Rheumatology key message.
People with chronic pain are at higher risk of developing COVID-19 and subsequent adverse outcomes.
Dear Editor, Individuals with chronic pain—a characteristic symptom of FM—have increased mortality, including >5-fold higher risk of respiratory disease–related deaths [1]. Whether excess mortality is driven by acute respiratory infections or chronic respiratory diseases has not been examined. This distinction is important, as people with chronic pain may represent a vulnerable population for COVID-19. Traditional observational study designs are limited by confounding and reverse causation, for example, obesity and smoking are both bidirectionally associated with chronic pain and are also strong risk factors for poor outcomes in COVID-19. Mendelian randomization (MR) uses genetic variants as instrumental variables to estimate the causal effect of an exposure on an outcome that is less susceptible to these biases [2]. We aimed to estimate the influence of chronic pain on COVID-19, and vice versa, in univariable and multivariable MR accounting for BMI and smoking.
We used genome-wide association data from 112 612 any-COVID-19 cases (lab-confirmed, physician-diagnosed or self-reported) and 2 474 079 population controls, 24 274 hospitalized COVID-19 cases and 2 061 529 controls, and 8779 very severe COVID-19 cases (respiratory support or death) and 1 001 875 controls [3]. To avoid the limitations of using binary exposures in MR [4], we used a related trait—multisite chronic pain (MSCP)—from 387 649 UK biobank participants [5]. MSCP represents the sum of seven chronic pain sites across the body (head, face, neck/shoulder, back, stomach/abdomen, hip, knee) and is strongly correlated with chronic widespread pain [5]. MSCP was instrumented using 39 independent genome-wide significant (P < 5 × 10−8) variants. A standard and conditional F statistic of >10 was taken to suggest adequate instrument strength in univariable and multivariable MR, respectively. We employed the inverse variance weighted method for the primary analysis and sensitivity methods (e.g. MR-Egger) to explore potential pleiotropy. We applied multivariable MR [6] to additionally account for effects from BMI and smoking. Sensitivity methods were used to account for weak instruments when required [7], which attenuates estimates to the null where populations are non-overlapping, as in this analysis. Finally, ‘reverse’ MR was performed to estimate the effect of COVID-19 on MSCP. Additional methods are presented in the Supplementary Materials, available at Rheumatology online.
The F statistic for MSCP was 17. Each additional site of chronic pain increased the risk of any COVID-19 [odds ratio (OR) 1.18; 95% CI 1.06, 1.32; P = 0.003] and hospitalization (OR 1.58; 95% CI 1.17, 2.13; P = 0.003) (Fig. 1). The results from MR-Egger (any COVID: OR 2.73; 95% CI 1.68, 4.44; P = 2.49 × 10−4; hospitalization: OR 5.57; 95% CI 1.27, 24.43; P = 0.029) and other univariable sensitivity analyses were consistent with the primary analysis (shown in the Supplementary Materials, available at Rheumatology online).
Fig. 1.
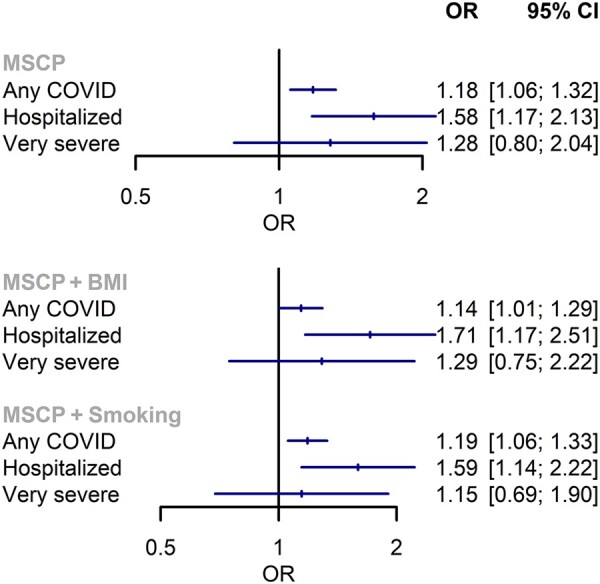
Mendelian randomization estimates of the influence of multisite chronic pain on COVID-19, using univariable (top) and multivariable (bottom) models
MSCP: multisite chronic pain; OR: odds ratio.
In multivariable MR, conditional F statistics for MSCP and BMI (9 and 30) and MSCP and smoking (19 and 9) suggested that analyses could be potentially influenced by weak instrument bias. Estimates accounting for BMI or smoking were not meaningfully different from univariable analyses, whether using weak instruments (Fig. 1) or with additional analyses accounting for weak instruments (Supplementary Materials, available at Rheumatology online).
Genetic liability to COVID-19 was positively associated with MSCP (any COVID: beta = 0.04, P = 0.048; hospitalized: beta = 0.02, P = 0.02; severe: beta = 0.01, P = 0.02). Additional results are presented in the Supplementary Materials, available at Rheumatology online.
The results of this study support a causal effect of chronic pain on the risk of developing and being hospitalized for COVID-19. These findings are consistent with prior observational data that demonstrate excess any-cause and respiratory-related mortality [1], and indicate that people with chronic pain are a vulnerable ‘at-risk’ group. We also showed that genetic liability for COVID-19 increases the risk of chronic pain. Recent studies have shown small-fibre pathology and autonomic dysfunction in ‘long COVID’, suggestive of a possible temporal association between infection and the nervous system [8]. The main limitation of this analysis is the as-yet unclear genetic basis of MSCP, which may pick up the genetic architecture of multiple diseases proximal to MSCP and could lead to misspecification of the primary phenotype in this analysis. However, the findings were robust to multivariable MR, even when accounting for potentially confounding traits such as smoking and BMI. The results were also supported by multiple pleiotropy robust methods. Further studies are needed to replicate these findings in cohorts with non-European ancestries. In conclusion, genetic analysis supports the hypothesis that chronic pain patient groups are at higher risk of and developing COVID-19 and subsequent adverse outcomes, which could potentially inform public health policy.
Supplementary Material
Acknowledgements
All authors were involved in the conception and design of the study, and analysis and interpretation of data. All authors contributed to the writing of the article and approved the final manuscript. Neither patients nor the public were involved in the design, conduct, reporting, or dissemination plans of this research. This analysis used publicly available summary statistics for which ethical approval had already been obtained.
Funding: S.S.Z. is supported by a National Institute for Health Research Clinical Lectureship. M.V.H. works in a unit that receives funding from the UK Medical Research Council and is supported by a British Heart Foundation Intermediate Clinical Research Fellowship (FS/18/23/33512) and the National Institute for Health Research Oxford Biomedical Research Centre. This research was funded by the Pain Relief Foundation.
Disclosure statement: M.V.H. has collaborated with Boehringer Ingelheim in research, and in adherence to the University of Oxford’s Clinical Trial Service Unit and Epidemiological Studies Unit (CSTU) staff policy, did not accept personal honoraria or other payments from pharmaceutical companies. All authors have declared no conflicts of interest that could bias this work.
Contributor Information
Sizheng Steven Zhao, Centre for Epidemiology Versus Arthritis, Division of Musculoskeletal and Dermatological Sciences, School of Biological Sciences, Faculty of Biology Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester; Institute of Life Course and Medical Sciences, University of Liverpool, Liverpool.
Michael V Holmes, MRC Population Health Research Unit, University of Oxford, Oxford.
Uazman Alam, Institute of Life Course and Medical Sciences, University of Liverpool, Liverpool; Department of Diabetes and Endocrinology, Liverpool University Hospital NHS Foundation Trust, Liverpool; Division of Diabetes, Endocrinology and Gastroenterology, Institute of Human Development, University of Manchester, Manchester, UK.
Data availability statement
All data used in this study are publicly available and accessible through relevant citations detailed.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Macfarlane GJ, Barnish MS, Jones GT.. Persons with chronic widespread pain experience excess mortality: longitudinal results from UK Biobank and meta-analysis. Ann Rheum Dis 2017;76:1815–22. [DOI] [PubMed] [Google Scholar]
- 2. Smith GD, Ebrahim S.. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 3.COVID-19 Host Genetics Initiative. Mapping the Human Genetic Architecture of COVID-19: an Update. 2021. https://www.medrxiv.org/content/10.1101/2021.11.08.21265944v1 (29 December 2021, date last accessed).
- 4. Burgess S, Labrecque JA.. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol 2018;33:947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston KJA, Adams MJ, Nicholl BI. et al. Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genetics 2019;15:e1008164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanderson E, Davey Smith G, Windmeijer F, Bowden J.. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol 2019;48:713–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanderson E, Spiller W, Bowden J.. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat Med 2021;40:5434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bitirgen G, Korkmaz C, Zamani A. et al. Corneal confocal microscopy identifies corneal nerve fibre loss and increased dendritic cells in patients with long COVID. Br J Ophthalmol 2021; Advance Access published , doi: 10.1136/bjophthalmol-2021-319450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are publicly available and accessible through relevant citations detailed.


