recovered by centrifugation, resuspended in 10 μl of fixer, and placed on a polystyrene surface for confocal microscopic analysis along with the attached conidia. Values are averages of results from three experiments.
DISCUSSION
Inhibition of germination and differentiation of fungal conidia by chemicals present on their surfaces (self-inhibition) has been known for a long time (19). Self-inhibitors have been found in more than 60 fungal species and are an ecological adaptation to ensure spatial and temporal distribution of the fungal species (28). They are also often lipophilic. Plant surface waxes or other hydrophobic materials were found to relieve self-inhibition in the conidia of M. grisea. Thus, it was suggested that when conidia land on a plant cuticle, self-inhibition might be relieved by the diffusion of the self-inhibitors into the plant cuticles (12). However, how the self-inhibitors exert their effect is not known.
The contact of conidia with a host surface induces expression of a set of early genes that is required for the conidia to respond to further host signals. Among the early genes is the cam gene, whose transcription was found to be induced by hard-surface contact maximally in a few hours and then to decline in C. gloeosporioides (15) and in C. trifolii (3). Subsequently, the conidia respond to host signals that cause the transcriptional activation of another set of genes, leading to the induction of germination and appressorium formation (13). At what stage in this progression of events the self-inhibitors exert their effects is not known. To test whether self-inhibitors suppress the expression of the early genes involved in conidium differentiation, cam was chosen as a representative, as it is the earliest gene yet found to be expressed during the process. Since the cam gene from M. grisea had not been studied previously, we first cloned calmodulin cDNA and genomic DNA from M. grisea. At the amino acid level, CAMMg is identical to other fungal calmodulins and highly homologous (>90%) to plant and animal calmodulins. Plants and animals have multiple calmodulins in their genomes with distinct patterns of expression in different organs (2). In the genome of M. grisea, camMg is present as a single-copy gene, and the positions of the five introns in this fungal gene are also highly conserved. Therefore, the cloned cam gene is suitable for testing if self-inhibitors inhibit early gene expression. Since direct measurement of the level of cam transcripts in a few conidia would be difficult, the cam promoter was fused to a GFP reporter gene whose expression can readily be measured (26, 29). GFP fluorescence reached a maximum at a fourfold level in 2 to 3 h of surface contact, just as cam expression was found to be an early event in the germination and differentiation processes of M. grisea, C. gloeosporioides (15), and C. trifolii (3). Thus, the increase in GFP fluorescence was found to be suitable for measuring cam gene expression.
Our results showed that cam gene expression as indicated by GFP fluorescence was much lower when the conidial density was high than when the conidial density was low and that the addition of plant surface waxes could prevent this inhibition. The addition of plant surface waxes also prevented the inhibition of appressorium formation resulting from a high conidial density, as observed previously (12). These results suggested that cam gene expression in M. grisea was inhibited by self-inhibitors, whose effects could be prevented by plant surface waxes. Exogenous self-inhibitors, added to conidia at a low density, were found to inhibit the development of GFP fluorescence. It was difficult to directly measure the transcript levels under the condition of low conidial density. However, with a high conidial density we could measure the transcript level and thus validate the theory that the inhibition of cam gene expression caused by self-inhibitors could be relieved by plant surface wax. Our results strongly suggest that self-inhibitors exert their effects at an early stage in the process of germination and differentiation.
Hard-surface contact is known to be required for appressorium formation (8). The attachment of fungal conidia to the plant host is one of the early steps in plant-pathogen interaction. The adhesive materials in conidial mucilage such as glycoproteins are mainly responsible for attaching the spores to the substratum. Mucilage, commonly present in fungi, may be carried externally on the spore during dispersal or may be internal and secreted within minutes of contacting the host surface or during germination (10, 11). We tested whether conidial attachment to a surface is necessary for early cam gene expression, using ConA to block the conidial attachment. When the unattached conidia were recovered by pipetting, the fluorescence of the conidia that were in contact with the surface but not attached to it did not increase, as did that of the conidia attached to the hard surface. Our approach of using a GFP reporter driven by the cam gene promoter allowed us to distinguish between the effect of conidial contact and that of conidial attachment to the surface on early cam gene expression. Our results showed that ConA inhibited surface attachment of conidia, GFP expression, and appressorium formation without affecting germination, suggesting that conidium attachment to, and not mere contact with, the surface is required for induction of cam expression as well as appressorium formation.
A small portion of the conidia did attach to the surface even in the presence of high concentrations of ConA. This may have been due to the heterogeneity in the conidial population with respect to the mucilage content and/or composition. Lectins (including ConA) are highly specific for the saccharide haptens they bind, and some conidia might not have at their surfaces the mucilage that has the specific types of ConA-binding saccharides. The possibility of the presence of conidia that lack any mucilage but can still attach to the surface cannot be ruled out in view of the report that certain isolated M. grisea conidia lacked spore tip mucilage (14). Those conidia that attached to the surface even at high concentrations of ConA germinated but were incapable of forming appressoria. Without ConA they would have formed appressoria, as virtually all conidia we used formed appressoria under normal conditions. Thus, for these conidia, attachment alone is not enough to allow appressorium formation and ConA can block appressorium formation by some mechanisms other than by merely blocking attachment. This observation raises the possibility that the conclusion, based on the correlation of ConA inhibition of attachment and appressorium formation, that attachment is absolutely required for appressorium formation may need to be reexamined.
Expression of the cam gene in M. grisea is induced in the early stages in conidial differentiation. Calmodulin is known to be required for cell cycle progression during G1 and mitosis (23). As a calcium-binding protein, calmodulin is a primary transducer of intracellular calcium signals. For example, by activating calmodulin-dependent kinases, calmodulin can affect transcription factors and regulate the transcription of many genes (25). Through the signal transduction cascade, slight changes in calmodulin level can have significant effects on cell growth and cell cycle progression. Calmodulin is required for the polar movement of chromosomes during mitosis as it regulates microtubule disassembly (or depolymerization); it is known to interact with microtubule-associated proteins (24). Appressorium formation may require cytoskeleton reorganization (5). It was reported that microtubules and actin filaments become depolymerized during appressorium formation in Uromyces appendiculatus (17). Calmodulin may be required for nuclear division in the conidium and the subsequent migration of the nucleus and cytoplasm into the appressorium since these processes involve microtubule function. As we demonstrate in this paper, the self-inhibition of fungal conidial differentiation involves early events in plant-pathogen interaction. Prevention of conidial germination and differentiation by interfering with an early event in this process would be an effective way for a self-inhibitor to ensure that the conidium embarks on further development only in a favorable environment. Such early events may also be very effective targets of antifungal strategies to protect plants.
ACKNOWLEDGMENTS
This work was supported in part by National Science Foundation grants IBN-9816868 and IBN-9318544.
We thank Linda Rogers and Nichole R. Gierat for assistance in preparing the manuscript and Daoxin Li and Yeon-ki Kim for helpful discussions.
REFERENCES
- 1.Bajar A, Podila G K, Kolattukudy P E. Identification of a fungal cutinase promoter that is inducible by a plant signal via a phosphorylated trans-acting factor. Proc Natl Acad Sci USA. 1991;88:8208–8212. doi: 10.1073/pnas.88.18.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braam J, Davis R W. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- 3.Buhr T L, Dickman M B. Gene expression analysis during conidial germ tube and appressorium development in Colletotrichum trifolii. Appl Environ Microbiol. 1997;63:2378–2383. doi: 10.1128/aem.63.6.2378-2383.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.Dean R A. Signal pathways and appressorium morphogenesis. Annu Rev Phytopathol. 1997;35:211–234. doi: 10.1146/annurev.phyto.35.1.211. [DOI] [PubMed] [Google Scholar]
- 6.Dickman M B, Buhr T L, Warwar V, Truesdell G M, Huang C X. Molecular signals during the early stages of alfalfa anthracnose. Can J Bot. 1995;73(Suppl. 1):S1169–S1177. [Google Scholar]
- 7.Dobbeling U, Boni R, Haffner A, Dummer R, Burg G. Method for simultaneous RNA and DNA isolation from biopsy material, culture cells, plants and bacteria. BioTechniques. 1997;22:88–90. doi: 10.2144/97221bm19. [DOI] [PubMed] [Google Scholar]
- 8.Emmet R W, Parberry D G. Appressoria. Annu Rev Phytopathol. 1975;13:147–167. [Google Scholar]
- 9.Fowler T, Berka R, Ward M. Regulation of the glaA gene of Aspergillus niger. Curr Genet. 1990;18:537–545. doi: 10.1007/BF00327025. [DOI] [PubMed] [Google Scholar]
- 10.Griffen D H. Fungal physiology. New York, N.Y: Wiley-Liss; 1994. [Google Scholar]
- 11.Hamer J E, Howard R J, Chumley F G, Valent B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science. 1988;239:288–290. doi: 10.1126/science.239.4837.288. [DOI] [PubMed] [Google Scholar]
- 12.Hedge Y, Kolattukudy P E. Cuticle waxes relieve self-inhibition of germination and appressorium formation by the conidia of Magnaporthe grisea. Physiol Mol Plant Pathol. 1997;51:75–84. [Google Scholar]
- 13.Hwang C-S, Kolattukudy P E. Isolation and characterization of genes expressed uniquely during appressorium formation by Colletotrichum gloeosporioides conidia induced by the host surface wax. Mol Gen Genet. 1995;247:282–294. doi: 10.1007/BF00293196. [DOI] [PubMed] [Google Scholar]
- 14.Jelitto T C, Page H A, Read N D. Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus. Planta. 1994;194:471–477. [Google Scholar]
- 15.Kim Y-K, Li D, Kolattukudy P E. Induction of Ca2+-calmodulin signaling by hard-surface contact primes Colletotrichum gloeosporioides conidia to germinate and form appressoria. J Bacteriol. 1998;180:5144–5150. doi: 10.1128/jb.180.19.5144-5150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolattukudy, P. E., Y. Kim, D. Li, Z. M. Liu, and L. Rogers. Early molecular communication between Colletotrichum gloeosporioides and its host. In Host specificity, pathology and host pathogen interaction of Colletotrichum, in press. The American Phytopathological Society, St. Paul, Minn.
- 17.Kwon Y H, Hoch H C, Staples R C. Cytoskeletal organization in Uromyces urediospore germling apices during appressorium formation. Protoplasma. 1991;165:37–50. [Google Scholar]
- 18.Liu Z M, Kolattukudy P E. Identification of a gene product induced by hard-surface contact of Collectotrichum gloeosporioides conidia as a ubiquitin-conjugating enzyme by yeast complementation. J Bacteriol. 1998;180:3592–3597. doi: 10.1128/jb.180.14.3592-3597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macko V. Inhibitors and stimulants of spore germination and infection structure formation in fungi. In: Turian G, Holh H R, editors. The fungal spore morphogenetic controls. New York, N.Y: Academic Press; 1981. pp. 565–584. [Google Scholar]
- 20.Melnick M B, Melnick C, Lee M, Woodward D O. Structure and sequence of the calmodulin gene from Neurospora crassa. Biochim Biophys Acta. 1993;117:334–336. doi: 10.1016/0167-4781(93)90079-s. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell T K, Dean R A. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenicity by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7:1869–1878. doi: 10.1105/tpc.7.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunberg J, Meade J, Cole G, Lawyer F, McCabe P, Schweickart V, Tal T, Wittman V, Flatgaard J, Innis M. Molecular cloning and characterization of the glucoamylase gene of Aspergillus awamori. Mol Cell Biol. 1984;4:2306–2315. doi: 10.1128/mcb.4.11.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen C D, Means A R. Calmodulin is required for cell cycle progression during G1 and mitosis. EMBO J. 1989;8:73–82. doi: 10.1002/j.1460-2075.1989.tb03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts D M, Harmon A C. Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:375–414. [Google Scholar]
- 25.Schulman H. The multifunctional Ca2+/calmodulin-dependent protein kinases. Curr Opin Cell Biol. 1993;5:247–253. doi: 10.1016/0955-0674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 26.Spellig T, Bottin A, Kahmann R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- 27.Staben C, Jensen B, Singer M, Pollosk J, Schechtman M, Linsey J, Selker E. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet Newsl. 1989;36:79–81. [Google Scholar]
- 28.Tsurushima T, Ueno T, Fukami H, Irie H, Inoue M. Germination self-inhibitors from Colletotrichum gloeosporioides f. sp. jussiaea. Mol Plant-Microbe Interact. 1995;8:652–657. [Google Scholar]
- 29.Wymelenberg A J V, Cullen D, Spear R N, Schoenike B, Andrews J H. Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surface. BioTechniques. 1997;23:686–690. doi: 10.2144/97234st01. [DOI] [PubMed] [Google Scholar]
- 30.Yasui K, Kitamoto K, Gomi K, Kumagai C, Ohya Y, Tamura G. Cloning and nucleotide sequence of the calmodulin-encoding gene (cmdA) from Aspergillus oryzae. Biosci Biotechnol Biochem. 1995;59:1444–1449. doi: 10.1271/bbb.59.1444. [DOI] [PubMed] [Google Scholar]