Abstract
The glutamyl-tRNA synthetase (gltX) gene from Pseudomonas aeruginosa was identified. A plasmid containing a 2.3-kb insert complemented the temperature-sensitive gltX mutation of Escherichia coli JP1449, and GltX activity was demonstrated. The inferred amino acid sequence of this gene showed 50.6% identity with GltX from Rhizobium meliloti.
The nature of the genetic code is defined by the combined action of the 20 aminoacyl-tRNA synthetases of the cell. The specific interaction of these enzymes with their cognate tRNA and amino acid substrates is essential for proper translation of the nucleic acid sequence to proteins. Currently, several hundred tRNA synthetase sequences have been reported from archaebacteria, eubacteria, mitochondria, chloroplasts, and eukaryotic cells. Initial sequence alignments revealed that these enzymes may be divided into two distinct structural classes (8), each containing 10 enzymes. The class I enzymes are distinguishable by the presence of the amino-terminal amino acid motifs HIGH and KMSKS (23). The crystal structures of the aminoacyl-tRNA synthetases for class I synthetases, such as glutamine from Escherichia coli (17) and glutamate from Thermus thermophilus (15), revealed that these sequences correlate with the presence of the classic α/β nucleotide binding domain termed the Rossmann fold. In contrast, the catalytic domain of class II enzymes, as exemplified by the crystal structure for the seryl-tRNA synthetase from E. coli (4), is composed of an antiparallel β pleated sheet. These enzymes may also be classified mechanistically; class I enzymes aminoacylate their cognate tRNAs at the 3′ hydroxyl, whereas class II enzymes typically aminoacylate the tRNA at the 2′ hydroxyl (8).
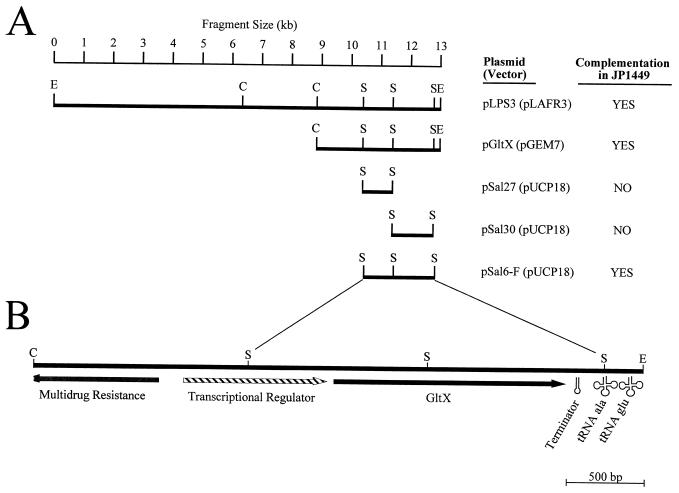
The Pseudomonas aeruginosa gltX gene, encoding glutamyl-tRNA synthetase, was recognized during sequence analysis of the cloned DNA in plasmid pLPS3, which contains genes for the synthesis of the lipopolysaccharide O antigen from the serogroup O11 P. aeruginosa strain PA103 (9). The gltX gene was found approximately 7.8 kb downstream of and transcribed in the same direction as genes of the lipopolysaccharide locus. Plasmid pLPS3 was able to complement the temperature-sensitive gltX351 mutation in E. coli JP1449 (18). Further subcloning (Fig. 1A), by standard, previously described techniques (3), localized the gltX gene to a 2.3-kb SalI fragment on the recombinant plasmid pSal6F. In this construct, the gltX gene was in the same orientation as the plasmid-encoded lac promoter. A similar construct with the plasmid-borne promoter in the opposite orientation was also able complement JP1449, suggesting that the insert DNA has its own promoter that is recognized in E. coli.
FIG. 1.
(A) Complementation of the temperature-sensitive gltX mutation in E. coli JP1449 with various subclones of pLPS3 (9). Vectors used in cloning were pLAFR3 (22), pGEM7 (Promega Corp., Madison, Wis.), and pUCP18 (20). Complementation was detected as the ability of E. coli JP1449 containing recombinant plasmids to grow at 42°C. The smallest subclone capable of complementing this gene was a 2.3-kb SalI fragment in pUCP18. (B) Genetic organization of the 3.9-kb DNA insert from pGltX. Two complete ORFs, coding for a hypothetical transcriptional regulator and the glutamyl-tRNA synthetase (GltX), were detected. Upstream of these genes is a divergently transcribed partial ORF with similarity to multidrug resistance gene products. Directly downstream of the gltX gene is a rho-independent terminator; distal to this terminator is a short transcript encoding two tRNA molecules, alanyl-tRNA (GGC) and glutamyl-tRNA (TTC), indicated as cloverleaf structures. Restriction endonuclease recognition sites are abbreviated as follows: C, ClaI, E, EcoRI; S, SalI.
Nucleotide sequence determination of the 3,892-bp insert from pGltX was performed by standard techniques, as previously described (5), and compared to the P. aeruginosa codon usage table (2). Analysis of this region revealed a 1,482-bp open reading frame (ORF) potentially encoding a 494-amino-acid protein with a predicted molecular mass of 56,777 daltons. This protein has 50.6% identity with GltX from Rhizobium meliloti and contains all of the class I motifs in the catalytic domain (Fig. 2). The overall G+C content of the insert, 64.3%, is typical for genes from this organism. The codon usage of gltX conforms to that of other well-expressed proteins from P. aeruginosa. A putative ORF was also found in the same position on the opposite strand from GltX. This predicted protein did not show significant similarity to any entries in GenBank, and codon usage was not consistent with that of other P. aeruginosa genes.
FIG. 2.
Multiple-sequence alignment of GltX. The predicted protein product from P. aeruginosa gltX was compared to five other known glutamyl-tRNA synthetases by using the Pileup program from the Genetics Computer Group (Madison, Wis.) package (Wisconsin Package, version, 9.1). Comparisons were made with the blosum 62 matrix, a gap weight of 12, and a gap length weight of 4. The GltX sequences used were as follows: P_aeru, P. aeruginosa PA103 (this study); R_meli, R. meliloti (GenBank accession no. P15189); T_aqua, Thermus aquaticus (GenBank accession no. P27000); B_ster, Bacillus stearothermophilus (GenBank accession no. P43818); B_subt, B. subtilis (GenBank accession no. P22249); E_coli, E. coli (GenBank accession no. P04805). Residues which were identical in all six sequences are printed in white on black, while those conserved in at least four of the six sequences are shaded. The positions of the Rossmann fold, including the HIGH and KMSKS motifs, and the zinc-binding domain are indicated. The locations of the four residues implicated in coordinating zinc in the E. coli glutamyl-tRNA synthetase are marked with asterisks.
Upstream of P. aeruginosa gltX is a potential gene product with sequence similarity to putative transcriptional regulators (Fig. 1B). However, the presence of this ORF was not required for complementation of the gltX351 mutation. Between the stop codon of this activator and the potential start codon of GltX is a potential ribosome binding site consistent with the orientation-independent complementation of E. coli JP1449.
It is interesting that downstream of the P. aeruginosa gltX gene and its rho-independent terminator are two tandemly organized tRNA genes, encoding tRNAAla and tRNAGlu (Fig. 1B). The alanyl-tRNA is 94% identical to that of E. coli and contains the invariant G-U base pair at positions 3 and 70 that discriminates tRNAAla (1). The glutamyl-tRNA is 96% identical to that of E. coli. A large stem-loop structure, reminiscent of RNase P substrates, is predicted between these tRNAs. The frequencies of the use of codons specified by the two tRNAs by P. aeruginosa are 56% (Ala-GCC) and 40% (Glu-GAA) (2).
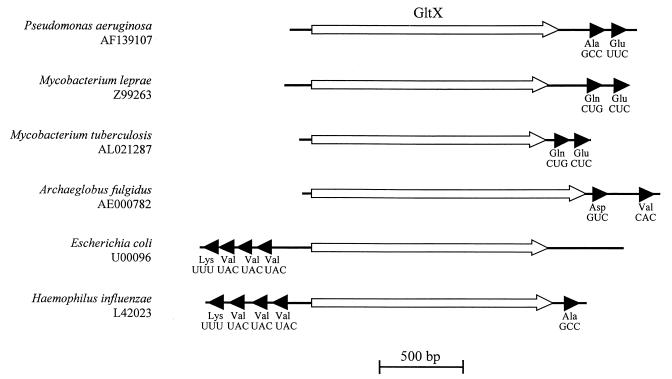
tRNAs are not always found adjacent to their respective tRNA synthetases. Of 29 bacterial gltX genes inspected, only the Mycobacterium leprae and Mycobacterium tuberculosis gltX genes have an adjacent tRNAGlu (Fig. 3). In E. coli and Haemophilus influenzae, GltX is expressed divergently from an operon containing tRNA genes, none of which encode tRNAGlu. In the case of the P. aeruginosa gene, it is clear that complementation of the gltX mutation in E. coli JP1449 is not simply due to pseudoreversion (i.e., increased levels of the cognate tRNA), since the pSalF construct does not contain the tRNAGlu locus.
FIG. 3.
Proximity of tRNA genes to glutamyl-tRNA synthetases. The gltX loci reported for 29 bacterial species were examined for the presence of adjacent tRNA genes; accession numbers are indicated. Species containing tRNAs adjacent to gltX are shown. Open arrows represent GltX, and solid arrows represent tRNA with amino acids and anticodons, as indicated. Species lacking tRNAs adjacent to gltX included the following, with accession numbers: Aquifex aeolicus, AE000657; Azospirillum brasilense, X99587; Bacillus stearotherophilus, M55072; B. subtilis, AL009126; Borrelia burgdorferi, AE00783; Chlamydia psittaci, U41758; Chlamydia trachomatis, AE001273; Helicobacter pylori, AE001439; Methanobacterium thermoautotrophicum, AE000666; Methanococcus jannaschii, L77117; Mycoplasma genitalium, L43967; Mycoplasma pneumoniae, U00089; Mycoplasma pulmonis, L25415; Neisseria gonorrhoeae, U76418; Pyrococcus horikoshii, pyro_h; R. meliloti, M27221; Rickettsia prowazekii, AJ235269; Staphylococcus xylosus, Y07614; Streptococcus coelicolor, AL031124; Synechocystis sp., AB001339; T. thermophilus, X64557; Treponema pallidum, AE000520; Vibrio cholerae, AF030977.
Glutamyl-tRNA synthetase activity was determined in E. coli JP1449 containing recombinant plasmids by a standard aminoacyl transferase assay (12) (Table 1). In brief, 1 liter of E. coli was grown in Luria-Bertani medium, with ampicillin (100 μg/ml) when needed, for 16 h at 30°C. Cells were pelleted and resuspended in 10 ml of 50 mM HEPES (pH 7.2) and then broken by one pass through a French pressure cell (10,000 psi). Cell debris was removed by low-speed centrifugation (4,000 × g, 4°C, 10 min), and cell membranes were pelleted by ultracentrifugation (100,000 × g, 4°C, 60 min).
TABLE 1.
Quantitation of GltX activity in E. coli JP1449
| Plasmida | Temp (°C)b | GltX sp act (nmol/min/mg of protein)c | Relative activity (%)d |
|---|---|---|---|
| None | 30 | 1.15 ± 0.28 | 100 |
| None | 42 | 0.18 ± 0.06 | 15.6 |
| pUCP18 | 30 | 0.99 ± 0.13 | 86.1 |
| pUCP18 | 42 | 0.17 ± 0.05 | 14.8 |
| pSal6F | 30 | 7.57 ± 0.01 | 673.9 |
| pSal6F | 42 | 7.77 ± 0.37 | 675.6 |
Cell extracts were made from E. coli JP1449 containing the indicated plasmids.
Reaction incubation temperature.
Results are averages of five experiments ± standard deviations.
Activity compared to E. coli JP1449 cell extracts assayed at 30°C.
The cell extract was then decanted and placed on ice for immediate use. The assay for GltX function contained the following components in a final volume of 100 μl: 50 mM HEPES (pH 7.2), 10% (vol/vol) glycerol, 25 mM MgCl2, 0.5 mM dithiothreitol, 1 mM ATP, 3 mg of crude E. coli tRNA (Sigma Chemical Co., St. Louis, Mo.) per ml, and 0.1 mM [14C]glutamic acid (10 mCi/mmol) (ICN Inc., Costa Mesa, Calif.). Reactions were initiated by the addition of cell extracts, were allowed to proceed at either 30 or 42°C, and were terminated after 5 min by the addition of 20 μl of 100% (wt/vol) trichloroacetic acid (TCA). Incorporation of [14C]glutamic acid into the TCA-precipitable pool was determined by filtering onto glass filters, followed by two 5-ml washes of 5% (wt/vol) TCA and 95% ethanol. Dried filters were placed in 10 ml of Scintosafe EconoF LSC fluid and counted for 2 min on a Wallac 1409 liquid scintillation counter. Protein concentrations were determined with the Bradford dye-binding assay from Bio-Rad Laboratories (Hercules, Calif.) with bovine serum albumin as a standard.
E. coli JP1449 extracts contained a low, but detectable, level of GltX activity at 30°C. This activity was abolished upon incubation of the reaction mixture at the nonpermissive temperature (42°C). Cells containing plasmid pSal6F expressed about sixfold more activity than JP1449 alone; this elevated activity was retained at the nonpermissive temperature.
The presence of tightly associated zinc has been reported for some tRNA synthetases from both class I and class II enzymes (13). Glutamyl-tRNA synthetases from E. coli and Bacillus subtilis, but not T. thermophilus, have been shown to contain one zinc atom per molecule. Removal of this ion with the metal chelator o-phenanthroline resulted in a conformational change and a concomitant loss of enzymatic activity (14). Sequence alignments revealed that the zinc-containing enzymes possessed two cysteine- and histidine-rich motifs, CXC and CRHSHEHH, in the tRNA acceptor domain. Extended X-ray absorption fine structure analysis of the E. coli glutamyl-tRNA synthetase demonstrated that the zinc atom was coordinated by three cysteine and one histidine residues. Site-directed mutagenesis of these motifs resulted in four variants—C98S, C100S, C125S, and H127Q—which no longer complemented the gltX351 defect of E. coli JP1449 (13). Inspection of the multiple-sequence alignment (Fig. 2) revealed that P. aeruginosa GltX contains cysteine and histidine residues corresponding to those involved in zinc coordination. As expected, preincubation of P. aeruginosa cell extracts with either 1 or 10 mM o-phenathroline diminished enzymatic activity at 42°C (Table 2).
TABLE 2.
Effects of o-phenanthroline on GltX activity
| Concn (mM)a | GltX sp act (nmol/min/mg of protein)b | Relative activity (%)c |
|---|---|---|
| 0 | 8.58 ± 0.68 | 100.0 |
| 1 | 0.79 ± 0.49 | 9.3 |
| 10 | 0.10 ± 0.11 | 1.2 |
GltX reactions were performed at 42°C with 20 μg of E. coli JP1449(pSal6F) extracts after preincubation of cell proteins with the indicated concentrations of o-phenanthroline for 10 min.
Results are averages of five experiments ± standard deviations.
Activity compared to assays with no added o-phenanthroline.
Given both the degree of sequence similarity and the conservation of functional domains between the glutamyl-tRNA synthetases from E. coli and P. aeruginosa, it is not surprising that the Pseudomonas gene can complement the gltX351 mutation of E. coli JP1449. Interestingly, similar attempts to complement E. coli with gltX from B. subtilis were unsuccessful (16); recombinant plasmids containing an intact B. subtilis gltX gene were found to be lethal for E. coli. It has been postulated that this toxic effect is due to the misacylation of tRNAGln with glutamate. B. subtilis, like most other organisms, uses one tRNA synthetase, GltX, to charge both tRNAGlu and tRNAGln with glutamate; the latter is subsequently converted to glutamine by a specific amidotransferase (11). Given the lack of lethality of P. aeruginosa gltX, we reasoned that Pseudomonas, like E. coli, may possess separate glutamyl- and glutaminyl-tRNA synthetases. A search of the incomplete P. aeruginosa genomic sequencing project (16a) revealed the presence of a glutaminyl-tRNA synthetase homolog that showed 60 and 59% identity with those of E. coli and H. influenzae, respectively.
Because of the crucial role that aminoacyl-tRNA synthetases play in protein biosynthesis, and their high degree of sequence conservation, these enzymes are of interest from several perspectives. They are enticing targets for novel antimicrobials. Indeed, pseudomonic acid, a natural product of Pseudomonas fluorescens, acts by blocking the action of isoleucyl-tRNA synthetases (10). Recently, several aminoadenylate analogs have been synthesized, some of which have been found to be potent antimicrobials (6, 19). Other researchers have used sequence comparisons of tRNA synthetases to investigate the phylogeny of prokaryotic cells (7, 21). In many cases, the phylogenetic relationships generated by these analyses differ from those obtained by analysis of rRNA genes. In such cases, horizontal gene transfer between organisms and/or gene duplications have been invoked. Although extensive, the database of tRNA synthetase genes is not yet sufficient to provide a single satisfactory model for the dissemination of these enzymes from a common ancestral gene. Thus, future discovery of aminoacyl-tRNA synthetases during genomic sequencing of organisms not only will lead to increased understanding of protein biosynthesis but also will provide possible leads to new antibiotics and clues to the origins of the diversity of life.
Nucleotide sequence accession number.
The nucleotide sequence of gltX has been deposited in GenBank and assigned accession no. AF139107.
Acknowledgments
We are grateful to Amy Staab, Yan Ren, and Betty Shiberu for excellent technical assistance and to Charles Dean for helpful discussions. We are indebted to Tim Bender and his group for help with GltX assays.
This research was supported by a grant from the NIH (R01 AI35674) to J.B.G.
REFERENCES
- 1.Atilgan T, Nicholas H B, Jr, McClain W H. A statistical method for correlating tRNA sequence with amino acid specificity. Nucleic Acids Res. 1986;14:375–380. doi: 10.1093/nar/14.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne M J, Jr, Goldberg J B. Cloning and characterization of the gene (rfc) encoding O-antigen polymerase of Pseudomonas aeruginosa PAO1. Gene. 1995;167:81–86. doi: 10.1016/0378-1119(95)00595-1. [DOI] [PubMed] [Google Scholar]
- 3.Coyne M J, Jr, Russell K S, Coyle C L, Goldberg J B. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J Bacteriol. 1994;176:3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusack S, Berthet-Colominas C, Hartlein M, Leberman R R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature (London) 1991;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 5.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. Characterization of the O antigen locus from the serogroup O11 Pseudomonas aeruginosa strain PA103: identification of the O antigen polymerase gene. Submitted for publication.
- 6.Desjardins M, Desgagnes J, Lacoste L, Yang F, Morin M-P, Lapointe J, Chenevert R. Synthesis of inhibitors of glutamyl-tRNA synthetase. Bioorg Med Chem Lett. 1997;7:2363–2366. [Google Scholar]
- 7.Doolittle R F, Handy J. Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr Opin Genet Dev. 1998;8:630–636. doi: 10.1016/s0959-437x(98)80030-0. [DOI] [PubMed] [Google Scholar]
- 8.Eriani G, Delarue M, Poch O, Gangloff J, Dino M. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 9.Evans D J, Pier G B, Coyne M J, Jr, Goldberg J B. The rfb locus from Pseudomonas aeruginosa strain PA103 promotes the expression of O antigen by both LPS-rough and LPS-smooth isolates from cystic fibrosis patients. Mol Microbiol. 1994;13:427–434. doi: 10.1111/j.1365-2958.1994.tb00437.x. [DOI] [PubMed] [Google Scholar]
- 10.Fuller A T, Mellows G, Woolford M, Banks G T, Barrow K D, Chain E B. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature (London) 1971;234:416–417. doi: 10.1038/234416a0. [DOI] [PubMed] [Google Scholar]
- 11.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNA1Glu in vitro. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Brisson A, Liu J, Roy P H, Lapointe J. Higher specific activity of the Escherichia coli glutamyl-tRNA synthetase purified to homogeneity by a six-hour procedure. Protein Expr Purif. 1992;3:71–74. doi: 10.1016/1046-5928(92)90058-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Gagnon Y, Gauthier J, Furenlid L, L’Heureux P-J, Auger M, Nureki O, Yokoyama S, Lapointe J. The zinc-binding site of Escherichia coli glutamyl-tRNA synthetase is located in the acceptor-binding domain. J Biol Chem. 1995;270:15162–15169. doi: 10.1074/jbc.270.25.15162. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Lin S-X, Blochet J-E, Pezolet M, Lapointe J. The glutamyl-tRNA synthetase of Escherichia coli contains one atom of zinc essential for its native conformation and its catalytic activity. Biochemistry. 1993;32:11390–11396. doi: 10.1021/bi00093a016. [DOI] [PubMed] [Google Scholar]
- 15.Nureki O, Vassylyev D G, Katayanagi K, Shimizu T, Sekine S, Kigawa T, Miyazawa T, Yokoyama S, Morikawa K. Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science. 1995;267:1958–1965. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- 16.Pelchat M, Lacoste L, Yang F, Lapointe J. Overproduction of the Bacillus subtilis glutamyl-tRNA synthetase in its host and its toxicity to Escherichia coli. Can J Microbiol. 1998;44:378–381. [PubMed] [Google Scholar]
- 16a.Pseudomonas Genome Project. 15 March 1999, revision date. [Online.] PathoGenesis Corp., Seattle, Wash. http://www.pseudomonas.com. [20 April 1999, last date accessed.]
- 17.Rould M A, Perona J J, Soll D, Steitz T A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 18.Russell R R B, Pittard A J. Mutants of Escherichia coli unable to make protein at 42°C. J Bacteriol. 1971;108:790–798. doi: 10.1128/jb.108.2.790-798.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmel P, Tao J, Hill J. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J. 1998;12:1599–1609. [PubMed] [Google Scholar]
- 20.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 21.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family: rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur J Biochem. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 22.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster T, Tsai H, Kula H T M, Mackie G A, Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984;226:1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]