Abstract
Salmonella typhimurium possesses two outer membrane receptor proteins, IroN and FepA, which have been implicated in the uptake of enterobactin. To determine whether both receptors have identical substrate specificities, fepA and iroN mutants and a double mutant were characterized. While both receptors transported enterobactin, the uptake of corynebactin and myxochelin C was selectively mediated by IroN and FepA, respectively.
Iron is essential for the multiplication of enterobacteria, since it is a component of enzymes (e.g., ribonucleotide reductase) which are required for the biosynthesis of macromolecules (e.g., DNA) and energy-generating electron transport processes. In order to obtain iron from insoluble Fe(III) complexes present under aerobic growth conditions, enterobacteria release low-molecular-weight compounds, designated siderophores, which bind this metal ion with high affinity (4). The Fe(III)-siderophore complexes are then internalized by iron-regulated outer membrane receptor proteins which display substrate specificity. The primary siderophore produced by enterobacteria is enterobactin, a cyclic trimer of N-(2,3-dihydroxybenzoyl)-l-serine (DBS). In Escherichia coli, transport of enterobactin across the outer membrane is mediated by the FepA outer membrane receptor protein (13).
An orthologue of fepA is present in Salmonella enterica, but the phenotype of a strain carrying a mutation in this gene has not been described to date (7, 19, 21). Recently, iroN, a gene absent from the E. coli K-12 genome, has been shown to encode an outer membrane siderophore receptor of S. enterica serotype Typhimurium (S. typhimurium) (3). When introduced into an E. coli fepA aroB mutant, the cloned iroN gene mediates the utilization of enterobactin as the sole source of iron (20a). However, an S. typhimurium iroN aroA mutant is not deficient in enterobactin uptake, presumably because it is still able to utilize this siderophore via FepA. These data suggest that S. typhimurium possesses two enterobactin receptor proteins, IroN and FepA.
The ability to produce siderophores is one of the main strategies by which microbes acquire Fe(III) in the environment. In addition to receptors for the utilization of enterobactin, S. typhimurium possesses several outer membrane receptors, including FhuA, FhuE, and FoxA, which are involved in the utilization of siderophores that are not produced by this organism (12, 21). Thus, a second strategy apparently used by S. typhimurium to acquire Fe(III) is to steal siderophores produced by other microbes. This siderophore piracy (6) may explain why S. typhimurium possesses two receptors for the utilization of enterobactin. The presence of these two outer membrane enterobactin receptors may be advantageous under conditions when the available siderophores can be utilized only through either FepA or IroN. An important assumption in this line of reasoning is that although FepA and IroN both serve as enterobactin receptors, each receptor may in addition mediate the uptake of substrates that are not recognized and transported by any other siderophore receptor in S. typhimurium. To test this hypothesis, we constructed S. typhimurium strains lacking FepA or IroN or both receptors. Analysis of the siderophore profile utilized by these mutants enabled us to determine that the enterobactin receptors IroN and FepA differ in their substrate specificities.
Isolation of an S. typhimurium fepA mutant.
The fepA gene is located at 14 min on the E. coli genetic map in a DNA region containing enterobactin biosynthesis (ent) genes (11, 13–15). According to a recent analysis of the 5′ end of the fepA gene and the adjacent DNA region, this genetic organization is conserved in S. typhimurium with regard to gene order and map location (21). Since it is transcribed as a monocistronic messenger, mutational inactivation of fepA has no effect on the biosynthesis of enterobactin in E. coli. Based on these data, we reasoned that a selectable marker in fepA could be used to move enterobactin biosynthesis genes by cotransduction (16). Our strategy to identify a fepA mutant was therefore to characterize transposon insertions, which are cotransduced with enterobactin biosynthesis genes. A phage lysate of a random bank of S. typhimurium Tn10dTc mutants was transduced into an S. typhimurium mutant (TA2700) which is defective for enterobactin production (17). Transductions were performed by using the high-frequency generalized transducing phage mutant P22HT 105/1 int 201. Transductants were selected on chrome azurol S (CAS) agar plates, on which colonies producing enterobactin can be readily detected by their ability to form a halo (20). A total of 130 colonies producing a halo on CAS agar plates were isolated, and enterobactin production was confirmed in a cross-feeding test with the S. typhimurium ent mutant (TA2700) (18). Outer membranes were isolated from these mutants as described previously by Hantke (8), and proteins were separated by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein bands were visualized by Coomassie blue staining. One mutant, WR1316, lacked an 83-kDa outer membrane protein, the size reported for FepA (data not shown). The Tn10dTc insertion in WR1315 was mapped to the enterobactin biosynthesis region by cotransduction with entB::MudJ (cotransduction frequency, 2%) and ent::MudJ from strain AR8439 (cotransduction frequency, 5%) (21). To confirm transposon insertion in fepA, a Southern blot of EcoRI-restricted genomic DNA of strains TA2700 and WR1315 was probed with the labeled (nonradioactive labeling kit from NEN) insert of pFT17, a plasmid carrying the 5′ ends of the S. typhimurium fepA gene (21). The probe hybridized with restriction fragments of 10 and 3 kb in TA2700 and WR1315, respectively. Thus, inactivation of fepA in strain WR1315 was suggested by Southern hybridization and the lack of an 83-kDa outer membrane protein.
Utilization of siderophores by FepA and IroN.
In order to study siderophore utilization of the fepA mutant, the Tn10dTc insertion in WR1315 was transduced into TA2700 (ent), and a transductant (WR1316) which was tetracycline resistant and unable to produce enterobactin was selected on CAS agar plates. The ability of strain WR1316 to utilize enterobactin and structurally related siderophores was tested. For this purpose, enterobactin was prepared and purified from E. coli AN311 according to a protocol reported previously (22). Myxochelin derivatives were synthesized and characterized by W. Trowitzsch-Kienast and H. D. Ambrosi, Technische Fachhochschule Berlin, Berlin, Germany (1, 10). The siderophore corynebactin was isolated from Corynebacterium glutamicum (5) and kindly provided by H. Budzikiewicz, Institute of Organic Chemistry, University of Cologne, Cologne, Germany. Utilization of siderophores was detected by an agar diffusion assay (18). To create iron-limiting growth conditions, 0.15 mM 2,2′-dipyridyl was added to Vogel Bonner medium (VBD) (18). The strain to be tested was poured into 3 ml of VBD top agar (VBD with 2% agar)-Noble agar onto a VBD agar plate. Filter paper disks impregnated with 5 μl of a 1-mg/ml solution of the respective siderophores were laid onto the top agar and, after incubation overnight at 37°C, growth stimulation around the filter disk was recorded. Comparison of the profiles of the siderophores utilized by WR1316 (ent fepA) and its parent, TA2700 (ent), revealed that myxochelin C utilization was abolished in the fepA mutant (Table 1). To quantify growth stimulation by siderophores, strains TA2700 (ent) and WR1316 (ent fepA) were cultured in VBD broth, and bacterial growth was recorded by measuring the absorbance at 620 nm. Myxochelin C stimulated the growth of strain TA2700, but not that of strain WR1316, in VBD broth.
TABLE 1.
Siderophore utilization of S. typhimurium mutants
| Siderophore | Growth zone (mm) produced by indicated strain (genotype)
|
||||
|---|---|---|---|---|---|
| TA2700 (ent) | WR1316 (ent fepA) | WR1316 pITS449 (ent fepA fepA+ on plasmid) | WR1223 (ent iroN) | WR1332 (ent fepA iroN) | |
| Enterobactin | 25 | 25 | 25 | 25 | 0 |
| DBS | 30 | 30 | 30 | 30 | 0 |
| Myxochelin A | 40 | 40 | 40 | 40 | 24 |
| Myxochelin C | 35 | 0 | 25 | 35 | 0 |
| Corynebactin | 35 | 35 | NDa | 0 | 0 |
ND, not determined.
We next tested whether the defect in myxochelin C utilization in WR1316 could be restored by introducing the cloned fepA gene from E. coli. A plasmid (pITS449) (2) carrying the E. coli fepA gene was kindly provided by Phil E. Klebba, University of Oklahoma. Introduction of plasmid pITS449 (E. coli fepA) restored the ability of strain WR1316 to utilize myxochelin C as an iron source (Table 1). These data showed that FepA can mediate the uptake of myxochelin C in S. typhimurium. Furthermore, the inability of WR1316 (ent fepA) to utilize myxochelin C suggested that FepA is the only outer membrane receptor involved in the transport of this substrate in S. typhimurium.
Unlike an E. coli fepA mutant, the S. typhimurium fepA mutant (WR1316) was able to utilize enterobactin, presumably because of the presence of a second enterobactin receptor (Table 1). To test the hypothesis that S. typhimurium possesses two outer membrane enterobactin receptors, strain TA2700 (ent) was used to generate isogenic ent iroN (WR1223) and ent iroN fepA (WR1332) mutants. A iroN allele which carries an insertion of suicide vector pGP704 (9) was moved by P22 transduction from S. typhimurium AJB64 (3). The parent (TA2700, ent), the ent fepA mutant (WR1316), and the ent iroN mutant (WR1223) were able to utilize enterobactin. However, utilization of enterobactin and DBS were abolished in the ent iroN fepA mutant (WR1332), suggesting that both IroN and FepA can mediate uptake of these siderophores in S. typhimurium (Table 1).
In contrast to the ent mutant (TA2700) and the ent fepA mutant (WR1316), strains WR1223 (ent iroN) and WR1332 (ent iroN fepA) were unable to utilize corynebactin, suggesting that this siderophore is transported via IroN in S. typhimurium (Table 1). To quantify growth stimulation by siderophores, bacterial strains were cultured in VBD broth supplemented with corynebactin, myxochelin C, or enterobactin. As a control, VBD broth was supplemented with myxochelin B, a siderophore, which is not transported by FepA or IroN. VBD broth supplemented with corynebactin did not support the growth of strain WR1223 (ent iroN) or WR1332 (ent iroN fepA). Furthermore, strain WR1332 was unable to grow in VBD broth supplemented with myxochelin C. While strain TA2700 and its derivatives (WR1332 and WR1223) were unable to grow in VBD broth lacking siderophore supplements, all strains were able to grow in VBD broth supplemented with myxochelin B. Enterobactin promoted the growth of strains TA2700 (ent), WR1223 (ent iroN), and WR1316 (ent fepA) but not that of strain WR1332 (ent iroN fepA). While cultures of strains TA2700 (ent) and WR1223 (ent iroN) reached the same density (optical density at 620 nm [OD620] = 0.5), the growth of strain WR1316 (ent fepA) was reduced (OD620 = 0.4), suggesting that enterobactin is utilized more effectively by FepA than by IroN.
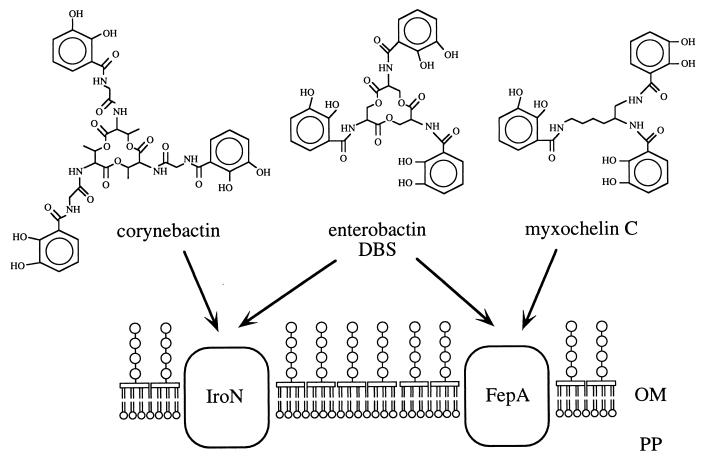
Our data show that S. typhimurium possesses two outer membrane enterobactin receptor proteins, IroN and FepA. These receptor proteins differ in their specificities for other substrates, such as myxochelin C and corynebactin (Fig. 1). Corynebactin is excreted by Corynebacterium glutamicum, an organism found in soil, an environment frequently encountered during the fecal-oral transmission of S. typhimurium. Growth in this environment may require the ability to utilize a wide spectrum of siderophores produced by the bacteria in the soil. It could therefore be speculated that IroN and FepA facilitate growth in soil, since the presence of these receptor proteins may increase the capability of S. typhimurium to obtain iron by siderophore piracy.
FIG. 1.
Substrate specificity of the S. typhimurium IroN and FepA outer membrane (OM) receptor proteins. PP, periplasmic space. The structures of enterobactin, corynebactin, and myxochelin C are shown above.
Acknowledgments
We thank K. E. Sanderson for providing S. typhimurium strains and K. Hantke and J. R. Roth for helpful discussions and support of the genetic work. We thank P. E. Klebba for kindly providing us with the fepA plasmid. Ute Strutz is gratefully acknowledged for her excellent technical assistance.
REFERENCES
- 1.Ambrosi H D, Hartmann V, Pistorius D, Reissbrodt R, Trowitzsch-Kienast W. Myxochelins B, C, D, E and F: a new structural principle for powerful siderophores imitating nature. Eur J Org Chem. 1998;98:541–551. [Google Scholar]
- 2.Armstrong S K, Francis C L, McIntosh M A. Molecular analysis of the Escherichia coli ferric enterobactin receptor FepA. J Biol Chem. 1980;265:14536–14543. [PubMed] [Google Scholar]
- 3.Bäumler A J, Norris T L, Lasco T, Voigt W, Reissbrodt R, Rabsch W, Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun V, Hantke K. Genetics of bacterial iron transport. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press; 1991. pp. 107–138. [Google Scholar]
- 5.Budzikiewicz H, Bössenkamp A, Taraz K, Pandey A, Meyer J M. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC14067 (Brevibacterium sp. DSM 20411) Z Naturforsch Sect C. 1997;52:551–554. [Google Scholar]
- 6.Cornelissen C N, Sparling P F. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Beros M E, Gonzalez C, McIntosh M, Cabello F C. Immune response to the iron-deprivation induced proteins of Salmonella typhi in typhoid fever. Infect Immun. 1989;57:1271. doi: 10.1128/iai.57.4.1271-1275.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hantke K. Regulation of the ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 9.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 10.Kunze B, Bedorf N, Kohl W, Höfle G, Reichenbach H. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales) J Antibiot. 1989;42:14–17. doi: 10.7164/antibiotics.42.14. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Duncan K, Walsh C T. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dihydrogenase. J Bacteriol. 1989;171:791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luckey M, Pollack J R, Wayne R, Ames B N, Neilands J B. Iron uptake in Salmonella typhimurium: utilization of exogenous siderophores as iron carriers. J Bacteriol. 1972;111:731–738. doi: 10.1128/jb.111.3.731-738.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 14.Nahlik M S, Brickman T J, Ozenberger B A, McIntosh M A. Nucleotide sequence and transcriptional organization of the Escherichia coli enterobactin synthesis cistrons entB and entA. J Bacteriol. 1989;171:784–790. doi: 10.1128/jb.171.2.784-790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettis G S, Brickman T J, McIntosh M A. Transcriptional mapping and nucleotide sequence of the Escherichia coli fepA-fes enterobactin region. Identification of a unique iron-regulated bidirectional promoter. J Biol Chem. 1988;263:18857–18863. [PubMed] [Google Scholar]
- 16.Pollack J R, Ames B N, Neilands J B. Iron transport in Salmonella typhimurium: mutants blocked in the biosynthesis of enterobactin. J Bacteriol. 1970;104:635–639. doi: 10.1128/jb.104.2.635-639.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabsch W. Characterization of the catecholate indicator strain S. typhimurium TA2700 as an ent fhuC double mutant. FEMS Microbiol Lett. 1998;163:79–84. doi: 10.1111/j.1574-6968.1998.tb13029.x. [DOI] [PubMed] [Google Scholar]
- 18.Reissbrodt R, Rabsch W. Further differentiation of Enterobacteriaceae by means of siderophore-pattern analysis. Zentbl Bakteriol Hyg. 1988;A268:306–317. doi: 10.1016/s0176-6724(88)80015-4. [DOI] [PubMed] [Google Scholar]
- 19.Rutz J M, Abdullah T, Singh S P, Kalve V I, Klebba P E. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol. 1991;173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 20a.Thulasiraman P, Newton S M, Xu J, Raymond K N, Mai C, Hall A, Montague M A, Klebba P E. Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J Bacteriol. 1998;180:6689–6696. doi: 10.1128/jb.180.24.6689-6696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsolis R M, Bäumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young I G, Gibson F. Isolation of enterochelin from Escherichia coli. Methods Enzymol. 1979;56:394–398. doi: 10.1016/0076-6879(79)56037-6. [DOI] [PubMed] [Google Scholar]