Abstract
The majority (591 of 791, or 76%) of Streptococcus pneumoniae clinical isolates examined showed the presence of two or more chromosomal SmaI fragments that hybridized with the lytA-specific DNA probe. Only one of these fragments, frequently having an approximate molecular size of 90 kb, was shown to carry the genetic determinant of the pneumococcal autolysin (N-acetylmuramic acid-l-alanine amidase). Strains carrying multiple copies of lytA homologues included both antibiotic-susceptible and -resistant isolates as well as a number of different serotypes and strains recovered from geographic sites on three continents. Mitomycin C treatment of strains carrying several lytA-hybridizing fragments caused the appearance of extrachromosomal DNA hybridizing to the lytA gene, followed by lysis of the bacteria. Such lysates contained phage particles detectable by electron microscopy. The findings suggest that the lytA-hybridizing fragments in excess of the host lytA represent components of pneumococcal bacteriophages. The high proportion of clinical isolates carrying multiple copies of lytA indicates the widespread occurrence of lysogeny, which may contribute to genetic variation in natural populations of pneumococci.
As part of a study of the expression of the autolysin gene in clinical isolates of Streptococcus pneumoniae, we probed SmaI digests of total DNA separated by pulsed-field gel electrophoresis (PFGE) with a probe specific for lytA. The sequence of lytA has no SmaI recognition site (16). Therefore, it was surprising that the majority of clinical isolates tested had more than one lytA-hybridizing band, indicating that there were two or more highly homologous genes in the chromosome.
In this communication, we present evidence that the supernumerary lytA-hybridizing fragments detected signal the presence of prophages and that the incidence of prophage carriage in natural isolates of pneumococci is high.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The S. pneumoniae isolates were obtained from The Rockefeller University collection. S. pneumoniae strains were grown in semisynthetic medium (21) at 37°C without aeration or in tryptic soy agar (Difco, Detroit, Mich.) supplemented with 3% sterile sheep blood incubated at 37°C.
Escherichia coli HB101 (pGL80) (17) was used as a source of DNA for the lytA gene probe. E. coli cells were grown in Luria-Bertani medium (Difco) with aeration or in Luria-Bertani agar (Difco) solid medium incubated at 37°C and supplemented with 75 μg of ampicillin/ml.
Probes for the lytA gene.
The probe for the lytA gene was obtained from plasmid pGL80 (17). Plasmid DNA was prepared by using the Wizard Midiprep kit (Promega, Madison, Wis.), digested with HindIII (New England Biolabs, Beverly, Mass.), and separated by agarose gel electrophoresis. The 1.2-kb fragment from pGL80 contained the entire lytA gene (957 nucleotides [nt]), a fragment of upstream sequence (199 nt), and 51 nt downstream of the gene. The 1.2-kb fragment containing the lytA gene was purified from the gel by using the Wizard DNA cleanup kit (Promega). A second, PCR-generated DNA probe included an internal, 890-nt lytA fragment lacking the first 57 and the last 10 nt of the gene. The 890-bp product was generated with primers Lytd-1 and Lytr-1 (Table 1), with DNA from strain R36A as the template. The PCR product was purified by using the Wizard DNA cleanup kit (Promega) before being labeled with either the ECL random prime labeling kit or the ECL direct labeling kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
TABLE 1.
Selected primers and probes used in this study
| Primer | Sequence (5′ to 3′) | Accession no. Z34303
|
|
|---|---|---|---|
| Positions | Amplified fragment | ||
| Lytd-1 | TATAGGCAAGTACACGC | 4602–4618 | 4602–5491 |
| Lytr-1 | GTAATCAAGCCATCTGGCTC | 5472–5491 | 4602–5491 |
| RecAd | AAGGATCAATCATGCGTTTGGG | 1432–1453 | 1432–2047 |
| RecAr | TTACCACGAACATCCAAGCGG | 2027–2047 | 1432–2047 |
Probe for the recA gene.
The recA probe was an internal fragment of 616 bp obtained by PCR with primers RecAd and RecAr (Table 1) and with DNA from strain R36A as the template. The PCR product was purified by using the Wizard DNA cleanup kit (Promega), and labeling was performed by using the ECL direct labeling kit (Amersham).
PFGE.
Chromosomal DNA fragments, generated by SmaI digestion, were separated and analyzed as previously described (40). SmaI PFGE patterns were assigned arbitrary numbers. For the visualization of extrachromosomal phage DNA, cultures were grown and induced with mitomycin C as described below. The optical density of the culture at 620 nm (OD620) was monitored. Cells were harvested immediately after a decrease in OD620 or just before lysis. Agarose disks for PFGE were prepared as previously described (40). The unrestricted DNA was separated in a 1% agarose gel, with 0.5× Tris-borate-EDTA as the buffering agent. The gel was run on a Chef DRII apparatus (Bio-Rad, Hercules, Calif.) under the following conditions: 6 V/cm, ramping of the pulse between 5 and 35 s, and a total running time of 23 h. The buffer was maintained at 7°C during the run.
Southern blot hybridization.
DNA fragments separated by PFGE were transferred to nylon membranes (Hybond N+; Amersham) by using the vacuum gene system (Pharmacia LKB Biotech, Uppsala, Sweden), according to the manufacturer’s instructions. Membranes were hybridized to specific DNA probes labeled with the ECL system (Amersham), as described above. Hybridization conditions for the ECL direct labeling system were as recommended by the manufacturer, using a sodium chloride concentration of 0.5 M. Hybridization conditions for the ECL random prime system were as recommended by the manufacturer. The molecular weights of the hybridization signal(s) (39, 41) and the corresponding SmaI fragments were determined.
Induction of the lytic cycle by mitomycin C in putative lysogenic strains.
Cultures were grown at 37°C until the OD620 reached approximately 0.2 to 0.3. Mitomycin C was then added to a final concentration of 0.1 μg/ml to induce the putative lysogenic strains. Incubation was continued, and growth was monitored by optical density.
Effect of high concentrations of choline on lysis of induced cultures.
Cultures were grown at 37°C until the OD620 reached approximately 0.2 to 0.3. Mytomicin C and choline were then added to final concentrations of 0.1 μg/ml and 2% (wt/vol), respectively. As a control for the prevention of lysis by high choline concentrations, penicillin was added at a concentration 10 times the MIC, together with choline. Incubation was continued, and growth was monitored by OD.
Preparation of samples for electron microscopy.
Cells for electron microscopy were harvested by centrifugation, washed twice with 0.9% sodium chloride, and suspended in 10 times the volume of the pellet in a solution composed of 100 mM sodium cacodylate, 100 mM sodium arsenate, and 2.5% glutaraldehyde (pH 7.4) (CAG). After embedding and sectioning, the preparations were stained with uranyl acetate for electron microscopic observation as described previously (44).
Phage particles were prepared for electron microscopy by allowing induced cells to lyse until the reduction in OD620 in 1 h was less than 10%. Cellular debris was removed by centrifugation at 3,750 rpm for 15 min. The supernatant was centrifuged in a Beckman ultracentrifuge model L5-50 with rotor model SW50.1 at 42,000 rpm for 90 min. The pellet was suspended in 100 μl of 0.9% NaCl, diluted in CAG, and prepared for negative staining as described previously (23).
RESULTS
Multiple copies of lytA in clinical isolates of S. pneumoniae.
Each one of the 791 clinical isolates of S. pneumoniae tested showed at least one chromosomal SmaI fragment that hybridized to the lytA DNA probe. Also, each of the 791 clinical isolates showed lysis by deoxycholate, a property involving the activity of the autolytic amidase, indicating that at least one of the lytA copies present in all pneumococcal isolates must be the determinant of the autolysin. The ubiquitousness of lytA, the genetic determinant of the major pneumococcal autolysin, an N-acetylmuramic acid-l-alanine amidase, among pneumococci, was demonstrated earlier by dot blot hybridization (31).
The presence of multiple copies of lytA in the majority of the pneumococcal isolates examined was an unexpected finding of these hybridization studies (Table 2). The frequency of S. pneumoniae strains carrying one to four lytA-hybridizing SmaI bands is shown for isolates collected from seven different countries (Table 2).
TABLE 2.
Frequency of isolates with multiple lytA-hybridizing fragments in seven countries
| No. of bands | Frequency (%) of isolates from indicated country
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Brazil (n = 81) | Bulgaria (n = 94) | Hungary (n = 79) | Mexico (n = 44) | U.S. (n = 240) | Iceland (n = 129) | Sweden (n = 124) | Total (n = 791) | |
| 1 | 12.3 | 19.1 | 21.5 | 2.3 | 23.8 | 14.0 | 55.6 | 24.0 |
| 2 | 67.9 | 61.7 | 58.2 | 70.5 | 57.9 | 51.2 | 40.3 | 56.3 |
| 3 | 19.8 | 19.1 | 17.7 | 25.0 | 17.1 | 33.3 | 4.0 | 18.7 |
| 4 | 0.0 | 0.0 | 2.5 | 2.3 | 1.2 | 1.6 | 0.0 | 1.0 |
Variation in the number and molecular size of SmaI fragments hybridizing with the lytA DNA probe.
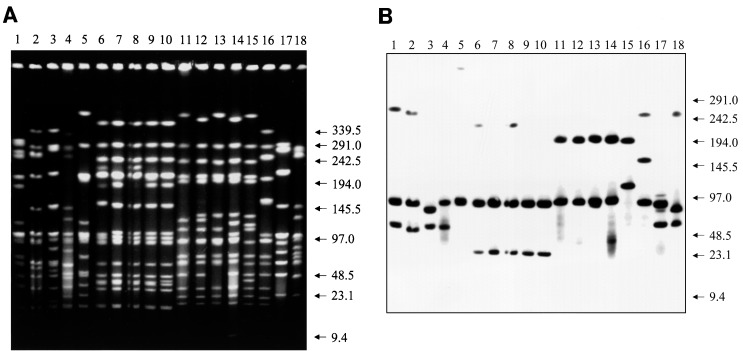
Figure 1A shows PFGE profiles of SmaI-restricted total DNA isolated from a group of 18 S. pneumoniae clinical isolates selected to illustrate the variability of the lytA hybridization pattern (Fig. 1B). The isolates tested were from the United States (Alaska and Ohio) and Iceland and included penicillin-susceptible and penicillin-resistant isolates and multidrug-resistant isolates along with three different serotypes and several clonal types, as defined by PFGE pattern. The properties of these strains are listed in the legend to Fig. 1. Some parallels between PFGE patterns and the corresponding lytA hybridization patterns are apparent in a comparison of Fig. 1A and B.
FIG. 1.
Multiple patterns of lytA hybridization among clinical isolates of S. pneumoniae. Total DNA was isolated from a group of pneumococcal clinical isolates. After digestion with SmaI and separation by PFGE (A), Southern hybridization was done to identify fragments hybridizing with lytA (B). Pneumococcal strains originated in Iceland (Ic), Alaska (Ala50, intermediate level resistance to penicillin), and Cleveland (Clev2, serotype 23F, resistance to penicillin, tetracycline, and chloramphenicol). Except for strain Ic189, which belongs to serogroup 19, and strains Ic165, Ic183, Ic202 (resistant to penicillin, erythromycin, tetracycline, chloramphenicol, and sulfamethoxazole-trimethoprim), and Ala50, which belong to serogroup 6, all strains belong to serogroup 23. The Icelandic isolates with serogroup 23 showed intermediate level resistance to penicillin. Lanes 1 to 18, respectively, Ic165, Ic183, Ala50, Ic202, Ic162, Clev2, Ic107, Ic118, Ic130, Ic155, Ic226, Ic134, Ic173, Ic189, Ic204, Ic156, Ic191, and Ic221. Arrows indicate molecular sizes in kilobases.
Induction of lysis by mitomycin C.
Mitomycin C is known to induce the lytic cycle of some prophages (29), although not all prophages respond to this inducing agent (2). A group of 18 pneumococcal strains selected for variation in genetic background and lytA hybridization pattern was tested for lysis induction by mitomycin C. The strains, from Hungary and the United States, included 14 different clonal types (as defined by PFGE pattern), nine serotypes, and both penicillin-susceptible and antibiotic-resistant strains. Mitomycin C treatment was shown to induce lysis in 11 of the 18 isolates (Table 3).
TABLE 3.
Relevant properties and response to mitomycin C in selected pneumococcal isolates
| Strain | Serogroup or type | PFGE patterna | lytA (kb)c | recA (kb)c | Pen MIC (μg/ml)d | Resistancee | Mitomycin induction |
|---|---|---|---|---|---|---|---|
| MA42 | 19 | 2(2) | 145, 90, 70 | 90 | 16 | E, T, Cm | Yes |
| MA47 | 19 | 18(11) | 280, 90–80 | 90 | 16 | E, T, Cm | Yes |
| MA49 | 19 | 18(13) | 190, 160, 90 | 90 | 16 | E, T, Cm | Yes |
| MA62 | 19 | 2(8) | 245, 90, 70 | 90 | 16 | E, T, Cm | Yes |
| MA75 | 19 | 18(9) | 280, 90 | 90 | 8 | E, T, Cm | Yes |
| MA81 | 22 | 27 | 260, 80 | 80 | 0.5 | T, Cm | No |
| SVMC12 | NDb | 11 | 200, 90 | ND | 0.3 | E, T | Yes |
| SVMC17 | 23F | 1(2) | 90, 30 | ND | 6 | E, T, Cm | No |
| SVMC23 | 16 | 16 | 90, 85 | ND | S | — | No |
| SVMC27 | 23 | 8 | 225, 85 | ND | 1 | T | Yes |
| SVMC28 | 23 | 17 | 95, 60 | ND | S | — | Yes |
| SVMC31 | 9 | 5(2) | 255, 95 | ND | 2 | T | No |
| SVMC32 | 14 | 6 | 115 | ND | 2 | E, T | No |
| SVMC33 | 18 | 3 | 225, 85, 70 | ND | S | — | Yes |
| SVMC35 | 23F | 1(2) | 90, 30 | ND | 2 | E, T, Cm | No |
| SVMC36 | 3 | 1(10) | 90, 30 | ND | 3 | E, T, Cm | No |
| SVMC52 | 19 | 23 | 95, 85 | ND | S | — | Yes |
| SVMC54 | 6 | 13 | 80, 60 | ND | 0.1 | E, T | Yes |
The pattern numbers given are arbitrary. The first number denotes the PFGE pattern type (defined by the number and size of the bands). Patterns that differ by fewer than four fragments were classified as subtypes and assigned a different number in parentheses.
ND, not determined.
Molecular sizes of fragments hybridizing to probes for the gene indicated.
Penicillin MIC. S, sensitive.
E, erythromycin; T, tetracycline; Cm, chloramphenicol. The dash indicates susceptibility.
Prevention of lysis by high concentrations of choline.
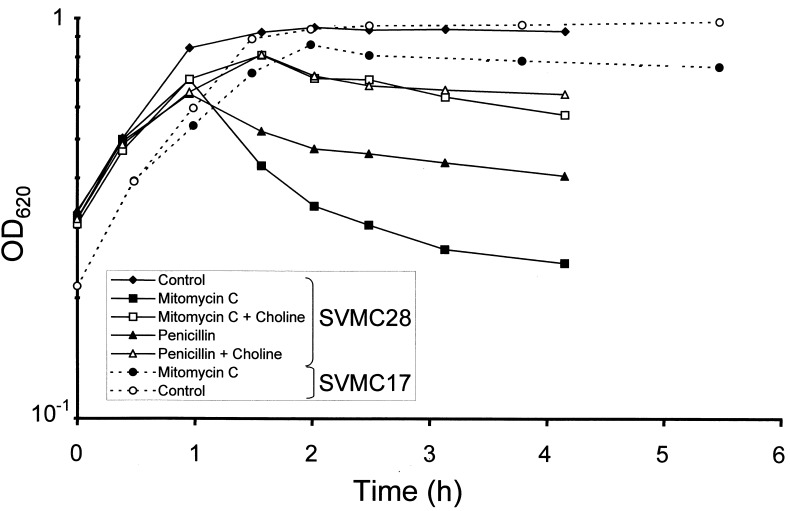
High concentrations of choline are known to inhibit lysis of pneumococcal autolysin, the product of lytA (8, 45). The dependence of the lytic activity of several pneumococcal bacteriophages on the presence of choline residues in the cell wall of the host bacterium was also established (13). Cultures of two S. pneumoniae strains, SVMC28, responding to mitomycin C treatment with culture lysis, and SVMC17, which did not lyse upon treatment with mitomycin C, were tested for the effect of high concentrations of choline on culture lysis induced by mitomycin C or penicillin. Antibiotics and choline were added to the cultures at time 0 in Fig. 2. The addition of high concentrations of choline inhibited both the mitomycin-induced and the penicillin-induced lysis in strain SVMC28. A high concentration of choline was found to inhibit each one of six additional cultures from lysing during mitomycin treatment (data not shown). The culture of SVMC17 showed no lysis during mitomycin treatment in spite of the fact that it possessed two SmaI fragments positive for the lytA gene probe (Table 3).
FIG. 2.
Lysis of S. pneumoniae induced by mitomycin C. Cultures of strains SVMC28 and SVMC17 received mitomycin C (0.1 μg/ml) or penicillin (0.1 μg/ml) with or without the addition of choline (2%) at time zero, and the OD of cultures was monitored at 620 nm, as described in Materials and Methods.
Electron microscopic detection of phage particles in culture supernatants and in thin sections of cells induced with mitomycin C.
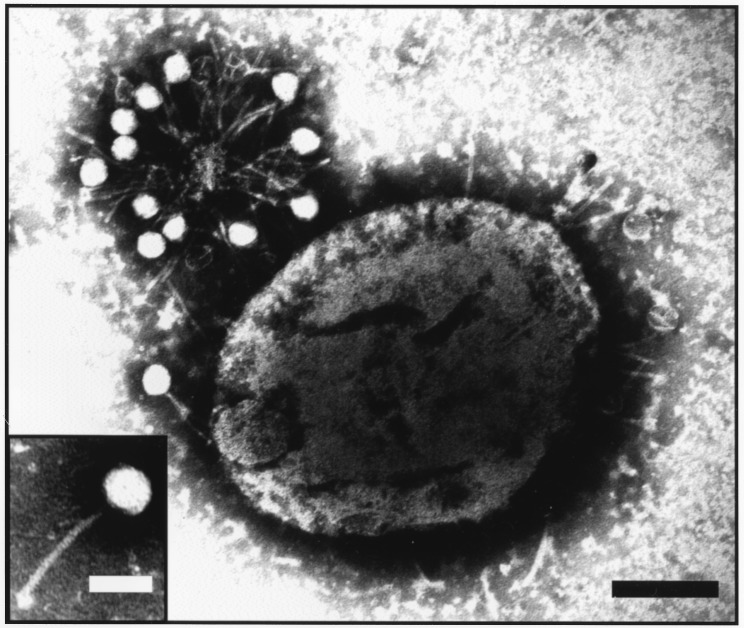
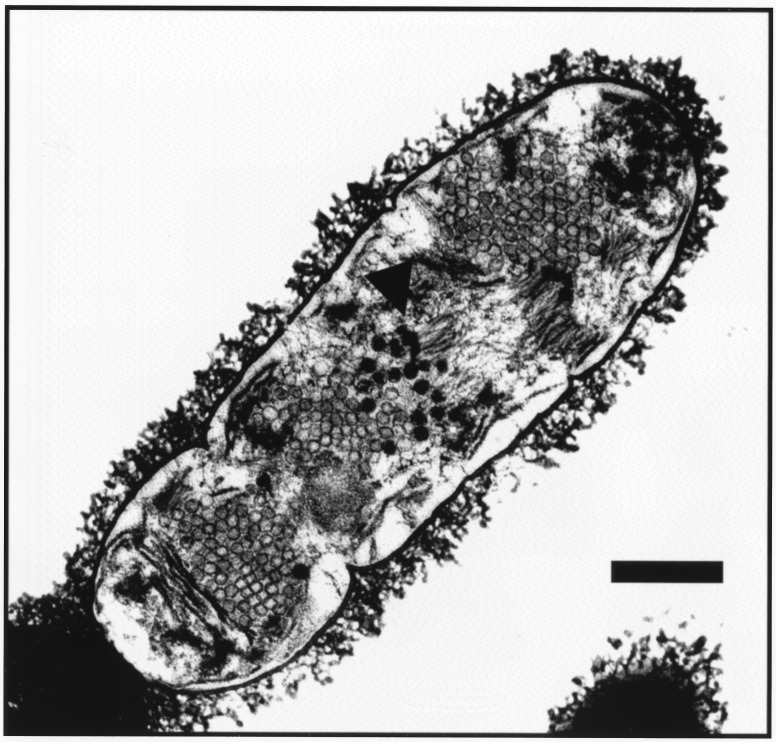
Mitomycin-treated cultures harvested before lysis as well as supernatants of mitomycin-lysed cultures were examined by electron microscopy for the presence of phage particles. Phage particles were detected in all three strains examined either by negative staining with phosphotungstic acid or by thin sectioning of the mitomycin-treated bacteria (Fig. 3). Mature phage particles were also detected in thin sections of SVMC28 in which the mitomycin C-induced lysis was blocked by the presence of high concentrations of choline in the medium (Fig. 4).
FIG. 3.
Detection of phage particles in the supernatant of strain SVMC28 induced to lyse by mitomycin C. The preparation of the sample and negative staining for electron microscopy were performed as described in Materials and Methods. The larger electron micrograph shows a pneumococcal cell with multiple phage tails attached and a cluster of phages and phage tails in the top left corner. This cluster is probably held together by a cellular fragment containing phage receptors. The inset shows a complete phage with a morphology that is typical of the Siphoviridae family and similar to that of the pneumococcal phage HB-3 (3). Black bar, 0.3 μm; white bar (inset), 0.1 μm.
FIG. 4.
Intracellular phage particles in pneumococci induced with mitomycin C but protected from lysis by a high concentration of choline in the medium. Samples for electron microscopy were prepared as described in Materials and Methods. The strain used in this experiment was SVMC28. Collapse of the membrane is evident from the white background in the cytoplasmic space. Escape of phage particles is prevented by the integrity of the cell wall. Arrowheads indicate fully assembled phage particles. Bar, 0.3 μm.
Production of extrachromosomal phage DNA in mitomycin C-treated bacteria.
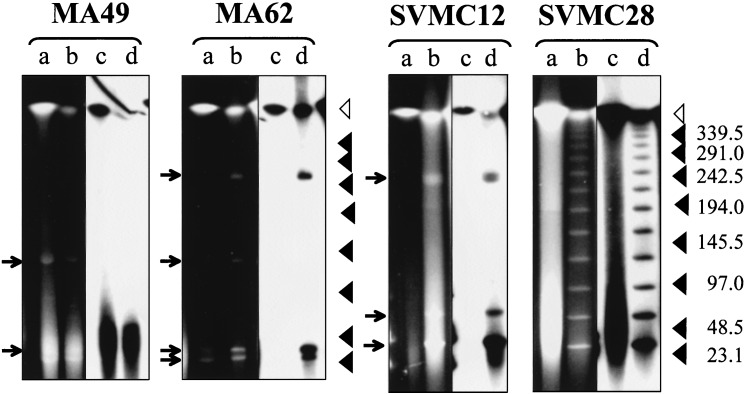
Four S. pneumoniae clinical isolates showing induction of lysis during mitomycin C treatment were tested for the presence of extrachromosomal phage DNA after exposure to mitomycin C. The chromosome of the bacterium is about 2.2 Mb (19) and migrates in the compression zone of the gel under the running conditions of PFGE. In most but not all strains, unrestricted total DNA from control cultures (not treated with mitomycin) showed no DNA fragments smaller than the chromosome (Fig. 5, lanes a). In contrast, DNA fragments generated by mitomycin treatment were smaller than the chromosome and separable by PFGE. Particularly striking was the appearance of the ladder-like pattern of DNA fragments in mitomycin-treated cultures of SVMC28. Like the bacterial chromosome, all mitomycin-induced DNA fragments hybridized with the lytA gene probe, suggesting that they correspond to extrachromosomal phage DNA.
FIG. 5.
Extrachromosomal phage DNA in S. pneumoniae induced with mitomycin C. Total DNA was isolated from cultures of strains SVMC12, SVMC28, MA49, and MA62 treated with mitomycin C or left untreated. Preparations were separated by PFGE without prior treatment with restriction enzymes. Lanes a and b show PFGE profiles in the UV, and lanes c and d show hybridization with the lytA probe. Lanes a and c are from control (untreated) cultures, and lanes b and d are from mitomycin C-treated cultures. Open triangles indicate the position of the bacterial chromosome that migrates in the compression zone of the gel. Arrowheads on the right indicate molecular sizes in kilobases. Arrows on the left indicate DNA fragments smaller than the bacterial chromosome.
DISCUSSION
Incidence of multiple bands hybridizing to the lytA probe and localization of the lytA gene.
A surprisingly large proportion (76%) of the 791 pneumococcal clinical isolates examined showed more than one SmaI DNA fragment hybridizing with the DNA probe specific for the pneumococcal autolysin gene. The number and molecular sizes of the lytA homologues were independent of the geographic origin, serotype, or antibiotic resistance of the isolates (Table 2). Since lytA does not have an SmaI cutting site (16), the observation raised questions concerning the nature of the multiple lytA-hybridizing bands.
All pneumococcal isolates had at least one lytA-hybridizing fragment, as expected from the ubiquitousness of the autolytic enzyme in pneumococci (31). The laboratory strain R6 had a single lytA-hybridizing band, and strain M31 (38), a lytA deletion mutant derivative of R6 selected for defective autolytic activity, gave no hybridization signal with the lytA DNA probe. Moreover, it was shown that the pneumococcal recA gene (accession no. Z34303) is located approximately 2 kb upstream of lytA and that, at least in some circumstances, recA is transcribed together with lytA (24). Each of a randomly selected group of 79 pneumococcal isolates probed with recA showed a single band that always colocalized with a lytA-hybridizing band (results not shown). In most cases, the size of this band was about 90 kb (Table 3), although fragments in the range of 80 to 130 kb were also observed. These findings indicate that the lytA-hybridizing band representing the genetic determinant of the host autolytic activity was associated in most, if not all, of the isolates with a SmaI fragment of approximately 90 kb.
Two possibilities could explain the multiplicity of lytA homologues in most of the clinical isolates. The lytA gene may have one or more SmaI recognition sites in some clinical isolates. Alternatively, the chromosome of pneumococcal isolates may carry multiple genes with a high degree of identity to lytA. The sequence of the lytA gene of the isolates presenting multiple positive fragments was not determined, and thus the first possibility cannot be formally excluded. However, several observations described in this communication strongly suggest an alternative explanation, namely, that the supernumerary lytA-hybridizing bands represent pneumococcal prophages. Consistent with this hypothesis is the observed variation in the molecular size and number of the lytA-hybridizing bands in different isolates, suggesting independent acquisition of the lytA-hybridizing genes.
Lysis induction by mitomycin C and detection of phage particles.
The majority (11 of 17) of the pneumococcal isolates tested carrying multiple copies of lytA lysed within a 5-h period following the addition of mitomycin C to the medium. Conditions that inhibit the activity of the pneumococcal amidase (the lytA gene product) are known to prevent phage-mediated lysis of pneumococcal cells (37). The inhibition of mitomycin-induced lysis by choline suggests that the phage lytic enzyme has biochemical properties similar to those of the host autolytic amidase. Electron microscopy of thin sections of mitomycin C-induced cells just before lysis or of cells in which mitomycin-induced lysis was blocked by high concentrations of choline revealed the presence of phage particles inside the cells. Phage particles were also detected in supernatants of mitomycin-lysed cultures. The morphology of the phage observed in the culture supernatant of strain SVMC28 was characteristic of the Podoviridae family (1), similar to the previously described pneumococcal temperate phage HB-3 (3) (Fig. 3).
Detection of extrachromosomal phage DNA.
Total DNA prepared from cells induced with mitomycin C was separated by PFGE, in order to confirm prophage induction. In strains that lysed upon mitomycin C induction, PFGE analysis allowed detection of discrete DNA fragments with molecular sizes smaller than that of the chromosome, and all extrachromosomal bands hybridized with the lytA probe. These results are compatible with the excision of the phage genome from the bacterial chromosome induced by mitomycin C.
In the mitomycin-treated strain SVMC28, PFGE identified a ladder-like pattern of DNA fragments, all of which hybridized to the lytA probe. These fragments appeared to be multiples of the smallest fragment (30 kb), suggesting that, like E. coli λ phage, the prophage infecting SVMC28 has a genome with cohesive ends (42). The estimated 30-kb genome size of the hypothetical prophage in SVMC28 is similar to the genome sizes of other pneumococcal phages (18). The multiple lytA-hybridizing fragments seen upon induction with mitomycin C in strain MA62 could correspond to multiple prophages. However, the possibility that the larger bands correspond to closed circular forms of one of the smaller bands cannot be clarified, since circular molecules are known to have unusual migration properties in PFGE (7).
The diversity of molecular sizes of DNA fragments obtained by mitomycin C induction and the fact that more than one fragment was obtained from several strains suggest that clinical isolates of pneumococci are lysogenic for more than one phage. This conclusion is supported by the fact that a number of isolates have more than two SmaI fragments positive for the lytA gene probe. In some cases (for instance, MA49 in Fig. 5), signals were also detected in uninduced controls with the same molecular size as that observed in mitomycin C-treated cells. This finding is compatible with a low-level spontaneous induction of prophage, similar to what was described previously in other systems (12, 14, 29).
Pneumococcal isolates that did not lyse upon mitomycin C treatment in spite of having multiple lytA-hybridizing fragments may carry defective prophages. Large fragments related to phage DNA have already been identified in the E. coli chromosome (34) as well as in S. pneumoniae (35). The phage-related fragment described in S. pneumoniae 8R1 encompassed the phage lytic genes precisely and provided the insertion site for temperate phage HB-746 (35). Pneumococci carrying defective prophages may represent bacterial hosts that survived cycles of infection and lysogenization or have lost part of the phage genome, as proposed for E. coli K12 (34).
Frequency of prophage carriage.
The incidence of prophage carriage in bacteria varies from 4% to almost 100%, depending on the species, origin of the isolates, and method used to evaluate carriage rates (9, 12, 20, 28). Early studies on S. pneumoniae that used mitomycin C for induction and relied on plaque formation for phage detection found that 8 of 12 (4) or 58 of 139 (3) pneumococcal isolates were lysogenic. The incidence of prophage based on lytA hybridization of PFGE-separated SmaI fragments determined in this study (76%) is higher than earlier estimates (combined average of 42.4%) (3, 4). Either one of these two values may be realistic, since the incidence of prophage in bacteria depends on many factors. Our method may overestimate the incidence of functional prophages, since it can also detect defective prophages. Conversely, the method of Bernheimer (3, 4), which relies on the sensitivity of an indicator strain, probably underestimates the incidence of prophage.
Lysogeny and competence.
Functional as well as defective prophages may promote genetic variation and may contribute to the structure of pneumococcal populations in their natural environment. The high incidence of lysogeny observed among clinical strains raises the possibility that some of the exchange of genetic information occurring in S. pneumoniae in vivo proceeds through transduction or is assisted by phage function(s). Some temperate phages are known to carry virulence-related genes (6, 15, 27, 46); an analogous process could be of selective advantage for pneumococci.
It has been suggested that lysogeny may inhibit DNA-mediated genetic transformation (25), based on a study with the temperate bacteriophage 304 and a single streptococcal isolate (R6X). However, tests of this proposition with a large number of clinical strains and their infecting prophages showed no correlation between transformability and prophage carriage. The results described in Table 4 demonstrate that strains that carried an inducible prophage transformed at a high level in response to competence-stimulating peptide (CSPα) (e.g., SVMC54), whereas others did not transform at all (e.g., SVMC52). Isolates presenting two lytA-hybridizing bands but not responding to mitomycin C induction also showed a high-level response to CSPα (e.g., SVMC23) or did not respond at all (e.g., SVMC35).
TABLE 4.
Relation between prophage carriage and competence development in selected isolates
| Strain | Molecular size (kb)a | Presence
of:b
|
Mitomycin induction | CSPα inductionc | |
|---|---|---|---|---|---|
| comA | comC | ||||
| SVMC12 | 200, 90 | + | + | + | I |
| SVMC17 | 90, 30 | + | + | − | L |
| SVMC23 | 90, 85 | + | + | − | H |
| SVMC27 | 225, 85 | + | + | + | H |
| SVMC28 | 95, 60 | + | + | + | H |
| SVMC31 | 255, 95 | + | + | − | H |
| SVMC32 | 115 | + | + | − | H |
| SVMC33 | 225, 85, 70 | + | + | + | I |
| SVMC35 | 90, 30 | + | + | − | None |
| SVMC36 | 90, 30 | + | + | − | None |
| SVMC52 | 95, 85 | + | + | + | None |
| SVMC54 | 80, 60 | + | + | + | H |
A process similar to transduction, but requiring competence development, was described previously in pneumococci (30). In this system, the host DNA seems to be packed into the phage heads but enters the cells through the same route as transforming DNA. Moreover, Porter and coworkers (30) concluded that the efficiency of pseudotransduction of a deletion of more than 15 kb is 10-fold more efficient than transformation of the same deletion. This result was obtained in spite of the median 16-kb fragment size in the DNA preparation. Porter et al. argue that protection of the DNA from excessive cuts by the DNA binding sites in the surface of the bacteria could account for this phenomenon. The possibility of in vivo DNA exchange through this mechanism is attractive, because the higher efficiency of pseudotransduction of large fragments of DNA compared to transformation could favor this route over transformation for the observed in vivo capsular switch events (10, 11, 26, 33).
The unexpectedly high incidence of phage infection in natural isolates of S. pneumoniae could have an impact on the structure of natural populations of pneumococci in their ecological niche. Phages capable of lysing nonencapsulated (nonlysogenic) indicator strains have been readily isolated from carriers or patients (23, 36, 43). On the other hand, it was shown that the presence of capsular polysaccharide protects pneumococci against infection by phages of the ω group (5). The overwhelming majority of pneumococcal clinical isolates express antiphagocytic capsules, and it is conceivable that defense against phage infection may be another selective pressure that affects the structure and maintenance of capsular polysaccharides in pneumococci.
The population structure of S. pneumoniae colonizing the nasopharynx may be modulated by the phage immunity pattern of the resident flora, which would provide a strong selective pressure against incoming strains. Superinfection immunity (22), restricted to closely related phages or associated with phages of a broader scope (2), is a frequent property of prophages.
ACKNOWLEDGMENTS
Partial support for these studies was received from grant no. RO1 AI37275 from the National Institutes of Health and from the Irene Diamond Fund. M.R. received partial support from the Gulbenkian Foundation (PGDBM) and the Fundação Luso Americana para o Desenvolvimento (FLAD).
We thank Sigurdur Vilhelmsson, Cristina Brandileone, Gabriela Aviles, and Lena Setchanova for technical assistance.
REFERENCES
- 1.Ackermann H W, Cantor E D, Jarvis A W, Lembke J, Mayo J A. New species definitions in phages of gram-positive cocci. Intervirology. 1984;22:181–190. doi: 10.1159/000149550. [DOI] [PubMed] [Google Scholar]
- 2.Barksdale L, Arden S B. Persisting bacteriophage infections, lysogeny, and phage conversions. Annu Rev Microbiol. 1974;28:265–299. doi: 10.1146/annurev.mi.28.100174.001405. [DOI] [PubMed] [Google Scholar]
- 3.Bernheimer H P. Lysogenic pneumococci and their bacteriophages. J Bacteriol. 1979;138:618–624. doi: 10.1128/jb.138.2.618-624.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernheimer H P. Lysogeny in pneumococci freshly isolated from man. Science. 1977;195:66–68. doi: 10.1126/science.12565. [DOI] [PubMed] [Google Scholar]
- 5.Bernheimer H P, Tiraby J G. Inhibition of phage infection by pneumococcus capsule. Virology. 1976;73:308–309. doi: 10.1016/0042-6822(76)90085-4. [DOI] [PubMed] [Google Scholar]
- 6.Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 7.Birren B, Lai E. Pulsed field gel electrophoresis: a practical guide. San Diego, Calif: Academic Press, Inc.; 1993. [Google Scholar]
- 8.Briese T, Hakenbeck R. Interaction between choline and the N-acetyl-muramyl-alanine-amidase of Streptococcus pneumoniae. In: Hakenbeck R, Holtje J-V, editors. The target of penicillin—the murein sacculus of bacterial cell walls architecture and growth. Berlin, Germany: Walter de Gruyter; 1983. pp. 173–178. [Google Scholar]
- 9.Coetzee J N, de Klerk H C. Lysogeny in the genus Lactobacillus. Nature. 1962;194:505. doi: 10.1038/194505a0. [DOI] [PubMed] [Google Scholar]
- 10.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 11.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berron S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 12.Davidson B E, Powell I B, Hillier A J. Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol Rev. 1990;7:79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x. [DOI] [PubMed] [Google Scholar]
- 13.Díaz E, López R, García J L. EJ-1, a temperate bacteriophage of Streptococcus pneumoniae with a Myoviridaemorphotype. J Bacteriol. 1992;174:5516–5525. doi: 10.1128/jb.174.17.5516-5525.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas J. Bacteriophages. 1st ed. London, England: Chapman and Hall Ltd.; 1975. [Google Scholar]
- 15.Freeman V J. Studies on the virulence of the bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 1951;61:675–688. doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia E, Garcia J L, Ronda C, Garcia P, Lopez R. Cloning and expression of the pneumococcal autolysin gene in Escherichia coli. Mol Gen Genet. 1985;201:225–230. doi: 10.1007/BF00425663. [DOI] [PubMed] [Google Scholar]
- 17.Garcia P, Garcia J L, Garcia E, Lopez R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 18.Garcia P, Martin A C, Lopez R. Bacteriophages of Streptococcus pneumoniae: a molecular approach. Microb Drug Resist. 1997;3:165–176. doi: 10.1089/mdr.1997.3.165. [DOI] [PubMed] [Google Scholar]
- 19.Gasc A M, Kauc L, Barraille P, Sicard M, Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991;173:7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang S C, Paul J H. Significance of lysogeny in the marine environment—studies with isolates and a model of lysogenic phage production. Microb Ecol. 1998;35:235–243. doi: 10.1007/s002489900079. [DOI] [PubMed] [Google Scholar]
- 21.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 22.Marsh P, Wellington E M H. Phage-host interactions in soil. FEMS Microbiol Ecol. 1994;15:99–108. [Google Scholar]
- 23.McDonnell M, Lain R, Tomasz A. ”Diplophage”: a bacteriophage of Diplococcus pneumoniae. Virology. 1975;63:577–582. doi: 10.1016/0042-6822(75)90329-3. [DOI] [PubMed] [Google Scholar]
- 24.Mortier-Barriere I, de Saizieu A, Claverys J P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 25.Moynet D J, Tiraby G J. Inhibition of transformation in Streptococcus pneumoniaeby lysogeny. J Bacteriol. 1980;141:1298–1304. doi: 10.1128/jb.141.3.1298-1304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 27.O’Brien A D, Newland J W, Miller S F, Holmes R K, Smith H W, Formal S B. Shiga-like toxin-converting phages from Escherichia colistrains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 28.Ogunseitan O A, Sayler G S, Miller R V. Application of DNA probes to analysis of bacteriophage distribution patterns in the environment. Appl Environ Microbiol. 1992;58:2046–2052. doi: 10.1128/aem.58.6.2046-2052.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuji N, Sekiguchi M, Iijima T, Takagi Y. Induction of phage formation in the lysogenic Escherichia coliK-12 by mitomycin C. Nature. 1959;184:1079–1080. doi: 10.1038/1841079b0. [DOI] [PubMed] [Google Scholar]
- 30.Porter R D, Shoemaker N B, Rampe G, Guild W R. Bacteriophage-associated gene transfer in pneumococcus: transduction or pseudotransduction? J Bacteriol. 1979;137:556–567. doi: 10.1128/jb.137.1.556-567.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pozzi G, Oggioni M R, Tomasz A. DNA probe for identification of Streptococcus pneumoniae. J Clin Microbiol. 1989;27:370–372. doi: 10.1128/jcm.27.2.370-372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez M, Morrison D A, Tomasz A. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:39–52. doi: 10.1089/mdr.1997.3.39. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez, M., and A. Tomasz. Acquisition of new capsular genes among clinical isolates of antibiotic resistant Streptococcus pneumoniae. Submitted for publication. [DOI] [PubMed]
- 34.Redfield R J, Campbell A M. Structure of cryptic lambda prophages. J Mol Biol. 1987;198:393–404. doi: 10.1016/0022-2836(87)90289-0. [DOI] [PubMed] [Google Scholar]
- 35.Romero A, Lopez R, Garcia P. The insertion site of the temperate phage HB-746 is located near the phage remnant in the pneumococcal host chromosome. J Virol. 1992;66:2860–2864. doi: 10.1128/jvi.66.5.2860-2864.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronda C, Lopez R, Garcia E. Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae. J Virol. 1981;40:551–559. doi: 10.1128/jvi.40.2.551-559.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronda-Lain C, Lopez R, Tapia A, Tomasz A. Role of the pneumococcal autolysin (murein hydrolase) in the release of progeny bacteriophage and in the bacteriophage-induced lysis of the host cells. J Virol. 1977;21:366–374. doi: 10.1128/jvi.21.1.366-374.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Puelles J M, Ronda C, Garcia J L, Garcia P, Lopez R, Garcia E. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytAgene. Eur J Biochem. 1986;158:289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 39.Schaffer H E, Sederoff R R. Improved estimation of DNA fragment lengths from agarose gels. Anal Biochem. 1981;115:113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- 40.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniaefrom Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 41.Southern E M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979;100:319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- 42.Taylor K, Wegrzyn G. Replication of coliphage lambda DNA. FEMS Microbiol Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 43.Tiraby J G, Tiraby E, Fox M S. Pneumococcal bacteriophages. Virology. 1975;68:566–569. doi: 10.1016/0042-6822(75)90300-1. [DOI] [PubMed] [Google Scholar]
- 44.Tomasz A, Jamieson J D, Ottolenghi E. The fine structure of Diplococcus pneumoniae. J Cell Biol. 1964;22:453–467. doi: 10.1083/jcb.22.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasz A, Waks S. Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc Natl Acad Sci USA. 1975;72:4162–4166. doi: 10.1073/pnas.72.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]