ABSTRACT
Formation of highly unique and complex facial structures is controlled by genetic programs that are responsible for the precise coordination of three-dimensional tissue morphogenesis. However, the underlying mechanisms governing these processes remain poorly understood. We combined mouse genetic and genomic approaches to define the mechanisms underlying normal and defective midfacial morphogenesis. Conditional inactivation of the Wnt secretion protein Wls in Pax3-expressing lineage cells disrupted frontonasal primordial patterning, cell survival and directional outgrowth, resulting in altered facial structures, including midfacial hypoplasia and midline facial clefts. Single-cell RNA sequencing revealed unique transcriptomic atlases of mesenchymal subpopulations in the midfacial primordia, which are disrupted in the conditional Wls mutants. Differentially expressed genes and cis-regulatory sequence analyses uncovered that Wls modulates and integrates a core gene regulatory network, consisting of key midfacial regulatory transcription factors (including Msx1, Pax3 and Pax7) and their downstream targets (including Wnt, Shh, Tgfβ and retinoic acid signaling components), in a mesenchymal subpopulation of the medial nasal prominences that is responsible for midline facial formation and fusion. These results reveal fundamental mechanisms underlying mammalian midfacial morphogenesis and related defects at single-cell resolution.
KEY WORDS: Wnt, Pax, Frontonasal hypoplasia, Midline facial clefts, Single-cell RNA-seq, Gene regulatory network (GRN)
Summary: Single-cell genomic approaches reveal transcriptomic atlases of midfacial primordia, and a mesenchymal gene regulatory network modulated by Wls during midline facial formation and fusion.
INTRODUCTION
Mammalian midfacial development is a complex and dynamic process, involving precisely controlled three-dimensional tissue growth and fusion of three paired midfacial primordia: the lateral nasal prominence (LNP), the medial nasal prominence (MNP) and the maxillary prominence (MxP) (Chai and Maxson, 2006; Cox, 2004; Helms et al., 2005; Ji et al., 2020; Jiang et al., 2006; Song et al., 2009; Suzuki et al., 2016; Szabo-Rogers et al., 2010). These midfacial primordia generate the upper jaw (maxilla), upper lip, palate, nose and associated structures. Defective midfacial morphogenesis can cause various malformations, such as orofacial clefts, which are among the most common structural birth defects in humans (Ji et al., 2020; Jugessur and Murray, 2005; Reynolds et al., 2019; Schutte and Murray, 1999). Many studies have examined upper lip and primary palate formation at the bilateral junction zones where the LNP, MNP and MxP merge and fuse (Abramyan and Richman, 2015; Ji et al., 2020). The fusion process involves a rapid expansion of neural crest-derived mesenchymal cells within the midfacial primordia and epithelial seam formation and apoptosis at the fusion site. Defective primordial growth and/or fusion at the junction zones may cause bilateral or unilateral cleft lip with or without cleft palate (CL/P) (Ji et al., 2020; Mossey et al., 2009). In contrast, the two adjacent MNPs merge at the midline (which does not involve epithelial fusion) to form the philtrum, nasal septum and associated structures (Abramyan and Richman, 2015). Although it is relatively rare, midline orofacial clefts, including median cleft lip and bifid nose, also occur in human newborns with unknown pathogenesis (Eppley et al., 2005).
Midfacial morphogenesis is regulated by several developmentally essential signaling pathways, including Wnt, Fgf, Tgfβ/Bmp, Shh, retinoic acid pathways, and Pdgf families and related transcription factors (Dudas and Kaartinen, 2005; Graf et al., 2016; He and Chen, 2012; Parada and Chai, 2012; Reynolds et al., 2019; Reynolds et al., 2020; Stanier and Pauws, 2012; Suzuki et al., 2016; Xavier et al., 2016). The Wnt family comprises 19 secreted glycoproteins that play crucial roles in development and disease (Nusse and Clevers, 2017). In particular, Wnt signaling interacts with other signaling pathways and is required for neural crest cell and midfacial development (Ji et al., 2019; Reynolds et al., 2019). Multiple Wnt signaling genes are associated with orofacial clefts in both human patients and mutant mouse models (Reynolds et al., 2019). For example, homozygous nonsense mutation in WNT3 is associated with CL/P and tetra-amelia in a large consanguineous family (Niemann et al., 2004); Wnt9b and its putative co-receptor Lrp6 in the canonical Wnt/β-catenin signaling pathway have been demonstrated to play a crucial role in the formation and fusion of the upper lip and primary palate at the midfacial primordial junction zones (Jin et al., 2012; Song et al., 2009). Wnt5a is required for outgrowth of craniofacial and other distal structures (Yamaguchi et al., 1999), and Wnt5a/Ror2 signaling in the non-canonical Wnt pathway is required for secondary palatogenesis (He et al., 2008).
Wnt proteins are post-translationally modified by lipidation or fatty acylation by Porcn (porcupine O-acyltransferase) in the endoplasmic reticulum (ER) (Berthiaume, 2014; Herr and Basler, 2012), and they are transported by Wls (Wntless or Gpr177) from the ER to the Golgi body and plasma membrane for their secretion to the extracellular space (Bänziger et al., 2006). Wls is required for body axis formation and early embryonic survival, which is identical to Wnt3 functions during early embryogenesis (Fu et al., 2011, 2009; Liu et al., 1999). Wls has been demonstrated to play cell lineage-specific roles during craniofacial development. Conditional ablation of either Wls or β-catenin with Foxg1-Cre in facial ectodermal and neuroepithelial cells arrests the formation of orofacial primordia (Wang et al., 2011; Zhu et al., 2016). This suggests a key role of Wls upstream of the canonical Wnt signaling pathway during early facial patterning. In contrast, conditional knockout of Wls with Wnt1-Cre in dorsal neuroepithelial cells and neural crest cells results in mid-hindbrain patterning defects and cleft palate, which is similar to the phenotypes of Wnt1/Wnt3a double knockouts or conditional ablation of β-catenin with Wnt1-Cre, respectively (Fu et al., 2011; Ikeya et al., 1997; Brault et al., 2001). Moreover, cranial ectodermal and mesenchymal Wls play distinct roles in osteoblast and dermal fibroblast formation (Goodnough et al., 2014). Nevertheless, the role of Wls in the fusion or merging process of the midline facial primordia remains undetermined. The present study employs conditional gene targeting of Wls in Pax3-expressing midfacial mesenchymal cells derived from both neural crest and mesoderm (Kasberg et al., 2013) in combination with single-cell RNA sequencing (scRNA-seq) and related bioinformatics approaches to define the transcriptomic atlases and gene regulatory mechanisms governing normal and defective midfacial primordia. Our results demonstrate a crucial role of Wls in modulating genetic programs and a gene regulatory network (GRN) required for the dorsal nasal primordial patterning, survival, directional outgrowth, and the midline merging of the medial nasal prominences during midfacial development.
RESULTS
Genetic fate-mapping and conditional gene-targeting analyses of Pax3Cre;Wlsflox during midfacial morphogenesis
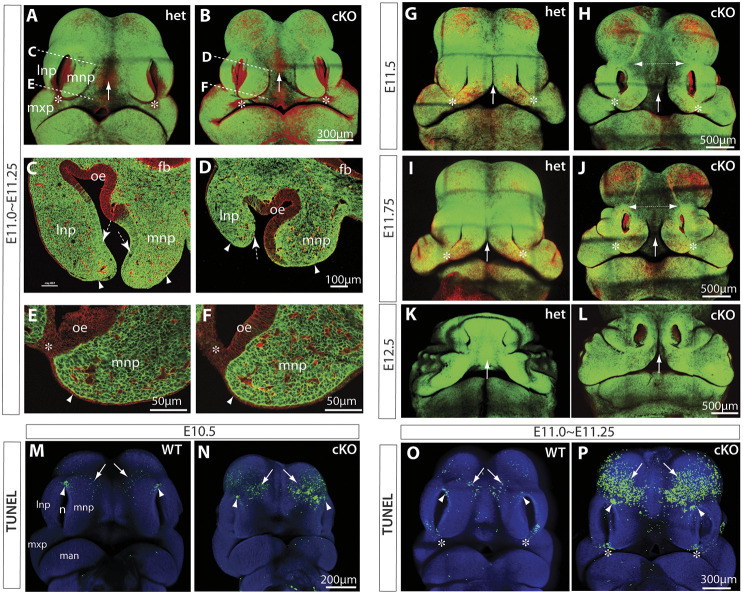
To extend our understanding of midfacial morphogenesis and its regulation by Wls, we combined genetic fate-mapping and Cre/loxP conditional gene-targeting approaches through serial mating of Pax3Cre/+ knock-in mice (Engleka et al., 2005), Wlsflox/flox mice (Carpenter et al., 2010), and the Cre reporter Rosa26mT/mG mice (Muzumdar et al., 2007). At embryonic day (E) 10.5, the enhanced green fluorescent protein (EGFP)-positive Pax3-expressing lineage cells were widely distributed in the midfacial primordia, including LNP, MNP and MxP, in the littermate heterozygous control of E10.5 Pax3Cre/+;Wlsflox/+;Rosa26mT/mG (abbreviated as Wls-het) embryos (Fig. S1A). There were no obvious morphological or fate-mapping alterations in the E10.5 Pax3Cre/+;Wlsflox/flox;Rosa26mT/mG conditional knockout (abbreviated as Wls-cKO) embryos (Fig. S1B). By E11.0-11.25, EGFP-positive cells occupied the midfacial mesenchyme and were also found in the distal portion of the non-neural nasal epithelium and adjacent surface ectoderm in the dorsal nasal primordia (Fig. 1A,C,E), which were not apparently altered in the Wls-cKOs (Fig. 1B,D,F), in line with published fate mapping results of Pax3Cre during midfacial development (Kasberg et al., 2013). By E11.5, the dorsal components of the paired MNPs grew toward each other and initiated midline merging (Fig. 1G,I), a process that was completed before E12.5 (Fig. 1K) in the heterozygous control embryos. These conformational rearrangements were severely disrupted in E11.0-11.5 Wls-cKO embryos, as evidenced by wider midline gaps between the paired MNPs (distance I in Fig. S1C-E and arrows in Fig. 1H,J) and between the borders of the dorsal nasal primordia and the forebrain vesicles (Fig. 1H,J, dashed double-arrow), and the midline facial clefts in E12.5 mutant embryos (Fig. 1L, arrow). It is important to note that the epithelial cells in the bilateral junction zones were EGFP negative (indicating no Pax3-Cre recombination activities) in both heterozygous control and conditional knockout (cKO) embryos (arrowheads in Fig. 1A,B,E,F), and that the fusion of the upper lip and primary palate around the MNP, LNP and MxP was not affected in Wls-cKO embryos (Fig. 1H,J) as a consequence of conserved Wls in the bilateral junction zone epithelial cells. The distances between the dorsal nasal pits (II), the bilateral junction zones (III) or the lateral edges of the LNPs (IV) were slightly increased in the Wls-cKOs but this was not statistically significant (Fig. S1C-E). Together, these results demonstrate that Wls is required in Pax3-expressing lineage cells (including all facial mesenchymal cells and a subpopulation of non-neural epithelial cells) for midfacial formation and merging at the midline, but not at the bilateral junction zones during midfacial morphogenesis.
Fig. 1.
Genetic fate mapping and apoptosis of Pax3Cre;Wls-cKO midfacial primordia. (A-L) EGFP expression, representing Pax3Cre recombination patterns in the midfacial primordia of E11-E12.5 Wls-het (A,C,E,G,I,K) and Wls-cKO (B,D,F,H,J,L) embryos. Dashed lines in A,B indicate respective section regions shown in C-F. Arrows indicate the facial midline between the paired MNPs during or after merging. Dashed arrows and arrowheads in C,D indicate EGFP-positive nasal epithelial cells and adjacent surface ectodermal cells around the dorsal nasal tips. Arrowheads in E,F indicate EGFP-negative surface ectodermal cells. Double-headed arrows in H,J show an enlarged midline gap in Wls-cKO embryos. Asterisks indicate the bilateral junction zone. (M-P) Whole-mount TUNEL assays for normal controls and Pax3Cre;Wls-cKOs at E10.5 and E11.0-E11.25. Arrows indicate apoptotic cells in the forehead region adjacent to the dorsal nasal prominences. Arrowheads indicate apoptotic cells in the dorsal junction zone between the LNP and LMP. Asterisks in O,P indicate apoptotic cells at the junction zone during bilateral upper lip fusion. fb, forebrain neuroepithelia; lnp, lateral nasal prominence; man, mandibular prominence; mnp, medial nasal prominence; mxp, maxillary prominence; n, nasal pit; oe, olfactory epithelia.
Ectopic apoptosis in forehead regions of Wls mutants
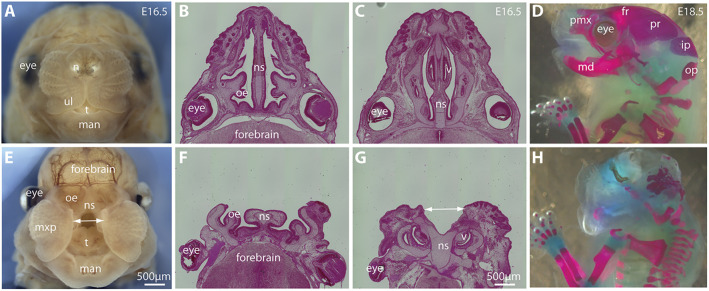
Apoptosis plays important roles during lip and palatal fusion (Ji et al., 2020). To test whether apoptosis also plays a role in the defective conformational changes of the Wls-deficient midfacial primordia, we conducted whole-mount terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays and found a cluster of apoptotic cells around the dorsal junction zone between the LNP and MNP in the normal control, which was not significantly altered in the Wls-cKO at E10.5 and E11.0-11.25 (Fig. 1M-P, arrowheads; Fig. S2C,F). We also found apoptotic cells scattered in the forehead region next to the dorsal nasal primordia of the normal control, but these apoptotic cells were significantly increased by 2.93-fold in the forehead region of the Wls-cKOs at E10.5 and increased by 7.42-fold in the mutant forehead at E11.0-11.25 before the paired MNPs merged at the midline (Fig. 1M-P, arrows; Fig. S2C,F). This ectopic apoptosis may contribute to the severe frontonasal hypoplasia and midline facial clefts observed in Wls-cKOs at later gestational ages (Fig. 2), which include a large open gap between the paired maxillary structures (Fig. 2E), shortened and split nasal septum (Fig. 2F,G), and lack of orofacial bone formation (Fig. 2H). These results demonstrate that Wls is required in Pax3-expressing lineage cells to prevent ectopic apoptosis during midfacial formation and midline merging, which provides a cellular basis for the severe frontonasal hypoplasia of the Wls mutants.
Fig. 2.
Midfacial clefts and frontonasal hypoplasia in Wls-deficient mice. (A,E) Front facial views of control (A) and Wls-cKO (E) embryos at E16.5. The double-headed arrow shows the gap between MNPs in the Wls-deficient embryo. (B,C,F,G) Histological transverse sections of control (B,C) and Wls-cKO (F,G) embryos at E16.5. The Wls-cKO embryo exhibits a split nasal septum (double-headed arrow in G). (D,H) Lateral views of skeletal preparation of control (D) and Wls-cKO (H) embryos at E18.5, showing the short and defective craniofacial bones and cartilages. fr, frontal bone; ip, interparietal bone; man, mandibular prominence; md, mandible; mxp, maxillary prominence; n, nasal pit; ns, nasal septum; oe, olfactory epithelium; op, occipital bone; pr, parietal bone; pmx, premaxilla; t, tongue; ul, upper lip; v, vomeronasal organ.
Single-cell RNA-seq analyses of the midfacial primordia of normal and Wls-cKO mouse embryos
To explore the molecular mechanisms underlying normal and defective midfacial morphogenesis as described above, we performed scRNA-seq of microdissected bilaterally paired midfacial primordia (MNPs, LNPs and MxPs) at E11.5 (Fig. 3). Following tissue dissociation, genotyping and fluorescence-activated cell sorting, isolated live cells were pooled from three wild-type (WT) littermates and two Wls-cKO embryos. RNA sequencing generated approximately 37,778 reads per cell and 350 million reads per lane. In total 378.9 million trimmed reads (161.8 million WT and 217.1 million Wls-cKO) were obtained, of which 91.50% of WT and 92.50% of cKO reads were mapped to the mouse genome, with 88.20% of WT and 89.3% of Wls-cKO exhibiting unique alignments. After quality control checks (Fig. S3) and filtering, 4284 and 3441 cells, 15,122 and 15,156 genes were retained from the raw results (Table S1) of WT and cKO samples, respectively, for subsequent analyses. Additional sequencing information is summarized in Table S1.
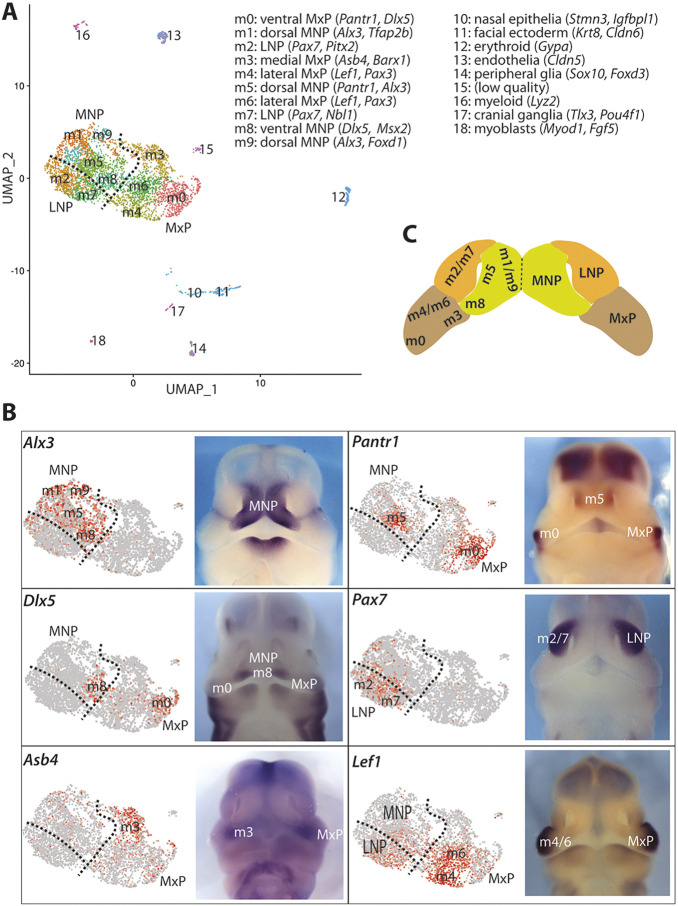
Fig. 3.
scRNA-seq identifies major cell types at the facial primordia in control samples. (A) UMAP plot of 4284 cells from WT samples showing 18 unique cell populations. Cluster-specific marker genes used for mapping the cluster onto the midfacial primordia are listed at the top right. (B) Identification of the mesenchymal clusters by whole-mount in situ hybridization. Left: Feature plots for marker genes. Right: Whole-mount in situ hybridization for the indicated genes at E11.5. (C) Schematic showing the identified anatomical locations of midfacial mesenchymal subpopulations at E11.5.
We used a recently developed algorithm named uniform manifold approximation and projection (UMAP) to generate graph-based cell clustering plots (Becht et al., 2018). Based on multiple conserved marker genes in WT control embryos, we were able to identify 18 unique cell populations, as demonstrated by UMAP (Fig. 3) and heatmaps with top 5 marker genes in each cluster (Figs S4 and S5). These include ten major subpopulations of mesenchymal cells (m0-m9), one cluster of nasal epithelial cells, one cluster of facial ectodermal cells, one cluster of erythrocytes, one cluster of myeloid cells, one cluster of endothelial cells, one cluster of peripheral glia, one cluster of cranial ganglia neurons and one cluster of myoblasts. The UMAP plot revealed highly organized mesenchymal subpopulations that resemble in vivo anatomical relationships among the three paired midfacial primordia: MNPs (m1/m5/m8/m9), LNPs (m2/m7) and MxP (m0/m3/m4/m6) (Fig. 3A,C), which were validated by feature plots and the expression patterns of multiple marker genes by whole-mount in situ hybridization (Fig. 3B). For instance, Alx3 was highly expressed in m1/m5/m8/m9 clusters, suggesting that these cells are from the MNPs. Clusters of m1/m5/m9 denote the dorsal MNPs and predominately express Tfap2b, with m5 expressing Pantr1, m9 expressing Shox2, and m8, which demarcates the ventral MNPs, expressing Dlx5. Clusters of m2/m7 demarcate the LNPs and express Pax7. Cluster m0 defines the ventral MxP and expresses Pantr1 and Barx1, whereas m3 from medial MxP expresses Asb4, and m4/m6 from the lateral MxP express Lef1 (Fig. 3B). These results provide a transcriptome atlas of unique cell populations in the midfacial primordia and also reveal an unexpected positional correlation between the UMAP and anatomical organization of the mesenchymal subpopulations. The latter suggests that the physical localizations of mesenchymal subpopulations are intrinsically determined by their unique transcriptome profiles.
Cell numbers and developmental processes are altered in Wls-deficient midfacial primordia
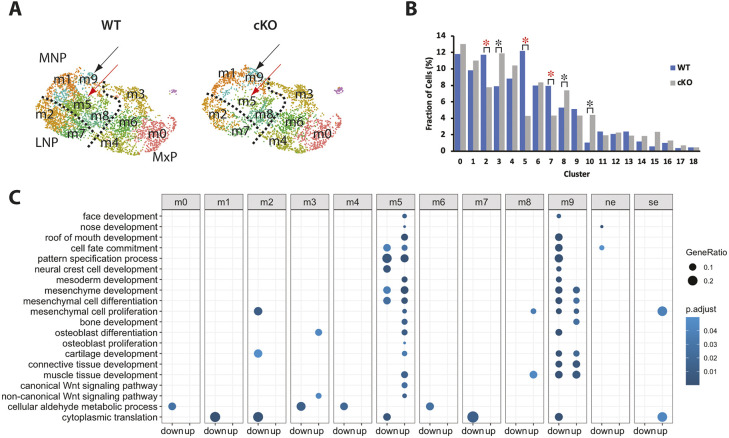
The UMAP for Wls-cKO samples indicated that cell number in the MNP, particularly in the m5 mesenchymal subpopulation, is significantly reduced compared with that in the WT control (Fig. 4A,B). In WT samples, cluster m5 contributed 37.58% of all MNP mesenchymal cells, but it only contributed 15.82% of all MNP cells in Wls-cKO samples, suggesting that m5 cells are preferentially affected by ectopic apoptosis in Wls-deficient MNPs. Cell numbers in mutant m2 and m7 of LNPs were also reduced (Fig. 4B). The reduced cell numbers in m2, m5 and m7 may be responsible for the increase in the proportion of other clusters, such as m3, m8 and cluster 10 (nasal epithelia) in the cKO. To explore additional reasons for the notably increased proportion of cells in the mutant cluster 10, we re-clustered it and found no significant alterations of cell cycle status but significantly fewer cells in the WT compared with the cKO (Fig. S6). This unusual phenomenon is likely caused by technical limitations during microdissections of midfacial primordia and scRNA-seq preparations due to altered anatomical structures and significantly reduced cell numbers in the cKO embryos.
Fig. 4.
scRNA-seq reveals the two most altered mesenchymal subpopulations in Wls-cKO midfacial primordia. (A) UMAP comparison of WT and Wls-cKO mesenchyme showing a drastically reduced m5 subpopulation (red arrows) and relatively conserved m9 subpopulation (black arrows) in the mutant MNP. (B) Comparisons of cell fractions (percentage) between WT and Wls-cKO clusters. Red asterisks indicate significant reduction (P<0.001; two-proportion Z-test, two-sided). Black asterisks indicate the clusters with a relatively increased ratio in the cKOs, which would be impacted by the ratio reductions in other clusters. (C) Gene ontology enrichment analysis of DEGs relevant to midfacial development revealed m5 (with mostly upregulated pathways) and m9 (with mostly downregulated pathways) as the two most affected mesenchymal subpopulations in the mutant midfacial primordia. The bottom two terms in C are not directly relevant to midfacial development and were used for visualizing all mesenchymal clusters (m0-m9) in the gene ontology chart. Nasal epithelia (ne, cluster 10) and surface ectoderm (se, cluster 11) are also included for reference.
To address systematically the impact of Wls deficiency on gene expression profiles within the midfacial primordia, we compared control and Wls-cKO samples to identify differentially expressed genes (DEGs). Gene ontology analyses of the developmentally relevant DEGs revealed that m5 and m9 mesenchymal subpopulations in the MNP have the greatest number of altered developmental processes, including cell fate commitment, pattern specification, mesenchymal cell development, and face, nose and roof of mouth development, with mostly downregulated processes in m9 and mostly upregulated in m5 of the mutants (Fig. 4C, Table S2). Regulation of Wnt signaling was also altered in m5 and m9 clusters, and regulation of apoptotic signaling involved in development was upregulated in m5, which had the greatest reduction in cell numbers in the mutants.
Wnt signaling alterations in Wls-deficient midfacial primordia
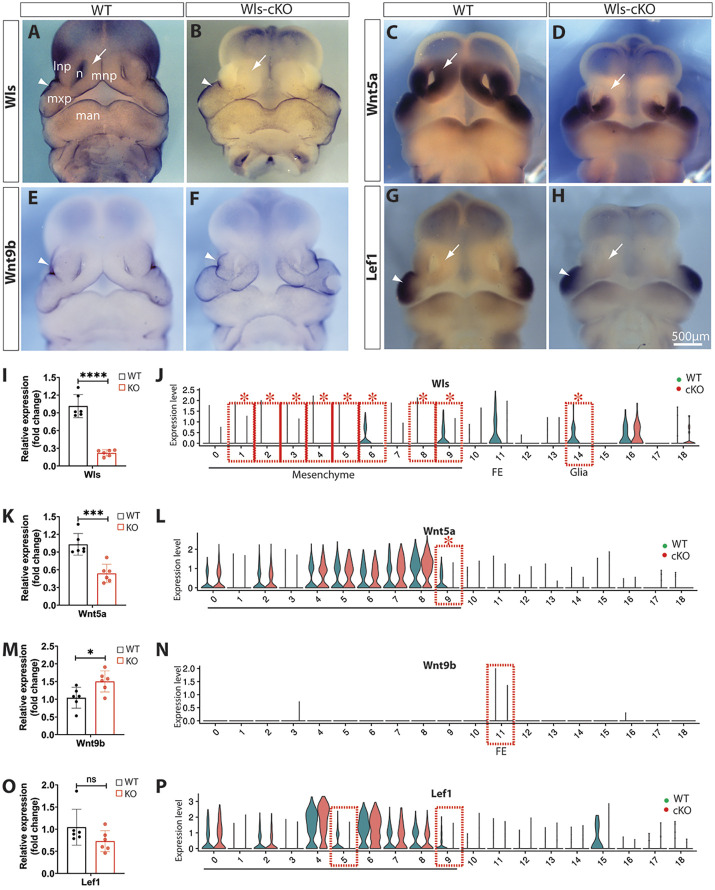
To investigate the molecular mechanisms underlying the severe frontonasal hypoplasia and midline facial clefts in Wls-cKO embryos, we analyzed the expression patterns of selected DEGs and related genes that are crucial for midfacial development. We validated significant candidates using whole-mount in situ hybridization and real-time reverse-transcription PCR (RT-qPCR) and through comparison with their single-cell transcriptomic profiles for cell cluster-specific alterations. Because Wls is required for the transportation and secretion of Wnt proteins that play vital roles in craniofacial development, we focused on several representative Wnt signaling genes and compared them with their violin plots generated from scRNA-seq analyses. Wls was widely expressed in mesenchymal cells in the orofacial primordia (Fig. 5A). Whole-mount in situ hybridization clearly demonstrated that Wls is abolished in orofacial mesenchymal cells, but it is relatively conserved in facial ectodermal cells in the E11.5 Wls-cKO embryo (Fig. 5B). The overall reduction of Wls in the mutant midfacial primordia is supported by RT-qPCR results (Fig. 5I) and cell type-specific reductions of Wls were further demonstrated by scRNA-seq, which identified Wls as a DEG in most mesenchymal subpopulations of the mutant midfacial primordia (Fig. 5J). These results demonstrate the effective ablation of Wls by Pax3Cre in midfacial mesenchymal cells.
Fig. 5.
Alterations of Wnt signaling genes in Wls-cKO midfacial primordia at E11.5. (A-H) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia showing altered and conserved expression patterns of Wls (A,B), Wnt5a (C,D), Wnt9b (E,F) and Lef1 (G,H). Arrows indicate diminished expression in the dorsal MNPs of the mutants (B,D,H), and arrowheads indicate conserved expression in the mutant surface ectoderm (B,F) or MxP (H) of the mutants. (I,K,M,O) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). *P<0.05; ***P<0.001; ****P<0.0001; ns, no statistical significance (P>0.05; t-test). (J,L,N,P) Violin plots of scRNA-seq showing cell cluster-specific alterations of representative Wnt signaling genes. Dashed boxes indicate cell clusters with diminished expression, and asterisks indicate statistical significance (P_adj<0.05; bimod, likelihood-ratio test). 0-9, mesenchymal subpopulations; 11, facial ectoderm (FE); 14, peripheral glia. lnp, lateral nasal prominence; man, mandibular prominence; mnp, medial nasal prominence; mxp, maxillary prominence; n, nasal pit.
Our scRNA-seq detected 13 out of 19 Wnt genes in the midfacial primordia (Fig. S7). Among these, Wnt5a, a key orofacial developmental gene, was shown by whole-mount in situ hybridization to be significantly diminished in the dorsal parts of the mutant MNPs (Fig. 5C,D) and this overall change was supported by RT-qPCR (Fig. 5K). scRNA-seq identified Wnt5a as a DEG in cluster m9 (Fig. 5L). In contrast, the cleft lip gene Wnt9b, which is exclusively expressed in the facial ectoderm, was slightly upregulated in the Wls-cKO mutants (Fig. 5E,F,M,N). Furthermore, the canonical Wnt signaling transcription factor Lef1 was predominantly expressed in the WT MxPs and also moderately expressed in the LNPs and MNPs and was clearly diminished in the mutant MNPs particularly in m5 and m9, but was not a DEG determined by scRNA-seq or RT-qPCR (Fig. 5G,H,O,P). Together, these results indicate that mesenchymal Wls ablation may block both canonical and noncanonical Wnt signaling pathways, especially Wnt5a in dorsal MNP mesenchymal cells, which could contribute to midfacial malformation.
Region-specific downregulation of facial cleft-associated genes in Wls-deficient midfacial mesenchyme
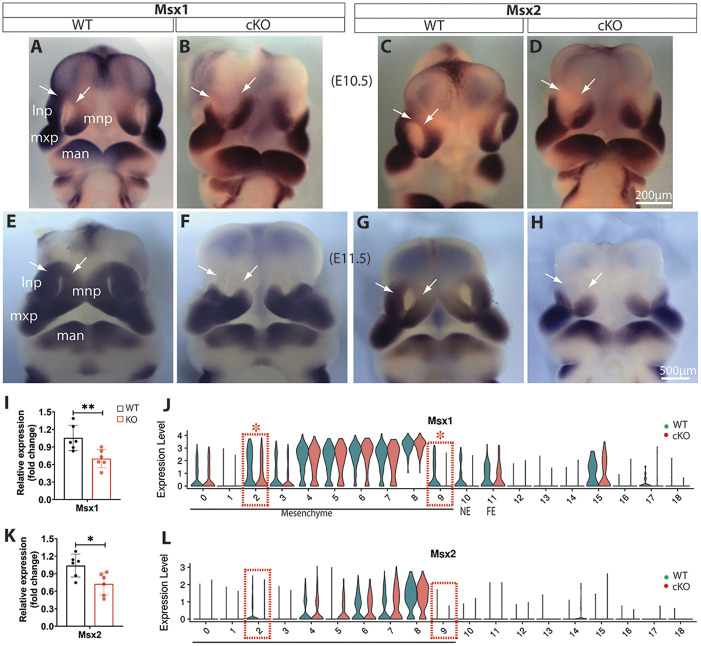
The muscle segment homeobox Msx genes play crucial roles in craniofacial development (Alappat et al., 2003). Whole-mount in situ hybridization demonstrated that Msx1 and Msx2 are clearly diminished in the dorsal MNPs and LNPs of Wls-cKO embryos at E10.5 and E11.5 (Fig. 6A-H), which is further supported by RT-qPCR results at E11.5 (Fig. 6I,K). Single-cell transcriptomics indicated that Msx1 is a significant DEG that is diminished primarily in mesenchymal clusters m2 in LNPs and m9 in MNPs at E11.5 (Fig. 6J), which matches well with their expression in the dorsal LNPs and MNPs. Msx2 was expressed at a lower level in the dorsal MNPs of E11.5 WT embryos and it was altered similarly, but to a lesser extent than, Msx1 in Wls-cKO mutants (Fig. 6G,H,K,L).
Fig. 6.
Alterations of Msx1 and Msx2 in Wls-cKO midfacial primordia. (A-H) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia at E10.5 (A-D) and E11.5 (E-H) showing altered and conserved expression patterns of Msx1 and Msx2. Arrows indicate diminished expression in the dorsal MNPs and LNPs of the mutants. (I,K) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). *P<0.05; **P<0.01 (t-test). (J,L) Violin plots of scRNA-seq showing cell cluster-specific alterations of Msx1 and Msx2. Dashed boxes indicate cell clusters with diminished expression in m2 (LNP) and m9 (MNP) subpopulations, and asterisks indicate statistical significance (P_adj<0.05; bimod test). lnp, lateral nasal prominence; man, mandibular prominence; mnp, medial nasal prominence; mxp, maxillary prominence.
The paired box genes Pax3 and Pax7 play a synergetic role in midline facial formation and clefts (Zalc et al., 2015). Pax3 was widely expressed in midfacial primordia of E11.5 WT embryos, including the LNP, MxP, and the dorsal and ventral MNP flanking a lower expression domain (Fig. 7A). It was significantly diminished in the dorsal MNP as demonstrated by whole-mount in situ hybridization and RT-qPCR (Fig. 7B,C), and was differentially expressed in m5 and m9 clusters of Wls-cKO MNPs as determined by single-cell transcriptomics (Fig. 7D). In contrast, Pax7 was predominately expressed in the LNPs and moderately expressed in the dorsal MNPs of E11.5 WT embryos (Fig. 7E), but it was downregulated in both regions of Wls-cKO embryos (Fig. 7F,G) and was differentially expressed in cluster m9 of the mutant MNPs (Fig. 7H).
Fig. 7.
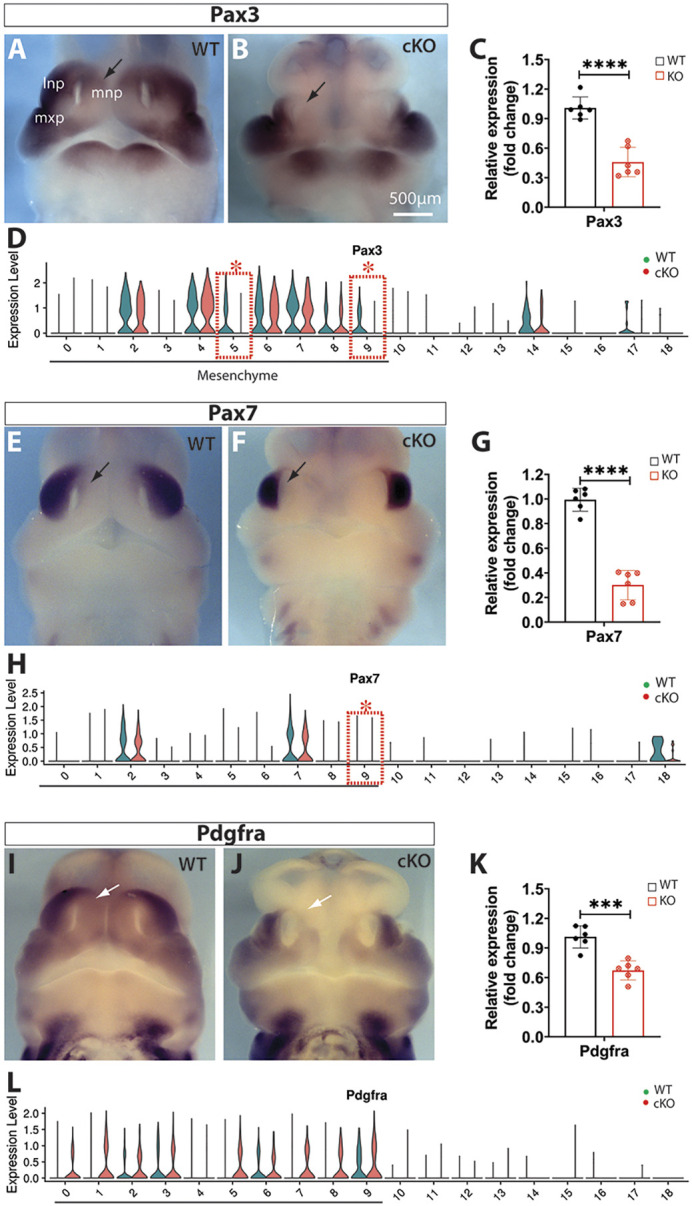
Alterations of Pax3, Pax7 and Pdgfra in Wls-cKO midfacial primordia at E11.5. (A,B,E,F,I,J) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia showing altered expression patterns. Arrows indicate diminished expression in the dorsal MNPs of the mutants. (C,G,K) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). ***P<0.001; ****P<0.0001 (t-test). (D,H,L) Violin plots of scRNA-seq showing consistently diminished expression of Pax3 in mutant m5 and m9, and Pax7 in mutant m9. Pdgfra alterations in the mutants were not detected by scRNA-seq. Dashed boxes indicate cell clusters with diminished gene expression in the mutants. Asterisks indicate statistical significance (P_adj<0.05; bimod test). lnp, lateral nasal prominence; mnp, medial nasal prominence; mxp, maxillary prominence.
Other than the aforementioned Msx and Pax transcription factors, platelet-derived growth factor receptor A (Pdgfra) signaling also plays an indispensable role in MNP development as conditional ablation of Pdgfra with Wnt1-Cre in neural crest lineage cells has been shown to result in midline facial clefts (He and Soriano, 2013). Whole-mount in situ hybridization and RT-qPCR demonstrated that Pdgfra is significantly downregulated in the midfacial primordia, particularly in the dorsal MNPs, of E11.5 Wls-cKOs (Fig. 7I-K), but single-cell transcriptomics showed no significant change in Pdgfra expression in mutant mesenchymal subpopulations (Fig. 7L).
Together, these results suggest that downregulation of Msx1/Msx2 and Pax3/Pax7 as well as Pdgfra in the dorsal nasal prominences is a primary contributor, at the molecular level, to the frontonasal hypoplasia and midline facial clefts in Wls-deficient mutants.
Complex alteration patterns of homeobox Alx genes in Wls-deficient midfacial mesenchyme
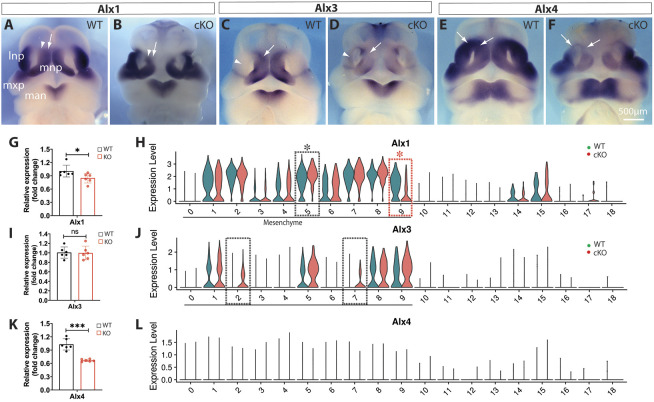
Mutations in the aristaless-like homeobox Alx genes are associated with midfacial clefts and frontonasal dysplasia in humans (Uz et al., 2010). An Alx1 mutation is associated with midline facial defects in cats (Lyons et al., 2016), and Alx3;Alx4 double mutants exhibit incomplete penetrant midline facial clefts in mice (Beverdam et al., 2001). Alx1 was diminished in cluster m9, but upregulated in m5 of Wls-cKO MNPs (Fig. 8A,B,G,H). Alx3 was diminished in MNPs, but was also upregulated in the LNPs of Wls-cKOs, as demonstrated by whole-mount in situ hybridization (Fig. 8C,D). However, the overall change in Alx3 in the mutant midfacial primordia was not detectable by RT-qPCR (Fig. 8I). Alx3 upregulation in m2 and m7 clusters of the mutant LNPs was supported by single-cell transcriptomics (Fig. 8J). Moreover, Alx4 expression was significantly diminished in dorsal MNPs and LNPs of Wls-cKOs, as demonstrated by whole-mount in situ hybridization and RT-qPCR (Fig. 8E,F,K), but not by single-cell transcriptomics (Fig. 8L). Some of these inconsistent results generated by different approaches may reflect the complex alteration patterns of Alx genes in Wls-deficient midfacial primordia. Nevertheless, these results suggest that altered Alx gene activity may also contribute to midline facial clefts in Wls-cKO mutants.
Fig. 8.
Alterations of Alx family genes in Wls-cKO midfacial primordia at E11.5. (A-F) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia showing altered expression patterns of Alx1, Alx3 and Alx4. Arrows indicate diminished expression, and arrowheads indicate upregulated expression in the mutant primordia. (G,I,K) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). *P<0.05; ***P<0.001; ns, no statistical significance (t-test). (H,J,L) Violin plots of scRNA-seq showing downregulated expression of Alx1 in m9 and concurrent upregulation in m5 of the mutant MNPs and Alx3 upregulation in m2 and m7 of the mutant LNPs. Alx4 alterations were not detected by scRNA-seq. Dashed boxes indicate cell clusters with altered gene expression in the mutants (black, upregulated; red, downregulated). Asterisks indicate statistical significance (P_adj<0.05; bimod test). lnp, lateral nasal prominence; man, mandibular prominence; mnp, medial nasal prominence; mxp, maxillary prominence.
Alterations of other crucial midfacial developmental genes in Wls-deficient midfacial primordia
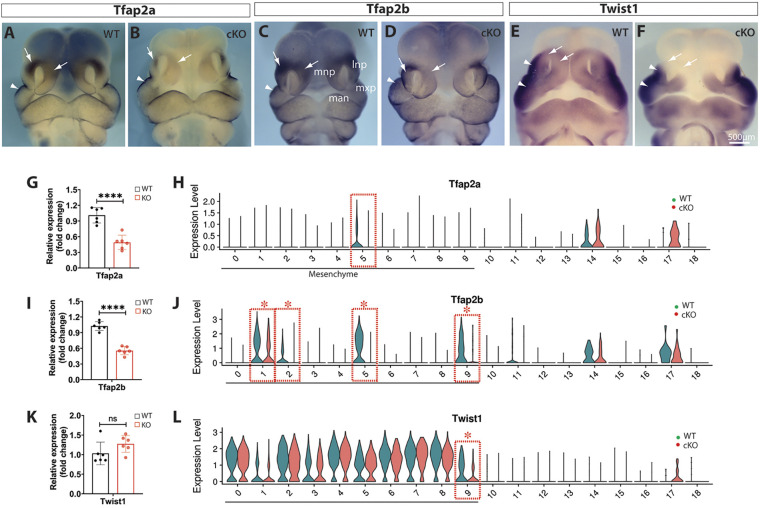
Tfap2a, which encodes a transcription factor required for craniofacial development and midline facial closure (Schorle et al., 1996), was significantly downregulated in the dorsal MNPs and LNPs, as demonstrated by whole-mount in situ hybridization and RT-qPCR (Fig. 9A,B,G). Tfap2a was also downregulated in the mutant MNP mesenchymal cluster m5 but was not identified as a DEG in any cell clusters by single-cell transcriptomics (Fig. 9H). Tfap2b was also significantly downregulated, as demonstrated by in situ hybridization and PCR approaches, and was identified as a DEG in multiple mesenchymal clusters, including m1, m2, m5 and m9 of Wls-deficient midfacial primordia (Fig. 9C,D,I,J). Twist1, which encodes a basic helix-loop-helix (bHLH)-containing transcription factor required for cranial closure (Chen and Behringer, 1995), was predominantly expressed in the lateral regions of LNPs and MxPs, and moderately expressed in the dorsal nasal primordia of E11.5 WT embryos. The latter expression domains were clearly diminished in Wls-cKOs and Twist1 was identified as a DEG in cluster m9 of MNPs, but RT-qPCR detected no significant change between WT and cKO midfacial samples at E11.5 (Fig. 9E,F,K,L).
Fig. 9.
Alterations of Tfap2a, Tfap2b and Twist1 in Wls-cKO midfacial primordia at E11.5. (A-F) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia showing altered expression patterns of the indicated genes. Arrows indicate diminished expression, and arrowheads indicate no obvious changes in the mutant primordia. (G,I,K) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). ****P<0.0001; ns, no statistical significance (t-test). (H,J,L) Violin plots of scRNA-seq showing downregulated expression of Tfap2a in m5, Tfap2b in m1, m2, m5 and m9, and Twist1 in m9 mesenchymal subpopulations of the mutants. Dashed boxes indicate cell clusters with diminished gene expression in the mutants. Asterisks indicate statistical significance (P_adj<0.05; bimod test). lnp, lateral nasal prominence; man, mandibular prominence; mnp, medial nasal prominence; mxp, maxillary prominence.
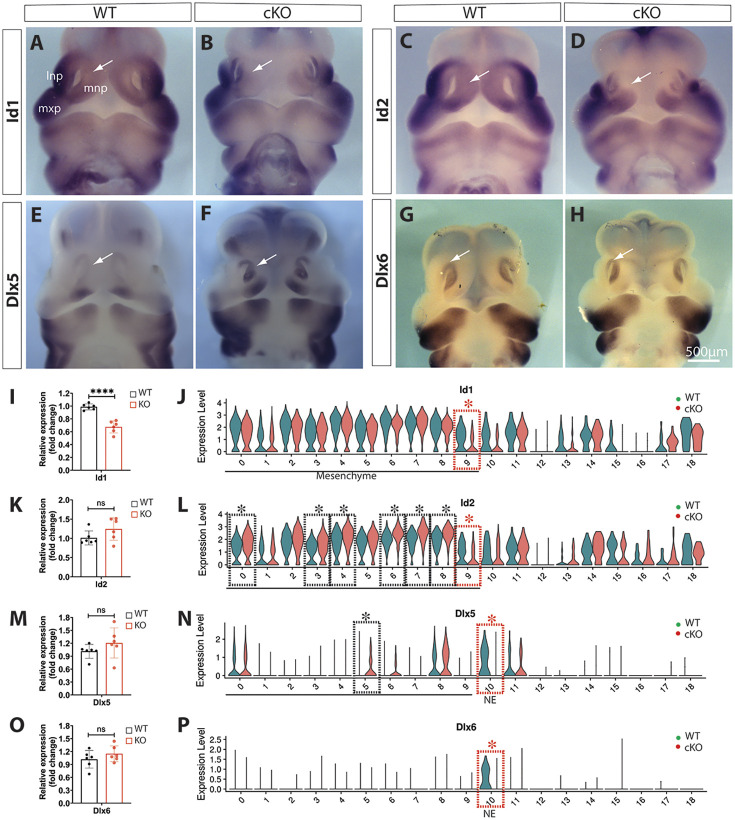
Moreover, Id1, which encodes a bHLH-containing inhibitor of DNA binding and cell differentiation, was downregulated in dorsal MNPs and identified as a DEG in m9 of Wls-cKOs at E11.5 (Fig. 10A,B,I,J). In contrast, Id2 was downregulated in m9 and upregulated in several other mesenchymal clusters in Wls-cKOs (Fig. 10C,D,K,L). The distal-less gene Dlx5 is required for craniofacial development (Acampora et al., 1999) and it was identified as a DEG being upregulated in m5 but downregulated in nasal epithelia (cluster 10) of the mutants (Fig. 10E,F,M,N). Dlx6 was predominantly expressed in the nasal epithelia of the WT midfacial primordia and showed no significant changes by in situ hybridization and RT-qPCR, but was identified as a DEG in the mutant nasal epithelia by single-cell transcriptomics (Fig. 10G,H,O,P).
Fig. 10.
Alterations of Id1, Id2, Dlx5 and Dlx6 in Wls-cKO midfacial primordia at E11.5. (A-H) Front views of whole-mount in situ hybridization of control and Wls-cKO facial primordia showing altered expression patterns of the indicated genes. Arrows indicate dorsal MNPs or nasal epithelia. (I,K,M,O) RT-qPCR validation of the indicated genes in WT and Wls-cKO embryos (n=6 per group). ****P<0.0001; ns, no statistical significance (t-test). (J,L,N,P) Violin plots of scRNA-seq showing downregulated expression of Id1 and Id2 in m9, upregulation of Id1 in m0, m3, m4, m6, m7 and m8, downregulation of Dlx5 in nasal epithelia (NE) and concurrent upregulation of Dlx5 in m5, and Dlx6 downregulation in nasal epithelia (NE) of the mutants. Dashed boxes indicate cell clusters with altered gene expression in the mutants (black, upregulated; red, downregulated). Asterisks indicate statistical significance (P_adj<0.05; bimod test). lnp, lateral nasal prominence; mnp, medial nasal prominence; mxp, maxillary prominence.
Together, these results suggest that Wls systematically modulates various transcription factors required for midfacial morphogenesis.
Alterations of Fgf8 and non-neural epithelial marker genes in Wls-deficient midfacial primordia
Because Pax3Cre is also activated in a subset of non-neural epithelia in the distal nasal pit and adjacent surface ectoderm (Fig. 1A-D), we examined several genes in the region by whole-mount in situ hybridization and scRNA-seq. The cell survival factor Fgf8 was predominantly expressed in the distal nasal epithelia at E10.5 and E11.5 in WT control embryos (Fig. S8A,C). This expression was diminished specifically in the dorsal domains of the distal nasal epithelia, but conserved in the ventral domains adjacent to LNPs and/or MNPs and in other facial ectodermal regions in the Wls-cKOs at E10.5 and E11.5 (Fig. S8B,D). The region-specific loss of Fgf8 in the mutant nasal epithelia was not confirmed by scRNA-seq (Fig. S9). Fgfr2 and Sp8 were not altered in Wls-cKO nasal epithelia at E11.5 by either approach (Figs S8E-H and S9). In contrast, no alteration in expression of Cldn6 and Krt8 was detected by in situ hybridization (Fig. S8I-L), but both were identified as DEGs in Wls-cKO cluster 10 by scRNA-seq (Fig. S9). These results suggest that Fgf8 and related epithelial genes are regulated by Wls in a subset of nasal epithelial cells with Pax3Cre activities.
Functional interaction and GRNs in an MNP mesenchymal subpopulation responsible for midline facial formation and clefts
Our results indicate that among multiple mesenchymal subpopulations m1 and m9 are uniquely located in the dorsal regions of the paired MNPs, which start to merge at the midline from E11.5 of WT embryos (Fig. 3) and that m9, but not m1, has the greatest number of downregulated and functionally relevant DEGs in Wls-deficient midfacial primordia (Fig. 4). Notably, these downregulated DEGs, which have been validated by at least two experimental approaches (whole-mount in situ hybridization, RT-qPCR or scRNA-seq) form an interactive network, which also involves Fgf8 (secreted by adjacent nasal epithelia) at the protein level (Fig. 11A), suggesting their coherent function in midline facial formation and clefts.
Fig. 11.
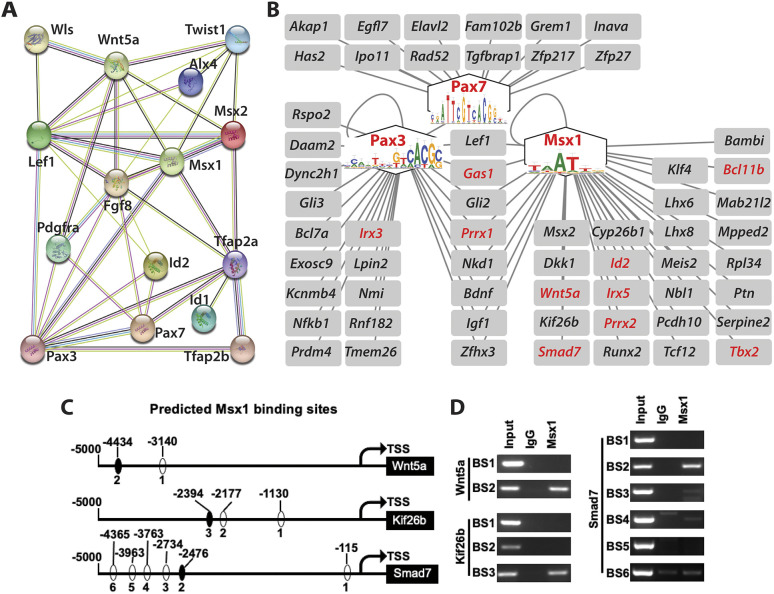
Functional interaction and GRN modulated by Wls in a MNP mesenchymal subpopulation during midline facial formation and fusion. (A) A string interaction network consists of crucial proteins encoded by Wls and downregulated DEGs in m9 that are validated in the current study. Fgf8 in the dorsal nasal tip (a regional signaling center) is also included for its functional relevance. Edges represent protein-protein associations and colors represent the following: light blue, from curated databases; purple, experimentally determined; red, gene fusions; green, gene neighborhood; blue, gene co-occurrence; yellow, from text mining, black, co-expression. (B) A significant GRN modular structure revealed by SCENIC consisting of three core regulons (Msx1, Pax3 and Pax7) and their predicated downstream target genes in m9. Cis-regulatory elements are shown in the respective regulons that are required for midfacial formation and fusion. The downstream target genes include components in multiple morphogenetic signaling pathways and many others with undefined functions in midfacial development. The genes in red are also DEGs downregulated in Wls-deficient m9. (C) Predicted Msx1-binding sites in the respective promoter regions of Wnt5a, Kif26b and Smad7. TSS, transcription start site. (D) ChIP demonstrates recruitment of the transcription factor Msx1 to a specific binding site of each of the selected downstream target genes. BS, binding site.
To investigate further the regulatory mechanisms within m9, we performed single-cell regulatory network inference and clustering (SCENIC) analyses (Aibar et al., 2017) to map the GRN activities in WT and Wls-cKO cells. A regulatory transcription factor co-expressed with its target genes in the same cells is defined as a regulon. SCENIC analysis revealed that a subcluster of WT m9 is strongly driven by Msx1, Pax3 and Pax7, which are inactivated in Wls-cKO m9 (Fig. S10). Based on co-expression and cis-regulatory motif analyses, SCENIC predicted 34 Msx1 targets, 24 Pax3 targets and 13 Pax7 target genes (Fig. 11B). Msx1 and Pax3 have eight common targets, including Lef1 (canonical Wnt signaling pathway), Gas1 (a Shh co-receptor), Gli2 (a Shh transcription factor) and Prrx1 (a paired related homeobox gene). Other notable Msx1 targets include its own family members (Msx1 and Msx2), Wnt signaling genes (Dkk1, Wnt5a and Kif26b), the Tgfβ signaling transcription factor Smad7, the retinoic acid signaling gene Cyp26b1, the mesenchymal transcription factors Id2, Irx5, Prrx2 and Runx2, and the cell cycle regulator Klf4. Other notable Pax3 targets include its own family members (Pax3 and Pax7), Wnt signaling genes (Rspo2 and Daam2), the dynin-2 complex member Dync2h1 and the Shh transcription factor Gli3. Nine of the predicted Msx1 targets (Bcl11b, Gas1, Id2, Irx5, Prxx1, Prxx2, Smad7, Tbx2 and Wnt5a) and three of the predicted Pax3 targets (Gas1, Irx3 and Prrx1) were downregulated in mutant m9, indicating robust regulon activities of Msx1 and Pax3 modulated by Wls.
To demonstrate, partially, the direct regulation of candidate downstream target genes by upstream transcription factors in this GRN module, we searched the presumptive upstream promotor regions of Wnt5a, Kif26b and Smad7, and found multiple Msx1-binding sites in the respective genes (Fig. 11C). Chromatin immunoprecipitation (ChIP) assays using WT midfacial primordial cells at E11.5 demonstrated that antibodies against Msx1 can pull down the binding site 2 (BS2) in Wnt5a, BS3 in Kif26b and BS2 in Smad7 promoter regions (Fig. 11D), suggesting a direct regulation by Msx1 of these candidate downstream target genes.
Together, these results provide an exemplary regulatory network mechanism underlying MNP mesenchymal morphogenesis in which Wls modulates a core GRN consisting of crucial regulons that further integrate Wnt, Tgfβ, Shh and retinoic acid signaling pathways and relevant components participating in midline facial formation and merging.
DISCUSSION
Cellular mechanisms of midline facial formation and clefts
During craniofacial development, midfacial primordia undergo dynamic morphological changes involving precisely regulated three-dimensional tissue growth, apoptosis, and fusion processes in a short period of time (Abramyan and Richman, 2015; Chai and Maxson, 2006; Ji et al., 2020; Jiang et al., 2006; Suzuki et al., 2016). Our study shows that E11 to E12 is a critical time window for midfacial morphogenesis in mice. During this time, mesenchymal cells from paired MNPs merge to the midline. The merging of mesenchyme occurs at the dorsal MNPs around E11.5 and finishes completely in less a day. The cellular and molecular mechanisms underlying midline merging remain poorly understood. Our results demonstrate that mesenchymal Wls deficiency disrupts mesenchymal convergence in the midline, which prevents normal development of the nasal septum, philtrum and associated orofacial structures, and results in midline facial clefts.
Excessive apoptosis was detected in bilateral forehead regions adjacent to the dorsal nasal primordia in Wls-deficient mutants, which clearly contributes to frontonasal hypoplasia as demonstrated by missing forehead tissues in the mutants. However, it is unclear whether apoptosis directly contributes to the midline clefts. Although the mutant primordia were much shorter and failed to fuse in the anterior midline, the nasal septum and its associated vomeronasal organs are still formed, suggesting that no excessive cell death occurs in the mutant midline mesenchyme, which generates the nasal septum and associated structures. Pax3Cre elicits recombination activities in both mesoderm- and neural crest-derived mesenchymal cells (Engleka et al., 2005). The missing bone and cartilage elements in Pax3Cre;Wls-cKO mutants further confirmed a crucial role for Wls in skeletal development and homeostasis (Fu et al., 2011; Goodnough et al., 2014; Maruyama et al., 2013; Wan et al., 2013; Zhong et al., 2012, 2015). Pax3Cre is also active in the surface ectodermal lineage cells of the dorsal nasal pit, which is considered a regional signaling center regulated by Fgf8 during nasal development (Kawauchi et al., 2005). Our results demonstrate that Fgf8 is specifically diminished in this regional signaling center, which may contribute to the excessive apoptosis observed in the adjacent tissues in Wls-cKO mutants. It has also been shown that cranial ectodermal Wls is required for specification and differentiation of osteoblast and dermal progenitors and surface ectoderm Wls mutants display severe craniofacial malformations (Goodnough et al., 2014). Nevertheless, it remains unclear whether surface ectodermal Wls also contributes to midline facial fusion and clefts.
Single-cell transcriptomics of midline facial formation and clefts
In addition to genetic fate mapping, conditional gene targeting and phenotypic analyses, the current study employed scRNA-seq to study transcriptomic mechanisms underlying normal and defective midline facial formation and fusion at the single-cell level. Transcriptomic profiles faithfully revealed nine unique cell types, including eight relatively small populations consisting of facial ectodermal cells, nasal epithelial cells, peripheral glia, cranial ganglion neurons, myoblasts, endothelial cells, erythrocytes and myeloid cells, and a large mesenchymal population (more than 87% of WT cells and 82% of cKO cells), which is consistent with the fact that the midfacial primordia mainly consist of neural crest-derived mesenchymal cells (Chai and Maxson, 2006; Helms et al., 2005).
The midfacial mesenchymal cells isolated from the whole structures of MNPs, LNPs and MxPs can be further divided into ten subpopulations based on enriched markers in both WT and Wls-cKO at E11.5, which is different from a published scRNA-seq study that revealed nine mesenchymal subclusters in the bilateral junction zones by excluding the dorsal MNPs and LNPs and ventral MxPs (Li et al., 2019). Based on marker gene expression patterns, we were able to locate the anatomical positions of these mesenchymal subpopulations, including m1, m5, m8 and m9, which are represented by strong Alx3 expression and are located in the MNP. The m2 and m7 clusters, represented by strong Pax7 expression, were located in the LNP, and m0, m3, m4 and m6 were located in the MxP based on marker gene expression patterns. The positional relationships among these mesenchymal subpopulations, visualized by UMAP, were highly correlated with their anatomical structures but with undefined mechanisms. One possible explanation is that the patterning of cranial neural crest cells can be influenced by the microenvironment, for example signal cues from the cranial mesoderm, ectoderm or endoderm during or post-migration (Trainor and Krumlauf, 2000; Trainor et al., 2003). Recent scRNA-seq studies have revealed unique properties of pre-migratory cranial neural crest cells by reactivation of the pluripotent gene Oct4 (Zalc et al., 2021) or prerequisite activation of the bHLH transcription factor Twist1 (Soldatov et al., 2019). However, it remains unclear when and how the cranial neural crest-derived mesenchymal cells gained the unique transcriptomic profiles that define their anatomical locations and related functional properties during midfacial development.
Notably, the number of cells of the m5 cluster, located in the dorsal MNP adjacent to the nasal pit, was drastically decreased from 12.18% in WT to 4.27% in cKO, suggesting that many apoptotic cells were removed from Wls-cKO m5 subpopulation during sample preparation by fluorescence-activated cell sorting for scRNA-seq, which used live cells only for the 10x pipeline. As a potential consequence, we detected predominantly upregulated DEGs in the mutant m5 mesenchymal cells, including genes involved in orofacial development, but there was no evidence for these upregulated genes in the pathogenesis of midline facial clefts in the mutants. However, a few downregulated DEGs in the m5 cluster, including Pax3, which is also downregulated in the m9 cluster, may play a role in MNP morphogenesis and midline fusion.
By contrast, m1 and m9, the other two mesenchymal subpopulations located in the dorsal MNPs adjacent to the midline, exhibited no major changes in the cell numbers between WT and Wls-cKO samples, suggesting that they are less impacted by the ectopic apoptosis occurring in the mutants. The m1 subpopulation had no significant DEGs relevant to midfacial formation, but m9 exhibited the greatest number of downregulated DEGs that are highly relevant to midline facial formation and fusion (which will be discussed in detail below), suggesting that m9 is a key mesenchymal subpopulation with unique transcriptomics regulated by Wls for midline facial formation and merging. It is worth mentioning that our scRNA-seq results did not detect all crucial DEGs, such as Alx3 and Alx4, which were clearly diminished in dorsal MNPs of the mutants detected by whole-mount in situ hybridization and RT-qPCR, implying a technical limitation of scRNA-seq that we cannot explain.
Molecular mechanisms and GRNs underlying midline facial formation and clefts
At the molecular level, our combined approaches using in situ hybridization, RT-qPCR and scRNA-seq revealed a panel of significant DEGs in a mesenchymal subpopulation of Wls-deficient MNPs, which encode crucial proteins that are interactively connected within the same network required for midfacial development (Fig. 11A). Among these functionally relevant DEGs, Wnt5a is required for outgrowth of craniofacial and other distal structures and Wnt5a-null embryos display a flattened face (Yamaguchi et al., 1999) that is similar to the significantly shortened snout and slightly widened face in the Wls-cKO embryos with diminished Wnt5a expression in the dorsal nasal primordia. These unique facial deformations can be a result of disrupted non-canonical Wnt/planar cell polarity signaling and convergent extension, which are required for directional tissue morphogenesis (Gao, 2012), but which have not been clearly addressed in midfacial morphogenesis. In contrast, Lef1 is a key transcription factor regulating downstream target genes in the canonical Wnt/β-catenin signaling pathway (Eastman and Grosschedl, 1999). Msx1 and Msx2 are downstream target genes of Lrp6-mediated canonical Wnt/β-catenin signaling during midfacial formation and fusion (Song et al., 2009), and Msx1/Msx2 double null mutants display severely disrupted craniofacial structures with orofacial clefts (Ishii et al., 2005). Pax3 is a direct target of Wnt/β-catenin signaling during neural tube closure (Zhao et al., 2014). Dominant-negative Pax3 knock-in mutants or double-knockout mutants of Pax3 and Pax7 exhibit severe frontonasal dysplasia, including midline facial clefts (Zalc et al., 2015). These results suggest that Msx1, Msx2 and Pax3 are key downstream effectors of Wls-modulated canonical Wnt signaling in midline facial formation and fusion. It remains unknown whether other genes, including Alx4, Pdgfra, Tfap2a and Twist1, which encode interactive proteins in the same network, are directly regulated by canonical Wnt/β-catenin signaling. The current study demonstrates that these molecules form a functionally interactive network regulated by Wls in midline facial formation and fusion.
The current study also demonstrates that the dorsal regions (which can be defined as the posterior regions of the body axis) of the medial and lateral nasal primordia are mostly affected, and the ventral (or anterior) regions of these primordia are less impacted by Wls deficiency, which may reflect an evolutionarily conserved role of Wnt signaling in body axis specification (Hikasa and Sokol, 2013). It may be also linked with the defective Fgf8 expression in the dorsal regions of the distal nasal pits in the Wls-cKO mutants. Fgf8 is a downstream target gene of Wnt/β-catenin signaling pathway during facial development, as demonstrated in our previous study (Wang et al., 2011). Intriguingly, Sp8 and Fgf8 induce one another in cortical patterning (Sahara et al., 2007), and conditional inactivation of Sp8 with Pax3Cre also causes midline facial clefts (Kasberg et al., 2013). However, we did not detect significant alterations of Sp8 in Pax3Cre;Wls-cKO mutants, suggesting that Sp8 is not a downstream effector of Wls in midline facial formation and fusion. Three-dimensional tissue growth is regulated by complex gene expression patterns that can be coordinated through GRNs that consist of transcription factors and cis-regulatory elements in modular structures for specific transcriptional outputs (Martin et al., 2016; Materna and Davidson, 2007). Our SCENIC analysis revealed a specific GRN modular structure in the mesenchymal subpopulation m9 regulated by three core transcription factors, Msx1, Pax3 and Pax7, which are all downregulated in Wls-deficient MNP mesenchymal cells and are required for midline facial formation and fusion, further demonstrating their functional relevance and molecular regulatory mechanisms in the same developmental process. Their downstream target genes include components in Wnt, Tgfβ, hedgehog and retinoic acid signaling pathways. These morphogenetic signaling pathways play interactive roles in midfacial development and related disorders, particularly in lip/palate formation and orofacial clefts (Reynolds et al., 2019; Reynolds et al., 2020; Suzuki et al., 2016). For instance, disrupting hedgehog and Wnt signaling interactions promotes cleft lip pathogenesis (Kurosaka et al., 2014). The effects and mechanisms underlying signaling interactions between Wnt pathway and other morphogenetic pathways in midfacial formation and clefts warrant further investigations.
Several signaling genes including Wnt5a, Lef1 and Smad7 are downregulated in the Wls-deficient MNP mesenchymal cells. Interestingly, Wnt5a/Ror signaling regulates Kif26b at the protein level by inducing its degradation by the ubiquitin-proteasome system (Susman et al., 2017). Kif26b is required for kidney development (Uchiyama et al., 2010), but its role in craniofacial development has not been defined. Also, the roles of many other target genes in midfacial development have not been defined. Our GRN analysis revealed that Kif26b, along with many others are predicted downstream target genes of Msx1. We demonstrate that Msx1 can directly bind to specific motifs in the promoter regions of Wnt5a, Smad7 and Kif26b in the midfacial primordial cells. Together, these results provide another layer of mechanistic insights into the modulation and integration of multiple signaling pathways by Wls through a core GRN in the MNP. This GRN module does not include several significantly altered genes in Wls-deficient mutants, such as Alx4, Pdgfra, Tfap2a and Twist1, suggesting that they may act in parallel or independently of this active GRN in the same region at the same age or they may act in different GRN modules at different ages during midfacial development. This study also provides future directions for investigations into the roles of new GRNs and downstream target genes in midfacial development and related disorders.
MATERIALS AND METHODS
Mouse lines
All animal studies were approved by UC Davis Animal Care and Use Committee and conformed to NIH guidelines. Pax3Cre knock-in (Engleka et al., 2005), Wlsflox (Carpenter et al., 2010) and Rosa26mT/mG reporter (Muzumdar et al., 2007) mouse lines were acquired from The Jackson Laboratory. Pregnant, timed-mated mice were euthanized prior to cesarean section. Noon of the day of conception was designated as E0.5.
Genetic fate-mapping and conditional gene-targeting analyses
Pax3Cre/+;Wlsflox/+ mice were mated with Rosa26-mT/mG;Wlsflox/flox mice for genetic fate mapping of the Pax3Cre lineage cells and conditional gene-targeting analyses of Wls in facial mesenchymal cells. Heterozygous Pax3Cre/+;Wlsflox/+ and cKO Pax3Cre/+;Wlsflox/flox embryos containing a Rosa26-mT/mG reporter were sampled at E10.5-E12.5 for genetic fate mapping and morphological analyses by confocal microscopy using a Nikon A1 confocal laser microscope. The three-dimensional frontal views of embryonic heads were reconstructed from z-stacks and large images using NIS-Elements C software.
Skeletal preparation
The whole-mount skeletal staining was carried out as previously described (Rigueur and Lyons, 2014). Briefly, E18.5 cKO embryos and littermate controls were dissected out and fixed in 80% ethanol overnight, followed by removal of the skin and internal organs. The samples were then dehydrated in 95% ethanol for 24 h, followed by acetone wash for 2-3 days. The embryos were then stained with 0.03% Alcian Blue (prepared in ethanol/glacial acetic acid) for 2 days, followed by dehydration in 70-95% ethanol overnight. The embryos were then cleared with 1% KOH and stained with 0.1% Alizarin Red for 6 h. The stained embryos were cleared in 50% glycerol with 1% KOH solution for several weeks until ready for brightfield stereo imaging using a Lumar V12 stereomicroscope (Zeiss).
Whole-mount in situ hybridization and TUNEL assays
Whole-mount in situ hybridization was performed as previously described (Song et al., 2009). Briefly, embryos fixed in 4% paraformaldehyde overnight at 4°C were processed for whole-mount in situ hybridization using digoxigenin-labeled antisense RNA probes. Primers for all probes were referenced to Allen Brain Atlas (https://mouse.brain-map.org/). At least two mutants and two littermate control embryos were used for each in situ experiment for consistent results. Whole-mount TUNEL assays were carried out as previously described (Kanzler et al., 2000) with minor modifications. The DeadEnd Fluorometric TUNEL system (Promega) was used for labeling apoptotic cells, and embryos were counterstained with DAPI. A series of z-stacks generated using a Nikon A1 confocal microscope were used for reconstruction of whole-mount images using NIS Elements software (Nikon). Three mutant embryos and three littermate controls were examined at each age. The automatic cell counting function of ImageJ (Schneider et al., 2012) was used to count numbers of TUNEL-positive cells in equal areas of the left and right nasal junction zones or forehead regions for statistical analyses. Two-tailed, unpaired Student's t-test was used for statistical comparisons when appropriate and differences were considered significant at P<0.05.
Total RNA isolation and RT-qPCR
Mouse embryonic midfacial primordia consisting of the maxillary, lateral and medial nasal prominences at E11.5 were microdissected in RNase-free PBS for total RNA extraction using a RNeasy kit (QIAGEN). Samples from six Wls-cKOs and six littermate control embryos were collected. RNA concentrations were measured using a NanoDrop One spectrophotometer (Thermo Fisher Scientific). RT-qPCR was performed as previously described (Song et al., 2009) with minor modifications. The iScript cDNA synthesis kit (Bio-Rad) was used to synthesize cDNAs from 400 ng of total RNA per sample for subsequent qPCR analysis. qPCR reactions were duplicated using TB Green Advantage qPCR Premix (Takara Bio) with the AriaMx real-time PCR system (Agilent Technologies). The relative expression level of target genes was calculated using the ΔCT method. The housekeeping gene Gapdh was used to normalize qPCR results, which are shown as mean±s.d. The calculation of mean±s.d. and statistical analysis (two-tailed, unpaired Student's t-test adjusted with Welch's correlation) was performed using GraphPad Prism V9.1 software. A P-value less than 0.05 was considered significant. The primers used in this study are included in Table S3.
Single-cell preparation and scRNA-seq analyses
E11.5 mouse midfacial primordia were microdissected in cold PBS and dissociated in cold protease solution modified from a published protocol (Adam et al., 2017). The freshly prepared single-cell suspensions were filtered and processed for fluorescent-activated cell sorting to remove the dead cells labeled by DAPI. After PCR genotyping, two cKOs and three littermate controls were respectively pooled for subsequent sequencing preparation. Barcoded 3′ single cell libraries were prepared from single-cell suspensions using the Chromium Single Cell 3′ Library and Gel Bead kit (10x Genomics) for sequencing according to the recommendations of the manufacturer. The cDNA and library fragment size distributions were verified by micro-capillary gel electrophoresis on a Bioanalyzer 2100 (Agilent Technologies). Libraries were quantified by fluorometry on a Qubit instrument (Life Technologies) and by qPCR with a Kapa Library Quant kit (Kapa Biosystems-Roche) prior to sequencing. Libraries were sequenced on the HiSeq 4000 sequencer (Illumina) with paired-end 100 bp reads.
Mapping
FASTQ files for all samples were generated by the DNA Technologies and Expression Analysis Core Lab of the UC Davis Genome Center. The Cell Ranger Single Cell software suite from 10x Genomics was used to align reads and generate feature-barcode matrices. Raw reads were processed using the Cell Ranger count program using default parameters. This program uses STAR to map cDNA reads to the transcriptome using mouse reference package (mm10).
Quality control and pre-processing
Seurat_3.0.3 (Butler et al., 2018) was used for all following analyses. All genes that were not detected in at least three single cells were excluded. Cells in which fewer than 300 unique genes were detected and for which the unique molecular identifier was greater than 60,000 were excluded to remove possible low-quality cells, doublets and multiplets. Mitochondrial quality control metrics were calculated using the ‘PercentageFeatureSet’ function to filter out cells with >20% mitochondrial counts, avoiding low-quality and dying cells. After filtering, 15,610 genes in total across 7725 cells (4284 from WT samples and 3441 from cKO samples) in the midfacial dataset remained for downstream analysis. Normalization was performed by the global-scaling normalization method ‘LogNormalize’ in Seurat, then the results were log-transformed for downstream analysis. Then the ‘FindVariableFeatures’ function was used to calculate a subset of highly variable features (default: 2000 genes) for each sample to help highlight biological signals in future analyses.
Integration analysis
The ‘FindIntegrationAnchors’ and ‘IntegrateData’ functions in Seurat_3.0.3 were employed to integrate the two datasets, WT and cKO (Stuart et al., 2019). Then a linear transformation as a standard pre-processing step was performed on the combined dataset and the effects of cell cycle heterogeneity in this combined dataset was regressed out during data scaling. Principal component analysis (a dimensional reduction technique) was conducted to determine the dimensionality of the dataset and UMAP (McInnes et al., 2018 preprint) was chosen to visualize the combined dataset. The ‘FindNeighbors’ and ‘FindClusters’ functions were applied to cluster cells, cluster-specific markers conserved across conditions were identified with the ‘FindConservedMarkers’ function and clusters were assigned to known cell types based on their specific markers. After all the cells were aligned, comparative analysis was performed to identify DEGs induced by gene knockout. Differential expression was assessed with ‘Bimod’ (likelihood-ratio test) and genes for which P_adj <0.05 and |logFC|>0.2 were defined as significant DEGs. All marker genes and DEGs were visualized with ‘FeaturePlot’, ‘DotPlot’ and ‘VlnPlot’ functions in Seurat and the R package ‘ggplot2’ (https://ggplot2.tidyverse.org). Specifically, for DEGs selected for pathway analysis, two R packages, ‘clusterProfiler’ and ‘GOplot’ (Walter et al., 2015; Yu et al., 2012), were used for gene set enrichment analysis and visualization. Only functional categories related to midfacial development functions were chosen.
SCENIC analysis
SCENIC analysis was run as described (Aibar et al., 2017) (SCENIC version 1.1.2.2, which corresponds to GENIE3 1.4.3, RcisTarget 1.2.1 and AUCell 1.4.1) using the 24,453 motifs database for RcisTarget (mm9-500bp-upstream-7species.mc9nr.feather; mm9-tss-centered-10kb-7species.mc9nr.feather). The input matrix was the raw/non-normalized expression counts matrix (including WT and cKO) from which 9651 genes passed the default filtering. From these genes, only the genes that were available in RcisTarget databases were kept (8787 genes) in the co-expression modules from GENIE3 and analyzed for motif enrichment with RcisTarget. Regulators (‘Regulon’ defined by SCENIC) for different cell types in two different conditions (WT and cKO) were selected based on the ‘RelativeActivity’ score.
ChIP assay
ChIP assays were carried out using a ChIP assay kit (Sigma-Aldrich, 17-295) according to the manufacturer's instruction as previously described (Wang et al., 2011). Briefly, midfacial primordia were microdissected from E11.5 WT embryos and homogenized in ice-cold PBS then incubated with 1% formaldehyde at 37°C for 10 min to allow crosslinking to occur. Following centrifugation (425 g for 4 min at 4°C), the pellet was re-suspended in lysis buffer and sonicated. Salmon sperm DNA/Protein A-agarose was then used to preclear the fragmented chromatin. The precleared protein-DNA complexes were incubated with a monoclonal anti-Msx1 antibody (Thermo Fisher Scientific, H00004487-M11) or a control mouse IgG2a (Cell Signaling Technology, 61656S). After washing, samples were reverse cross-linked. To determine Msx1-binding sites (TAATTG or CAATTA) on gene promoter regions, we retrieved promoter sequences of Wnt5a, Kif26b and Smad7 (−5000 to +100 bp) from the Eukaryotic Promoter Database (https://epd.epfl.ch//index.php). For protein-DNA interaction validation, we used GoTaq® G2 DNA Polymerase (Promega) for PCR amplification (35-40 cycles) and electrophoresed the product on 2% agarose gels. The separated bands in agarose gels were then observed under ultraviolet light. Primer sets used for ChIP assays are shown in Table S4.
Supplementary Material
Acknowledgements
We are grateful to Arjun Stokes, Huan Zhao, Rebecca Donham, Taylor Imai, Yue Liu, Michael Garland, Sarwat Amina, Santosh Kumar, Sunil Rai, Vui Doan, and other Zhou lab members for their efforts or discussions; and Bridget Mclaughlin and Jonathan Van Dyke (UC Davis Flow Cytometry Shared Resource) for conducting fluorescence-activated cell sorting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.J.Z.; Methodology: R.G., S.Z., S.K.S., Y.J., B.S., C.J.Z.; Software: S.Z.; Validation: R.G., S.Z., S.K.S., C.J.Z.; Formal analysis: S.Z., S.K.S., C.J.Z.; Investigation: R.G., S.Z., S.K.S., C.J.Z.; Data curation: R.G., S.Z., S.K.S., Y.J., K.R., M.M., B.S., M.I., D.B.-W., C.J.Z.; Writing - original draft: R.G., S.Z., S.K.S., C.J.Z.; Writing - review & editing: K.R., P.A.T., Y. Chen, Y.X., Y. Chai, C.J.Z.; Visualization: R.G., S.Z., S.K.S., C.J.Z.; Supervision: C.J.Z.; Project administration: C.J.Z.; Funding acquisition: C.J.Z.
Funding
This work was supported by grants from the National Institutes of Health (R01DE026737, R01NS102261 and R01DE021696 to C.J.Z.) and the Shriners Hospitals for Children (85105 to C.J.Z. and postdoctoral fellowships to R.G. and B.S.). Research in the Trainor lab is supported by the Stowers Institute for Medical Research; the Chen lab by the National Institutes of Health; the Xu lab by the Ministry of Science and Technology and the National Natural Science Foundation of China; and the Chai lab by the National Institutes of Health. Deposited in PMC for release after 12 months.
Data availability
The sequencing data for this project are accessible from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA579176.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200533.
References
- Abramyan, J. and Richman, J. M. (2015). Recent insights into the morphological diversity in the amniote primary and secondary palates. Dev. Dyn. 244, 1457-1468. 10.1002/dvdy.24338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acampora, D., Merlo, G. R., Paleari, L., Zerega, B., Postiglione, M. P., Mantero, S., Bober, E., Barbieri, O., Simeone, A. and Levi, G. (1999). Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126, 3795-3809. 10.1242/dev.126.17.3795 [DOI] [PubMed] [Google Scholar]
- Adam, M., Potter, A. S. and Potter, S. S. (2017). Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144, 3625-3632. 10.1242/dev.151142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar, S., González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H., Hulselmans, G., Rambow, F., Marine, J.-C., Geurts, P., Aerts, J.et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083-1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alappat, S., Zhang, Z. Y. and Chen, Y. P. (2003). Msx homeobox gene family and craniofacial development. Cell Res. 13, 429-442. 10.1038/sj.cr.7290185 [DOI] [PubMed] [Google Scholar]
- Bänziger, C., Soldini, D., Schütt, C., Zipperlen, P., Hausmann, G. and Basler, K. (2006). Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509-522. 10.1016/j.cell.2006.02.049 [DOI] [PubMed] [Google Scholar]
- Brault, V., Moore, R., Kutsch, S., Ishibashi, M., Rowitch, D. H., McMahon, A. P., Sommer L., Boussadia, O. and Kemler, R. (2001). Inactivation of the (β)-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253-1264. 10.1242/dev.128.8.1253 [DOI] [PubMed] [Google Scholar]
- Becht, E., McInnes, L., Healy, J., Dutertre, C.-A., Kwok, I. W. H., Ng, L. G., Ginhoux, F. and Newell, E. W. (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38-44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Berthiaume, L. G. (2014). Wnt acylation: seeing is believing. Nat. Chem. Biol. 10, 5-7. 10.1038/nchembio.1414 [DOI] [PubMed] [Google Scholar]
- Beverdam, A., Brouwer, A., Reijnen, M., Korving, J. and Meijlink, F. (2001). Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975-3986. 10.1242/dev.128.20.3975 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E. and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. C., Rao, S., Wells, J. M., Campbell, K. and Lang, R. A. (2010). Generation of mice with a conditional null allele for Wntless. Genesis 48, 554-558. 10.1002/dvg.20651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y. and Maxson, R. E.Jr. (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353-2375. 10.1002/dvdy.20833 [DOI] [PubMed] [Google Scholar]
- Chen, Z. F. and Behringer, R. R. (1995). twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9, 686-699. 10.1101/gad.9.6.686 [DOI] [PubMed] [Google Scholar]
- Cox, T. C. (2004). Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin. Genet. 65, 163-176. 10.1111/j.0009-9163.2004.00225.x [DOI] [PubMed] [Google Scholar]
- Dudas, M. and Kaartinen, V. (2005). Tgf-beta superfamily and mouse craniofacial development: interplay of morphogenetic proteins and receptor signaling controls normal formation of the face. Curr. Top. Dev. Biol. 66, 65-133. 10.1016/S0070-2153(05)66003-6 [DOI] [PubMed] [Google Scholar]
- Eastman, Q. and Grosschedl, R. (1999). Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol. 11, 233-240. 10.1016/S0955-0674(99)80031-3 [DOI] [PubMed] [Google Scholar]
- Engleka, K. A., Gitler, A. D., Zhang, M., Zhou, D. D., High, F. A. and Epstein, J. A. (2005). Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396-406. 10.1016/j.ydbio.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Eppley, B. L., van Aalst, J. A., Robey, A., Havlik, R. J. and Sadove, A. M. (2005). The spectrum of orofacial clefting. Plast. Reconstr. Surg. 115, 101e-114e. 10.1097/01.PRS.0000164494.45986.91 [DOI] [PubMed] [Google Scholar]
- Fu, J., Jiang, M., Mirando, A. J., Yu, H.-M. I. and Hsu, W. (2009). Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc. Natl. Acad. Sci. USA 106, 18598-18603. 10.1073/pnas.0904894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., Ivy Yu, H.-M., Maruyama, T., Mirando, A. J. and Hsu, W. (2011). Gpr177/mouse Wntless is essential for Wnt-mediated craniofacial and brain development. Dev. Dyn. 240, 365-371. 10.1002/dvdy.22541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B. (2012). Wnt regulation of planar cell polarity (PCP). Curr. Top. Dev. Biol. 101, 263-295. 10.1016/B978-0-12-394592-1.00008-9 [DOI] [PubMed] [Google Scholar]
- Goodnough, L. H., Dinuoscio, G. J., Ferguson, J. W., Williams, T., Lang, R. A. and Atit, R. P. (2014). Distinct requirements for cranial ectoderm and mesenchyme-derived wnts in specification and differentiation of osteoblast and dermal progenitors. PLoS Genet. 10, e1004152. 10.1371/journal.pgen.1004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, D., Malik, Z., Hayano, S. and Mishina, Y. (2016). Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor. Rev. 27, 129-139. 10.1016/j.cytogfr.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. and Chen, Y. P. (2012). Wnt signaling in lip and palate development. Front. Oral Biol. 16, 81-90. 10.1159/000337619 [DOI] [PubMed] [Google Scholar]
- He, F. and Soriano, P. (2013). A critical role for PDGFRalpha signaling in medial nasal process development. PLoS Genet. 9, e1003851. 10.1371/journal.pgen.1003851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., Xiong, W., Yu, X., Espinoza-Lewis, R., Liu, C., Gu, S., Nishita, M., Suzuki, K., Yamada, G., Minami, Y.et al. (2008). Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development 135, 3871-3879. 10.1242/dev.025767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J. A., Cordero, D. and Tapadia, M. D. (2005). New insights into craniofacial morphogenesis. Development 132, 851-861. 10.1242/dev.01705 [DOI] [PubMed] [Google Scholar]
- Herr, P. and Basler, K. (2012). Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev. Biol. 361, 392-402. 10.1016/j.ydbio.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Hikasa, H. and Sokol, S. Y. (2013). Wnt signaling in vertebrate axis specification. Cold Spring Harb. Perspect. Biol. 5, a007955. 10.1101/cshperspect.a007955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya, M., Lee, S. M., Johnson, J. E., McMahon, A. P. and Takada, S. (1997). Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389, 966-970. 10.1038/40146 [DOI] [PubMed] [Google Scholar]
- Ishii, M., Han, J., Yen, H.-Y., Sucov, H. M., Chai, Y. and Maxson, R. E.Jr. (2005). Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 132, 4937-4950. 10.1242/dev.02072 [DOI] [PubMed] [Google Scholar]
- Ji, Y., Hao, H., Reynolds, K., McMahon, M. and Zhou, C. J. (2019). Wnt signaling in neural crest ontogenesis and oncogenesis. Cells 8, 1173. 10.3390/cells8101173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., Garland, M. A., Sun, B., Zhang, S., Reynolds, K., McMahon, M., Rajakumar, R., Islam, M. S., Liu, Y., Chen, Y.et al. (2020). Cellular and developmental basis of orofacial clefts. Birth Defects Res 112, 1558-1587. 10.1002/bdr2.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R., Bush, J. O. and Lidral, A. C. (2006). Development of the upper lip: morphogenetic and molecular mechanisms. Dev. Dyn. 235, 1152-1166. 10.1002/dvdy.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.-R., Han, X. H., Taketo, M. M. and Yoon, J. K. (2012). Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development 139, 1821-1830. 10.1242/dev.075796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur, A. and Murray, J. C. (2005). Orofacial clefting: recent insights into a complex trait. Curr. Opin. Genet. Dev. 15, 270-278. 10.1016/j.gde.2005.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler, B., Foreman, R. K., Labosky, P. A. and Mallo, M. (2000). BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development 127, 1095-1104. 10.1242/dev.127.5.1095 [DOI] [PubMed] [Google Scholar]
- Kasberg, A. D., Brunskill, E. W. and Steven Potter, S. (2013). SP8 regulates signaling centers during craniofacial development. Dev. Biol. 381, 312-323. 10.1016/j.ydbio.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi, S., Shou, J., Santos, R., Hébert, J. M., McConnell, S. K., Mason, I. and Calof, A. L. (2005). Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development 132, 5211-5223. 10.1242/dev.02143 [DOI] [PubMed] [Google Scholar]
- Kurosaka, H., Iulianella, A., Williams, T. and Trainor, P. A. (2014). Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. J. Clin. Invest. 124, 1660-1671. 10.1172/JCI72688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Jones, K. L., Hooper, J. E. and Williams, T. (2019). The molecular anatomy of mammalian upper lip and primary palate fusion at single cell resolution. Development 146, dev174888. 10.1242/dev.174888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Wakamiya, M., Shea, M. J., Albrecht, U., Behringer, R. R. and Bradley, A. (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Gen. 22, 361-365. 10.1038/11932 [DOI] [PubMed] [Google Scholar]
- Lyons, L. A., Erdman, C. A., Grahn, R. A., Hamilton, M. J., Carter, M. J., Helps, C. R., Alhaddad, H. and Gandolfi, B. (2016). Aristaless-Like Homeobox protein 1 (ALX1) variant associated with craniofacial structure and frontonasal dysplasia in Burmese cats. Dev. Biol. 409, 451-458. 10.1016/j.ydbio.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., Organista, M. F. and de Celis, J. F. (2016). Structure of developmental gene regulatory networks from the perspective of cell fate-determining genes. Transcription 7, 32-37. 10.1080/21541264.2015.1130118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, T., Jiang, M. and Hsu, W. (2013). Gpr177, a novel locus for bone mineral density and osteoporosis, regulates osteogenesis and chondrogenesis in skeletal development. J. Bone Miner. Res. 28, 1150-1159. 10.1002/jbmr.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna, S. C. and Davidson, E. H. (2007). Logic of gene regulatory networks. Curr. Opin. Biotechnol. 18, 351-354. 10.1016/j.copbio.2007.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy, J. and Melville, J. (2018). UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv 1802.03426. 10.48550/arXiv.1802.03426 [DOI] [Google Scholar]
- Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J. and Shaw, W. C. (2009). Cleft lip and palate. Lancet 374, 1773-1785. 10.1016/S0140-6736(09)60695-4 [DOI] [PubMed] [Google Scholar]
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Niemann, S., Zhao, C., Pascu, F., Stahl, U., Aulepp, U., Niswander, L., Weber, J. L. and Müller, U. (2004). Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am. J. Hum. Genet. 74, 558-563. 10.1086/382196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse, R. and Clevers, H. (2017). Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985-999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Parada, C. and Chai, Y. (2012). Roles of BMP signaling pathway in lip and palate development. Front. Oral Biol. 16, 60-70. 10.1159/000337617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, K., Kumari, P., Sepulveda Rincon, L., Gu, R., Ji, Y., Kumar, S. and Zhou, C. J. (2019). Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis. Model. Mech. 12, dmm037051. 10.1242/dmm.037051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, K., Zhang, S., Sun, B., Garland, M. A., Ji, Y. and Zhou, C. J. (2020). Genetics and signaling mechanisms of orofacial clefts. Birth Defects Res. 112, 1588-1634. 10.1002/bdr2.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigueur, D. and Lyons, K. M. (2014). Whole-mount skeletal staining. Methods Mol. Biol. 1130, 113-121. 10.1007/978-1-62703-989-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara, S., Kawakami, Y., Izpisua Belmonte, J. C. and O'Leary, D. D. M. (2007). Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev. 2, 10. 10.1186/1749-8104-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle, H., Meier, P., Buchert, M., Jaenisch, R. and Mitchell, P. J. (1996). Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235-238. 10.1038/381235a0 [DOI] [PubMed] [Google Scholar]
- Schutte, B. C. and Murray, J. C. (1999). The many faces and factors of orofacial clefts. Hum. Mol. Genet. 8, 1853-1859. 10.1093/hmg/8.10.1853 [DOI] [PubMed] [Google Scholar]
- Soldatov, R., Kaucka, M., Kastriti, M. E., Petersen, J., Chontorotzea, T., Englmaier, L., Akkuratova, N., Yang, Y., Haring, M., Dyachuk, V.et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536. 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Song, L., Li, Y., Wang, K., Wang, Y.-Z., Molotkov, A., Gao, L., Zhao, T., Yamagami, T., Wang, Y., Gan, Q.et al. (2009). Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development 136, 3161-3171. 10.1242/dev.037440 [DOI] [PubMed] [Google Scholar]
- Stanier, P. and Pauws, E. (2012). Development of the lip and palate: FGF signalling. Front. Oral Biol. 16, 71-80. 10.1159/000337618 [DOI] [PubMed] [Google Scholar]
- Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., III, Hao, Y., Stoeckius, M., Smibert, P. and Satija, R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888-1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susman, M. W., Karuna, E. P., Kunz, R. C., Gujral, T. S., Cantu, A. V., Choi, S. S., Jong, B. Y., Okada, K., Scales, M. K., Hum, J.et al. (2017). Kinesin superfamily protein Kif26b links Wnt5a-Ror signaling to the control of cell and tissue behaviors in vertebrates. eLife 6, e26509. 10.7554/eLife.26509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, A., Sangani, D. R., Ansari, A. and Iwata, J. (2016). Molecular mechanisms of midfacial developmental defects. Dev. Dyn. 245, 276-293. 10.1002/dvdy.24368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo-Rogers, H. L., Smithers, L. E., Yakob, W. and Liu, K. J. (2010). New directions in craniofacial morphogenesis. Dev. Biol. 341, 84-94. 10.1016/j.ydbio.2009.11.021 [DOI] [PubMed] [Google Scholar]
- Trainor, P. and Krumlauf, R. (2000). Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat. Cell Biol. 2, 96-102. 10.1038/35000051 [DOI] [PubMed] [Google Scholar]
- Trainor, P. A., Melton, K. R. and Manzanares, M. (2003). Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int. J. Dev. Biol. 47, 541-553. [PubMed] [Google Scholar]
- Uchiyama, Y., Sakaguchi, M., Terabayashi, T., Inenaga, T., Inoue, S., Kobayashi, C., Oshima, N., Kiyonari, H., Nakagata, N., Sato, Y.et al. (2010). Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc. Natl. Acad. Sci. USA 107, 9240-9245. 10.1073/pnas.0913748107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uz, E., Alanay, Y., Aktas, D., Vargel, I., Gucer, S., Tuncbilek, G., von Eggeling, F., Yilmaz, E., Deren, O., Posorski, N.et al. (2010). Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 86, 789-796. 10.1016/j.ajhg.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, W., Sánchez-Cabo, F. and Ricote, M. (2015). GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912-2914. 10.1093/bioinformatics/btv300 [DOI] [PubMed] [Google Scholar]
- Wan, Y., Lu, C., Cao, J., Zhou, R., Yao, Y., Yu, J., Zhang, L., Zhao, H., Li, H., Zhao, J.et al. (2013). Osteoblastic Wnts differentially regulate bone remodeling and the maintenance of bone marrow mesenchymal stem cells. Bone 55, 258-267. 10.1016/j.bone.2012.12.052 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Song, L. and Zhou, C. J. (2011). The canonical Wnt/beta-catenin signaling pathway regulates Fgf signaling for early facial development. Dev. Biol. 349, 250-260. 10.1016/j.ydbio.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Xavier, G. M., Seppala, M., Barrell, W., Birjandi, A. A., Geoghegan, F. and Cobourne, M. T. (2016). Hedgehog receptor function during craniofacial development. Dev. Biol. 415, 198-215. 10.1016/j.ydbio.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T. P., Bradley, A., McMahon, A. P. and Jones, S. (1999). A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 126, 1211-1223. 10.1242/dev.126.6.1211 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang, L.-G., Han, Y. and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc, A., Rattenbach, R., Auradé, F., Cadot, B. and Relaix, F. (2015). Pax3 and Pax7 play essential safeguard functions against environmental stress-induced birth defects. Dev. Cell 33, 56-66. 10.1016/j.devcel.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Zalc, A., Sinha, R., Gulati, G. S., Wesche, D. J., Daszczuk, P., Swigut, T., Weissman, I. L. and Wysocka, J. (2021). Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 371, eabb4776. 10.1126/science.abb4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, T., Gan, Q., Stokes, A., Lassiter, R. N. T., Wang, Y., Chan, J., Han, J. X., Pleasure, D. E., Epstein, J. A. and Zhou, C. J. (2014). beta-catenin regulates Pax3 and Cdx2 for caudal neural tube closure and elongation. Development 141, 148-157. 10.1242/dev.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z., Zylstra-Diegel, C. R., Schumacher, C. A., Baker, J. J., Carpenter, A. C., Rao, S., Yao, W., Guan, M., Helms, J. A., Lane, N. E.et al. (2012). Wntless functions in mature osteoblasts to regulate bone mass. Proc. Natl. Acad. Sci. USA 109, E2197-E2204. 10.1073/pnas.1120407109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. A., Zahatnansky, J., Snider, J., Van Wieren, E., Diegel, C. R. and Williams, B. O. (2015). Wntless spatially regulates bone development through β-catenin-dependent and independent mechanisms. Dev. Dyn. 244, 1347-1355. 10.1002/dvdy.24316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.-J., Liu, Y., Yuan, X., Wang, M., Zhao, W., Yang, X., Zhang, X., Hsu, W., Qiu, M., Zhang, Z.et al. (2016). Ectodermal Wnt controls nasal pit morphogenesis through modulation of the BMP/FGF/JNK signaling axis. Dev. Dyn. 245, 414-426. 10.1002/dvdy.24376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.