Abstract
The structure of the membrane protein MntB, a component of a manganese transporter system in Synechocystis sp. strain PCC 6803, was examined with a series of fusions to the reporter proteins alkaline phosphatase and β-galactosidase. The results support a topological model for MntB consisting of nine transmembrane segments, with the amino terminus of the protein being in the periplasm and the carboxyl terminus being in the cytoplasm.
The first high-affinity transporter system for the transition metal manganese was identified in the cyanobacterium Synechocystis sp. strain PCC 6803 (1, 2). This system is a member of the ABC transporter (10) or traffic ATPase (5) superfamily of transporters that mediate movements of numerous diverse substrates across various biological membranes from microbes to humans (17, 19). In gram-negative bacteria, a typical uptake ABC transporter consists of five components: a substrate-binding protein located in the periplasm, two transmembrane proteins that form the pathway for the substrate, and two ATP-binding proteins peripherally located at the cytoplasmic face of the inner membrane. Except with the ATP-binding domains in the last-named polypeptides, sequence similarities between the different ABC transporters are usually not high. However, the overall structures of all ABC transporters are believed to be similar. The two hydrophobic transmembrane domains of the majority of the ABC transporters were originally thought to span the membrane 12 times (six transmembrane segments per domain) (10), a prediction that has since been experimentally confirmed in a number of cases. An exception was found for the MalF protein of the maltose transporter, which was predicted, and then experimentally shown, to have eight transmembrane segments (7).
In Synechocystis sp. strain PCC 6803, the ABC transporter system for manganese is encoded by an operon of three closely linked genes: mntC, which encodes the substrate-binding protein; mntA, which encodes the ATP-binding protein; and mntB, which encodes the transmembrane protein (1). The MntB protein is predicted to be extremely hydrophobic, with a molecular mass of 33.4 kDa. Hydropathy analysis with the Kyte and Doolittle algorithm in the TopPred II program (4) suggested the presence of eight putative transmembrane segments in the MntB protein (1). However, using the same program but a different algorithm developed by Engelman and coworkers (6), we found that the MntB protein may have up to 10 membrane-spanning segments (data not shown). To determine the actual number of transmembrane spans in this protein, we used a reporter gene fusion approach to examine the membrane topology of MntB. This method is based on the observation that the enzyme activities of certain reporter proteins translationally fused to a membrane protein can indicate the subcellular locations of the fusion sites in the hybrid protein (9, 20). In the present study, we selected two such reporters that have been widely used to study topologies of many bacterial proteins expressed in Escherichia coli cells. The first of these, alkaline phosphatase, encoded by the phoA gene, is enzymatically active in the periplasm but not in the cytoplasm (11, 14). In contrast, the second reporter protein β-galactosidase, encoded by the lacZ gene, is active in the cytoplasm but not in the periplasm (7).
Construction of MntB fusion proteins.
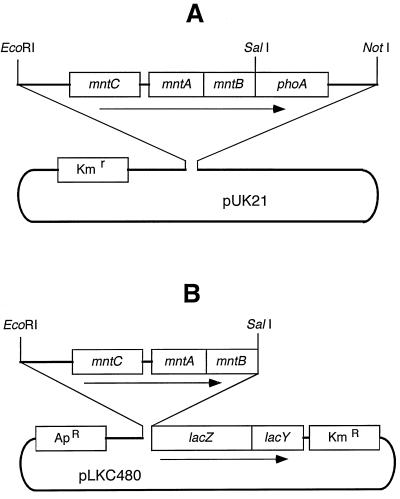
We used synthetic oligonucleotides to generate various fusion proteins. First, SalI restriction sites were introduced throughout the mntB gene by using a site-directed mutagenesis procedure (15). For this purpose, DNA sequences from the mntB gene were amplified by PCR with one universal forward primer, 5′-TTTTGGTGAGTTGCGAAGGAGCGTTTTCCT-3′, and several reverse mutagenic primers (Table 1). All of these PCR products were sequenced to ensure that no undesired mutation was introduced. Next, for the MntB-PhoA fusions, the mntB gene present in an EcoRI-SalI fragment of Synechocystis sp. strain PCC 6803 DNA (containing mntC, mntA, and various 5′-fragments of mntB) as well as the phoA gene present in a SalI-NotI DNA fragment (containing the promoterless phoA gene from the pPHO7 plasmid [8]) was cloned in the pUK21 vector (21) (Fig. 1A). Alternatively, for the MntB-LacZ fusions, the same EcoRI-SalI fragments of Synechocystis sp. strain PCC 6803 DNA were cloned into the polylinker region of the pLKC480 vector, which carries the lacZ gene (18) (Fig. 1B). The reporter fusions were selectively created at a number of sites present in various predicted cytoplasmic and periplasmic loops of the MntB protein.
TABLE 1.
Oligonucleotides used for the construction of SalI sites in the mntB gene of Synechocystis sp. strain PCC 6803
| Oligonu-cleotide | Sequence (5′ to 3′)a | Site of fusion in MntB protein |
|---|---|---|
| B1 | CAAAGGCTCCGTCGACCAATGCCAAAA | 13W |
| B2 | AGGCGCTCACCCAAGTCGACCGGATTAA | 25R |
| B3 | CCATTAAGTCGACCAACCTTTGAGA | 49W |
| B4 | GAATAGTCGACGCATAGGCCAGCACTA | 69A |
| B5 | TGCCAATGACTGCAGTCGACTTTAACCG | 99K |
| B6 | TGGAATAGGTCGACTTACTAGGAA | 124S |
| B7 | CAAAACAAGAGCAGGTCGACTTGCG | 163K |
| B8 | TGCGTAGTCGACGCCTTGGCATGGTTT | 177A |
| B9 | TTGTAAAGTCGACACAATGGTGAGG | 198V |
| B10 | AAGCATAGTCGACTCAAAGCGATCGCT | 227F |
| B11 | CCGGTCGACACATCAAAGTGATAGC | 254V |
| B12 | CCACAGTCGACTTTTCATCGTCTTCC | 297E |
SalI sites are underlined. Boldface nucleotides indicate introduced mutations.
FIG. 1.
Schematic illustrations of the plasmid constructions used for the expression of MntB-PhoA (A) and MntB-LacZ (B) fusion proteins in E. coli CC118.
Expression of MntB-PhoA fusions.
The fusion proteins were expressed in E. coli CC118, which lacks the phoA and lacZ genes (13). The mntCAB operon is expressed in E. coli cells from its own promoter, as was detected by immunoblot analysis with antibodies raised against the MntC protein (data not shown).
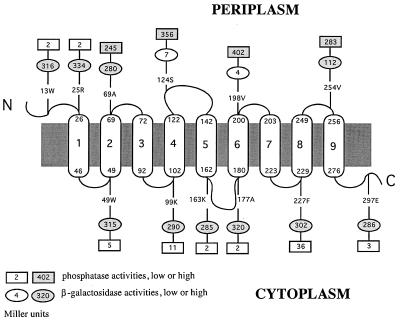
Alkaline phosphatase activities of cells expressing MntB-PhoA fusions were determined by measuring the rates of hydrolysis of the substrate p-nitrophenyl phosphate (13). The PhoA activities (in Miller units per minute per milligram of total cellular protein) corresponding to various fusion sites are shown in Fig. 2. The activities were relatively high for fusions at amino acid positions 69, 124, 198, and 254 of the MntB protein, indicating that these sites are located in the periplasm. In comparison, PhoA fusions at amino acid positions 49, 99, 163, 177, 227, and 297 of MntB demonstrated significantly lower activities, suggesting that these residues are located in the cytoplasm. It is noteworthy that despite the low activities of the fusions at positions 13 and 25, we have reasoned that they belong to a periplasmic domain, because during TopPred II analysis (4), all algorithms strongly predicted the first transmembrane segment to be between residues 26 and 46. Reporter genes fused to periplasmic N termini of proteins that lack signal peptides display a cytoplasmic phenotype, since without a transmembrane segment following them, the reporters are not translocated through the membrane (3, 12).
FIG. 2.
Experimentally derived model for the structure of the MntB protein with nine transmembrane domains (numbered capsules) and locations of alkaline phosphatase and β-galactosidase fusions (rectangles and ovals, respectively). The small numbers at the tops and bottoms of the capsules correspond to positions of amino acid residues in MntB. Sites of fusions are indicated by similar numbers followed by the one-letter designations of the corresponding amino acids.
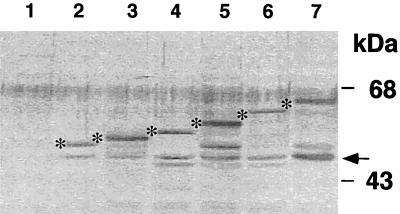
To demonstrate that differences in the enzymatic activities of various fusion proteins are not a result of different levels of protein expression, we performed immunoblot analysis. Whole-cell extracts of exponentially growing E. coli cultures were fractionated on a denaturing sodium dodecyl sulfate–12% polyacrylamide gel, transferred to nitrocellulose filters, and immunostained with a rabbit anti-PhoA immunoglobulin G preparation (5 Prime → 3 Prime, Inc.). The filters were presoaked with cellular extracts of untransformed E. coli CC118 to decrease nonspecific hybridization. As shown in Fig. 3, for the first six fusions, there was no significant difference in the levels of expression of the hybrid proteins irrespective of whether they exhibited low or high alkaline phosphatase activities. However, we could not detect the last six hybrid proteins, even for fusions with high activities (data not shown). It is possible that the larger fusion proteins were relatively unstable. Similar effects have been observed in other studies of different membrane proteins (reviewed in reference 13). Indeed, in all of the fusion-bearing strains, we observed a band at 47 kDa (Fig. 3), which corresponds to the size of the normal PhoA protein and which was not present in the control sample (extracts from E. coli CC118 without a plasmid [Fig. 3, lane 1]). We suggest that in all of these samples, the presence of the native PhoA protein results from the degradation of the hybrid proteins in these cells.
FIG. 3.
Western blot analysis of MntC-PhoA fusion proteins expressed in E. coli CC118. The bands corresponding to the predicted molecular masses of various fusion proteins are marked by asterisks. The arrow denotes a 47-kDa band corresponding to the size of the normal PhoA protein. Lane 1, no plasmid; lanes 2 to 7, fusion constructions created with primers B1 to B6 (Table 1), respectively.
Expression of MntB-LacZ fusions.
To examine the topology of MntB by a second method, we used an alternative reporter protein, β-galactosidase. β-Galactosidase activities of cells expressing MntB-LacZ fusions were analyzed by measuring the rates of hydrolysis of o-nitrophenyl-β-d-galactopyranoside according to the method described in reference 16. As explained above, at any given fusion site, the β-galactosidase fusion is expected to display levels of activity opposite to those of the PhoA fusion. This was found to be true for 10 of the 12 MntB-LacZ fusions (Fig. 2). However, two fusions at amino acid positions 69 and 254 had higher than the expected levels of β-galactosidase activity, based on the analysis of the corresponding MntB-PhoA fusions. It is known that β-galactosidase is a less reliable reporter than PhoA (9). It has been suggested that fused to periplasmic domains, β-galactosidase sometimes exhibits high activity because of disruption of the membrane integration of the fusion protein by this reporter (9). In contrast, high activity of a PhoA fusion requires active translocation of the reporter enzyme moiety through the cytoplasmic membrane. Therefore, in a case where both reporters at the same fusion site displayed high activities, we concluded that the site was periplasmic.
Membrane topology of the MntB protein.
Considering together the predictions of theoretical analysis and the experimental results with reporter gene fusions, we have derived a model of the topology of the MntB protein in the cytoplasmic membranes of bacterial cells (Fig. 2). According to this model, MntB has nine membrane-spanning segments and has its amino terminus in the periplasm and its carboxyl terminus in the cytoplasm. The extent of each transmembrane segment shown in Fig. 2 was as predicted by the TopPred II program (4) with the algorithm of Engelman et al. (6). The major difference between this experimentally derived nine-span model and the previous theoretically predicted eight-span model (1) of this protein is the identification of the domain between residues 49 and 69 as the second membrane span. We have previously reported that MntB has a high degree of sequence similarity with the membrane protein components of a number of putative ABC transporters (1). More recent analysis has revealed that the MntABC transporter is the representative member of a newly identified subfamily of ABC-type metal transporters present in a number of bacterial species (17, 19). Since the hydrophobicity profiles of the membrane protein components of all members if this subfamily are highly similar to that of MntB, we suggest that these proteins also have nine transmembrane segments.
Acknowledgments
This work was supported by grants from the U.S. Department of Agriculture (NRI 9501081) and the International Human Frontier Science Program to H.B.P. V.V.B. was partially supported by a fellowship from Monsanto Co. to the Plant Biology Program at Washington University.
REFERENCES
- 1.Bartsevich V V, Pakrasi H B. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsevich V V, Pakrasi H B. Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1996;271:26057–26061. doi: 10.1074/jbc.271.42.26057. [DOI] [PubMed] [Google Scholar]
- 3.Cartwright C P, Tipper D J. In vivo topological analysis of Ste2, a yeast plasma membrane protein, by using β-lactamase gene fusions. Mol Cell Biol. 1991;11:2620–2628. doi: 10.1128/mcb.11.5.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. CABIOS. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 5.Doige C A, Ames G F-L. ATP-dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug-resistance. Annu Rev Microbiol. 1993;47:291–319. doi: 10.1146/annurev.mi.47.100193.001451. [DOI] [PubMed] [Google Scholar]
- 6.Engelman D M, Steitz T A, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 7.Froshauer S, Green G N, McGovern D B K, Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:1989. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessey E S, Broome-Smith J K. Gene-fusion techniques for determining membrane-protein topology. Curr Opin Struct Biol. 1993;3:524–531. [Google Scholar]
- 10.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman J C, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis M J, Chang J A, Simoni R D. A topological analysis of subunit A from Escherichia coli F1F0-ATP synthase predicts eight transmembrane segments. J Biol Chem. 1990;256:10541–10550. [PubMed] [Google Scholar]
- 13.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 14.Manoil C, Beckwith J. TnPhoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 17.Saier M H. Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv Microb Physiol. 1998;40:81–136. doi: 10.1016/s0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- 18.Tiedeman A A, Smith J M. lacZY gene fusion cassettes with Kanr resistance. Nucleic Acids Res. 1988;8:3587. doi: 10.1093/nar/16.8.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomii K, Kanehisha M. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 1998;8:1048–1059. doi: 10.1101/gr.8.10.1048. [DOI] [PubMed] [Google Scholar]
- 20.Traxler B, Boyd D, Beckwith J. The topological analysis of integral cytoplasmic membrane proteins. J Membr Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 21.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Methods Enzymol. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]