Abstract
Atopic dermatitis (AD) is a chronic, itchy skin condition that affects 15–20% of children but may occur at any age. It is estimated that 16.5 million US adults (7.3%) have AD that initially began at age >2 years, with nearly 40% affected by moderate or severe disease. Therefore, a quantitative measurement that tracks the evolution of AD severity could be extremely useful in assessing patient evolution and therapeutic efficacy. Currently, SCOring Atopic Dermatitis (SCORAD) is the most frequently used measurement tool in clinical practice. However, SCORAD has the following disadvantages: (i) time consuming—calculating SCORAD usually takes about 7–10 minutes per patient, which poses a heavy burden on dermatologists and (ii) inconsistency—owing to the complexity of SCORAD calculation, even well-trained dermatologists could give different scores for the same case. In this study, we introduce the Automatic SCORAD, an automatic version of the SCORAD that deploys state-of-the-art convolutional neural networks that measure AD severity by analyzing skin lesion images. Overall, we have shown that Automatic SCORAD may prove to be a rapid and objective alternative method for the automatic assessment of AD, achieving results comparable with those of human expert assessment while reducing interobserver variability.
Abbreviations: AD, atopic dermatitis; AI, artificial intelligence; ASCORAD, Automatic SCOring Atopic Dermatitis; AUC, area under the curve; CADx, Computer-Aided Diagnosis; FAR, full agreement rate; IoU, intersection over union; PAR, partial agreement rate; RMAE, relative mean absolute error; RSD, relative SD; SCORAD, SCOring Atopic Dermatitis
Introduction
Atopic dermatitis (AD) is a multifaceted, chronic relapsing inflammatory skin disease that is commonly associated with other atopic manifestations such as allergic conjunctivitis, allergic rhinitis, and asthma (Berke et al., 2012; Bieber, 2008; Drucker et al., 2017). It is the most common skin disease in children, affecting approximately 15‒20% of children and 1‒3% of adults (Eichenfield et al., 2014; Nutten, 2015). The onset of the disease is most common by age 5 years, and early diagnosis and treatment are essential to avoid complications of AD and improve QOL (Eichenfield et al., 2014).
The European Task Force on Atopic Dermatitis developed the SCOring Atopic Dermatitis (SCORAD) index (Stalder et al., 1993) to create a consensus on assessment methods for AD. The SCORAD index consists of the interpretation of the extent of the disorder, that is, the intensity, composed of six visual items (erythema, edema/papules, effect of scratching, oozing/crust formation, lichenification, and dryness), and two subjective symptoms (itch and sleeplessness); the maximum score is 103 points. If the subjective symptoms (itch and sleeplessness) of the SCORAD are not assessed, the maximum score is 83 points, and it is known as the objective SCORAD. The SCORAD index is influenced by subjective ratings that may be affected by social and cultural factors, and therefore, the European Task Force on Atopic Dermatitis recommends the objective SCORAD. One of the advantages of using the objective SCORAD system is that it is based on a European consensus of experts on pediatric dermatology. The system is representative and well-evaluated (Schmitt et al., 2013) but shows, as with all other systems, intraobserver and interobserver disagreements. However, it is currently widely used in clinical practice to assess patient evolution and measure the effectiveness of treatments (Butler et al., 2020; Nahm et al., 2020; Panahi et al., 2012; Silverberg et al., 2020; Yoo et al., 2020).
Much work has been done in the development of a better scoring system to reach a more objective and quicker-to-fill method. Novel tools for patients, such as the patient-oriented validated scoring system patient-oriented-SCORAD (Stalder et al., 2011) detect changes in signs and symptoms without the intervention of doctors. Likewise, the Three Item Severity score is a simple method to determine the severity of AD, and takes about 43 seconds per patient. The Eczema Area and Severity Index (Hanifin et al., 2001) showed a good interobserver and intraobserver variability, but it is a complex and time-consuming index to fill. However, all these scoring systems still suffer from the same variability problem because they share similarities with SCORAD (Chopra et al., 2017).
In recent years, artificial intelligence (AI) has achieved human-expert-like performance in a wide variety of tasks such as skin cancer classification, detection, and lesion segmentation. Extensive work has been done in the detection of AD with different imaging methods, including multiphoton tomography (Guimarães et al., 2020), clinical image (Wu et al., 2020), and even electronic health records (Gustafson et al., 2017). Skin pathologies such as psoriasis have also attracted the attention of researchers for the same reasons as AD, because the main scoring system, PASI, is a time-consuming and highly subjective scoring method. Dash et al. (2019) proved that convolutional neural networks are able to segment psoriasis with high accuracy, sensitivity, and specificity, outperforming existing methods. Pal et al. (2016) showed the effectiveness of convolutional neural networks in visual sign classification, a key task to automatic severity grading. Dash et al. (2020) combined both segmentation and severity grading, creating a computer-aided diagnosis (CADx) system for psoriasis lesion grading.
Creating a more objective and practical scoring system for AD assessment is key to improving evidence-based dermatology. In this study, we introduce the Automatic SCORAD (ASCORAD), an automatic version of the SCORAD that provides a quick, accurate, and fully automated scoring method.
Results
Annotation
Firstly, we calculated the variability among the expert dermatologists across the three datasets. This provided a baseline that made possible the appraisal of the results of the Legit. Health-SCORADNet algorithm. We found out that the lesion segmentation annotation was very consistent across datasets, with an accuracy of 81.0‒91.3%, area under the curve (AUC) of 0.91, F1 of 0.86‒0.91, and relative SD (RSD) of 8.6‒9.1%. It can also be seen that Legit.Health-AD-FPK-IVI had the largest disagreement, if we look at the intersection over union (IoU) metric, with 0.80 against 0.86 and 0.91 on light-skin datasets. Note that the F1 score is also the lowest for the light-skin dataset. In regard to visual sign severity assessment, Legit.Health-AD had more disagreement among the specialists, but the other datasets had more positively skewed distributions, meaning that the majority of the intensity values were close to 0.
Lesion surface segmentation
We compared the difference at pixel-level because there was no physical reference on the images to obtain the real size of the lesions. As shown in Table 1, the annotations of the three datasets had an RSD close to 9%, Cohen’s kappa of 0.79, and AUC around 0.90. Despite the similarity in the results on the previously mentioned metrics, Legit.Health-AD-FPK-IVI seemed to have more discrepancies among the annotators because it showed the lowest IoU and F1 values, 0.80 and 0.86, respectively.
Table 1.
Annotator’s Performance in Lesion Surface Segmentation
| Datasets | ACC | AUC | IoU | F11 | RSD | Cohen’s Kappa |
|---|---|---|---|---|---|---|
| Legit.Health-AD | 86.9 | 0.91 | 0.91 | 0.88 | 8.6 | 0.78 |
| Legit.Health-AD-Test | 81.0 | 0.91 | 0.86 | 0.91 | 9.1 | 0.79 |
| Legit.Health-AD-FPK-IVI | 91.3 | 0.91 | 0.80 | 0.86 | 9.0 | 0.80 |
Abbreviations: ACC, accuracy; AUC, area under the curve; IoU, intersection over union; RSD, relative SD.
These results provide the background for comparing with the results of Legit.Health-SCORADNet.
F1 denotes F1 score.
Visual sign severity assessment
The results presented in Table 2, Table 3, Table 4 provide the baseline to appraise the results of Legit.Health-SCORADNet in the visual sign severity-assessment task. All the values are below random RSD and above random full agreement rate (FAR), partial agreement rate (PAR) 1, and PAR2 for all visual signs. Erythema was the visual sign that obtained the best Cohen’s kappa value in general, and lichenification (0.06) in Legit.Health-AD and excoriations (0.08) and dryness (0.09) in Legit.Health-AD-FPK-IVI had values very close to 0. The six visual signs constitute a maximum of 63 points of the SCORAD because the sum of the intensities was multiplied by (equation 2). Given the RSD results in terms of SCORAD points, the variability of Legit.Health-AD was around 11 points ( = 17%), and both Legit.Health-AD-Test and Legit.Health-AD-FPK-IVI had the same variability, on average, of 8 points ( = 12%).
Table 2.
Annotator’s Performance in Legit.Health-AD Visual Sign Severity Assessment
| Visual Signs | RSD | RMAE (Mean) | RMAE (Median) | FAR | PAR1 | PAR2 | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|
| Erythema | 11.5 | 10.7 | 8.3 | 33.1 | 92.0 | 94.5 | 0.34 |
| Edema | 16.2 | 14.7 | 11.9 | 21.3 | 74.1 | 84.9 | 0.15 |
| Oozing | 20.0 | 18.2 | 14.8 | 18.0 | 59.6 | 79.3 | 0.19 |
| Excoriations | 17.4 | 15.9 | 12.9 | 22.6 | 66.5 | 81.2 | 0.17 |
| Lichenification | 20.3 | 18.3 | 15.1 | 10.7 | 59.1 | 74.6 | 0.06 |
| Dryness | 18.7 | 16.9 | 12.8 | 20.0 | 69.3 | 82.3 | 0.14 |
| Average | 17.4 | 15.8 | 13.8 | 14.4 | 64.7 | 79.3 | 0.17 |
Abbreviations: FAR, full agreement rate; PAR, partial agreement rate; RMAE, relative mean absolute error; RSD, relative SD.
These results provide the baseline to appraise the results of Legit.Health-SCORADNet.
Table 3.
Annotator’s Performance in Legit.Health-AD-Test Visual Sign Severity Assessment
| Visual Signs | RSD | RMAE (Mean) | RMAE (Median) | FAR | PAR1 | PAR2 | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|
| Erythema | 12.1 | 11.2 | 8.8 | 34.0 | 88.0 | 91.5 | 0.35 |
| Edema | 7.9 | 7.3 | 5.6 | 55.8 | 93.1 | 96.7 | 0.22 |
| Oozing | 10.3 | 9.5 | 7.5 | 44.4 | 89.9 | 93.1 | 0.39 |
| Excoriations | 12.7 | 11.6 | 9.4 | 39.7 | 79.0 | 87.1 | 0.20 |
| Lichenification | 10.1 | 9.3 | 7.4 | 46.8 | 88.0 | 92.9 | 0.21 |
| Dryness | 16.5 | 14.9 | 12.2 | 20.4 | 72.4 | 80.3 | 0.19 |
| Average | 11.6 | 10.6 | 8.5 | 40.2 | 85.0 | 90.3 | 0.26 |
Abbreviations: FAR, full agreement rate; PAR, partial agreement rate; RMAE, relative mean absolute error; RSD, relative SD.
These results provide the baseline to appraise the results of Legit.Health-SCORADNet.
Table 4.
Annotator’s Performance in Legit.Health-AD-FPK-IVI Visual Sign Severity Assessment
| Visual Signs | RSD | RMAE (Mean) | RMAE (Median) | FAR | PAR1 | PAR2 | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|
| Erythema | 11.9 | 10.8 | 8.8 | 42.3 | 80.1 | 88.2 | 0.23 |
| Edema | 8.6 | 8.0 | 6.3 | 54.0 | 90.9 | 94.5 | 0.13 |
| Oozing | 12.7 | 11.6 | 9.4 | 35.1 | 81.9 | 87.3 | 0.27 |
| Excoriations | 9.7 | 9.0 | 7.0 | 45.0 | 92.7 | 95.5 | 0.08 |
| Lichenification | 13.3 | 12.2 | 9.7 | 27.9 | 85.5 | 90.9 | 0.27 |
| Dryness | 18.2 | 16.4 | 13.4 | 10.8 | 70.2 | 81.0 | 0.09 |
| Average | 12.4 | 11.3 | 9.1 | 35.9 | 86.6 | 89.6 | 0.18 |
Abbreviations: FAR, full agreement rate; PAR, partial agreement rate; RMAE, relative mean absolute error; RSD, relative SD.
These results provide the baseline to appraise the results of Legit.Health-SCORADNet.
Legit.Health-SCORADNet
Legit.Health-SCORADNet was validated through two experiments in which the network was trained on several data splits because we applied a k-fold cross-validation technique: 6-fold for the first experiment and 3-fold for the second experiment. All the results presented in Table 5, Table 6, Table 7, Table 8, Table 9 were obtained by averaging the results of the network’s performance on the different data splits and were measured using the same metrics as the annotation, with the purpose of making a direct comparison of both.
Table 5.
Legit.Health-SCORADNet’s Results in Light Skin Lesion Surface Segmentation
| Clinical Sign | ACC, % (95% CI) | AUC (95% CI) | IoU (95% CI) | F11 (95% CI) |
|---|---|---|---|---|
| Lesion surface | 84.6 (80.9‒88.3) | 0.93 (0.90‒0.96) | 0.64 (0.59‒0.69) | 0.75 (0.71‒0.79) |
Abbreviations: ACC, accuracy; AUC, area under the curve; CI, confidence interval; IoU, intersection over union.
F1 denotes F1 score.
Table 6.
Legit.Health-SCORADNet’s Results in Dark Skin Lesion Surface Segmentation
| XXX | Experiment 1 |
Experiment 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| ACC, % (95% CI) | AUC (95% CI) | IoU (95% CI) | F11 (95% CI) | ACC, % (95% CI) | AUC (95% CI) | IoU (95% CI) | F11 (95% CI) | |
| Lesion surface | 74.0 | 0.83 | 0.32 | 0.42 | 79.2 | 0.87 | 0.45 | 0.55 |
| (65.9‒82.1) | (0.76‒0.90) | (0.23‒0.41) | (0.33‒0.51) | (66.3‒92.1) | (0.76‒0.98) | (0.29‒0.61) | (0.39‒0.71) | |
Abbreviations: ACC, accuracy; AUC, area under the curve; CI, confidence interval; IoU, intersection over union.
Results are divided by experiment. The algorithm in experiment 1 was trained solely on light-skinned patient images, and the algorithm in experiment 2 was trained on mixed data containing 8% of dark-skinned patient images.
F1 denotes F1 score.
Table 7.
Legit.Health-SCORADNet’s Results in Visual Sign Severity Assessment
| Range | Training GT | Legit.Health-AD-Test |
Legit.Health-AD-FPK-IVI |
||
|---|---|---|---|---|---|
| RMAE 11.(95% CI) | RMAE 22 (95% CI) | RMAE 1 (95% CI) | RMAE 2 (95% CI) | ||
| 0‒3 | Median | 13.6 (9.7‒17.5) | 14.3 (10.4‒18.2) | 21.2 (17.3‒25.0) | 20.8 (16.9‒24.7) |
| 0‒10 | Median | 14.3 (10.4‒18.2) | 13.2 (9.3‒17.0) | 22.8 (18.9‒26.7) | 20.0 (16.0‒23.9) |
| 0‒100 | Median | 14.4 (10.5‒18.3) | 13.0 (9.1‒16.9) | 22.6 (18.7‒26.5) | 19.8 (15.9‒23.7) |
| 0‒100 | Mean | 13.5 (9.6‒17.4) | 13.4 (9.5‒17.3) | 21.1 (17.2‒25.0) | 19.9 (16.0‒23.8) |
Abbreviations: CI, confidence interval; DEX, Deep EXpectation; RMAE, relative mean absolute error.
The models were trained on Legit.Health-AD using a different range and ground truth method and tested on Legit.Health-AD-Test and Legit.Health-AD-FPK-IVI.
RMAE 1 is obtained by applying the argmax function to the prediction.
RMAE 2 is obtained by applying the DEX method to the prediction.
Table 8.
Legit.Health-SCORADNet’s Results in Light-Skin Visual Sign Severity Assessment
| Visual Signs | RMAE 11 (95% CI) | RMAE 22 (95% CI) |
|---|---|---|
| Erythema | 14.1 (10.2‒18.0) | 13.3 (9.4‒17.2) |
| Edema | 16.1 (12.2‒20.0) | 16.0 (12.1‒19.9) |
| Oozing | 22.3 (18.4‒26.2) | 19.4 (15.5‒23.3) |
| Excoriations | 11.5 (7.6‒15.4) | 9.6 (5.7‒15.4) |
| Lichenification | 10.3 (6.4‒14.2) | 8.7 (4.8‒12.6) |
| Dryness | 12.4 (8.5‒16.3) | 11.3 (7.4‒15.2) |
| Average | 14.4 (10.5‒18.3) | 13.0 (9.1‒16.9) |
Abbreviations: CI, confidence interval; DEX, Deep EXpectation; RMAE, relative mean absolute error.
RMAE 1 is obtained by applying the argmax function to the prediction.
RMAE 2 is obtained by applying the DEX method to the prediction.
Table 9.
Legit.Health-SCORADNet’s Results in Dark Skin Visual Sign Severity Assessment
| Visual Signs | Experiment 1 |
Experiment 2 |
||
|---|---|---|---|---|
| RMAE 11 (95% CI) | RMAE 22 (95% CI) | RMAE 1 (95% CI) | RMAE 2 (95% CI) | |
| Erythema | 17.8 (13.9‒21.7) | 15.7 (11.8‒19.6) | 16.2 (12.2‒20.2) | 14.3 (10.3‒18.3) |
| Edema | 16.8 (12.9‒20.7) | 18.6 (14.7‒22.5) | 18.1 (14.1‒22.0) | 15.4 (11.4‒19.4) |
| Oozing | 24.9 (21.0‒28.8) | 22.7 (18.8‒26.6) | 9.3 (5.3‒13.3) | 9.0 (5.0‒13.0) |
| Excoriations | 10.1 (6.2‒14.0) | 9.6 (5.7‒13.5) | 10.2 (6.2‒14.2) | 8.0 (4.0‒12.0) |
| Lichenification | 25.9 (22.0‒29.8) | 20.6 (16.7‒24.5) | 24.0 (20.0‒28.0) | 19.8 (15.8‒23.8) |
| Dryness | 39.9 (36.0‒43.8) | 31.7 (27.8‒35.6) | 26.0 (22.0‒30.0) | 19.3 (15.3‒23.3) |
| Average | 22.6 (18.7‒26.5) | 19.8 (15.9‒23.7) | 17.3 (13.3‒21.3) | 14.3 (10.3‒18.3) |
Abbreviations: CI, confidence interval; DEX, Deep EXpectation; RMAE, relative mean absolute error.
Results are divided by experiment. The algorithm in experiment 1 was trained solely on light-skinned patient images, and the algorithm in experiment 2 was trained on mixed data containing 8% of dark-skinned patient images.
RMAE 1 is obtained by applying the argmax function to the prediction.
RMAE 2 is obtained by applying the DEX method to the prediction.
Legit.Health-SCORADNet showed a good performance at visual sign severity assessment, obtaining a relative mean absolute error (RMAE) of 13.0% and AUC of 0.93 at surface estimation. The total execution time of Legit.Health-SCORADNet for a single image was 0.34 seconds, running on an Intel Xeon Platinum 8260 CPU at 2.40 GHz (Intel, Santa Clara, CA).
Lesion surface segmentation
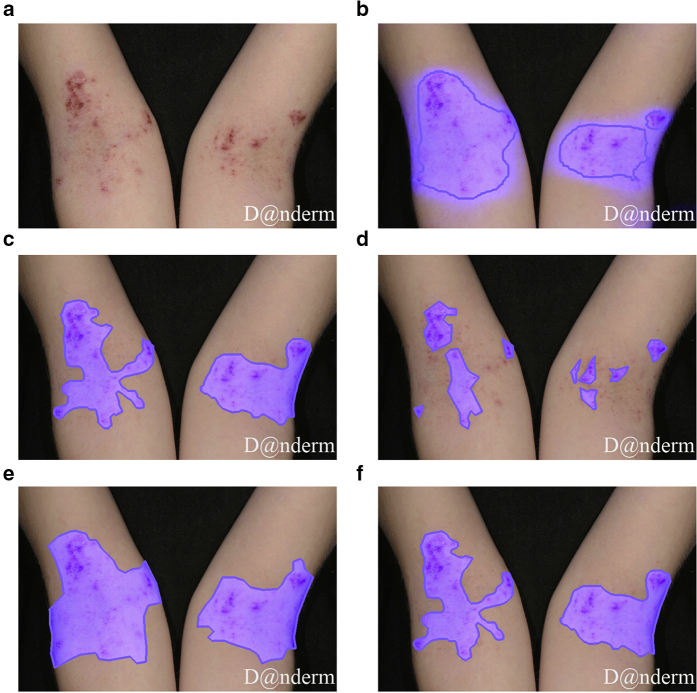
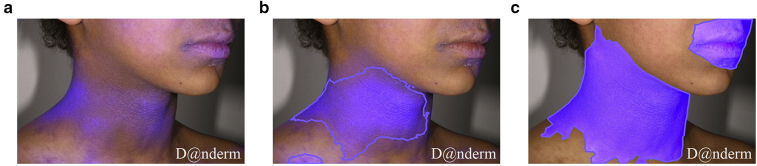
Legit.Health-SCORADNet’s lesion surface segmentation results are presented in Tables 5 and 6. The AUC, IoU, and F1 for light skin were 0.93, 0.64, and 0.75, respectively, whereas the results on those metrics were 0.83, 0.32, and 0.42, respectively, for dark skin. However, when training in a small subset of dark skin images (experiment 2), the results significantly improved (0.41 for IoU and 0.33 for F1), as shown in Table 6. Figures 1 and 2 show the ground truth and the prediction for a sample case of Legit.Health-AD-Test and Legit.Health-AD-FPK-IVI, respectively.
Figure 1.
Lesion surface segmentation masks. (a) Original image. (b) Legit.Health-SCORADNet’s prediction. (c) Ground truth. (d) Mask drawn by the first specialist. (e) Mask drawn by the second specialist. (f) Mask drawn by the third specialist. Legit.Health-AD-Test sample image gathered from Danderm Dermatology Atlas with the owner's permission.
Figure 2.
Results of experiments 1 and 2 models on a dark skin image. (a) The predicted surface mask of the model trained on light skin. (b) The predicted surface mask of the model trained on both light and dark skin. (c) The ground truth mask. Legit.Health-AD-FPK-IVI sample image gathered from Danderm Dermatology Atlas with the owner's permission.
Visual sign severity assessment
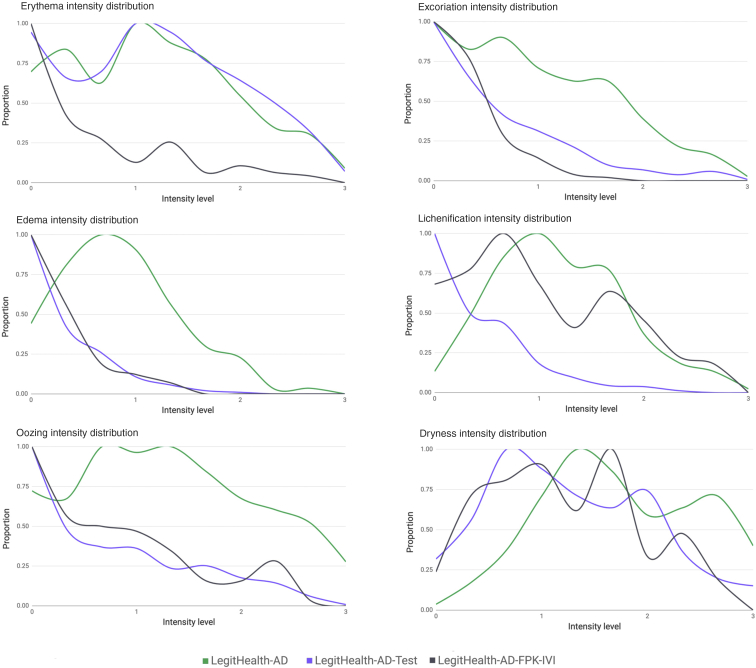
On average, we achieved the best performance when we trained the network with the ground truth that resulted from applying the median and normalizing the outcome into the 0‒100 range (Table 7). Using that configuration, we ran experiments 1 and 2, and we got an RMAE of 13.0% in Legit.Health-AD-Test, which had an interobserver RMAE of 10.6%, having trained Legit.Health-SCORADNet on a dataset with 15.8% RMAE (Table 8). The RMAE on Legit.Health-AD-FPK-IVI was slightly higher: 14.3% (Table 9) when including dark skin images in the training set, and 19.8%, without including dark skin images. The visual sign with the worst performance on light skin was oozing (19.4%), followed by edema (16.0%). Lichenification (19.8%) and dryness (19.3%) were the most difficult visual signs for the algorithm to correctly predict on dark skin, with edema (15.4%) also having a value above the average. Interestingly, oozing got a much lower RMAE on Legit.Health-AD-FPK-IVI, whereas both test datasets had the same oozing intensity distribution. The distribution of predicted intensity values was plotted next to the ground truth distributions (Figure 3) to show that Legit.Health-SCORADNet was able to predict values in the whole range and not only the mean of the distribution.
Figure 3.
Legit.Health-AD-Test visual sign intensity distribution of ground truth labels and predictions. The horizontal axis is in the range 0‒100 because the results are given using the best performing model, which was trained with ground truth labels in that range.
Discussion
ASCORAD shows promise as an automatic scoring system that might enable a more objective and quicker evaluation. Indeed, a deep learning algorithm could simplify the assessment of AD, a very common skin disease that affects 15‒20% of children (Asher et al., 2006) and 1‒3% of adults worldwide. Scoring systems such as SCORAD and Eczema Area and Severity Index have high interobserver variability and are time-consuming. An AI-automated approach may help to reduce such bias and therefore be a more precise and objective criterion for evaluation in pharmaceutical studies and routine clinical practice.
Our results show that deep learning may be noticed as a fast and objective alternative method for the automatic assessment of AD with great potential, already achieving results comparable with those of human expert assessment, while reducing interobserver variability and being more time-efficient. ASCORAD could also be used in situations where face-to-face consultations are not possible, providing an automatic assessment of clinical signs and lesion surface. It could also be a potential tool to reduce the time and effort of training clinical assessors for clinical trials and in clinical practice.
However, additional validation studies are needed in real-world settings and with diverse populations to ensure generalizability. Despite that the dataset used in this study captures the variability of a wide range of parameters, the algorithm should be tested on other datasets to prove its robustness and generalizability, in particular to dark skin tones. In the future, we intend to test ASCORAD in validation studies in which the objective part of the SCORAD will be assessed in person by the dermatologist. Comparing the result of the algorithm with those of face-to-face assessment is crucial because some visual signs such as edema, dryness, or oozing might present more difficulties in estimating the severity by image than in person. Furthermore, the AI Marker will be used in this study, helping the CADx system to correctly calculate the surface by converting lesion pixels into a metric unit of measurement.
To put our results into clinical context, the annotated lesion area was compared with the algorithm-predicted area. Because some photographs do not show the complete lesion area, live assessment method cannot be directly compared with the photograph assessment method. However, image-based area assessment by an expert and predicted area have the same basis for their analysis and are therefore directly comparable. Legit.Health-SCORADNet resulted in a good overall RMAE of 13.0% and an excellent AUC of 0.93 and IoU of 0.75 for lesion surface estimation on light skin.
Legit.Health-AD-Test and Legit.Health-AD-FPK-IVI datasets have strong positively skewed distributions for all the visual signs, which means that the most frequent intensities are 0 and 1. It seems that a vast majority of images are of mild AD or that the observers had a strong bias toward low-intensity values. If the majority of the visual signs are close to zero intensity, it is possible that the RSD reflected lower disagreement (9% vs. 17% in Legit.Health-AD). In fact, Oranje et al.(2007) found an RSD of 20%, which was very close to the interobserver variability found in Legit.Health-AD.
Looking at Cohen’s kappa values, it seems that some of the visual signs such as lichenification in Legit.Health-AD and excoriations and dryness in Legit.Health-AD-FPK-IVI have a null interobserver agreement. However, Cohen's kappa is a statistical measure for nominal classification problems, and metrics such as RSD, RMAE, FAR, PAR1, and PAR2 show that the annotation of the specialists is far from random. For example, the visual sign excoriations in Legit.Health-AD-FPK-IVI obtains a Cohen’s kappa value of 0.09 and PAR2 of 95.5%, far from the random value (62%).
In short, we have proved that a convolutional neural network trained with the observer’s average results can achieve an RMAE similar to that of one of the experts. Furthermore, our automatic method outputs a value in the range 0‒100 for each visual sign instead of the range 0‒3 as the usual SCORAD, broadening the spectrum of possible outputs and turning the discrete problem into more continuous.
We believe that our algorithm has the potential to reduce costs in dermatology by saving time while improving the documentation process of the evolution of the disease. This could be interesting for the application in pharmaceutical clinical trials, as well as in clinical practice.
Materials and Methods
Datasets and annotations
In this retrospective, noninterventional study, three new annotated datasets were constructed to train and validate the performance of the lesion surface segmentation and visual sign severity assessment algorithms. The first two datasets comprise solely light-skinned patients (Fitzpatrick I‒III) because it proved to be easier to gather datasets of such characteristics, whereas the third consists of images of IV‒VI skin types according to the Fitzpatrick scale. Demographic characteristics of each dataset are gathered in Table 10. Clinical images were collected from online public sources, and patient consent and ethics committee approval were not necessary. Published images belong to Danderm Dermatology Atlas, and the author gave his consent for publication.
Table 10.
Demographic Characteristics
| Datasets | Age Groups (%) |
Sex (%) |
Skin Type (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| <18 | 18‒29 | 30‒39 | 40‒49 | 50‒64 | >65 | Male | Female | Light | Dark | |
| Legit.Health-AD | 31 | 23 | 26 | 14 | 4 | 2 | 39 | 61 | 100 | 0 |
| Legit.Health-AD-Test | — | — | — | — | — | — | — | — | 100 | 0 |
| Legit.Health-AD-FPK-IVI | — | — | — | — | — | — | — | — | 0 | 100 |
Legit.Health-AD dataset
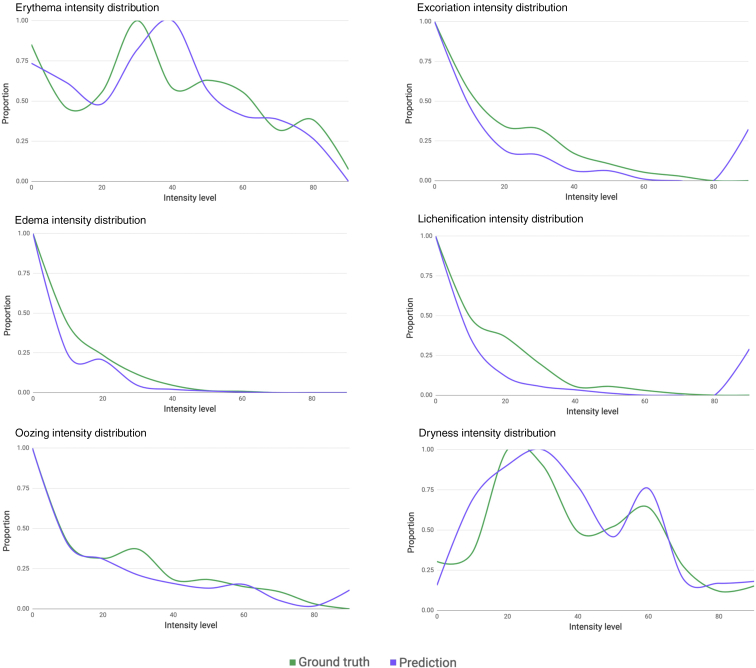
Legit.Health-AD is a dataset collected from online dermatological atlases that consist of 604 images that belong to light-skinned patients, of which one third are children (Table 10), suffering from AD, with lesions present on different body parts. The dataset contains the following percentage of body zones: head (22%), trunk (11%), arms (23%), hands (9%), legs (16%), feet (8%), genitalia (3%), full body (1%), and skin close-up (7%). The dataset contains a substantial variety of clinical images taken from different angles, distances, light conditions, body parts, and disease severity. Figure 4 depicts the normalized intensity distribution by visual sign. The images have a minimum size of 260 × 256 pixels, an average size of 667 × 563 pixels, and a maximum size of 1,772 × 1,304 pixels.
Figure 4.
Comparison of the intensity level distribution by a visual sign of the datasets used in the study.
Legit.Health-AD-Test dataset
A second dataset, Legit.Health-AD-Test, was built for testing purposes. The dataset was gathered from several dermatological atlases publicly available and contains a total number of 367 images that belong exclusively to light-skinned patients. The dataset is only characterized by skin type (Table 10), and basic demographic information such as age and sex is missing because the original sources do not provide that information. The images were downloaded one by one, and each of them was reviewed by a physician to approve the inclusion of the image in the dataset. Duplicates or very similar images were removed, and no other data sampling technique was applied. Similar to Legit.Health-AD, the dataset contains images of children and adults with great variability in angles, distances, light conditions, body parts, and disease severity. The dataset contains the following percentage of body zones: head (35%), trunk (20%), arms (18%), hands (7%), legs (13%), feet (2%), genitalia (2%), and skin close-up (3%). The visual sign intensity distribution of this dataset is different from that of Legit.Health-AD, having more cases of zero intensity for most of the visual signs (Figure 4). The images have a minimum size of 313 × 210 pixels, an average size of 574 × 537 pixels, and a maximum size of 2,848 × 3,252 pixels.
Legit.Health-AD-FPK-IVI dataset
Legit.Health-AD-FPK-IVI is a dataset collected from online dermatological atlases that contain photos of children and adult patients with Fitzpatrick IV‒VI skin types suffering from AD. The same manual procedure as that of Legit.Health-AD-Test was applied to gather the dataset, and basic demographic information such as age and sex is also missing (Table 10). It is composed of 112 images with a minimum size of 200 × 204 pixels, an average size of 766 × 695 pixels, and a maximum size of 3,024 × 4,032 pixels. The dataset contains the following percentage of body zones: head (41%), trunk (10%), arms (17%), hands (8%), legs (13%), feet (3%), and skin close-up (8%). The goal of including this dataset in the study was to gather preliminary evidence of the efficiency of the algorithms in dark skin.
Ground truth labels
The corresponding ground truths of each dataset were prepared by nine experts, three for each dataset, who treat patients with AD in their daily practice, to reduce variability by combining their results. The experts annotated the images without more context than the images. They had to draw a mask over the lesion and choose a score from 0 to 3 for each visual sign that comprise the SCORAD.
We obtained the ground truth labels for lesion segmentation and visual sign intensity classification by averaging the masks of the three annotators and by averaging the intensity levels. We chose the mean over the median because it is the statistical measure that gets the best results for generating ground truth labels from multiannotator ordinal data (Lakshminarayanan and Teh, 20131).
Data preprocessing
Images were resized to 512 × 512, and pixel values scaled between 0 and 1. In addition, images in which the disease was too small in the picture were cropped, focusing on the disease. Ground truth labels were obtained from averaging the results as explained in the previous section. However, we ran some additional experiments using an alternative ground truth only for the training set, consisting of the median visual intensity, instead of the mean. As a result of applying the mean and median, discrete visual sign intensity levels yielded real numbers, which had to be rounded to return to the discrete range 0‒3. To prevent information loss, we considered rescaling the values to 0‒10 and 0‒100 before rounding and compared these ranges with the original one.
With regard to lesion surface masks, the average mask was computed, resulting in a grayscale image in the range 0‒255. A pixel intensity threshold of 155 was applied to obtain a binary mask that was used as the ground truth. Images were finally normalized to the range 0‒1.
Deep learning model
The ASCORAD calculation can be divided into two parts: lesion surface segmentation and visual sign severity assessment. We trained two separated models, one for each task, and named Legit.Health-SCORADNet to the neural networks involved in the calculation of the ASCORAD (source code is available at github.com/Legit-Health/ASCORAD).
Lesion surface segmentation
For the lesion surface segmentation problem, we applied a U-Net, an architecture that was first designed for biomedical image segmentation and showed great results on the task of cell tracking (Ronneberger et al., 20152). The main contribution of this architecture was the ability to achieve good results even with hundreds of examples. The U-Net consists of two paths: a contracting path and an expanding path. The contracting path is a typical convolutional network where convolution and pooling operations are repeatedly applied. We decided to use the Resnet-34 (He et al., 20153) architecture, which is the typical backbone used in the contracting path.
Visual sign severity assessment
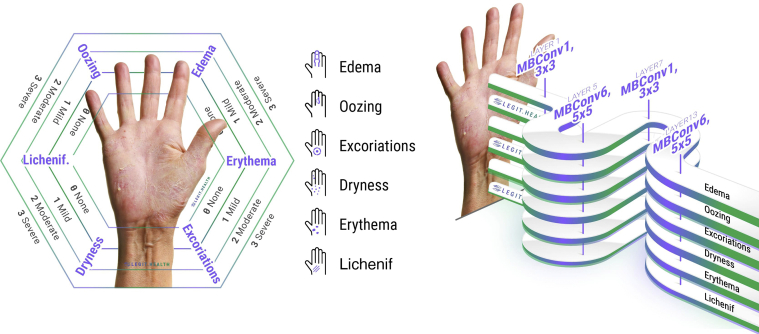
We trained a multioutput (Xu et al., 2020) classifier, with one softmax layer per visual sign (Figure 5). We used the EfficientNet-B0 network architecture (Tan and Le, 20194) that was pre-trained on approximately 1.28 million images (1,000 object categories) from the 2014 ImageNet Large Scale Visual Recognition Challenge (Russakovsky et al., 20145) and trained it on our dataset using transfer learning (Pan and Yang, 2010). EfficientNets achieve better accuracy and efficiency than previous convolutional neural networks with fewer parameters by applying a new scaling method that uniformly scales all dimensions of depth/width/resolution using a simple yet highly effective compound coefficient. There are eight versions, consisting of a different number of parameters, with the B0 being the smallest network that achieves state-of-the-art 77.1% top-1 accuracy on ImageNet for a network consisting of 5 million parameters.
Figure 5.
The visual signs that compose the SCORAD. Each visual sign can be classified into four intensity levels: none (0), mild (1), moderate (2), and severe (3). The multioutput EfficientNet-B0 network trained for visual sign intensity estimation has one head for each visual sign. Lichenf., lichenification; SCORAD, SCOring Atopic Dermatitis.
Visual sign severity grading can be seen as a piecewise regression or alternatively as a discrete classification with four discrete value labels for each visual sign intensity. In the case of multiple visual signs, a multilabel classification network can be used to solve the problem. However, to exploit methods such as Deep EXpectation (Rothe et al., 2015), one softmax layer per visual sign is needed. So, for the purpose of applying the Deep EXpectation method, we constructed a multioutput classifier with six softmax layers consisting of N neurons each, with N being 4, 11, or 101, depending on the range of the ground truth labels.
Deep EXpectation method proved to obtain better results on regression metrics by approaching a regression problem the same way that you would approach a classification problem, and therefore applying a softmax expected value:
| (1) |
where is the -dimensional output layer of each visual sign, representing softmax output probabilities , and are the discrete intensity levels corresponding to each class .
Evaluation metrics
Dermatologists may have a bias, a fixed effect where one observer consistently measures high or low. There may also be a random effect or heterogeneity, where the observer scores higher than others for some patients and lower for others. To measure interobserver variability, understand annotation quality in more detail, and compare it with the performance of the algorithms, we calculated the following set of metrics.
First of all, we computed the RSD and Cohen’s kappa for all the visual signs and lesion surface segmentation. In the case of the annotation of visual sign intensity, we also measured the times that the three observers gave the same result or the FAR. To complement FAR, two more metrics were calculated: the times that at least two observers gave the same result, whereas the third observer gave a result that deviated ±1 from the other observer’s or the PAR1. The same metrics without the ±1 condition for the third observer were called PAR2. Therefore, the metrics are ordered as follows in regard to their restrictiveness: FAR > PAR1 > PAR2. To assess the quality of the annotations and understand the results in more depth, we compared the results with an algorithm that randomly picked three intensity values for each visual sign. We ran this millions of times and found that RSD of a random visual sign evaluation tends to 27%, FAR tends to 6%, PAR1 tends to 34%, and PAR2 tends to 62%.
We also calculated the metrics that allowed a direct comparison of the Legit.Health-SCORADNet and the annotation, for both lesion segmentation and visual sign severity assessment. Pixel accuracy, AUC, IoU, and F1 score metrics were the preferred metrics for segmentation, whereas for the severity assessment of visual signs, we used RMAE.
Experimental setup
We ran two main experiments for each task: one with images containing only light skin and another adding a small number of dark skin images in the training set.
In the first experiment, we used Legit.Health-AD for training and Legit.Health-AD-Test and Legit.Health-AD-FPK-IVI for testing. We followed a six-fold cross-validation strategy to train the models. The models trained on the different folds were tested on both test sets, and the results were averaged over the folds to reduce the variance and bias.
The second experiment was built to better understand the performance of the network on dark skin when including a tiny fraction of dark-skinned patient images in the training set. In this experiment, we used Legit.Health-AD, Legit.Health-AD-Test, and a subset of Legit.Health-AD-FPK-IVI for training and the rest for testing. The training and test subsets of Legit.Health-AD-FPK-IVI were obtained with a three-fold cross-validation strategy. This means that the training set was composed of 971 light-skinned patient images, Legit.Health-AD and Legit.Health-AD-Test combined, and 75 dark-skinned patient images, which is a tiny fraction of the total images (8%). The dark skin test set was composed of the remaining 37 images. This split was done three times (three-fold), including different images in the training and test set, to obtain more reliable results.
In the case of visual sign severity assessment, we also ran experiments to find the optimal range, testing 0‒3, 0‒10, and 0‒100 ranges. In addition, we tested the mean and the median as the statistical measure for obtaining the ground truth of the training set. This project was entirely run on a single NVIDIA Tesla V100 (32 gigabytes) graphics processing unit (Nvidia, Santa Clara, CA).
CADx system
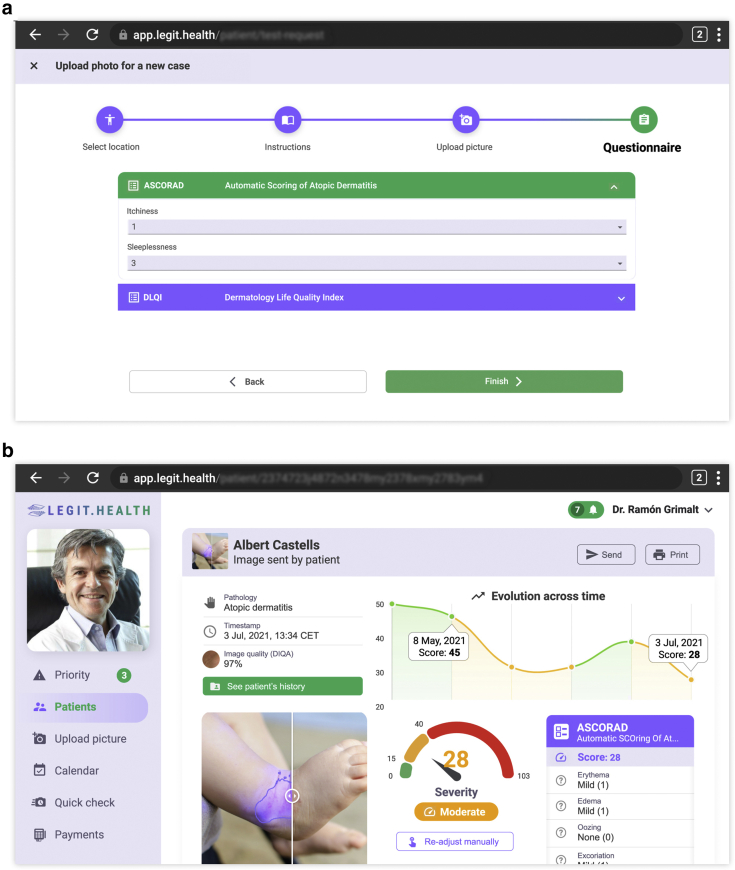
With the objective of making the algorithms accessible to the healthcare professional, we created a fully integrated CADx system, a web application that integrates the Legit.Health-SCORADNet algorithm and calculates the patient-based ASCORAD using clinical images. The CADx system includes three stages: uploading the images of the affected areas, processing the images, and reporting the ASCORAD.
In the first stage, images of affected areas are uploaded to the system using a simple user interface, depicted in Figure 6a. The user has to choose the body zone from the options defined in the original SCORAD (Stalder et al., 1993): head and neck, right upper limbs, left upper limbs, right lower limbs, left lower limbs, anterior trunk, back, and genitals. In some cases, such as children aged <2 years with all bodies affected, a full-body photograph can also be uploaded. In addition, the patient answers a simple questionnaire of two items: itchiness (0‒10) and sleeplessness (0‒10).
Figure 6.
CADx system. (a) Illustration of the questionnaire. (b) Illustration of the report generated by the CADx system. The report contains the evolution across the time of the ASCORAD, the last reported ASCORAD item by item, a picture of the lesion surface predicted by the algorithm, the final ASCORAD score with its translation to a category, and some additional information such as image quality. The example record shown is fictional. ASCORAD, Automatic SCOring Atopic Dermatitis; CADx, computer-aided diagnosis; CET, Central European Time; DIQA, Dermatology Image Quality Assessment; DLQI, Dermatology Life Quality Index; Jul, July.
In the second stage, the Legit.Health-SCORADNet algorithm processes the images and automatically calculates the severity of AD by calculating the intensity of each visual sign and the surface of the lesion. Finally, the output of the algorithm is shown in a user-friendly report containing an image with the estimated lesion surface and a chart with the evolution of the ASCORAD over time. The final report of the proposed CADx system is depicted in Figure 6b.
Computing the ASCORAD requires calculating the proportion of skin covered by the lesion. We solved this by including a small piece of hardware called AI Marker, a sticker with several shapes and colors that helps to translate pixels into a metric unit of measurement. The AI Marker should be kept close to the lesion, and it is automatically detected. In addition, the body surface area is calculated with the patient's height and weight using the Mosteller (Lee et al., 2008; Orimadegun and Omisanjo, 2014) formula. Once the surface of the lesion and body surface area are estimated, the percentage can be calculated by dividing the surface of the lesion by the body surface area (equation 2). This allows the CADx system to calculate the final value of ASCORAD. When the AI Marker is not used, lesion surface percentage is input by the user manually, although the CADx system is still capable of calculating the visual sign intensity values automatically.
When more than one image is uploaded, the surface of the images is summed, and the maximum (Dirschka et al., 2017) of each visual sign intensity is used for the ASCORAD calculation. Therefore, the final formula for N images of the whole body can be written as follows:
| (2) |
where stands for the lesion surface in a metric unit of measurement, stands for visual sign intensity, C stands for the sum of the symptoms input by the patient.
Software and statistical analysis
The models were implemented and trained using Pytorch (Paszke et al., 2019); Metrics and k-fold were calculated in Python using the SciKit-Learn package (Pedregosa et al., 20126) and plotted using MatPlotLib (Hunter, 2017).
Data availability statement
The images of Legit.Health-AD, Legit.Health-AD-Test, and Legit.Health-AD-FPK-IVI datasets related to this article can be found at http://www.atlasdermatologico.com.br/, hosted at Dermatology Atlas; http://www.danderm-pdv.is.kkh.dk/, hosted at Danderm; https://www.dermatlas.net/, hosted at Interactive Dermatology Atlas; https://www.dermis.net/dermisroot/en/home/index.htm, hosted at DermIS (Diepgen and Eysenbach, 1998); https://dermnetnz.org/, hosted at DermNet NZ; and http://www.hellenicdermatlas.com/en/, hosted at Hellenic Dermatological Atlas.
ORCIDs
Alfonso Medela: http://orcid.org/0000-0001-5859-5439
Taig Mac Carthy: http://orcid.org/0000-0001-5583-5273
S. Andy Aguilar Robles: http://orcid.org/0000-0003-0618-6179
Carlos M. Chiesa-Estomba: http://orcid.org/0000-0001-9454-9464
Ramon Grimalt: http://orcid.org/0000-0001-7204-8626
Author Contributions
Conceptualization: AM, RG; Data Curation: AM; Formal Analysis: AM, CMCE; Investigation: AM, TMC, SAAR, CMCE, RG; Methodology: AM, TMC, RG; Project Administration: SAAR; Visualization: TMC; Writing - Original Draft Preparation: AM; Writing - Review and Editing: AM, TMC, SAAR, CMCE, RG
Conflict of Interest
The authors state no conflict of interest.
Acknowledgments
The authors thank Fernando Alfageme Roldán for technical advice, BioCruces Bizkaia Health Research Institute for the academic support, and IBM for providing the computing infrastructure for the deep learning experiments.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2022;X:100107
Lakshminarayanan B, Teh YW. Inferring ground truth from multi-annotator ordinal data: a probabilistic approach. arXiv 2013.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. arXiv 2015.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. arXiv 2015.
Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. arXiv 2019.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. arXiv 2014.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. arXiv 2012.
References
- Asher M.I., Montefort S., Björkstén B., Lai C.K., Strachan D.P., Weiland S.K., et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional survey [published correction appears in Lancet 2007;370:1128] Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- Berke R., Singh A., Guralnick M. Atopic dermatitis: an overview. Am Fam Physician. 2012;86:35–42. [PubMed] [Google Scholar]
- Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- Butler É., Lundqvist C., Axelsson J. Lactobacillus reuteri DSM 17938 as a novel topical cosmetic ingredient: a proof of concept clinical study in adults with atopic dermatitis. Microorganisms. 2020;8:1026. doi: 10.3390/microorganisms8071026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R., Vakharia P.P., Sacotte R., Patel N., Immaneni S., White T., et al. Relationship between EASI and SCORAD severity assessments for atopic dermatitis. J Allergy Clin Immunol. 2017;140:1708–1710.e1. doi: 10.1016/j.jaci.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash M., Londhe N.D., Ghosh S., Raj R., Sonawane R.S. A cascaded deep convolution neural network based CADX system for psoriasis lesion segmentation and severity assessment. Appl Soft Comput. 2020;91:106240. [Google Scholar]
- Dash M., Londhe N.D., Ghosh S., Semwal A., Sonawane R.S. PsLSNet: automated psoriasis skin lesion segmentation using modified U-net-based fully convolutional network. Biomed Signal Process Control. 2019;52:226–237. [Google Scholar]
- Diepgen T.L., Eysenbach G. Digital images in dermatology and the dermatology online atlas on the World Wide Web. J Dermatol. 1998;25:782–787. doi: 10.1111/j.1346-8138.1998.tb02505.x. [DOI] [PubMed] [Google Scholar]
- Dirschka T., Pellacani G., Micali G., Malvehy J., Stratigos A.J., Casari A., et al. A proposed scoring system for assessing the severity of actinic keratosis on the head: actinic keratosis area and severity index. J Eur Acad Dermatol Venereol. 2017;31:1295–1302. doi: 10.1111/jdv.14267. [DOI] [PubMed] [Google Scholar]
- Drucker A.M., Wang A.R., Li W.Q., Sevetson E., Block J.K., Qureshi A.A. The burden of atopic dermatitis: summary of a report for the National Eczema Association. J Invest Dermatol. 2017;137:26–30. doi: 10.1016/j.jid.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Eichenfield L., Tom W., Chamlin S., Feldman S., Hanifin J., Simpson E., et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. J Am Acad Dermatol. 2014;70:338–351. doi: 10.1016/j.jaad.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães P., Batista A., Zieger M., Kaatz M., Koenig K. Artificial intelligence in multiphoton tomography: atopic dermatitis diagnosis. Sci Rep. 2020;10:7968. doi: 10.1038/s41598-020-64937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson E., Pacheco J., Wehbe F., Silverberg J., Thompson W. A machine learning algorithm for identifying atopic dermatitis in adults from electronic health records. IEEE Int Conf Healthc Inform. 2017;2017:83–90. doi: 10.1109/ICHI.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanifin J.M., Thurston M., Omoto M., Cherill R., Tofte S.J., Graeber M. The eczema area and severity index (EASI): assessment of reliability in atopic dermatitis. EASI Evaluator Group. Exp Dermatol. 2001;10:11–18. doi: 10.1034/j.1600-0625.2001.100102.x. [DOI] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- Lee J.Y., Choi J.W., Kim H. Determination of body surface area and formulas to estimate body surface area using the alginate method. J Physiol Anthropol. 2008;27:71–82. doi: 10.2114/jpa2.27.71. [DOI] [PubMed] [Google Scholar]
- Nahm D.H., Ye Y.M., Shin Y.S., Park H.S., Kim M.E., Kwon B., et al. Efficacy, safety, and immunomodulatory effect of the intramuscular administration of autologous total immunoglobulin G for atopic dermatitis: a randomized clinical trial. Allergy Asthma Immunol Res. 2020;12:949–963. doi: 10.4168/aair.2020.12.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. 2015;66(Suppl 1):8–16. doi: 10.1159/000370220. [DOI] [PubMed] [Google Scholar]
- Oranje A.P., Glazenburg E.J., Wolkerstorfer A., De Waard-van der Spek F.B. Practical issues on interpretation of scoring atopic dermatitis: the SCORAD index, objective SCORAD and the three-item severity score. Br J Dermatol. 2007;157:645–648. doi: 10.1111/j.1365-2133.2007.08112.x. [DOI] [PubMed] [Google Scholar]
- Orimadegun A., Omisanjo A. Evaluation of five formulae for estimating body surface area of Nigerian children. Ann Med Health Sci Res. 2014;4:889–898. doi: 10.4103/2141-9248.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Chaturvedi A, Garain U, Chandra A, Chatterjee R. Severity grading of psoriatic plaques using deep CNN based multi-task learning. Paper presented at: 23rd International Conference on Pattern Recognition (ICPR). 4‒8 December 2016; Cancun, Mexico.
- Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–1359. [Google Scholar]
- Panahi Y., Davoudi S.M., Sahebkar A., Beiraghdar F., Dadjo Y., Feizi I., et al. Efficacy of aloe vera/olive oil cream versus betamethasone cream for chronic skin lesions following sulfur mustard exposure: a randomized double-blind clinical trial. Cutan Ocul Toxicol. 2012;31:95–103. doi: 10.3109/15569527.2011.614669. [DOI] [PubMed] [Google Scholar]
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: an imperative style, high-performance deep learning library. Paper presented at: 33rd Conference on Neural Information Processing Systems (NeurIPS 2019). 8–14 December 2019; Vancouver, Canada.
- Rothe R, Timofte R, Gool L. DEX: Deep EXpectation of apparent age from a single image. Paper presented at: 2015 IEEE International Conference on Computability Vision Work (ICCVW). 7–13 December 2015; Santiago, Chile.
- Schmitt J., Langan S., Deckert S., Svensson A., von Kobyletzki L., Thomas K., et al. Assessment of clinical signs of atopic dermatitis: a systematic review and recommendation. J Allergy Clin Immunol. 2013;132:1337–1347. doi: 10.1016/j.jaci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Silverberg J.I., Simpson E.L., Thyssen J.P., Gooderham M., Chan G., Feeney C., et al. Efficacy and safety of abrocitinib in patients with moderate-to-severe atopic dermatitis: a randomized clinical trial. JAMA Dermatol. 2020;156:863–873. doi: 10.1001/jamadermatol.2020.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J.F., Barbarot S., Wollenberg A., Holm E.A., De Raeve L., Seidenari S., et al. Patient oriented SCORAD (PO-SCORAD): a new self assessment scale in atopic dermatitis, validated in Europe. Allergy. 2011;66:1114–1121. doi: 10.1111/j.1398-9995.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- Stalder J.F., Taieb A., Atherton D.J., Bieber P., Bonifazi E., Broberg A., et al. Severity scoring of atopic dermatitis: the SCORAD index. Consensus report of the European Task Force on Atopic Dermatitis. Dermatology. 1993;186:23–31. doi: 10.1159/000247298. [DOI] [PubMed] [Google Scholar]
- Wu H., Yin H., Chen H., Sun M., Liu X., Yu Y., et al. A deep learning, image based approach for automated diagnosis for inflammatory skin diseases. Ann Transl Med. 2020;8:581. doi: 10.21037/atm.2020.04.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Shi Y., Tsang I.W., Ong Y.S., Gong C., Shen X. Survey on multi-output learning. IEEE Trans Neural Netw Learn Syst. 2020;31:2409–2429. doi: 10.1109/TNNLS.2019.2945133. [DOI] [PubMed] [Google Scholar]
- Yoo J., Choi J.Y., Lee B.Y., Shin C.H., Shin J.W., Huh C.H., et al. Therapeutic effects of saline groundwater solution baths on atopic dermatitis: a pilot study. Evid Based Complement Alternat Med. 2020;2020 doi: 10.1155/2020/8303716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The images of Legit.Health-AD, Legit.Health-AD-Test, and Legit.Health-AD-FPK-IVI datasets related to this article can be found at http://www.atlasdermatologico.com.br/, hosted at Dermatology Atlas; http://www.danderm-pdv.is.kkh.dk/, hosted at Danderm; https://www.dermatlas.net/, hosted at Interactive Dermatology Atlas; https://www.dermis.net/dermisroot/en/home/index.htm, hosted at DermIS (Diepgen and Eysenbach, 1998); https://dermnetnz.org/, hosted at DermNet NZ; and http://www.hellenicdermatlas.com/en/, hosted at Hellenic Dermatological Atlas.