Abstract
Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic has adversely affected humankind and caused millions of deaths globally since January 2020. Robust and quick serological tests such as antibody detection assays for SARS-CoV-2 provide relevant information and aid in the process of vaccine development and diagnostics, as well as in sero-epidemiological monitoring of antibody response to the virus. The receptor-binding domain (RBD) of spike and nucleocapsid protein are specific targets for detecting SARS-CoV-2 antibodies. Here, we present the development of a stable spike (S) and nucleocapsid (N) protein-based ELISA antibody detection test “CoroSuchak,” with 99% sensitivity, 98% specificity, cost-effective, and detection in a minimum time for serodiagnosis and mass screening of the population for antibodies against SARS-CoV-2. Blood samples were analyzed from 374 SARS-CoV-2 reverse transcription-polymerase chain reaction (RT-PCR) positive, 772 negative and asymptomatic, and 874 random groups of subjects. We found that the antibody titer was significantly higher (p < 0.0001) in infected and vaccinated group compared to the only vaccinated and only infected group. Using enzyme-linked immunosorbent assay (ELISA), we detected SARS-CoV-2 immunoglobulin G (IgG) antibodies in 118/123 (96%) infected individuals, 570/653 (87%) non-infected but vaccinated individuals, 231/237 (97%) individuals who were both infected and vaccinated, and 499/874 (57%) from randomly selected individuals from the first and second waves of the pandemic. Similarly in the third wave, 14/14 (100%) infected and 16/20 (80%) RT-PCR-negative but symptomatic subjects were detected. Thus, the highly sensitive and specific in-house developed ELISA antibody detection kit “CoroSuchak” is extremely useful to determine the seroprevalence of SARS-CoV-2 antibodies in the coronavirus-exposed population.
Key points
•Indigenous kit using a combination of spike and nucleocapsid proteins and peptide sequences.
•High sensitivity and specificity to detect variants.
•Highly sensitive for mass screening.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-022-12113-8.
Keywords: COVID-19, Antibody detection, Human, IgG, ELISA, Mass screening
Introduction
Multiple human instances of new coronavirus infections were reported in the Huanan Seafood Wholesale Market (South China Seafood City Food Market) in Wuhan, China, at the end of 2019 and the beginning of 2020. The virus was identified as a novel coronavirus on January 7, 2020, and was officially named 2019nCoV, the new coronavirus, and the World Health Organization (WHO) declared it a pandemic on March 12, 2020 (WHO 2020a), as it caused unprecedented disruption on a global level. The first wave of infection caused by dominant circulating strain Alpha B.1.1.7 and Beta all over the world lasted for 5 to 6 months in 2020, claiming more than 640 thousand deaths globally (Giuliana Viglione 2020). Fortunately, there was a period of 6 to 7 months after the first wave when the infection rate was reduced worldwide. Before people could recover physiologically, psychologically, and economically from the devastating time of the year 2020, another potent strain of the coronavirus, the Delta strain, attacked the world with a second wave, causing havoc and claiming lives irrespective of age and gender for nearly 4 to 5 months from March to July 2021. Again, there was a lull period of 4 months to recover from the dreaded disease, but yet again, another strain Omicron appeared globally in the third wave from late December 2021. The virus spread across continents in a short time, infecting about 215 countries and territories. By early 2022, the global outbreak data logged more than 423 million (42,34,37,674) total cases and more than 5 million (58,78,328) deaths around the world. India itself has reported over 42 million cases and over 5.12 million deaths to date.
COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the Coronaviridae family. Coronaviruses (CoVs) are a large group of enclosed, single-stranded RNA viruses that mostly infect humans and animals, causing respiratory problems of varying severity (Chen et al. 2020). CoVs are enveloped, non-segmented viruses with a genome size ranging from 26 to 32 kilobases, the largest known viral RNA genome. The nucleocapsid region (N), made up of genomic RNA and phosphorylated nucleocapsid protein, is concealed inside phospholipid bilayers and protected by spike protein (S). SARS-CoV-2 infects human target cells with its viral trans-membrane S protein, a trimeric class-I fusion protein. The S1 makes it easier for the virus to attach to the host entry receptor angiotensin-converting enzyme 2 (ACE2), while the S2 subunit makes it possible for the viral and human cellular membranes to fuse (Hoffmann et al. 2020; Zhou et al. 2020). S protein is a crucial target for the development of specific medical therapies or vaccines because of its important role.
SARS-CoV-2 have a long incubation period of 2–14 days. Specific immunoglobulin M (IgM) and immunoglobulin A (IgA) antibodies are the early antibody responses that begin and peak within 7 to 10 days, followed by specific immunoglobulin G (IgG) a few days later (between 10 and 18 days) and are assumed to continue as protective antibodies for the rest of the life without much decline (Gao et al. 2021). IgG antibody detection against COVID-19 is expected to be more important in understanding antibody-mediated protection and vaccination trials than IgM or total antibodies (Theel et al. 2020; Wu et al. 2021). They are longer-lasting antibodies important for many basic immune responses such as viral neutralization, opsonization, and complement fixation (Huang et al. 2020). Thus, the detection of IgG antibodies against SARS-CoV-2 plays a significant role in this pandemic. As per the available data, IgG developed against SARS-CoV-2 antigens is detectable in patients after at least 8 to 10 days. Over 90% of patients demonstrate seropositivity by day 14 of illness. However, some patients may take longer to seroconvert depending on their immune status or may never seroconvert if severely immuno-suppressed (Long et al. 2020; Zhao et al. 2020). The fact that IgG production is prolonged, could mean that IgG is important in both the humoral immune response infection and the elimination of residual viral sources following recovery to acute SARS-CoV-2.
Molecular assays, as approved by the WHO, are currently the mainstay of COVID-19 diagnosis (WHO 2020b). Serological techniques such as ELISA are used to establish the true burden of illness and its transmission by detecting IgM and IgG antibodies against SARS-CoV-2 (Ye et al. 2020b; Bai et al. 2020; Long et al. 2020). Using immunofluorescence assays, Haveri et al. (Haveri et al. 2020) discovered the existence of serum IgM and IgG antibodies against SARS-CoV-2. Certifying agencies like Central Drugs Standard Control Organization (CDSCO), Indian Council of Medical Research (ICMR), Government of India (cdsco.gov.in), have certified IgM/IgG ELISA for COVID-19 for commercial purposes/research in India. These assays are simple to use and cost-effective for SARS-CoV-2 antibody detection and for surveillance in resource-constrained settings (Haveri et al. 2020).
The receptor-binding domain (RBD) has been employed as a target for the detection of anti-SARS-CoV-2 IgG antibodies in various studies. However, none of these studies has revealed any systematic optimization of the test parameters (Amanat et al. 2020; Premkumar et al. 2020). Other RBD-based IgG detection methods (Amanat et al. 2020; Whitman et al. 2020) are time-consuming and thus limiting daily throughput. In the present study, samples were tested for anti-S and anti-N SARS-CoV-2 IgG antibody levels in individuals categorized into different groups based on their infection and vaccination status. Considerable optimization was done for the designing of a cocktail of N and S peptides with N and S recombinant proteins for the detection of IgG antibodies in blood samples against SARS-CoV-2, to take care of multiple variants of SARS-CoV-2 and also minimize the chances of missing the low antibody titer. The cocktail-based IgG ELISA was developed with high sensitivity and specificity. The optimized ELISA was turned into a stabilized kit with less than 80-min assay time. A broad panel of samples from negatives (n = 772) was used to test the kit, including interference-prone samples and SARS-CoV-2 RT-PCR-positive cases (n = 374). The present kit “CoroSuchak” has high sensitivity, specificity, and stability with a long shelf-life and is cost-effective.
The kit, named as “CoroSuchak” is being used for mass screening of the population to understand whether the sequence of infection and/or vaccination or repeated exposures alters the specificity and magnitude of antibody generation in all three waves of SARS-CoV-2 infection.
Material and methods
Study subjects and ethical statement
A total of 2016 subjects (560 females and 1456 males) volunteered for the study. The age ranged from 19 to 80 years for both the genders, with a mean age of 38.85 years. For standardization of the kit, 509 samples were collected from a government-designated COVID-19 hospital. Before initiating the study, the volunteers were provided with detailed information regarding the study design and its importance. All the volunteers understood the nature of the study and provided informed written consent. After approval of the kit for mass screening, volunteers (n = 2016) were provided with a questionnaire consisting of demographic details of the individuals, including infection and vaccination status, any co-morbidity, medication for COVID-19 treatment, and date of positive and negative RT-PCR reports.
Blood sample collection
Samples were carefully collected following all the necessary protocols and guidelines issued by WHO and ICMR, Government of India, using biosafety level (BSL) facilities. Samples were randomly collected irrespective of the time of vaccination and sample collection. Three milliliters of blood sample was collected in clot activator vacutainers from all the volunteers after about 3 to 6 weeks of infection and 5 to 7 weeks after the second dose of vaccination. Samples were stored at room temperature for 1 h and centrifuged at 1200 rpm for 10–15 min for serum separation. To determine the duration of sustenance of antibodies, blood samples from some individuals were collected four times at an interval of 2 to 3 months. The collected serum samples were aliquoted and stored at – 40 °C until further analysis.
Coating of microtiter plate with a cocktail of proteins and peptides as antigen
Coating material
Recombinant protein
The S and N proteins were obtained from Abgenex Pvt. Ltd., Odisha, India (21–1008; 21–1003), for coating of plates. In brief, the S protein was expressed in the Chinese hamster ovary (CHO-K1) with the sequence (AA 14–683) of the SARS-CoV-2/COVID-19 spike S1 sequence fused with a 6xHis tag in the C-terminal. Similarly, the target nucleocapsid protein was expressed in E. coli with the sequence (Ser2-Ala419) of the SARS-CoV-2/COVID-19 nucleocapsid fused with a 6xHis tag in the N-terminal.
Synthetic peptide
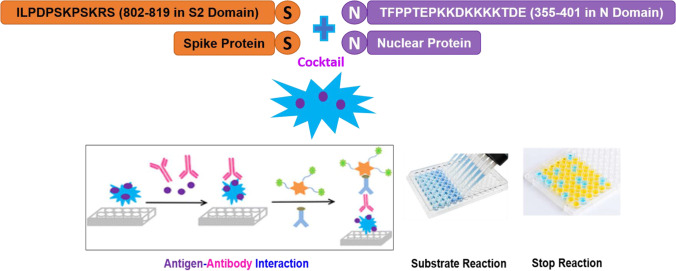
Highly antigenic nucleotide sequences were identified, synthesized, purified, and well characterized. The target sequence ILPDPSKPSKRS {(E5) residue 802–819 lie in the S2 domain)} was identified from the S-protein across the several structural proteins of SARS-CoV-2 (from Wuhan-Hu-1 variant). The selection of S protein was based on its exposure to the cell surface and advantage of sequence antigenicity and initiation of viral infection by binding to the receptor ACE2.
Another target sequence, TFPPTEPKKDKKKKTDE, {(E4) residue 355–401)}, was identified from the N protein immunogenic region. Interestingly, this region is highly conserved in the SARS and may effectively induce antibodies across several serotypes of SARS viruses.
The target sequences were selected based on the combined B cell immunogenicity scores from various prediction methods such as Immune Epitope Database (IEDB), ABCpred, and BcePred and combined to the linear B cell epitope candidate list across multiple variants of SARS-CoV-2. VaxiJen 2.0 was applied to evaluate the predicted antigenicity of the target epitopes. In all the selection methods, a stringent criterion was used to have epitopes with an antigenicity score of 0.9 and keep the window size to 7 in the prediction.
Coating of the plate with cocktail
An IgG capture ELISA was developed for the serological detection of SARS-CoV-2 infection from patients’ serum based on the principle of antigen–antibody interaction. 96-well polystyrene microtiter ELISA plates (Corning/Coster-92592 NY, USA) were coated with 275 µl of the indigenously prepared cocktail of multiplexing proteins and peptides combination (protein-N, 200 ng; protein-S, 50 ng; peptide-N, 15 ng; and peptide-S, 10 ng/well), in carbonate buffer (50–150 mM, pH 9.5–9.7), followed by incubation at 2–8 °C for 12 h (Fig. 1). The plates were washed 5–6 times using wash buffer, phosphate buffer saline, and Tween-20 {phosphate-buffered saline/Tween (PBST-100 Mm)}, with 250–350 µl/ well, followed by blocking with 10–100 mM phosphate buffer having 3.0–5.0% bovine serum albumin (BSA-Fr-V, Sigma, USA), and 0.5–1.0% Casein Hammarsten (Sigma-9000–71-9). The plates were kept at 2–8° C for 12 h, followed by washing as above. Plate stabilizing reagents using phosphate buffer having 3% sucrose was added to the plate followed by incubation at 2–8 °C for 12 h.
Fig. 1.
Depiction of principle of antigen antibody interaction in a cocktail of S and N peptide-protein coated on microtiter plate. A cocktailof multiplexing proteins (S and N) and peptides (ILPDPSKPSKRS, identified from S-protein and TFPPTEPKKDKKKKTDE, identified from the N-protein immunogenic regions) were used for coating the microtiter ELISA plates. This was followed by multiple rounds of incubation, washing, and blocking of the interaction complex
Standardization of SARS-CoV-2 IgG antibody detection by ELISA
Protein-peptide cocktail pre-coated plates were used for sample analysis for antibody detection against SARS-CoV-2. One hundred microliters of sample diluent was added to each well, followed by the addition of 10 µl negative control (inactivated and stabilized normal human serum), positive control (inactivated and stabilized SCoV-2 human serum), and test samples to respective wells. The plates were incubated at 37 °C for 30 min. After the incubation, plates were washed five times with 300 µl PBS. This was followed by the addition of 100 µl of horseradish peroxidase (HRP) conjugated to an anti-human IgG antibody into each well and kept for incubation at 37 °C for 30 min. The incubated plates were again washed five times with 300 µl PBS. One hundred microliters of 3,3,5,5′-tetramethylbenzidine (TMB) substrate was added to each well for color development. The addition of substrate started an enzyme-catalyzed color change, which was stopped within 15 min by the addition of 50 µl of 2 N sulfuric acid (H2SO4). Finally, the absorbance value was measured in terms of optical density (OD) using an ELISA reader at 450 nm (BioTek, Agilent, CA, USA).
Determination of cut-off values, index titer value (ITV) calculation
The cut-off value (COV) for qualitative estimation was determined by combining negative control mean (NCM) and threshold value (0.200).
A. Interpretation of results by the qualitative method:
Calculation of the cut-off value (COV):
• Calculate the negative control mean (NCX), e.g., negative control 1 absorbance = 0.009, negative control 2 absorbance = 0.010, negative control 3 absorbance = 0.011.
• Negative control mean (NCX) = (0.009 + 0.010 + 0.011)/3 = 0.010.
- • Cut-off value (COV) = NCX + 0.200 = 0.010 + 0.200 = 0.210.
Interpretation
• Samples with absorbance greater than or equal to the cut-off value are considered reactive or with presence of antibody to SARS-CoV-2 by the assay analysis.
• Samples with absorbance less than the cut-off value are considered as non-reactive or with absence of antibody to SARS-CoV-2 by the assay analysis.
• Gray zone results: samples whose absorbance falls in between ± 10% of cut-off value should be considered to be in the gray zone.
• The samples falling under the gray zone should be repeated to validate the results.
B. Interpretation of results by the semi-quantitative method
Semi-quantitative estimation of index titer value (ITV) for determination of the level of immune response was calculated using the formula:
Interpretation
| Results in ITV | Interpretation |
|---|---|
| < 10 | No immune response (absent) |
| > 10 to 20 | Low immune response |
| > 20 to 40 | Moderate immune response |
| > 40 | High immune response |
Validation and approval by Indian government agency
Once the method was designed, developed, and standardized into a kit form, an in-house blind study was performed on approximately 500 samples. This was followed by further third-party validation by a COVID-19 nominated center. Finally, three batches of the kit were submitted to ICMR and CDSCO, Government of India, for technical and regulatory approvals after obtaining perfect results. Both the agencies approved the kit for detecting COVID-19 IgG antibodies (license number: MFG/IVD/2020/000162).
Mass screening of population from different regions in India
The approved kit was used for mass screening of antibodies in different regions of India, viz., Delhi, Jodhpur, Ladakh, Udhampur, Mumbai, and 3496 samples were screened from June 2021 to December 2021. The study subjects were divided into five groups according to their infection and vaccination status {after two doses of vaccination (Table 1)}.
Table 1.
Categorization of subjects based on their infection and vaccination status. The study included COVID-19 RT-PCR positive and negative subjects from the three COVID-19 infection waves
| Group | Subjects distribution in different categories | No. of subjects included | Table |
|---|---|---|---|
| Group-1 |
COVID-19 infected 1 a: From first wave (April–August, 2020) 1 b: From second wave (April–August, 2021) |
18 105 |
2, a 2, b |
| Group-2 |
Non-infected but vaccinated 2a: Non-infected and vaccinated with vaccine-A 2b: Non-infected and vaccinated with vaccine-B 2c: Non-infected and vaccinated with vaccine-C 2d: Non-infected and vaccinated with vaccine-D |
143 504 02 04 |
3, a 3, b 3, c 3, d |
| Group-3 |
COVID-19 infected and vaccinated 3a: Infected and vaccinated with vaccine-A 3b: Infected and vaccinated with vaccine-B |
104 133 |
4, a 4, b |
| Group-4 |
COVID-19 non-infected and non-vaccinated Asymptomatic group |
95 | 5 |
| Group-5 |
Random group Had no proper details of infection or vaccination |
402 + 472 | 6, a + b |
| Group-6 |
Third wave subjects (December–January, 2022) |
34 | 7 |
Statistical analysis
The statistical analysis was performed using GraphPad Prism 7 software. For the continuous variables, scatter plots were drawn and compared using a one-way non-parametric analysis of variance (ANOVA) with no matching or pairing data. The Tukey post hoc test was used to provide a multiple comparison test for statistical hypothesis testing. The data are presented as mean ± standard deviation (S.D.), with a p value of 0.05 considered statistically significant.
Results
The combination of proteins and peptides is vital to bring broad coverage to the sequence across different members of the CoV-2 family. In addition, the N and S proteins are vital for viral replication and transcription of viral RNA. The selected epitopes’ physio-chemical properties were the best candidates for kit development on the basis of their accessibility, antigenic propensity, exposed surface, flexibility, hydrophilicity, polarity, and turn values. The presence of antibodies against N and S proteins suggests that the individual was potentially exposed to SARS-CoV-2. The presence of IgG expression confirms the infection status. Reports indicate that for any viral infections, IgG usually persists longer and provides immunity from re-infection, it is yet to be proved for COVID-19. The COVID-19 IgG antibody detection ELISA was performed by “CoroSuchak” on the Indian population, categorized into various groups depending on their infection and vaccination status (groups 1 to 5). The blood samples were collected after about 3–6 weeks of infection and 5–7 weeks after vaccination.
Analysis of samples from first and second wave–infected individuals—group 1
Group 1 included COVID-19-positive (RT-PCR confirmed), non-vaccinated individuals from the first wave (n = 18; 08 females and 10 males) and the second wave (n = 105; 35 females and 70 males) (Table 2). Among the first wave of infected individuals, 100% of females had CoV-2 IgG antibodies present (8/8) compared to 90% of males (9/10). Taken collectively, 94% (17/18) of subjects had the presence of CoV-2 antibodies, whereas one individual failed to show the presence of antibody after getting infected in the first wave (Table 2, a). Similarly, amongst the second wave–infected individuals, 94% of females (33/35) and 97% of males (68/70) had the presence of CoV-2 antibodies. Overall, 96% of individuals had the presence of antibodies, whereas two females and two males failed to show any antibody titer against the CoV-2 (Table 2, b). This analysis allowed us to estimate the average anti-S and anti-N antibodies in each individual at 3 to 6 weeks post-infection.
Table 2.
Group 1: COVID-19-infected individuals. 1a, first wave infected; 1b, second wave infected
| a | |||||
| Group-1a | N size | Abs present | Abs absent | % | |
| Female | 8 | 8 | 0 | 100 | |
| Male | 10 | 9 | 1 | 90 | |
| Total | 18 | 17 | 1 | 94 | |
| b | |||||
| Group-1b | N size | Abs present | Abs absent | % | |
| Female | 35 | 33 | 2 | 94 | |
| Male | 70 | 68 | 2 | 97 | |
| Total | 105 | 101 | 4 | 96 | |
Analysis of samples from non-infected but vaccinated subjects—group 2
Group 2 included 653 non-infected but vaccinated individuals. One hundred forty-three individuals (37 females and 106 males) received vaccine-A; 504 (78 females and 426 males) received vaccine-B; 02 females received vaccine-C; while 04 (02 females and 02 males) received vaccine-D (Table 3). Among vaccine-A-jabbed individuals, a total of 86% of individuals had antibodies present. Out of which, 86% were females, and 81% were males (Table 3, a). In vaccine-B-jabbed individuals, a total of 88% had antibodies, with 76% females and 90% males (Table 3, b). All subjects receiving vaccine-C (n = 02) and vaccine-D (n = 04) had the presence of CoV-2 antibodies (Table 3, c and d). This analysis allowed us to estimate the average anti-S and anti-N antibody levels in each individual at 5 to 7 weeks post-vaccination.
Table 3.
Group 2 subjects: Presence of SARS-CoV-2 antibodies in non-infected but vaccinated individuals. 2a, vaccine-A-jabbed individuals; 2b, vaccine-B-jabbed individuals; 2c, vaccine-C-jabbed individuals; 2d, vaccine-D-jabbed individuals
| a | |||||
| Group-2a | N size | Abs present | Abs absent | % | |
| Female | 37 | 32 | 5 | 86 | |
| Male | 106 | 86 | 20 | 81 | |
| Total | 143 | 118 | 25 | 83 | |
| b | |||||
| Group-2b | N size | Abs present | Abs absent | % | |
| Female | 78 | 59 | 19 | 76 | |
| Male | 426 | 382 | 44 | 90 | |
| Total | 504 | 441 | 63 | 88 | |
| c | |||||
| Group-2c | N size | Abs present | Abs absent | % | |
| Female | 2 | 2 | 0 | 100 | |
| Male | 0 | 0 | 0 | 0 | |
| Total | 2 | 2 | 0 | 100 | |
| d | |||||
| Group-2d | N size | Abs present | Abs absent | % | |
| Female | 2 | 2 | 0 | 100 | |
| Male | 2 | 2 | 0 | 100 | |
| Total | 4 | 4 | 0 | 100 | |
Analysis of samples from infected and vaccinated subjects—group 3
The group included individuals infected and vaccinated with either vaccine-A (group-3a) or vaccine-B (group-3b). Group-3a individuals vaccinated with vaccine-A consisted of 104 subjects comprising 37 females and 67 males (Table 4). Overall, 97% of individuals had the presence of antibodies, which included 97% of females (36/37) and 97% of males (65/67) (Table 4, a). Similarly, in group-3b, 133 individuals vaccinated with vaccine-B, including 24 females and 109 males, had a total 98% antibody presence, with 96% females (23/24) and 98% males (107/109) (Table 4, b). It was observed that vaccinated individuals who experienced an infection had higher antibody titer 30 days post-positive RT-PCR test. There was no reporting of any COVID-19-positive cases in individuals completely vaccinated with vaccines 3 and 4.
Table 4.
Group 3: Presence of SARS-CoV-2 antibodies in COVID-19-infected and vaccinated individuals. 3a, COVID-19-infected and vaccinated with vaccine-A; 3b, COVID-19-infected and vaccinated with vaccine-B
| a | |||||
| Group-3a | N size | Abs present | Abs absent | % | |
| Female | 37 | 36 | 1 | 97 | |
| Male | 67 | 65 | 2 | 97 | |
| Total | 104 | 101 | 3 | 97 | |
| b | |||||
| Group-3b | N size | Abs present | Abs absent | % | |
| Female | 24 | 23 | 1 | 96 | |
| Male | 109 | 107 | 2 | 98 | |
| Total | 133 | 130 | 3 | 98 | |
Analysis of samples from non-infected and non-vaccinated subjects—group 4
This group consisted of 95 individuals who were neither infected nor vaccinated, out of which 25 were females and 70 males. Though these individuals had never had any symptoms and, therefore, never got the RT-PCR test done, interestingly, 71% of individuals had the presence of CoV-2 antibodies. Out of which, 84% were females (21/25), and 66% were males (46/70) (Table 5).
Table 5.
Group 4: Presence of SARS-CoV-2 antibodies in non-infected and non-vaccinated individuals
| Group-4 | N size | Abs present | Abs absent | % | |
| Female | 25 | 21 | 4 | 84 | |
| Male | 70 | 46 | 24 | 66 | |
| Total | 95 | 67 | 28 | 71 |
Analysis of samples from the random group—group 5
Group 5 included samples of individuals selected randomly from the remote region of North India who had received vaccination but could not give any information about the type of vaccine or the date of vaccination nor about their infection status (Table 6). In group-5a, the number of randomly collected samples was 402: 240 females and 162 males. Among them, 48% of individuals had antibodies present, including 45% of females and 54% of males (Table 6, a). Group-5b included the largest number of randomly selected individuals with unknown infection as well as vaccination status (n = 472; 59 females and 413 males). The overall group represented a total 65% presence of CoV-2 antibodies, including 59% females and 65% males (Table 6, b).
Table 6.
Group 5 with randomly selected individuals. 5a, received vaccination but without any further details; 5b, randomly selected individuals of unknown infection or vaccination status
| a | |||||
| Group-5a | N size | Abs present | Abs absent | % | |
| Female | 240 | 107 | 133 | 45 | |
| Male | 162 | 87 | 75 | 54 | |
| Total | 402 | 194 | 208 | 48 | |
| b | |||||
| Group-5b | N size | Abs present | Abs absent | % | |
| Female | 59 | 35 | 24 | 59 | |
| Male | 413 | 270 | 143 | 65 | |
| 472 | 305 | 167 | 65 | ||
Analysis of samples from III wave infected subjects: group 6
This group included both RT-PCR-positive (n = 14; 04 females and 10 males) and RT-PCR-negative samples (n = 20; 09 females and 11 males) who were symptomatic during the third wave. Among RT-PCR-positive subjects, all 14 subjects (100%) had antibodies present in high titers. Among RT-PCR-negative and symptomatic individuals, 75% (8/9) females and 73% (8/11) males had antibodies present (Table 7).
Table 7.
Group 6: Samples collected from COVID-19-positive and COVID-19-negative individuals during the third wave
| Group-6 | N size | Abs present | Abs absent | % | |
| Female | 13 | 12 | 1 | 92 | |
| Male | 21 | 18 | 3 | 86 | |
| Total | 34 | 30 | 4 | 88 |
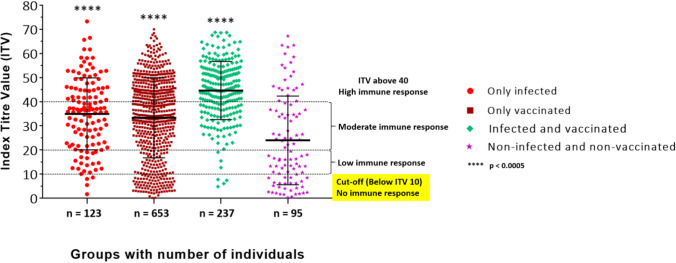
The overall comparison of antibody response against infection and vaccination in individuals (from different groups) analyzed for the SARS-CoV-2 antibody by “CoroSuchak” revealed that overall, 66.1% of females (371/561) and 78.4% of males (1144/1459) had the presence of CoV-2 IgG antibodies. For the 123 specimens which were COVID-19 infected, the mean ITV was 35 with a standard deviation of 14.88. Similarly, for only vaccinated (n = 653), infected and vaccinated (n = 237), and non-infected and non-vaccinated (n = 95) subjects, the mean ITV were 33.31, 44.67, and 24.5, respectively, with standard deviations of 16.61, 12.05, and 18.37 respectively (Fig. 2). Thus, the total Indian population included in this study revealed 75.0% of subjects with presence of CoV-2 antibodies. Evaluation of the IgG antibody titers (OD450) against recombinant proteins and peptides has been represented in Supplementary Figure S1.
Fig. 2.
Evaluation of the IgG antibody titers (index titer value, ITV) against recombinant S and N proteins and peptides using “CoroSuchak.” Serum samples from various groups of subjects were collected and analyzed for the presence of SARS-CoV-2 IgG antibodies. An ITV value of 10 was used as a cut-off value, i.e., ITV below 10 was considered as no immune response, between 10 and 20 was considered as a low immune response, between 20 and 40 was considered a moderate immune response, and ITV > 40 as a high immune response. The error bars represent standard deviation (mean ± S.D.)
Antibody status with regard to age
Female and male subjects of all the groups (1–5) were categorized in the age range of 18–40 and 40 + years. It was observed that 63–100% of females from 18–40 years and 46–100% of females from 40 + years included in this study had the presence of CoV-2 antibodies, whereas over 50% of males from both the age groups had CoV-2 antibodies present (Table 8). The present data of mass screening of the Indian population reveals a higher percentage of females with the presence of antibodies than males.
Table 8.
Table depicting different age groups and gender wise percentage for presence of SARS-CoV-2 antibodies
| Groups | Age (years) | Presence of CoV-2 antibody in females (n/%) | Presence of CoV-2 antibody (%) in males (n/%) |
|---|---|---|---|
| 1a (first wave) infected | 18–40 | 7 (100%) | 9 (90%) |
| 40 + | 1 (100%) | 0 | |
| 1b (second wave) infected | 18–40 | 20 (91%) | 46 (100%) |
| 40 + | 14 (93%) | 21 (91%) | |
|
2a Only vaccinated (Vaccine-A) |
18–40 | 11 (85%) | 26 (74%) |
| 40 + | 21 (88%) | 60 (85%) | |
|
2b Only vaccinated (Vaccine-B) |
18–40 | 31 (74%) | 163 (94%) |
| 40 + | 28 (77%) | 219 (87%) | |
|
2c Only vaccinated (Vaccine-C) |
18–40 | 1 (100%) | 0 |
| 40 + | 1 (100%) | 0 | |
|
2d Only vaccinated (Vaccine-D) |
18–40 | 0 | 2 (100%) |
| 40 + | 2 (100%) | 0 | |
|
3a (infected + vaccinated |
18–40 | 13 (100%) | 26 (96%) |
| 40 + | 23 (96%) | 39 (98%) | |
|
3b (infected + vaccinated) |
18–40 | 9 (90%) | 40 (95%) |
| 40 + | 14 (100%) | 67 (100%) | |
|
4 (not infected + not vaccinated) |
18–40 | 16 (84%) | 27 (66%) |
| 40 + | 5 (83%) | 24 (73%) | |
| 5a (random) | 18–40 | 29 (63%) | 234 (69%) |
| 40 + | 06 (46%) | 36 (50%) | |
| 5b (random) | 18–40 | 69 (40%) | 45 (51%) |
| 40 + | 38 (56%) | 42 (57%) |
Kit validation and comparative analysis
The performance validation of the “CoroSuchak” IgG antibody microwell ELISA detection kit was conducted at various sites using known positive and known negative serum/plasma samples (around 700 samples). The validation was carried out by both internal and external investigators, including Vanguard Diagnostics Private Limited, Delhi; Rajiv Gandhi Super Speciality Hospital, Delhi; Defence Institute of Physiology and Allied Sciences (DIPAS), Delhi; and ICMR/NARI, Pune (Table 9). Comparative analysis for the sensitivity and specificity of CoroSuchak was performed with two additional ICMR approved ELISA kits (kit-1 and kit-2), using clinically confirmed COVID-19-negative and COVID-19-positive serum/plasma samples (Table 10). The sensitivity of the kit was 98.7% and specificity 99.8%, with 24 months shelf life.
Table 9.
Validation of CoroSuchak at various test centers across India using clinically known positive and negative serum/plasma specimens
| Testing site | No. of samples | True positive | True negative | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Rajiv Gandhi Super Speciality Hospital, Delhi | 90 | 45 | 45 | 97.7% | 100% |
| DIPAS, Delhi | 71 | 20 | 51 | 100% | 100% |
| ICMR/NARI, Pune | 200 | 100 | 100 | 97% | 99% |
| Vanguard Diagnostics Private Limited, Delhi | 362 | 62 | 300 | 100% | 100% |
| Total | 723 | 227 | 496 | 98.7% | 99.8% |
Table 10.
Comparative analysis of CoroSuchak with other two ICMR approved COVID-19 ELISA kits
| Test parameters | CoroSuchak | Kit-1 | Kit-2 | |||
|---|---|---|---|---|---|---|
| Mfg. 01/2021 | Exp.-06/2022 | Mfg. 12/2020 | Exp. 11/2021 | Mfg. 07/2020 | Exp. 06/2021 | |
| Absorbance | Results | Absorbance | Results | Absorbance | Results | |
| Blank | 0.000 | - | - | - | - | - |
|
Negative control |
0.031 | Negative | 0.05 | Negative | 0.177 | Negative |
|
Negative control |
0.032 | Negative | 0.086 | Negative | 0.124 | Negative |
|
Negative control |
0.039 | Negative | 0.063 | Negative | 0.125 | Negative |
| Mean of NCx | 0.034 | - | 0.066 | - | 0.142 | - |
|
Positive control |
0.926 | Positive | 1.074 | Positive | 1.849 | Positive |
|
Positive control |
0.937 | Positive | 1.025 | Positive | 1.83 | Positive |
| Mean of PCx | 0.931 | - | 1.049 | - | 1.839 | - |
| Cut-off value | Mean of NCx + 0.200 | 0.234 | Mean of NCx + 0.200 | 0.266 | Mean of NCx + 0.200 | 0.342 |
| Known non-reactive samples/true negative samples | ||||||
| Sample ID No | Abs | Results | Abs | Results | Abs | Results |
| 9 | 0.049 | Negative | 0.08 | Negative | 0.153 | Negative |
| 12 | 0.042 | Negative | 0.051 | Negative | 0.075 | Negative |
| 14 | 0.050 | Negative | 0.047 | Negative | 0.094 | Negative |
| 15 | 0.052 | Negative | 0.081 | Negative | 0.193 | Negative |
| 17 | 0.050 | Negative | 0.062 | Negative | 0.095 | Negative |
| 19 | 0.049 | Negative | 0.036 | Negative | 0.100 | Negative |
| 21 | 0.042 | Negative | 0.046 | Negative | 0.105 | Negative |
| 24 | 0.038 | Negative | 0.063 | Negative | 0.094 | Negative |
| 25 | 0.039 | Negative | 0.056 | Negative | 0.116 | Negative |
| 27 | 0.052 | Negative | 0057 | Negative | 0.077 | Negative |
| 32 | 0.038 | Negative | 0.044 | Negative | 0.088 | Negative |
| 36 | 0.049 | Negative | 0.043 | Negative | 0.095 | Negative |
| 37 | 0.053 | Negative | 0.043 | Negative | 0.133 | Negative |
| 38 | 0.050 | Negative | 0.056 | Negative | 0.068 | Negative |
| 39 | 0.043 | Negative | 0.086 | Negative | 0.099 | Negative |
| 43 | 0.051 | Negative | 0.036 | Negative | 0.115 | Negative |
| 44 | 0.041 | Negative | 0.046 | Negative | 0.150 | Negative |
| 46 | 0.042 | Negative | 0.102 | Negative | 0.121 | Negative |
| 47 | 0.050 | Negative | 0.095 | Negative | 0.096 | Negative |
| 49 | 0.050 | Negative | 0.061 | Negative | 0.107 | Negative |
| Known reactive/true-positive samples | ||||||
| 16 | 0.384 | Positive | 2.155 | Positive | 1.208 | Positive |
| 18 | 0.532 | Positive | 2.363 | Positive | 3.356 | Positive |
| 20 | 0.309 | Positive | 1.124 | Positive | 1.919 | Positive |
| 22 | 0.429 | Positive | 0.705 | Positive | 3.322 | Positive |
| 272 | 0.478 | Positive | 0.715 | Positive | 3.271 | Positive |
| 30 | 0.501 | Positive | 0.356 | Positive | 2.904 | Positive |
| 31 | 0.492 | Positive | 0.85 | Positive | 2.05 | Positive |
| 83/II | 0.526 | Positive | 2.106 | Positive | 3.568 | Positive |
| 83/IV | 0.487 | Positive | 1.872 | Positive | 3.24 | Positive |
| 216 | 0.498 | Positive | 0.705 | Positive | 2.584 | Positive |
| 122 | 0.406 | Positive | 0.578 | Positive | 1.814 | Positive |
| 131 | 0.318 | Positive | 0.728 | Positive | 2.162 | Positive |
| 140/II | 0.394 | Positive | 1.453 | Positive | 2.797 | Positive |
| 140/III | 0.301 | Positive | 1.27 | Positive | 2.881 | Positive |
| 292/III | 0.392 | Positive | 2.249 | Positive | 3.653 | Positive |
| 292/IV | 0.365 | Positive | 2.316 | Positive | 3.641 | Positive |
| 314/III | 0.279 | Positive | 2.184 | Positive | 3.086 | Positive |
| 314/IV | 0.318 | Positive | 2.21 | Positive | 3.235 | Positive |
| 315/III | 0.406 | Positive | 1.23 | Positive | 3.097 | Positive |
| 375 | 0.462 | Positive | 2.08 | Positive | 3.252 | Positive |
Discussion
In the present challenging times, it is very important to know the number of people with antibodies present to indicate how many people are likely to have had the infection or have been vaccinated, to identify individuals who have had COVID-19 in the past, or who have developed antibodies after vaccination in both symptomatic and asymptomatic cases.
Antibody detection (serological assay) aids in monitoring and control of the disease in masses (Udugama et al. 2020; Cheng et al. 2020). In addition to molecular testing, serological assays for detecting antibodies against SARS-CoV-2 are getting prominent attention during the current pandemic time. Unlike molecular testing, detecting an individual’s immune response against a virus is a direct indicator of an infection or exposure to infection. Serological diagnosis is also becoming a valuable tool for determining the prevalence of COVID-19 in the population and identifying those who are immune and somewhat protected from going into the severity of the disease, especially crucial for patients with mild to severe sickness. The rise in the IgG antibody levels from the second week onwards after the symptoms develop makes it the most sensitive and early serological marker (Lou et al. 2020). So far, the US Food and Drug Administration (FDA) (www.fda.gov) has approved around 90 SARS-CoV-2 serological or antibody tests. As per the latest available reports, to date in India, 146 rapid antigen test kits (https://www.icmr.gov.in/pdf/-19/kits/List_of_rapid_antigen_kits_06012022.pdf), 207 RT-PCR kits, including 02 RT-PCR kits for detection of Omicron variant (https://www.icmr.gov.in/pdf/-19/kits/RT_PCR_Tests_Kits_Evaluation_Summ_06012022.pdf), and antibody detection kits have been approved by the ICMR, Government of India, for mass screening of the population. As vaccines are already in use, reproducible approaches to detect and quantify vaccine-mediated anti-SARS-CoV-2 antibodies are essential for assessing the extent and duration of immunogenicity and the efficacy of generating antibodies. The appropriate sensitivity and specificity levels for an antibody test are determined by the test’s purpose and must be considered.
There is lack of availability of indigenous, government-approved, and cost-effective kits. The present study describes the development of an ELISA kit for detecting anti-SARS-CoV-2 human IgG antibodies, the indigenously developed “CoroSuchak” kit. The kit has 98% sensitivity and 99% specificity and is tremendously useful, particularly in detecting immune-protected individuals who have been exposed previously without showing any symptoms. “CoroSuchak,” based on a cocktail of peptides and recombinant proteins from both N and S proteins, can be used to screen healthcare personnel, industry workers, and the general population. An antibody detection test is now regarded as a potential surrogate for immunity to reinfection (Adams et al. 2020). Confirmation of antibody status would thus alleviate anxiety, give authorities the confidence to relax social distancing measures, and guide policymakers in the gradual release of population lockdown, possibly in tandem with digital contact tracing (Ferretti et al. 2020). As a diagnostic tool, serology may be used in conjunction with RT-PCR testing to improve sensitivity, especially in instances that appear after the onset of symptoms (Zhao et al. 2020; Long et al. 2020).
Antibody responses may not consistently reach levels adequate to be detected by antibody tests in moderate and asymptomatic cases (Long et al. 2020). The magnitude and persistence of antibody responses in the context of illnesses ranging from asymptomatic to severe infections and across diverse populations, age, genetic backgrounds, and comorbidities are still being investigated. Antibodies generated against infection tend to decrease over time (Kellam and Barclay 2020; Lin et al. 2020), which may not always imply a loss of immunity. Despite reductions in titers, patients who recovered from Middle East respiratory syndrome (MERS-CoV) infections had measurable antibody titers for at least a year (Choe et al., 2017). However, research into the immunological response to SARS-CoV-2 has found that many asymptomatic cases roam around without getting detected as positive, which is a major threat to the spread of infection in the community during the convalescent phase of infection (Long et al. 2020). Such cases either do not get themselves tested, or antibody detection assays are not sensitive enough to catch the low antibody levels.
Detection of IgG antibodies against SARS-CoV-2 has a more prominent role to play during this pandemic. IgG is a longer-lasting antibody associated with viral neutralizing activity, essential for recovery from viral disease (Xiao et al. 2021; Casadevall and Pirofski 2020). Although positive IgM and IgG ELISA results have been reported as early as the fourth day following the onset of symptoms, larger levels are found in the second and third weeks of illness (Sethuraman et al. 2020). The majority of antibodies are often developed against the N protein. As a result, the most sensitive tests would be those that identify antibodies to N proteins. The RBD domain of S protein, on the other hand, is the host attachment protein. Hence, antibodies to RBD-S should be more specific and neutralizing. As a result, high sensitivity might be achieved by employing one or both antigens to detect IgG and IgM. Antibodies to SARS-CoV-2 and possibly other coronaviruses may, however, have cross-reactivity. Rapid point-of-care diagnostics for antibody detection have been widely developed and sold. However, their quality varies. Therefore, in “CoroSuchak,” we have used both N and S proteins to recognize a wide range of antibodies generated against SARS-CoV-2 and to avoid any cross-reactivity with other SARS family members.
In a very limited number of subjects, we observed 100% interaction between the vaccine type (vaccine-C and -D) and the generation of antibodies. The accuracy of IgG tests was 99% for 3–6 post-symptom onset. Thus, antibody tests provide a promise and a peril in the ongoing COVID-19 pandemic. Thus, understanding the severity and magnitude of immune responses following infections is critical for driving vaccination and pandemic planning activities (Collier et al. 2022). In animal trials, serum-neutralizing antibody titers best correlate with the immune protection against SARS-CoV-2 (Cromer et al. 2022; Lau et al. 2021; Khoury et al. 2021). We show that infected/vaccinated subjects are endowed with greater potency, breadth, and durability of antibodies relative to individuals who received two doses of COVID-19 vaccines only or those who were only infected by SARS-CoV-2 in the years 2020, 2021, and 2022. Serum antibodies in infected and vaccinated subjects were greater against the variants Alpha, Beta, Delta, and Omicron at all time points than those of only vaccinated individuals (after two doses) (Walls et al. 2022; Bates et al. 2022). Thus, it suggests that the kit “CoroSuchak” is sensitive and specific enough to indicate that vaccination enhances the immune response even to a variant to which the infected person had not been previously exposed.
A test report depends majorly on the sensitivity and specificity of the assay and its cut-off value. The absence of antibody results will occur if there are no antibodies or if antibody levels are too low to be detected in an assay system. Therefore, to obtain correct and true results, it is very important to develop a very robust assay method for detection. In the present study, amongst the total population analyzed, it was found that almost 75% of the adult community population showed the presence of antibodies to SARS-CoV-2, suggesting that either they were infected in the past or had been vaccinated. As our total data suggests, 3% (first wave, April–October 2020), 70% (second wave, April–August 2021), and 88% (third wave, December 2021–January 2022) of individuals showed the presence of SARS-CoV-2 antibodies.
The percentage of antibodies in adults continued to increase and remained high in those aged between 18 and 40 years (41.5%), whereas in those aged 40 and above, it was 33.3%. The presence of antibodies is directly proportional to the SARS-CoV-2 infection and vaccination but is not only responsible for protection against the disease. Having antibodies can help to prevent individuals from getting the infection, but it does not guarantee that an individual cannot be infected with COVID-19.
The marked enhancement of antibodies for vaccinated-only subjects after receiving two doses to levels comparable to infected/vaccinated cases suggests that the number of exposures and/or delay between exposures to SARS-CoV-2, whether through vaccination or infection, antibody responses as well as resilience to variants. It will be quite interesting to determine whether the number of exposures also correlates with the sustainability of antibody responses for vaccinated-only individuals as compared to infected/vaccinated subjects. However, evidence suggests that COVID-19 vaccination antibody responses prove to provide better neutralization of the virus or the variants than the natural exposure (Cavanaugh et al. 2021; León et al. 2022). Few epidemiologic studies support the benefit of vaccination for previously infected cases (Walls et al. 2022). Our findings suggest that COVID-19 disease and vaccination combinations elicit a magnitude of antibody response for better protection than only infected or vaccinated. According to a recent study, survivors of SARS-CoV-2 infection who received the COVID-19 vaccine had elevated/higher neutralizing antibody responses than those only exposed to the SARS-CoV-2 virus or vaccine (Tan et al. 2021). The findings strengthen and support the ongoing development of various sarbecovirus-specific vaccines (Martinez et al. 2021; Cohen et al. 2021; Walls et al. 2021) that elicit broader immunity and protection against SARS-CoV-2 variants and other zoonotic sarbecoviruses.
There is a difference between having immunity and the presence of antibodies. Modulation in the antibody tires occurs following infection or vaccination but still cannot be detected due to the low sensitivity of detection methods. But it certainly does not mean that a person has no protection against SARS-CoV-2 because only antibodies are not the deciding factors, T cells also play a major role in the generation of an immune response. An individual’s immune response is dominated by many factors, including health conditions and age. It is not yet clear or confirmed exactly how many antibodies are needed to rise to give protection. Therefore, data from different surveys are likely to differ depending on the number of individuals tested, method and time of collection, detection method protocols, tools for analyses, etc., and should not be used as a basis for the progress of the vaccination program. But at the same time, as the analysis develops at various places, the survey-based estimates will enable a possible future analysis of the population having received a vaccine with other characteristics collected in the survey.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all the volunteers for their voluntary participation and providing blood samples for this study. The authors also express their sincere thanks to Metabolome Hospital, Mumbai; Northern Command Hospital, Udhampur; 153 GH, Leh; and Rajiv Gandhi Super Speciality Hospital, Delhi, for performing COVID-19 antibody tests using CoroSuchak. AKS and AG would like to recognize the University Grants Commission (UGC) and Council of Scientific and Industrial Research (CSIR), respectively, for providing research fellowships.
Author contribution
LG, RS, RPT, VK designed the study. RCM, RV, LG monitor the progress. AKS, DC, LG wrote the manuscript. AG, RCM, RS, MRE, YS, RPT performed the assay development and evaluation experiments. RPT, VK, LG analyzed the experimental data. RV reviewed and approved the manuscript. AKS, DC and MRE performed experiments. DC, HG, K, SS, YS assisted in the collection of clinical samples. All authors approved the final manuscript.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during this study.
Declarations
Ethical statement
All procedures performed in this study were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki guidelines. The study was approved by the Ethical Committee of the hospital (IEC RGSSH/01/2020/03).
Consent to participate
A written informed consent was obtained from all the participants before beginning the study.
Conflict of interest
R P Tiwari and Veena Kohli are employed by Vanguard Diagnostics Private Limited, Delhi, India. All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams ER, Ainsworth M, Anand R, Andersson MI, Auckland K, Baillie JK, Barnes E, Beer S, Bell JI, Berry T, Bibi S, Carroll M, Chinnakannan SK, Clutterbuck E, Cornall RJ, Crook DW, de Silva T, Dejnirattisai W, Dingle KE, Dold C, Espinosa A, Eyre DW, Farmer H, Mendoza MF, Georgiou D, Hoosdally SJ, Hunter A, Jefferey K, Kelly DF, Klenerman P, Knight J, Knowles C, Kwok AJ, Leuschner U, Levin R, Liu C, Lopez-Camacho C, Martinez J, Matthews PC, McGivern H, Mentzer AJ, Milton J, Mongkolsapaya J, Moore SC, Oliveira MS, Pereira F, Perez E, Peto T, Ploeg RJ, Pollard A, Prince T, Roberts DJ, Rudkin JK, Sanchez V, Screaton GR, Semple MG, Slon-Campos J, Skelly DT, Smith EN, Sobrinodiaz A, Staves J, Stuart DI, Supasa P, Surik T, Thraves H, Tsang P, Turtle L, Walker AS, Wang B, Washington C, Watkins N, Whitehouse J, National COVID-19 Testing Scientific Advisory Panel Antibody testing for COVID-19: A report from the National -19 Scientific Advisory Panel [version 1; peer review: 2 approved] Wellcome Open Res. 2020;5:1–17. doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F, Stadlbauer D, Strohmeier S, Nguyen TH, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai SL, Wang JY, Zhou YQ, Yu DS, Gao XM, Li LL, Yang F. Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province. Zhonghua Yu Fang Yi Xue Za Zhi [Chin J Prev Med] 2020;54:E005–E005. doi: 10.3760/cma.j.issn.0253-9624.2020.0005. [DOI] [PubMed] [Google Scholar]
- Bates TA, McBride SK, Leier HC, Guzman G, Lyski ZL, Schoen D, Winders B, Lee JY, Lee DX, Messer WB, Curlin ME. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. 2022;7:eabn8014. doi: 10.1126/sciimmunol.abn8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh AM, Spicer KB, Thoroughman D, Glick C, Winter K. Reduced risk of reinfection with SARS-CoV-2 after COVID-19 vaccination—Kentucky, May–June 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1081–1083. doi: 10.15585/mmwr.mm7032e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, Libman M, Dittrich S, Yansouni CP. Diagnostic testing for severe acute respiratory syndrome–related coronavirus 2: a narrative review. Ann Intern Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe PG, Perera RAPM, Park WB, Song KH, Bang JH, Kim ES, Kim HB, Ko LWR, Park SW, Kim NJ, Lau EH, Poon LLM, Peiris M, Oh M. MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis. 2017;23:1079–1084. doi: 10.3201/eid2307.170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, Gnanapragasam PN, Lee YE, Hoffman PR, Ou S, Kakutani LM, Keeffe JR, Wu HJ, Howarth M, West AP, Barnes CO. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AR, Brown CM, McMahan KA, Yu J, Liu J, Jacob-Dolan C, Chandrashekar A, Tierney D, Ansel JL, Rowe M, Sellers D. Characterization of immune responses in fully vaccinated individuals after breakthrough infection with the SARS-CoV-2 delta variant. Sci Transl Med. 2022;14:1–7. doi: 10.1126/scitranslmed.abn6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer D, Steain M, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Kent SJ, Triccas JA, Khoury DS, Davenport MP. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. The Lancet Microbe. 2022;3:e52–e61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, Parker M, Bonsall D, Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368:1–7. doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Xu Y, Sun C, Wang X, Guo Y, Qiu S, Ma K. A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect. 2021;54:12–16. doi: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveri A, Smura T, Kuivanen S, Österlund P, Hepojoki J, Ikonen N, Pitkäpaasi M, Blomqvist S, Rönkkö E, Kantele A, Strandin T. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25:2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://cdsco.gov.in/opencms/opencms/system/modules/CDSCO.WEB/elements/download_file_division.jsp?num_id=ODA2OQ==
- https://www.fda.gov/medical-devices/coronavirus-COVID-19-and-medical-devices/COVID-19-tests-and-collection-kits-authorized-fda-infographic
- https://www.icmr.gov.in/pdf/-19/kits/List_of_rapid_antigen_kits_06012022.pdf
- https://www.icmr.gov.in/pdf/-19/kits/RT_PCR_Tests_Kits_Evaluation_Summ_06012022.pdf
- Huang AT, Garcia-Carreras B, Hitchings MD, Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon BD, Trimmer-Smith L, Etienne V. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Lau EH, Tsang OT, Hui DS, Kwan MY, Chan WH, Chiu SS, Ko RL, Chan KH, Cheng S, Perera RA, Cowling BJ. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12:1–7. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León TM, Dorabawila V, Nelson L, Lutterloh E, Bauer UE, Backenson B, Bassett MT, Henry H, Bregman B, Midgley CM, Myers JF. COVID-19 cases and hospitalizations by COVID-19 vaccination status and previous COVID-19 diagnosis—California and New York, May–November 2021. Morb Mortal Wkly Rep. 2022;71:125–131. doi: 10.15585/mmwr.mm7104e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zhu L, Ni Z, Meng H, You L. Duration of serum neutralizing antibodies for SARS-CoV-2: Lessons from SARS-CoV infection. J Microbiol Immunol Infect. 2020;53:821–822. doi: 10.1016/j.jmii.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, Yu F, Ge SX, Zou QD, Yuan Q, Lin S. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J. 2020;56:1–10. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DR, Schäfer A, Leist SR, De la Cruz G, West A, Atochina-Vasserman EN, Lindesmith LC, Pardi N, Parks R, Barr M, Li D. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann AJ, Cornaby C, Bartelt L, Weiss S, Park Y, Edwards CE. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Tan CW, Chia WN, Young BE, Zhu F, Lim BL, Sia WR, Thein TL, Chen MIC, Leo YS, Lye DC, Wang LF. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N Engl J Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol. 2020;58:e00797–e820. doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VY, Chen H, Mubareka S, Gubbay JB, Chan WC. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Viglione G. How many people has the coronavirus killed? Nature. 2020;585:22–24. doi: 10.1038/d41586-020-02497-w. [DOI] [PubMed] [Google Scholar]
- Walls AC, Miranda MC, Schäfer A, Pham MN, Greaney A, Arunachalam PS, Navarro MJ, Tortorici MA, Rogers K, O’Connor MA, Shirreff L. Elicitation of broadly protective Sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, Stewart C, Cameroni E, McCallum M, Goecker EA, Degli-Angeli EJ. SARS-CoV-2 breakthrough infections elicit potent, broad and durable neutralizing antibody responses. Cell. 2022;175:872–880. doi: 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020;38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020a) WHO Director-General’s opening remarks at the media briefing on COVID-19—11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-openingremarks-at-the-media-briefing-on-covid-19-11-march-2020
- World Health Organization (2020b) Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection: interim guidance, 10 January 2020. (No. WHO/2019-nCoV/laboratory/2020.1) World Health Organization
- Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, Yang X, Wang B, Chen L, Chen Q, Wu Y. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun. 2021;12:1–9. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Yang H, Liu B, Pang X, Du J, Liu M, Liu Y, Jing X, Chen J, Deng S, Zhou Z, Du J, Yin L, Yan Y, Mou H, She Q. Antibodies can last for more than one year after SARS-CoV-2 infection: a follow-up study from survivors of COVID-19. Front Med. 2021;8:1–10. doi: 10.3389/fmed.2021.684864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Xu S, Rong Z, Xu R, Liu X, Deng P, Liu H, Xu X. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020;94:133–138. doi: 10.1016/j.ijid.2020.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during this study.