Abstract
Objectives
To report an outbreak of hypervirulent Klebsiella pneumoniae (hvKp) in COVID-19 patients.
Methods
Prospective, observational study including consecutive COVID-19 patients with hvKp infections admitted to the University Hospital of Pisa (Italy). Clinical data and outcome of patients were collected. All patients were followed-up to 30 days from the diagnosis of infection. Mortality within 30 days of the diagnosis of hvKp infection was reported. The hypermucoviscous phenotype was determined by the ‘string test’. Molecular typing was performed on three strains collected during different periods of the outbreak. The strains underwent whole genome sequencing using the Illumina MiSeq instrument. The complete circular assemblies were also obtained for the chromosome and a large plasmid using the Unicycler tool.
Results
From November 2020 to March 2021, hvKp has been isolated from 36 COVID-19 patients: 29/36 (80.6%) had infections (15 bloodstream infections, 8 ventilator-associated pneumonias and 6 complicated urinary tract infections), while 7/36 (19.4%) had colonization (3 urine, 2 rectal and 2 skin). The isolates belonged to ST147 and their plasmid carried three replicons of the IncFIB (Mar), IncR and IncHI1B types and several resistance genes, including the rmpADC genes encoding enhancers of capsular synthesis. The hvKp isolates displayed an ESBL phenotype, with resistance to piperacillin/tazobactam and ceftolozane/tazobactam and susceptibility only to meropenem and ceftazidime/avibactam. The majority of patients were treated with meropenem alone or in combination with fosfomycin. Thirty-day mortality was 48.3% (14/29).
Conclusions
ST147 ESBL-producing hvKp is associated with high mortality in COVID-19 patients. Strict microbiological surveillance and infection control measures are needed in this population.
Introduction
Hypervirulent (hypermucoviscous) Klebsiella pneumoniae (hvKp) is a concerning organism because it may cause life-threatening diseases. Although not specific for hvKp, an atypical trait of hvKp strains is the hypermucoviscous phenotype defined by a positive string test.1
Bacterial superinfections have been increasingly reported in patients with COVID-19 and may lead to poor outcome in these patients.2 Prolonged hospitalization, widespread use of broad-spectrum antibiotics, use of steroids and immunomodulators are factors that increase the risk of multidrug-resistant organisms (MDRO) in these patients.2 Recently, a fatal case of hvKp infection in a Japanese patient with COVID-19 has been reported.3 Here, we describe a cluster of hvKp infections observed in COVID-19 patients in the University Hospital of Pisa (Italy).
Patients and methods
This single-centre, prospective, observational study included consecutive COVID-19 patients with hvKp infections admitted to the tertiary care University Hospital of Pisa (Italy) from November 2020 to March 2021. Epidemiological and demographic information, medical history, symptoms at admission and treatment details were collected. The use of steroids was classified as low- or high-dose based on the cutoff of 1 mg/kg/day of methylprednisolone or equivalents, as previously described.4 Patients were followed-up to 30 days from the diagnosis of infection. Thirty-day mortality was defined as death occurring within 30 days from the diagnosis of hvKp infection.
The hypermucoviscous phenotype was determined by the ‘string test’,5 a positive reaction being defined as a bacteriological loop being able to generate a viscous filament ≥5 mm in length by stretching bacterial colonies growth at 37°C by 18–24 h on a blood agar plate (see Figure S1, available as Supplementary data at JAC Online). Susceptibility tests were performed with Merlin microdilution panels on a Tecan automated platform.
Molecular typing was performed on three strains (arbitrarily named PA, ZO and VA, Bioproject PRJNA746575) representative of three different periods of the outbreak November 2020, January 2021 and March 2021). The strains were analysed by WGS performed on genomic DNA purified using the Macherey Nagel DNA extraction kit (Düren, Germany), preparing paired-end libraries sequenced using the Illumina MiSeq instrument (Illumina Inc). De novo assembly of Illumina reads was performed using the SPADES 3.8 software (https://w3.iss.it/site/aries/). The PA strain, selected as representative of the outbreak clone, was also subjected to nanopore sequencing on the MinION Flow Cell (R9.4.1) following SQK-RBK004 sequencing procedures on an Mk1C MinION platform.6
The complete circular assemblies (Bioproject PRJNA746592; Acc no.s CP084984–CP084985) were obtained for the chromosome (5 379 443 bp) and a large plasmid (382 377 bp) by using the Unicycler tool (https://usegalaxy.eu). In silico analysis of antimicrobial resistance, replicons, and virulence gene content was performed on the chromosome and the plasmid of the PA strain, using Plasmid Finder, ResFinder and Kleborate online tools, respectively.7–9
The study was approved by the local Ethics Committee (Number 17681).
Results
During the study period, 1037 patients with COVID-19 were admitted to our hospital. We isolated hvKp from 36 COVID-19 patients. Of these, 29 (80.6%) patients had infections caused by hvKp [15 with bloodstream infection (BSI), 8 with ventilator-associated pneumonia (VAP), and 6 with complicated urinary tract infection (cUTI)], while 7 (19.4%) had colonization (3 urine, 2 rectal and 2 skin). Demographics and clinical features of patients with infections caused by hvKp are reported in Table 1.
Table 1.
Demographics and clinical characteristics of COVID-19 patients (N = 29) with infection caused by hvKp
| Characteristic | Value |
|---|---|
| Age, years, median (IQR) | 73 (65.5–77.5) |
| Male sex, n (%) | 25 (86.2) |
| Coexisting comorbidities, n (%) | |
| COPD | 3 (10.3) |
| Diabetes mellitus | 5 (17.2) |
| Cardiovascular disease | 20 (68) |
| Solid cancer | 3 (10.3) |
| Ward of hospitalization (at time of infection), n (%) | |
| Intensive care unit | 21 (72.4) |
| Sub-intensive care unit | 3 (10.3) |
| Medical ward | 5 (17.2) |
| Charlson Comorbidity Index, median (IQR) | 3 (3–5) |
| SOFA score (at time of hvKp infection), median (IQR) | 8.5 (0.75–11.25) |
| Septic shock (at time of hvKp infection), n (%) | 8 (27.6) |
| Previous antibiotic therapy, n (%) | 26 (89.7) |
| Source of infection, n (%) | |
| Bloodstream infection | 15 (51.7) |
| Ventilator-associated pneumonia | 8 (27.6) |
| Urinary tract infection | 6 (20.7) |
| Concomitant medications, n (%) | |
| Remdesivir | 6 (20.7) |
| Steroids | 27 (93.1) |
| Dexamethasone 6 mg daily | 8 (27.6) |
| Steroids high dosea | 19 (65.5) |
| Low-molecular-weight heparin | 29 (100) |
| Immunomodulatory drugsb | 7 (24.1) |
| Respiratory support (at time of hvKp infection), n (%) | |
| Invasive mechanical ventilation | 21 (72.4) |
| Non-invasive mechanical ventilation | 3 (10.3) |
| Supplementary oxygen therapy | 5 (17.2) |
| Time from admission to hvKp infection, days, median (IQR) | 17 (9.5–20) |
| Time from hvKp infection to death, days, median (IQR) | 5.5 (2–22.5) |
| 30 day mortality, n (%)c | 14 (48.3) |
COPD, chronic obstructive pulmonary disease; hvKp, hypervirulent K. pneumoniae.
High dose of steroids was defined as use of 1 mg/kg/day of methylprednisolone or equivalents
Immunomodulatory drugs included baricitinib or tocilizumab.
30 day mortality was defined as occurrence of death within 30 days from diagnosis of hvKp infection.
Overall, 21/29 (72.4%) patients were cared for in ICU and underwent invasive mechanical ventilation (MV), 3/29 (10.3%) received non-invasive MV. The median time from admission to hvKp infection was 17 days (IQR 9.5–20). All patients received low molecular weight heparin and 27/29 (93.1%) steroids. Immunomodulatory drugs were administered in 7 (24.1%) patients. Twenty-six (89.7%) patients received intravenous antibiotics before the occurrence of hvKp infection: the majority received piperacillin/tazobactam with or without linezolid (16, 55.2%), followed by ceftriaxone with or without azithromycin (5, 17.2%), azithromycin alone (2, 6.9%), ceftobiprole (1, 3.4%), levofloxacin (1, 3.4%) and meropenem (1, 3.4%). Among the 26 patients who received antibiotics before the occurrence of hvKp infection, 18 (69%) had a previous documented infection: 2 patients had a documented respiratory coinfection (due to Streptococcus pneumoniae), 10 patients a documented VAP (4 Pseudomonas aeruginosa, 4 K. pneumoniae, 2 K. pneumoniae plus Staphylococcus aureus), 6 (23%) patients had a urinary tract infection. Eight patients received antibiotic therapy without a documented isolate.
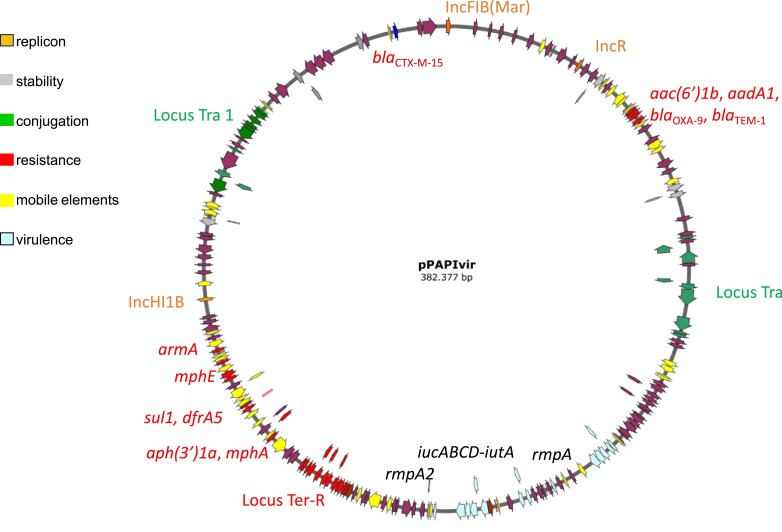
All hvKp strains (100%) were resistant to third-generation cephalosporins, aminoglycosides, and fluoroquinolones and the majority (94.4%) were resistant to piperacillin/tazobactam, ceftolozane/tazobactam, and trimethoprim/sulfamethoxazole. The hvKp isolates were fully susceptible to meropenem and ceftazidime/avibactam (Table S1). In silico WGS studies performed by Kleborate demonstrated that the three isolates were ST147. Single nucleotide polymorphism (SNP) analysis demonstrated that they were closely related to each other (3–4 SNPs of difference across 4283 core genes, data not shown).The plasmid carried three replicons of the IncFIB (Mar), IncR and IncHI1B types and several resistance genes [aac(6′)-Ib, aadA1, blaOXA-9, blaTEM-1A, aph(3′)-Ia, mph(A), sul1, dfrA5, mph(E), sul2], including armA, which encodes the 16S rRNA methyltransferase conferring resistance to all aminoglycosides, and the blaCTX-M-15 extended-spectrum β-lactamase gene. No carbapenemases genes were detected on the plasmid. However, the plasmid carried the iucA-D-iutA hydroxamate siderophore aerobactin virulence determinant, and the rmpADC (allele 27) genes (rmpA2 is frameshifted) encoding the enhancers of capsular synthesis, known to be responsible for the hypervirulent phenotype observed in this hvKp clone (Table 2 and Figure 1). On the chromosome, two other relevant virulence genetic determinants were identified, including the mrkA-H type 3 fimbriae synthesis cluster and the complete fyu-ybt-irp yersiniabactin biosynthetic operon. Two additional the blaCTX-M-15 genes were identified in the chromosome, contributing to resistance to cephalosporins (Table 2).
Table 2.
Genetic features deduced from whole genome sequencing of the hypervirulent ST147 K. pneumoniae isolates
| Locus | Locationa | Positionsb | Description/phenotype | |
|---|---|---|---|---|
| IncFIB(Mar) | Plasmid | 646 | 208 | Replicon |
| IncR | Plasmid | 35 322 | 35 072 | Replicon |
| aac(6’)-Ib | Plasmid | 52 273 | 52 878 | Amikacin, gentamicin, kanamycin resistance |
| aadA1 | Plasmid | 52 948 | 53 736 | Streptomycin resistance |
| bla OXA-9 | Plasmid | 53 781 | 54 620 | Ampicillin resistance |
| bla TEM-1A | Plasmid | 55 320 | 56 180 | Ampicillin resistance |
| rmpA | Plasmid | 162 998 | 163 630 | Enhancer of cps synthesis |
| iucA | Plasmid | 180 751 | 182 541 | Hydroxamate siderophore aerobactin |
| iucB | Plasmid | 182 542 | 183 489 | |
| iucC | Plasmid | 183 489 | 185 222 | |
| iucD | Plasmid | 185 226 | 186 503 | |
| iutA | Plasmid | 186 585 | 188 786 | |
| rmpA2 | Plasmid | 193 677 | 194 314 | Enhancer of cps synthesis |
| aph(3′)-Ia | Plasmid | 240 342 | 239 527 | Kanamycin resistance |
| mph(A) | Plasmid | 242 200 | 241 298 | Erythromycin, azithromycin resistance |
| sul1 | Plasmid | 249 689 | 248 850 | Sulfisoxazole resistance |
| dfrA5 | Plasmid | 250 687 | 250 214 | Trimethoprim resistance |
| mph(E) | Plasmid | 258 500 | 257 616 | Erythromycin, azithromycin resistance |
| msr(E) | Plasmid | 260 031 | 258 556 | Erythromycin, azithromycin resistance |
| armA | Plasmid | 263 103 | 262 330 | Amikacin, gentamicin, kanamycin, streptomycin resistance |
| sul2 | Plasmid | 265 585 | 266 400 | Sulfisoxazole resistance |
| IncHI1B | Plasmid | 279 395 | 278 826 | Replicon |
| bla CTX-M-15 | Plasmid | 369 187 | 370 062 | Ampicillin, ceftriaxone resistance |
| mrkA | Chromosome | 876 941 | 877 549 | Type 3 fimbriae synthesis |
| mrkB | Chromosome | 877 645 | 878 346 | |
| mrkC | Chromosome | 878 358 | 880 844 | |
| mrkD | Chromosome | 880 835 | 881 830 | |
| mrkF | Chromosome | 881 844 | 882 479 | |
| mrkJ | Chromosome | 882 514 | 883 230 | |
| mrkH | Chromosome | 883 964 | 884 668 | |
| bla CTX-M-15 | Chromosome | 1 597 014 | 1 597 889 | Ampicillin, ceftriaxone resistance |
| bla CTX-M-15 | Chromosome | 1 671 695 | 1 672 570 | Ampicillin, ceftriaxone resistance |
| fyuA | Chromosome | 1 863 698 | 1 865 719 | Yersiniabactin receptor resistance |
| ybtE | Chromosome | 1 865 850 | 1 867 427 | Yersiniabactin biosynthetic operon |
| ybtT | Chromosome | 1 867 431 | 1 868 234 | |
| ybtU | Chromosome | 1 868 231 | 1 869 331 | |
| irp1 | Chromosome | 1 869 328 | 1 878 819 | |
| irp2 | Chromosome | 1 878 907 | 1 885 014 | |
| ybtA | Chromosome | 1 885 205 | 1 886 164 | |
| ybtP | Chromosome | 1 886 421 | 1 888 133 | |
| ybtQ | Chromosome | 1 888 120 | 1 889 922 | |
| ybtX | Chromosome | 1 889 915 | 1 891 195 | |
| ybtS | Chromosome | 1 891 223 | 1 892 527 | |
Plasmid refers to location in the single large plasmid of 382 377 bp with FIB(Mar)-R-HI1B replicons, identified in the PA strain, carrying both virulence and resistance genes.
Nucleotide positions refers to start and stop codon of each gene of the plasmid and chromosome complete circular sequences, respectively (Bioproject PRJNA746592; Accession no.s CP084984–CP084985).
Figure 1.
Major structural features of the FIB(Mar)-R-HI1B resistance and virulence plasmid identified in the hypervirulent outbreak ST147 clone, Pisa 2020–2021. Predicted coding sequences are indicated by coloured arrows oriented in the direction of transcription of each respective gene. Important plasmid features (virulence, resistance, replicons, transfer loci) are coloured as indicated in the key.
Patients were treated with meropenem with or withou fosfomycin (17/29, 58.6%), ceftazidime/avibactam (4/29, 13.8%), and colistin/tigecycline (1/29, 3.4%). Seven patients did not receive any in vitro active therapy. Fourteen (48.3%) patients died within 30 days of hvKp infection: two had a cUTI, five a VAP and seven a BSI.
Discussion
We describe an outbreak of infections caused by hvKp in patients with SARS-CoV-2 pneumonia. To date, only one case of hvKp in a patient with COVID-19 has been reported.3 Remarkably, all the hvKp strains belonged to ST147 and produced some specific virulence factors. Our study highlights some concerns.
First, ST147 is a high-risk Kp clone with the potential to become a major threat to public health. ST147 consists of multiple clades/clusters associated with various carbapenemases (KPC, NDM, OXA-48-like, and VIM), and has been responsible for several outbreaks in India, Italy, Greece, North Africa and, more recently, Spain.10,11 In our hospital, NDM-producing Kp belonging to ST147 has been endemic since November 2018.11–13 In patients with rectal colonization by this strain, we observed an increased risk of developing bacteraemia caused by the same organism, a fact that highlights its high propensity to cause severe infections.12 The hypervirulent phenotype poses a serious therapeutic problem, since this hvKp remains fully susceptible only to meropenem and new β-lactams/β-lactamases inhibitors (ceftazidime/avibactam and meropenem/vaborbactam). Resistance of hvKp isolates to piperacillin/tazobactam and ceftolozane/tazobactam represents a major concern. Susceptibility to these two β-lactam/β-lactamase inhibitor combinations may be impaired in ESBL-producing organisms carrying the blaCTX-M-15 gene, especially in high-inoculum infections.14,15 In line with these findings we found that hvKp in our study carried blaCTX-M-15 both in plasmid and chromosomal locations. This MDR phenotype makes necessary the use of carbapenems or, alternatively, of drugs such as ceftazidime/avibactam with increased costs and risk of induction of resistance.16
The second important point is the high mortality associated with hvKp infections (48.3%), which is significantly higher than that we detected in COVID-19 patients who had superinfections in our hospital (18.8%) during the first wave.2 This finding suggests a role for the virulence factors associated with the hypervirulent phenotype, such as the aerobactin cluster, already found to be associated with increased virulence in mice,1,17 and with the hypermucoviscous phenotype, such as rmpA genes that reduce binding affinity of the strain to macrophage cells and cause resistance to phagocytosis.18
Finally, a significant proportion of patients in our cohort received broad-spectrum antibiotics before the occurrence of hvKp infections because of suspected or confirmed infection. It should be underlined that the majority of patients in our cohort were hospitalized in ICU and received antibiotics because of documented superinfections. In any case, the use of antibiotics remains unacceptably high in hospitalized patients with COVID-19. A recent study found that 37% and 85% of patients with COVID-19 received an antibiotic before they were admitted and during their hospital stay, respectively.19 Antimicrobial stewardship programmes and adherence to current guidelines should be implemented in COVID-19 patients.20
The spread of hvKp in our hospital reinforces the importance of adhering to standard infection prevention and control precautions in COVID-19 patients, to prevent the cross-transmission between patients and dissemination of MDRO. To this end we implemented strict surveillance cultures in COVID-19 wards, contact precautions and isolation procedures for infected/colonized patients, which has been followed by a decline of the isolation rate during the last 6 months (March to September 2021). As reported in a recent study, in our centre we performed a systematic surveillance for rectal colonization by carbapenem-resistant Enterobacteriaceae (CRE) but not by ESBL-producing organisms.12 Thus, we did not know the rectal colonization status for hvKp. After the outbreak of hvKp in COVID-19 patients, we implemented our surveillance system and we now search for hvKp on rectal swabs in specific high-risk patients, such as solid organ transplant recipients admitted to ICU.
In conclusion, hvKp superinfections may represent a significant cause of mortality in patients with severe COVID-19. Strict microbiological surveillance and infection control measures are needed.
Supplementary Material
Funding
This study was supported by internal funding.
Transparency declarations
M.F. received grants and/or speaker honoraria from MSD, Angelini, Shionogi, Pfizer, Menarini, Gilead and Nordic Pharma. F.M. has participated in advisory board and/or received speaker honoraria from Angelini, Correvio, Merck Sharp &Dohme (MSD), Nordic Pharma, Pfizer, Astellas, Gilead, Bristol-Myers Squibb (BMS), Janssen, ViiV, bioMérieux, Biotest, Becton Dickinson, Pfizer, and Shionogi. The remaining authors have none to declare.
Author contributions
M.F. conceived the study and wrote the manuscript; G.T. collected data and wrote the manuscript; G.A., G.B. and A.C. coordinated experimental design, performed data analysis and wrote the manuscript; A.L., C.G. and S.B. performed experiments; S.T. collected data; R.M. was involved in patient assistance; F.M. was involved in patient assistance and revised the manuscript.
Data availability
The three Klebsiella pneumoniae (PA, ZO and VA) genomes have been released under Bioproject PRJNA746575.
Supplementary data
Figure S1 and Table S1 are available as Supplementary data at JAC Online.
References
- 1. Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 2017; 8: 1111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falcone M, Tiseo G, Giordano Cet al. . Predictors of hospital-acquired bacterial and fungal superinfections in COVID-19: a prospective observational study. J Antimicrob Chemother 2021; 76: 1078–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hosoda T, Harada S, Okamoto Ket al. . COVID-19 and fatal sepsis caused by hypervirulent Klebsiella pneumoniae, Japan, 2020. Emerg Infect Dis 2021; 27: 556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tiseo G, Yahav D, Paul Met al. . What have we learned from the first to the second wave of COVID-19 pandemic? An international survey from the ESCMID Study Group for Infection in the Elderly (ESGIE) group. Eur J Clin Microbiol Infect Dis 2021, Nov 13:1–8. doi: 10.1007/s10096-021-04377-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisenmenger EF, Guajardo E, Finch Net al. . “String Test” for hypermucoviscous Klebsiella pneumoniae. Am J Med 2021; 134: e520–1. [DOI] [PubMed] [Google Scholar]
- 6. Carattoli A, Arcari G, Bibbolino Get al. . Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 2021; 65: e0057421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carattoli A, Zankari E, Garciá-Fernández Aet al. . In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zankari E, Hasman H, Cosentino Set al. . Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lam MMC, Wick RR, Watts SCet al. . A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 2021; 12: 4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peirano G, Chen L, Kreiswirth BNet al. . Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother 2020; 64: e01148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falcone M, Tiseo G, Antonelli Aet al. . Clinical features and outcomes of bloodstream infections caused by New Delhi Metallo-β-Lactamase-producing Enterobacterales during a regional outbreak. Open Forum Infect Dis 2020; 7: ofaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Falcone M, Tiseo G, Galfo Vet al. . Bloodstream infections in patients with rectal colonization by Klebsiella pneumoniae producing different type of carbapenemases: a prospective, cohort study (CHIMERA study). Clin Microbiol Infect 2021, Jun 28. doi: 10.1016/j.cmi.2021.06.031. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13. Falcone M, Giordano C, Barnini Set al. . Extremely drug-resistant NDM-9-producing ST147 Klebsiella pneumoniae causing infections in Italy, May 2020. Euro Surveill 2020; 25: 2001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Canovas J, Petitjean G, Chau Fet al. . Expression of CTX-M-15 limits the efficacy of ceftolozane/tazobactam against Escherichia coli in a high-inoculum murine peritonitis model. Clin Microbiol Infect 2020; 26: 1416.e5–9. [DOI] [PubMed] [Google Scholar]
- 15. Yang YM, Osawa K, Kitagawa Ket al. . Differential effects of chromosome and plasmid blaCTX-M-15 genes on antibiotic susceptibilities in extended-spectrum β-lactamase-producing Escherichia coli isolates from patients with urinary tract infection. Int J Urol 2021; 28: 623–8. [DOI] [PubMed] [Google Scholar]
- 16. Tiseo G, Falcone M, Leonildi Aet al. . Meropenem-Vaborbactam as salvage therapy for ceftazidime-avibactam-, cefiderocol-resistant ST-512 Klebsiella pneumoniae-producing KPC-31, a D179Y Variant of KPC-3. Open Forum Infect Dis 2021; 8: ofab141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng HY, Chen YS, Wu CYet al. . RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010; 192: 3144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu Q, Yang X, Chan EWCet al. . The hypermucoviscosity of hypervirulent K. pneumoniae confers the ability to evade neutrophil-mediated phagocytosis. Virulence 2021; 12: 2050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuehn BM. Antibiotic use in UK’s COVID-19 patients often unnecessary. JAMA 2021; 326: 214. [DOI] [PubMed] [Google Scholar]
- 20. Sieswerda E, de Boer MGJ, Bonten MMJet al. . Recommendations for antibacterial therapy in adults with COVID-19 - an evidence based guideline. Clin Microbiol Infect 2021; 27: 61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three Klebsiella pneumoniae (PA, ZO and VA) genomes have been released under Bioproject PRJNA746575.