Abstract
High-level expression of the major pilus subunit (PapA) of uropathogenic strains of Escherichia coli results in part from the unusually long lifetime of the mRNA that encodes this protein. Here we report that the longevity of papA mRNA derives in large measure from the protection afforded by its 5′ untranslated region. This papA RNA segment can prolong the lifetime of an otherwise short-lived mRNA to which it is fused. In vivo alkylation studies indicate that, in its natural milieu, the papA message begins with a stem-loop structure. This stem-loop is important for the stabilizing effect of the papA 5′ untranslated region, as evidenced by the significant acceleration in papA mRNA decay that results from its removal.
The pyelonephritis-associated pili (pap) genes of uropathogenic strains of Escherichia coli encode the surface fimbrial structures by which E. coli cells attach themselves to the epithelium of the upper urinary tract, a critical step in pathogenesis (11). The protein products of the pap gene cluster are involved in pilus assembly and in the regulation of pap gene expression. Two of these genes, specifying the major pilus subunit PapA and the transcription factor PapB, are cotranscribed from an inducible promoter as a dicistronic papBA operon transcript. Despite their coordinate transcription, the PapA protein is produced in substantial molar excess over PapB due to a posttranscriptional regulatory mechanism involving the differential stability of the papB and papA segments of the 1.2-kb papBA transcript (3). The 5′-terminal papB segment of this transcript is rapidly degraded with a half-life of 2 to 3 min, whereas the 3′ papA segment decays slowly, with a half-life of 20 to 30 min, after being liberated as a discrete 0.7-kb mRNA processing product by RNase E cleavage at an intercistronic site (3, 19). The greater longevity of the papA message allows this mRNA to accumulate to a large molar excess over its papBA mRNA precursor, thereby accounting for much of the difference in expression of the papB and papA genes.
We set out to investigate the basis for the longevity of papA mRNA. Our studies indicate that the lifetime of this mRNA is determined primarily by its 5′ untranslated region (UTR) and that a stem-loop near the 5′ end of this UTR helps to protect the message from degradation.
The papA 5′ UTR can stabilize a heterologous mRNA.
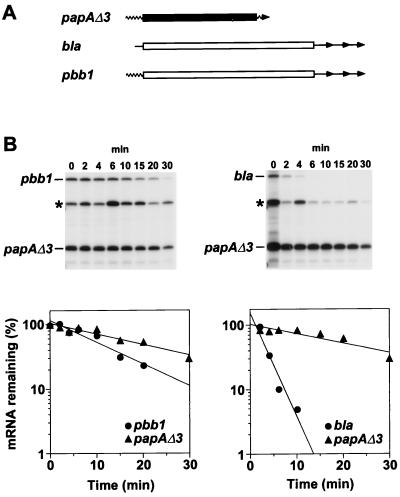
Earlier studies had shown that the unusual longevity of the E. coli ompA transcript is a consequence of its 5′ UTR, which functions as an mRNA stabilizer capable of prolonging the lifetime of a variety of heterologous messages to which it is fused (4, 10). To determine whether the 5′ UTR of papA can likewise function as an mRNA stabilizer, we decided to investigate whether the longevity of the short-lived bla message increases when its 5′ UTR is replaced with that of papA. To facilitate these studies, we first constructed a gene (papAΔ3) encoding a monocistronic papA message expressed from a constitutive promoter as a primary transcription product. Except for the absence of three nucleotides (AUU) from the 5′ end and the presence of a 5′-terminal triphosphate, this pseudo-wild-type transcript is identical to the papA message that arises by RNase E processing of the dicistronic papBA transcript in E. coli cells bearing the entire pap gene cluster. We then constructed a plasmid (pPBB1E) encoding a hybrid papAΔ3-bla transcript (pbb1) in which the 84-nucleotide papAΔ3 5′ UTR was joined precisely to the 286-codon protein-coding region and 3′ UTR of the short-lived bla message (Fig. 1A). As an internal control, this plasmid also bore a copy of the papAΔ3 gene. For comparison, we constructed an additional plasmid (pBLAE) bearing both the wild-type bla gene and the papAΔ3 gene.
FIG. 1.
Decay of a hybrid papA-bla mRNA. (A) The papA-bla hybrid transcript pbb1 is represented diagrammatically along with its mRNA progenitors papAΔ3 and bla. These mRNAs were expressed from plasmid pPBB1E or pBLAE, each a derivative of plasmid pBLA200 (8), by transcription from a bla promoter. Wavy lines, papA UTRs; straight lines, bla UTRs; solid rectangle, papA coding region: open rectangles, bla coding region; arrowheads, mRNA 3′ ends. There are three alternative sites of transcription termination downstream of the bla gene. (B) Cultures of a recA mutant derivative of E. coli MG1693 (2) harboring either pPBB1E (left) or pBLAE (right) were grown exponentially for several generations at 37°C in Luria-Bertani medium supplemented with glucose (0.4%) and Casamino Acids (0.5%). At time intervals after transcription inhibition with rifampin (200 μg/ml), total cellular RNA was isolated. Equal amounts of each RNA sample (2 μg) were then analyzed by S1 protection with a mixture of two 5′-end-labeled probes complementary to a 5′-terminal segment of papAΔ3 mRNA or bla mRNA. The radioactivity in bands that correspond to papAΔ3, pbb1, and bla mRNA was quantitated with a Molecular Dynamics PhosphorImager. Also marked is a band corresponding to the reannealed papA probe DNA (∗). Beneath each autoradiogram is a semilogarithmic plot of mRNA concentration versus time. Half-lives were calculated from the slope of each plot, and half-life errors were estimated from the standard deviation of the slopes. The measured half-lives were 8.9 ± 1.1 min for pbb1 mRNA and 18 ± 2 min for papAΔ3 mRNA (left) and 1.9 ± 0.4 min for bla mRNA and 20 ± 4 min for papAΔ3 mRNA (right). For procedural details concerning bacterial cell growth, RNA isolation, and S1 analysis, see reference 8.
E. coli cells containing either pPBB1E or pBLAE were grown exponentially at 37°C. Rifampin was added to halt further initiation of transcription, and total cellular RNA was extracted from culture samples withdrawn at time intervals thereafter. The decay of pbb1, bla, and papAΔ3 mRNA was then examined by S1 protection analysis of the extracted RNA samples (Fig. 1B) with a mixture of 5′-end-labeled DNA probes complementary to the first 0.3 kb of the papAΔ3 transcript or the first 0.8 kb of bla mRNA. These studies revealed that the half-life of bla mRNA increases more than fourfold when its 5′ UTR is replaced with the 5′ UTR of papAΔ3, rising from 1.9 ± 0.4 min (bla mRNA) to 8.9 ± 1.1 min (pbb1 mRNA). This half-life of the papAΔ3-bla mRNA hybrid is approximately half as long as that of the pseudo-wild-type papAΔ3 transcript in the same cells (18 ± 2 min, a value similar to the half-life reported for wild-type papA mRNA in E. coli [3]). These findings show that the papA 5′ UTR can act in cis to prolong the lifetime of a heterologous message to which it is fused, which in turn suggests that this 5′ RNA segment plays a key role in protecting the papA message from degradation in E. coli.
Secondary structure of the papAΔ3 5′ UTR in E. coli.
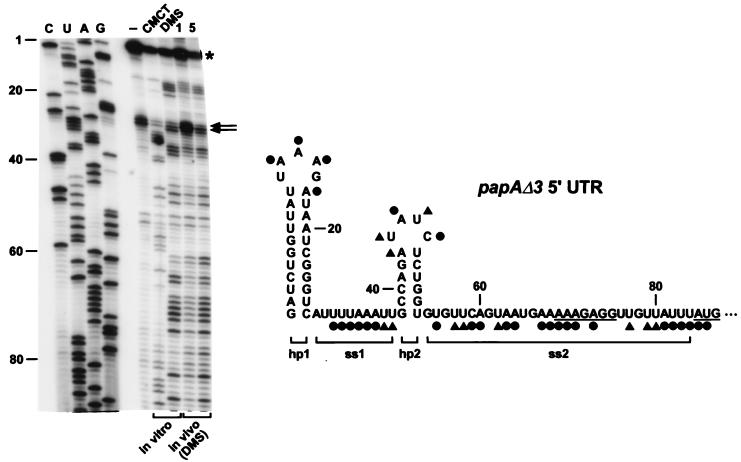
Previously, the secondary structure of the papBA intercistronic region was analyzed in vitro by cleavage with structure-specific ribonucleases (16). Because a variety of environmental perturbations in vitro (including the absence of bound ribosomes) can prevent mRNA from assuming its natural in vivo conformation, we decided to examine the secondary structure of the papA 5′ UTR in E. coli as a step toward understanding the basis for mRNA stabilization by this RNA segment. This was accomplished by chemical modification with dimethylsulfate (DMS), an alkylating agent whose reactivity is sensitive to base pairing (14). DMS alkylates unpaired adenosine residues at N1 and unpaired cytidine residues at N3. In contrast, 1-cyclohexyl-3-(2-morpholino-ethyl)carbodiimide metho-p-toluenesulfo-nate (CMCT) alkylates unpaired uridine residues at N3 and, to a lesser extent, unpaired guanosine residues at N1 (14). Nucleotides engaged in Watson-Crick base pairing are protected from alkylation by these two reagents. Sites of alkylation can be mapped readily, as these base modifications block primer extension with reverse transcriptase.
Being a small, uncharged molecule able to pass through cell membranes, DMS can be used to probe the secondary structure of mRNAs in their native conformation within the cytoplasm of living cells (6). It is also possible to treat extracted RNA with DMS to allow a comparison of RNA conformations in vivo and in vitro. In contrast, CMCT only can be used to alkylate RNA in vitro because this reagent does not penetrate cell membranes.
The secondary structure of the papAΔ3 5′ UTR was analyzed by alkylation with DMS and CMCT, followed by primer extension (Fig. 2, left). As a negative control, primer extension was also performed with an unalkylated RNA sample to identify sites where reverse transcriptase naturally pauses or terminates. The pattern of alkylation by DMS was similar in E. coli and in vitro, indicating that the conformation of this RNA segment does not change perceptibly upon extraction from cells. This finding validated the in vitro alkylation data that we obtained by using CMCT. Overall, the in vivo alkylation data were consistent with data from earlier studies based on RNase cleavage in vitro (16). Together, these data indicate that the papAΔ3 5′ UTR contains two stem-loops, one of which (hp1) is situated at the 5′ terminus (Fig. 2, right). These two stem-loops are separated by a short single-stranded RNA segment (ss1) and followed by a second single-stranded segment (ss2) that contains the signals for translation initiation.
FIG. 2.
Alkylation of the 5′ UTR of papAΔ3 mRNA. (Left) Primer extension analysis of alkylated papAΔ3 mRNA. Total cellular RNA was isolated from an exponential-phase culture of E. coli C600S (17) containing pPAPAΔ3 after treatment of aliquots of the culture with DMS (in vivo, 1 or 5 μl per ml of culture). In addition, samples of RNA extracted from an untreated culture were alkylated in vitro with DMS or CMCT. Sites of alkylation were mapped by primer extension with avian myeloblastosis virus reverse transcriptase and a 5′-end-labeled DNA primer (5′-AAGACACCACTGCCATAGCT-3′) complementary to the coding region of papA mRNA. The resulting extension products were then analyzed by gel electrophoresis beside sequencing ladders that were generated by extension of the same 5′-end-labeled primer on a papAΔ3 DNA template. Unalkylated RNA (lane −) served as a negative control to identify primer extension products unrelated to alkylation. Blockage of primer extension by an alkylated RNA base results in the production of a complementary DNA fragment one nucleotide shorter than that arising from incorporation of a dideoxynucleotide opposite the same base. In the experiment shown, CMCT did not react detectably with guanosine nucleotides, precluding a direct assessment of base pairing by those residues. The sequencing lanes (C, U, A, G) are labeled to indicate the sequence of the RNA, not the complementary DNA. Calibration is in nucleotides from the papAΔ3 5′ end. An asterisk marks the site of transcription initiation. The degree of chemical modification can be difficult to assess at sites where it is no greater than the basal level of termination by reverse transcriptase on an unalkylated RNA template. With the avian myeloblastosis virus enzyme, such sites often correspond to the 3′ boundary of secondary structure elements of significant thermodynamic stability (6), and in this case, the two major sites of premature termination on unalkylated RNA (marked by arrows) map to the foot of the 5′-terminal papAΔ3 stem-loop. For procedural details, see reference 9. (Right) Summary of the alkylation data for the papAΔ3 5′ UTR. ●, heavy alkylation; ▴, moderate alkylation. Brackets delineate the boundaries of the four structural domains (hp1, ss1, hp2, and ss2) within the papAΔ3 5′ UTR. The Shine-Dalgarno element and initiation codon are underlined.
A 5′-terminal stem-loop contributes to the stability of papA mRNA.
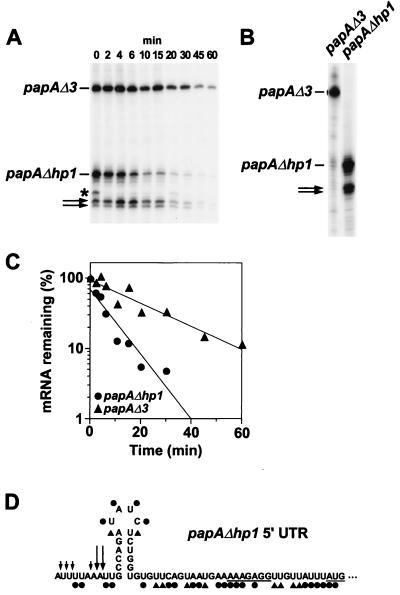
Like the papAΔ3 5′ UTR, the 5′ UTR of the long-lived E. coli ompA transcript functions as an mRNA stabilizer. Previous studies have shown that a stem-loop present at the 5′ terminus of the ompA 5′ UTR plays an important role in protecting the ompA message from degradation (1, 9). To determine whether the 5′-terminal stem-loop of papAΔ3 is similarly important for message longevity, we constructed a plasmid (pPAPAΔhp1E) encoding both papAΔ3 mRNA and a variant papA transcript (papAΔhp1) from which the 5′ stem-loop had been deleted. The rates of decay of these two plasmid-encoded mRNAs were monitored simultaneously in E. coli by primer extension analysis of RNA samples extracted at time intervals after rifampin addition (Fig. 3A).
FIG. 3.
Accelerated decay of a truncated papA transcript lacking the 5′ stem-loop. (A) A culture of E. coli JC10287 (7) harboring pPAPAΔhp1E was grown exponentially for several generations in supplemented Luria-Bertani medium at 37°C. At time intervals after transcription inhibition with rifampin (200 μg/ml), total cellular RNA was isolated. Equal amounts of each RNA sample (2 μg) were then analyzed by primer extension with avian myeloblastosis virus reverse transcriptase and a 5′-end-labeled DNA primer (5′-AAGACACCACTGCCATAGCT-3′) complementary to the coding region of papA mRNA. Bands that correspond to papAΔ3 and papAΔhp1 mRNA are indicated, as are two bands corresponding to a pair of apparent papA mRNA cleavage products (arrows). The origin of an additional band (∗) seen only at 0 min is not known. (B) That the two cleavage products are derived principally from papAΔhp1 mRNA and not significantly from papA mRNA is demonstrated by an additional primer extension experiment performed with RNA samples isolated from a pair of isogenic E. coli strains that expressed either papAΔ3 mRNA or papAΔhp1 mRNA. (C) Semilogarithmic plot of mRNA concentration versus time after rifampin addition. The measured half-lives were 6.6 ± 0.9 min for papAΔhp1 mRNA and 18 ± 2 min for papAΔ3 mRNA. (D) Summary of the DMS and CMCT alkylation data obtained in vivo and in vitro for the 5′ UTR of papAΔhp1 mRNA by the procedure described in Fig. 2. ●, heavy alkylation; ▴, moderate alkylation. Sites of premature termination by reverse transcriptase on an unalkylated RNA template are indicated by arrows: large arrows, major termination sites thought to represent the 5′ ends of RNase E cleavage products generated in vivo; small arrows, minor termination sites that preclude a direct assessment of base pairing by the preceding residue. The Shine-Dalgarno element and initiation codon are underlined.
Consistent with our previous experiments (Fig. 1), the pseudo-wild-type papAΔ3 transcript decayed slowly in these cells, with a half-life of 18 ± 2 min (Fig. 3C). The papAΔhp1 transcript, on the other hand, decayed more rapidly, with a half-life of 6.6 ± 0.9 min, corresponding to a degradation rate nearly three times faster than that of papAΔ3 mRNA (Fig. 3C). Structural analysis of the papAΔhp1 5′ UTR by chemical alkylation confirmed that deletion of the 5′ stem-loop had not disrupted the secondary structure of the remainder of the UTR (Fig. 3D). We conclude that this 5′-terminal stem-loop makes an important contribution to the stabilizing effect of the papAΔ3 5′ UTR. This finding raises the possibility that hp1 may help to shield papAΔ3 mRNA from degradation via an RNase E-dependent pathway (the principal pathway for mRNA decay in E. coli [2, 13, 15, 20]), as internal cleavage by this endonuclease in E. coli can be hindered by the presence of a stem-loop near the RNA 5′ end (5).
The protective effect of hp1 suggests a similar role for the nearly identical 5′ stem-loop of the wild-type papA message, an mRNA that arises by cleavage of the papBA precursor transcript at an intercistronic site two nucleotides upstream of this stem-loop (19). In this regard, it is noteworthy that the protective stem-loop at the 5′ end of ompA mRNA retains its stabilizing influence even when it is preceded by two unpaired nucleotides (9). The 5′ papA stem-loop might be particularly important for the longevity of the wild-type papA message if this mRNA is targeted for subsequent digestion by RNase E, as this 3′ RNA processing product is thought to begin with a 5′ monophosphate. Recent in vitro studies have shown that RNase E is especially aggressive at cleaving RNAs that begin with a 5′ monophosphate rather than a 5′ triphosphate, but that this accelerated cleavage can be significantly impeded by base pairing at or near the RNA 5′ terminus (12). Therefore, the contribution of papA hp1 to mRNA stability may be even greater for the natural papA message than it is for papAΔ3, which presumably begins with a 5′ triphosphate. We note that the presence of hp1 not only enhances the longevity of papAΔ3 mRNA, but also reduces the relative abundance of a pair of apparent degradation intermediates arising from cleavage in the vicinity of a known RNase E site (16) within the unpaired ss1 segment (see bands marked by arrows in Fig. 3A and B). Thus, by deterring RNase E from degrading the papA message following its liberation from the papBA operon transcript, hp1 may be critically important for the differential stability of the papB and papA segments of the papBA transcript and hence for the differential expression of these two cotranscribed genes. Consistent with this conclusion, a large deletion within the papBA intercistronic region that removes hp1 together with 85 flanking nucleotides (81 upstream and 4 downstream) reduces PapA protein production substantially, resulting in truncated cell surface fimbriae (18). The stem-loop structures present just downstream of endonucleolytic processing sites in various other mRNAs may have a similar protective function.
Acknowledgments
We thank David Low for providing a plasmid clone of the papBA operon.
This research was funded by a grant from the National Institutes of Health (GM35769) and by a Faculty Research Award (to J.G.B.) from the American Cancer Society (FRA-419).
REFERENCES
- 1.Arnold T E, Belasco J G. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 2.Babitzke P, Kushner S R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Båga M, Göransson M, Normark S, Uhlin B E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988;52:197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- 4.Belasco J G, Nilsson G, von Gabain A, Cohen S N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986;46:245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet P, Belasco J G. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen L-H, Emory S A, Bricker A L, Bouvet P, Belasco J G. Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J Bacteriol. 1991;173:4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csonka L N, Clark A J. Deletions generated by the transposon Tn10 in the srl recA region of the Escherichia coli K-12 chromosome. Genetics. 1979;93:321–343. doi: 10.1093/genetics/93.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emory S A, Belasco J G. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emory S A, Bouvet P, Belasco J G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 10.Hansen M J, Chen L-H, Fejzo M L S, Belasco J G. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol Microbiol. 1994;12:707–716. doi: 10.1111/j.1365-2958.1994.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 11.Hultgren S J, Normark S, Abraham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 12.Mackie G A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 13.Melefors Ö, von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991;5:857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 14.Moazed D, Stern S, Noller H F. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 15.Mudd E A, Krisch H M, Higgins C F. RNase E, an endoribonuclease, has a general role in the chemical decay of E. coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 16.Naureckiene S, Uhlin B E. In vitro analysis of mRNA processing by RNase E in the pap operon of Escherichia coli. Mol Microbiol. 1996;21:55–68. doi: 10.1046/j.1365-2958.1996.6121101.x. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson G, Belasco J G, Cohen S N, von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci USA. 1987;84:4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson P, Naureckiene S, Uhlin B E. Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J Bacteriol. 1996;178:683–690. doi: 10.1128/jb.178.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson P, Uhlin B E. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNase E-dependent endonucleolytic processing. Mol Microbiol. 1991;5:1791–1799. doi: 10.1111/j.1365-2958.1991.tb01928.x. [DOI] [PubMed] [Google Scholar]
- 20.Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]