Abstract
Analysis of the nucleotide sequence of an Escherichia coli colicin S4 determinant revealed 76% identity to the pore-forming domain of the colicin A protein, 77% identity to the colicin A immunity protein, and 82% identity to the colicin A lysis protein. The N-terminal region, which is responsible for the Tol-dependent uptake of colicin S4, has 94% identity to the N-terminal region of colicin K. By contrast, the predicted receptor binding domain shows no sequence similarities to other colicins. Mutants that lacked the OmpW protein were resistant to colicin S4.
Colicins are plasmid-encoded proteins that are synthesized by Escherichia coli and kill sensitive strains of E. coli and closely related species (3, 5, 10, 11). The narrow host range is determined by a highly specific uptake into sensitive cells. Colicins use proteins of the outer membrane to bind to the E. coli cell. These proteins include porins and receptors for vitamin B12, siderophores, and nucleosides (3, 5, 10, 11). Genetic studies have shown that, in addition to the receptor proteins, two different translocation systems are required for colicin import (6, 7). Group B colicins use the Ton system, which consists of the proteins TonB, ExbB, and ExbD (3), and group A colicins utilize the Tol system, which consists of the proteins TolA, TolB, TolQ, and TolR (21). Colicin S4 belongs to the group A colicins (7). However, Ferber et al. (8) described E. coli Φ mutants supposedly mutated in the tonB gene that were insensitive to colicin S4. A dependence on both import systems, Tol and Ton, has only been described, by us, for a colicin U mutant (17). For this reason and since the receptor protein of colicin S4 is unknown, we characterized the colicin S4 genes and the colicin S4 import proteins.
Sequencing of the colicin S4 genes.
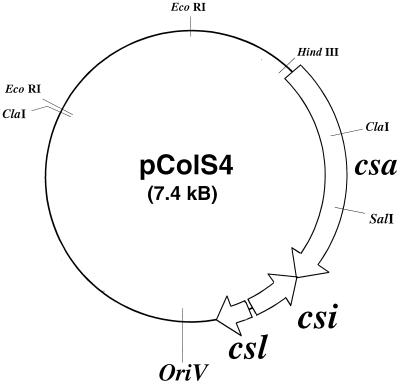
The colicin S4 plasmid pColS4 (Fig. 1) coding for the colicin S4 genes was isolated from strain E. coli K-12, which was originally obtained from a patient with an uncharacterized infection. E. coli K-12 was identified as a colicin S4-producing strain by showing cross-immunity to the colicin S4-producing reference strain Shigella dispar P15. The colicin S4 determinant was excised from plasmid pColS4 (Fig. 1) with EcoRI, and the 5.2-kb restriction fragment was cloned into the EcoRI site of pBCSK+. E. coli 5K transformed with the resulting plasmid pHP189 released colicin S4 and was immune to crude cell extracts obtained from E. coli K-12 (pColS4). Both strands of a 2,809-bp fragment were completely sequenced and revealed three open reading frames, which we designated csa (colicin S4 activity), csi (colicin S4 immunity), and csl (colicin S4 lysis). csa and csl have the same transcription polarity, and csi has the opposite polarity, an arrangement typical for genes of pore-forming colicins. csa, csi, and csl encode open reading frames of 499 (Mr = 54,085), 179 (Mr = 20,527), and 51 (Mr = 4,911) amino acid residues, respectively. The electrophoretic mobility of colicin S4 corresponded to an Mr of 52,000.
FIG. 1.
Arrangement of the csa, csi, and csl genes on the natural plasmid pColS4. The EcoRI restriction sites were used for cloning of the colicin S4 genes.
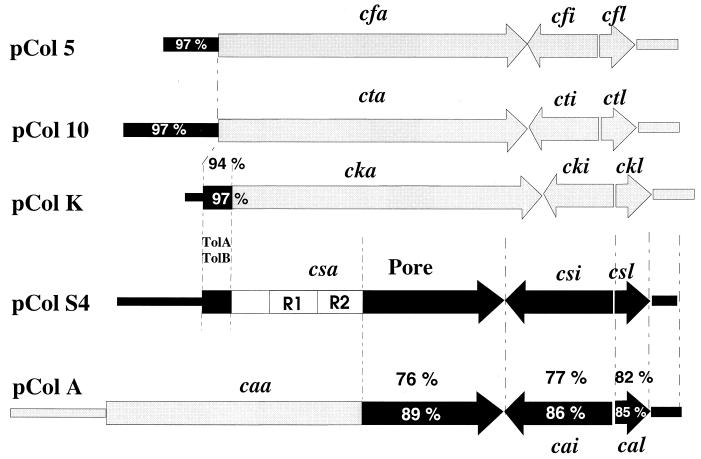
The nucleotide sequences flanking the genes of the colicin S4 operon exhibited high similarity to the sequences of other colicin determinants. The promoter region of csa is 97% identical to the corresponding region of colicin 10 (Fig. 2). The nucleotide sequences upstream of the colicin 5 and K genes display the same high degree of identity (Fig. 2). The sequence of the colicin S4 genes shows a mosaic-like structure (Fig. 2) like that of other colicins (15, 16, 20). Colicin S4 consists of three domains: an N-terminal domain, which is responsible for translocation through the outer membrane; a central domain, which binds to the receptor in the outer membrane; and a C-terminal domain, which contains the lethal activity. csa and the colicin K gene cka display 97% sequence identity in the 5′ region; the N-terminal 48 amino acids of colicin S4 and colicin K are 94% identical (Fig. 2). Since the N-terminal domain of the colicins is responsible for interaction with the proteins of the translocation systems (2), the N-terminal domains of colicins S4 and K could be responsible for interaction of these colicins with the Tol system proteins.
FIG. 2.
Comparison (percent sequence identity) of the activity, immunity, and lysis genes and the encoded proteins to the corresponding genes (within the bars) and proteins (above the bars) of colicin S4.
The region of the hydrophobic pore-forming domain of colicin S4 is nearly identical to that of colicin A (Fig. 3); in colicin A, it consists of a central hydrophobic hairpin (helices 8 and 9) surrounded by eight amphipathic helices (14). Our previous assignment of the immunity-specifying region to the hydrophobic hairpin (18) is supported by the complete cross-resistance to colicin S4 and colicin A of cells producing one of the two colicins (data not shown). The cross-immunity is also reflected by the high degree of sequence identity (77%) between the Csi and Cai immunity proteins, which stands in contrast to the low sequence similarities between the immunity proteins of other colicins of the colicin A family (18).
FIG. 3.
Sequence comparison of the C-terminal 200 amino acids of colicins S4 and A. Asterisks denote identical residues; dashes indicate similar residues. The hydrophobic segment is boxed, and the amino acids of the tip of the hydrophobic hairpin are indicated in boldface type.
The central part of colicin S4 shows no sequence similarities to any known proteins. This may reflect the unique receptor specificity of colicin S4, since in all colicins the receptor binding domain has been assigned to the central domain. Colicin S4 contains a pair of identical sequences in this central domain that might have evolved by duplication of a gene fragment (Fig. 4). The duplicated sequence may imply that colicin S4 binds to two copies of the receptor protein to enter cells. Colicin S4 represents another example that supports our previous proposal that colicins evolved by mixing DNA fragments that encode functional domains (15, 16, 20).
FIG. 4.
Sequence duplication in the putative receptor binding domain of colicin S4. Asterisks denote identical residues; dashes indicate similar residues.
Identification of the receptor protein of colicin S4.
To identify the colicin S4 receptor, colicin S4-resistant mutants of E. coli 5K were isolated from colonies in zones of growth inhibition on plates onto which a crude extract of colicin S4 had been applied. Mutations in the tol genes were excluded by testing the sensitivity of the colicin S4-insensitive mutants to colicins A and K. Ten colicin S4-insensitive mutants (HP151 to HP160) were highly sensitive to colicins A and K. To identify the receptor protein of colicin S4 in the outer membrane, mutant HP151 was transformed with an E. coli gene bank (2- to 6-kb fragments of the E. coli chromosome ligated into pACYC184; kindly provided by Silke Patzer of this institute). One out of 800 transformants contained a plasmid (pHP205) that restored colicin S4 sensitivity to E. coli HP151. The 3.6-kb fragment of pHP205 comprises yciB to trpB at 28.5 min of the E. coli chromosome. No functions have been assigned to the open reading frames of this region, except to trpA. By differential solubilization, two-dimensional gel electrophoresis, tryptic digestion, mass spectrometry of the isolated peptides, and comparison of the nucleotide sequence with the E. coli genome sequence, a protein was assigned to the gene product of the yciD gene and named OmpW (13) on the basis of its sequence similarity to the OmpW protein of Vibrio cholerae (9). The subcloned yciD gene (located on a 1-kb SspI fragment; pHP206) conferred colicin S4 sensitivity to all resistant mutants (HP151 to HP160), indicating that OmpW is the receptor or an essential part of the receptor. All E. coli strains tested with defined mutations in genes coding for outer membrane proteins were fully sensitive to colicin S4, including the BL21 omp8 mutant, whose genes (ompF, ompC, lamB, and ompA) encoding major outer membrane proteins are deleted (19).
The previously observed TonB dependence of colicin S4 uptake (8) probably resulted from a deletion that included tonB and yciD. Larger deletions in this region of the chromosome are frequently observed (4). E. coli H2300, with a deletion extending from trpB to tonB (8a), was insensitive to colicin S4. E. coli H2300 transformed with pHP206 was sensitive to colicin S4, which indicates that yciD alone complements the colicin S4-resistant phenotype, in contrast to a cloned tonB gene, which did not restore the colicin S4 sensitivity of E. coli H2300. These data demonstrate that uptake of colicin S4 into sensitive cells is not tonB dependent but requires the yciD gene product.
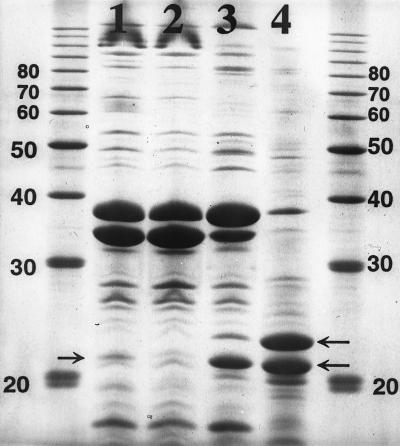
Comparison of outer membranes of the colicin S4-resistant E. coli HP151 with that of the colicin S4-sensitive E. coli 5K revealed that only a weak band present in the outer membrane fraction of E. coli 5K was missing in the outer membrane fraction of E. coli HP151 (Fig. 5, compare lanes 1 and 2). To increase the amounts of the OmpW protein, yciD was cloned downstream of the phage T7 promoter and transcribed by the T7 RNA polymerase (22). OmpW from the induced recombinant strain formed two strong bands; the band with the lower mobility was probably the OmpW precursor with the signal sequence (Fig. 5, lane 4). After induction of the RNA polymerase, more OmpW was synthesized, and synthesis of the OmpF and OmpC porins and the OmpA protein was strongly suppressed (Fig. 5, compare lanes 3 and 4). Like other outer membrane proteins, OmpW was heat modifiable, as revealed by the electrophoretic mobility of samples prepared at 37 and 95°C (data not shown).
FIG. 5.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of outer membranes of E. coli 5K (lane 1), HP151 (lane 2), BL21(pHP211) uninduced (lane 3), and BL21(pHP211) induced (lane 4). The arrows indicate the 21-kDa OmpW protein and its presumed 24-kDa precursor.
Colicin S4 binding to OmpW was assayed by mixing 1 ml of cell suspension (2 × 109 cells per ml) with 1 ml of colicin S4 solution (103 dilution titer). The mixture was incubated for 20 min at 37°C and centrifuged, and the amount of unbound colicin in the supernatant was determined. OmpW had to be overproduced to bind 90% of colicin S4 to cells of E. coli BL21(pHP211).
Sequence similarities of E. coli OmpW to other proteins.
OmpW of E. coli shows significant sequence similarities to OmpW of V. cholerae, a protein that is highly immunogenic (12). The biological function of the OmpW protein of V. cholerae is unknown. Like E. coli OmpW, V. cholerae OmpW is produced in minor amounts under laboratory conditions (12). Recently it has been shown that synthesis of Omp21 from Comamonas acidovorans, which is 30% identical to E. coli OmpW, is enhanced by oxygen depletion (1).
Thorne and Corwin (23) localized a gene locus between the trp genes and tonB of E. coli that is involved in the uptake of aromatic amino acids. Using indole acrylic acid, they isolated an E. coli mutant with a 60 to 80% decrease in tryptophan uptake. Therefore, we tested whether OmpW serves as a pore for tryptophan uptake across the outer membrane. The colicin S4-resistant E. coli mutant HP151 showed no reduction in the uptake of 3H-labeled tryptophan (data not shown), which suggests that the gene locus identified by Thorne and Corwin (23) is not yciD. Since OmpW belongs to a new family of outer membrane proteins with unknown functions (1), it will be interesting to investigate the biological function of OmpW for the E. coli cell, apart from its being the colicin S4 receptor protein.
Nucleotide sequence accession number.
The EMBL GenBank accession number for the colicin S4 sequence is Y18684.
Acknowledgments
We thank Karen A. Brune for critical reading of the manuscript.
We thank the Deutsche Forschungsgemeinschaft for financial support (SFB 323, project B1).
REFERENCES
- 1.Baldermann C, Lupas A, Lubieniecki J, Engelhardt H. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J Bacteriol. 1998;180:3741–3749. doi: 10.1128/jb.180.15.3741-3749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouveret E, Rigal A, Lazdunski C, Benedetti H. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol Microbiol. 1998;27:143–157. doi: 10.1046/j.1365-2958.1998.00667.x. [DOI] [PubMed] [Google Scholar]
- 3.Braun V, Pilsl H, Gross P. Colicins: structures, modes of action, transfer through membranes, and evolution. Arch Microbiol. 1994;161:199–206. doi: 10.1007/BF00248693. [DOI] [PubMed] [Google Scholar]
- 4.Conkell M B, Yanofsky C. Influence of chromosome structure on the frequency of tonB trp deletions in Escherichia coli. J Bacteriol. 1971;105:864–872. doi: 10.1128/jb.105.3.864-872.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Structure-function of the channel-forming colicins. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 6.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. J Bacteriol. 1975;123:96–101. doi: 10.1128/jb.123.1.96-101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferber D M, Fowler J M, Brubaker R R. Mutations to tolerance and resistance to pesticin and colicins in Escherichia coli φ. J Bacteriol. 1981;146:506–511. doi: 10.1128/jb.146.2.506-511.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Günther, K. Unpublished results.
- 9.Jalajakumari M B, Manning P A. Nucleotide sequence of the gene, ompW, encoding a 22 kDa immunogenic outer membrane protein of Vibrio cholerae. Nucleic Acids Res. 1990;18:25. doi: 10.1093/nar/18.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James R, Kleanthous C, Moore G R. The biology of E colicins: paradigms and paradoxes. Microbiology. 1996;142:1569–1580. doi: 10.1099/13500872-142-7-1569. [DOI] [PubMed] [Google Scholar]
- 11.Lazdunski C J, Bouveret E, Rigal A, Journet L, Lloubes R, Benedetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning P A, Bartowsky E J, Leavesly D I, Hackett J A, Heuzenroeder M W. Molecular cloning using immune sera of a 22-kDal outer membrane protein of Vibrio cholerae. Gene. 1985;34:95–103. doi: 10.1016/0378-1119(85)90299-9. [DOI] [PubMed] [Google Scholar]
- 13.Molloy M P, Herbert B R, Walsh B J, Tyler M I, Traini M, Sanchez J-C, Hochstrasser D F, Williams K L, Gooley A A. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- 14.Parker M W, Postma J P M, Pattus F, Tucker A D, Tsernoglou D. Refined structure of the pore-forming domain of colicin A at 2.4Å resolution. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 15.Pilsl H, Braun V. Novel colicin 10: assignment of four domains to the TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol Microbiol. 1995;16:57–67. doi: 10.1111/j.1365-2958.1995.tb02391.x. [DOI] [PubMed] [Google Scholar]
- 16.Pilsl H, Braun V. Strong function-related homology between the pore-forming colicins K and 5. J Bacteriol. 1995;177:6973–6977. doi: 10.1128/jb.177.23.6973-6977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilsl H, Braun V. The Ton system can functionally replace the TolB protein in the uptake of a mutated colicin U. FEMS Microbiol Lett. 1998;164:363–367. doi: 10.1111/j.1574-6968.1998.tb13110.x. [DOI] [PubMed] [Google Scholar]
- 18.Pilsl H, Šmajs D, Braun V. The tip of the hydrophobic hairpin of colicin U is dispensable for colicin U activity but is important for interaction with the immunity protein. J Bacteriol. 1998;180:4111–4115. doi: 10.1128/jb.180.16.4111-4115.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prilipov A, Phale P S, Van Gelder P, Rosenbusch J P, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 20.Roos U, Harkness R E, Braun V. Assembly of colicin genes from a few DNA fragments. Nucleotide sequence of colicin D. Mol Microbiol. 1989;3:891–902. doi: 10.1111/j.1365-2958.1989.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 21.Sun T P, Webster R E. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J Bacteriol. 1987;169:2667–2674. doi: 10.1128/jb.169.6.2667-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorne G M, Corwin L M. Mutations affecting aromatic amino acid transport in Escherichia coli and Salmonella typhimurium. J Gen Microbiol. 1975;90:203–216. doi: 10.1099/00221287-90-2-203. [DOI] [PubMed] [Google Scholar]