Abstract
Bacterial endospores are encased in a complex protein coat, which confers protection against noxious chemicals and influences the germination response. In Bacillus subtilis, over 20 polypeptides are organized into an amorphous undercoat, a lamellar lightly staining inner structure, and an electron-dense outer coat. Here we report on the identification of a polypeptide of about 30 kDa required for proper coat assembly, which was extracted from spores of a gerE mutant. The N-terminal sequence of this polypeptide matched the deduced product of the tasA gene, after removal of a putative 27-residue signal peptide, and TasA was immunologically detected in material extracted from purified spores. Remarkably, deletion of tasA results in the production of asymmetric spores that accumulate misassembled material in one pole and have a greatly expanded undercoat and an altered outer coat structure. Moreover, we found that tasA and gerE mutations act synergistically to decrease the efficiency of spore germination. We show that tasA is the most distal member of a three-gene operon, which also encodes the type I signal peptidase SipW. Expression of the tasA operon is enhanced 2 h after the onset of sporulation, under the control of ςH. When tasA transcription is uncoupled from sipW expression, a presumptive TasA precursor accumulates, suggesting that its maturation depends on SipW. Mature TasA is found in supernatants of sporulating cultures and intracellularly from 2 h of sporulation onward. We suggest that, at an early stage of sporulation, TasA is secreted to the septal compartment. Later, after engulfment of the prespore by the mother cell, TasA acts from the septal-proximal pole of the spore membranes to nucleate the organization of the undercoat region. TasA is the first example of a polypeptide involved in coat assembly whose production is not mother cell specific but rather precedes its formation. Our results implicate secretion as a mechanism to target individual proteins to specific cellular locations during the assembly of the bacterial endospore coat.
Bacterial endospores are encased within a complex multilayered protein structure known as the coat, which serves two main roles. First, it confers protection against bactericidal enzymes and chemicals, such as lysozyme and chloroform, thus contributing to the spore’s resistance properties and viability. Second, the coat influences the spore’s ability to monitor its environment and to germinate within minutes of exposure to appropriate germinants (1, 15). In Bacillus subtilis, the coat is composed of a heterogeneous group of over 20 polypeptides, ranging in size from about 6 to 69 kDa, which are arranged in three main structural layers: a diffuse undercoat, a laminated lightly staining inner layer, and a thick and electron-dense outer coat. The process of coat assembly spans a long developmental period, during which the sporangium undergoes profound cytological modifications (15, 43). Early in the process of sporulation, the rod-shaped cell is asymmetrically divided into a smaller prespore and a larger mother cell compartment. The septal membranes, which define a septal compartment, then migrate around the forespore, eventually engulfing it. The engulfed prespore is then enveloped by the cortex peptidoglycan, which fills the septal compartment, and later by the coat layers, which are assembled on the forespore outer membrane (15). A large number of genes have been directly implicated in coat assembly. These include genes for 18 coat structural proteins (cot genes), as well as genes encoding morphogenetic proteins that act by guiding the assembly of the structural components but need not be part of the final structure. Expression of all the coat genes is governed by a cascade of four transcription factors which appear in the mother cell compartment in the sequence ςE, SpoIIID, ςK, and GerE (23, 26, 43, 51). Thus, the process of coat assembly is normally seen as an exclusive function of the mother cell.
The initial stages in coat assembly occur soon after septation and involve functional interactions among at least three morphogenetic proteins, all of which are made under ςE control (4, 35, 41, 50, 52). First, the SpoIVA protein localizes at the outer forespore membrane. Second, SpoIVA directs the assembly of CotE in a ring-like structure that surrounds the forespore at a distance of about 75 nm from it (12). The gap defined by the localization of SpoIVA and CotE is thought to become the site of assembly of the inner coat components. Within this region, the undercoat may correspond to the more internal sector, adjacent to the SpoIVA protein. In contrast, the outer coat proteins are assembled on the outside of the CotE structure (12). Most of the coat components are made after engulfment of the forespore by the mother cell, when ςK is activated (2, 7, 16, 19, 27, 36, 44, 48, 50). Certain components such as CotT and CotS are targeted to the inner coat (6, 44), while others such as CotB, CotC, and CotG are directed to the outer coat (36, 52). Only one protein, CotJC, has been proposed to associate with the undercoat (39). A variety of posttranslational modifications, including protein-protein cross-linking and endoproteolytic processing, may in part be designed to reinforce the correct interactions among individual components or their targeting to specific layers within the maturing coat (15).
Our study focuses on a 30-kDa polypeptide which appears as the predominant product solubilized by NaOH treatment from spores of a gerE mutant. The N-terminal amino acid sequence analysis revealed that the 30-kDa polypeptide is the mature form of a protein encoded by the tasA gene (Fig. 1). We show that tasA is preceded by the yqxM and sipW genes in an operon whose expression is under ςH control and that the maturation of the TasA precursor is coupled to the expression of the sipW gene, encoding a type I signal peptidase (SPase) (46). TasA is detected in sporulating cells and in culture supernatants from 2 h of sporulation onward. TasA is also found in extracts enriched for coat material from gerE mutant spores or from wild-type spores purified 48 h after the onset of sporulation. Remarkably, disruption of tasA results in the production of asymmetric spores that accumulate misassembled material in the undercoat region. Inactivation of tasA also causes abnormal assembly of the outer coat layer. Our results implicate protein secretion as a mechanism in assembly of the spore coat. Moreover, our results suggest that coat assembly is initiated at septation by the synthesis of proteins that are secreted to the septal compartment. It is hypothesized that these components either are translocated across the forespore outer membrane or act from this position to nucleate undercoat formation.
FIG. 1.
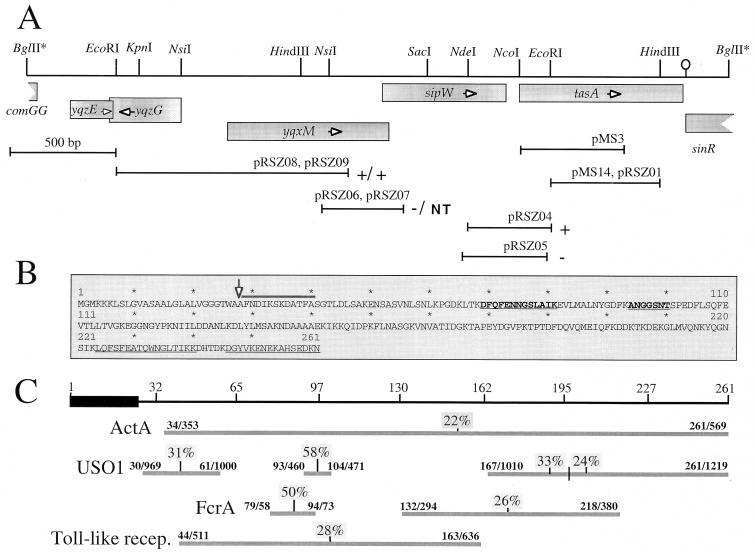
Chromosomal organization and structure of tasA. (A) Partial restriction map and genetic organization of the tasA region. The boxes below the restriction map indicate the extent and direction of transcription of the different cistrons in the region, as deduced from the analysis of the B. subtilis genome sequence (22). The stem-and-loop structure downstream of tasA indicates the position of a possible transcription terminator. The lines below the restriction map represent DNA fragments cloned into the indicated plasmids. The plus or minus sign to the right of a plasmid denotes the Lac phenotype of a B. subtilis strain carrying a transcriptional fusion of the corresponding fragment to the lacZ gene inserted in single copy at the amyE locus or at the tasA region, respectively (see text). pRSZ04 (integrating at the tasA region) and pRSZ05 (amyE integrational) are shown in separate lines, since the inserts in each plasmid differ slightly. “NT” indicates that no transformants of pRSZ07 were obtained. (B) Deduced primary structure of TasA. The thick line above the sequence denotes the N-terminal amino acid sequence determined by the Edman reaction of the coat-associated TasA polypeptide. The first 23 residues of TasA are thought to be a signal peptide, and the processing site is indicated by a vertical arrow. The internal segments underlined were determined by MALDI mass spectrometry analysis, after cleavage of a six-His-S.Tag-TasA fusion protein with Lys-C protease (see Materials and Methods). The sequences in boldface were determined by the Edman reaction. (C) Regions of sequence similarity between TasA and the indicated proteins. The numbers above the horizontal lines indicate the residues that delimit those regions in both proteins. The numbers above the vertical lines indicate the percentages of sequence identity for the indicated segments. The ActA, USO1, FcrA, and Toll-like protein sequences have the following database accession numbers: S20887, Z74106, S35760, and U88879, respectively. recep., receptor.
MATERIALS AND METHODS
Bacterial strains and media.
The B. subtilis strains used in this work are listed in Table 1. The wild-type strain MB24 (trpC2 metC3), and its congenic derivatives were used for the isolation of spore coat proteins (by the sodium dodecyl sulfate-dithiothreitol [SDS-DTT] method; see below), as well as the analysis of TasA production, processing, and assembly. Strain PY79 (prototrophic) and congenic derivatives bearing different developmental mutations were used for the analysis of β-galactosidase production driven by various lacZ fusions, for spore heat and lysozyme resistance tests, for germination assays, and for the extraction of coat proteins by treatment with NaOH. Cloning experiments were carried out with Escherichia coli DH5α. Strain BL21(DE3)pLysS (Novagen) was used for the production of a six-His–S.Tag-TasA fusion protein (see below). Yeast extract-tryptone (YT; 2×) medium was used for routine growth of E. coli and B. subtilis. Sporulation of B. subtilis was induced by exhaustion in Difco sporulation medium (DSM) (30). Antibiotics, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and isopropyl-β-d-thiogalactopyranoside (IPTG) were used as previously described (14, 27).
TABLE 1.
B. subtilis strains
| Strain | Genotype/phenotype | Origin |
|---|---|---|
| AZ393 | spo0H::sp/Spr Spo− | Laboratory stock |
| BK556 | spoIVCB23/Spo− | Laboratory stock (S. Cutting) |
| BK395 | spoIIID83/Spo− | Laboratory stock (S. Cutting) |
| KS450 | gerE36/Ger− | Laboratory stock (S. Cutting) |
| SC500 | spoIIIGΔ1/Spo− | Laboratory stock (S. Cutting) |
| SC1159 | spoIIAC1/Spo− | Laboratory stock (S. Cutting) |
| SC1163 | spoIIGB55/Spo− | Laboratory stock (S. Cutting) |
| PY17 | trpC2 SPβ | Laboratory stock |
| PY79 | Prototrophic | Laboratory stock |
| MB24 | trpC2 metC3 | Laboratory stock |
| AH17 | trpC2 spo0H81/Spo− | Laboratory stock |
| AH94 | trpC2 gerE36 | Laboratory stock |
| AH131 | trpC2 metC3 ΔamyE::erm | Laboratory stock |
| AH679 | trpC2 metC3 Pspac-spo0H/Cmr Spβ spoVE-lacZ/Cmr MLSra | A. O. Henriques (13a) |
| AH1700 | trpC2 metC3 tasA::pRSZ01/Cmr | This work |
| AH1802 | trpC2 metC3 Pspac-tasA/Cmr | This work |
| AH1822 | trpC2 metC3 tasA::cat | This work |
| AH1823 | trpC2 metC3 ΔcotE::erm | This work |
| AH1824 | trpC2 metC3 ΔcotE::erm tasA::cat | This work |
| AH1825 | trpC2 gerE36 ΔcotE::erm | This work |
| AH1826 | trpC2 gerE36 ΔcotE::erm tasA::sp | This work |
| AH1827 | trpC2 gerE36 tasA::cat | This work |
| AZ402 | tasA::pRSZ01(tasA-lacZ′ bla cat)/Cmr | This work |
| AZ403 | gerE36 tasA::pRSZ01(tasA-lacZ′ bla cat)/Cmr | This work |
| AZ404 | tasA::pRSZ04(tasA′-lacZ bla cat)/Cmr | This work |
| AZ405 | ΔamyE::tasA′-lacZ/Cmr | This work |
| AZ406 | ΔamyE::sipW′-′lacZ/Cmr | This work |
| AZ408 | ΔamyE::yqxM-′lacZ/Cmr | This work |
| AZ409 | yqxM::pRSZ07(yqxM′-lacZ bla cat)/Cmr | This work |
| AZ410 | tasA-lacZ spc | This work |
| AZ411 | Pspac-spo0H tasA::tasA′-lacZ/Cmr Spr | This work |
| AZ412 | gerE36 tasA::tasA′-lacZ/Cmr | This work |
| AZ413 | spo0H::sp tasA::tasA′-lacZ/Cmr | This work |
| AZ414 | spoIIID83 tasA::tasA′-lacZ/Cmr | This work |
| AZ415 | spoIVCB23 tasA::tasA′-lacZ/Cmr | This work |
| AZ416 | spoIIIGΔ1 tasA::tasA′-lacZ/Cmr | This work |
| AZ417 | spoIIAC1 tasA::tasA′-lacZ/Cmr | This work |
| AZ418 | spoIIGB55 tasA::tasA′-lacZ/Cmr | This work |
MLSr, macrolide-lincosamide-streptogramin B resistant.
Construction of tasA insertional mutants.
An 813-bp DNA fragment, internal to the tasA coding region, was amplified by PCR with oligonucleotides N1 (5′-CGATCAGCAGCGCCATTA-3′) and N4 (5′-TCGAATGAGAATTGAAGC-3′). The amplified product was purified and doubly digested with EcoRI and HindIII, and a 516-bp fragment was cloned between the EcoRI and HindIII sites of the integrational plasmids pER19 (34) and pUS19 (5), to yield plasmids pRSZ01 and pMS14, respectively (Fig. 1). Competent cells of strains PY17 and MB24 were transformed with pRSZ01, with selection for Cmr cells. These crosses produced the tasA insertion mutants AZ402 and AH1822, which were shown by PCR analysis to result from the integration of pRSZ01 into the tasA region of the chromosome by a single reciprocal crossover (a Campbell-type recombination event). Plasmid pRSZ01 was similarly transferred to the chromosomes of AH94 (gerE36) and AH1823 (ΔcotE::erm) to yield the double mutants AH1827 (gerE36 tasA::cat) and AH1824 (ΔcotE::erm tasA::cat). Mutant AH1825 (gerE36 ΔcotE::erm) was constructed by transforming AH94 with chromosomal DNA from AH1823 (absence of congression to Ger+ strain was verified with an SPβ cotC-lacZ-transducing phage). Finally, the triple mutant AH1826 (gerE36 ΔcotE::erm tasA::sp) was obtained by transformation of AH1825 with pMS14, with selection for spectinomycin resistance. Chromosomal DNA from AZ402 was also used to transform KS450 (gerE36) to Cmr, generating AZ403 (gerE36 tasA::cat) (Table 1).
Construction of a Pspac-tasA fusion.
A 495-bp DNA fragment carrying part of the tasA 5′ region and part of its coding sequence was amplified via PCR with the high-fidelity Pfu polymerase (Stratagene) and oligonucleotides N15 (5′-CTACTTAAGCTTCAGTTGTAAACCTGGC-3′) and N16 (5′-ACATCAAATACAGATCTTTAAGGTTCGC-3′). The 495-bp fragment was digested with BglII and HindIII and inserted into pDH88 that had been cut with the same enzymes (13). This produced pMS3, in which the tasA fragment is just downstream of the IPTG-inducible Pspac promoter (Fig. 1). Integration of pMS3 into the chromosome of wild-type host MB24 by Campbell-type recombination created the Cmr Pspac-tasA conditional mutant AH1802 (Table 1).
Transcriptional fusions of yqxM, sipW, and tasA to the lacZ gene.
The 5′ region of tasA was cloned by a chromosome walking step. First, we prepared chromosomal DNA from strain AZ402 and digested it with NdeI. Next, the digested DNA was religated in the presence of phage T4 DNA ligase, under conditions known to promote recircularization of the DNA. Finally, the ligated DNA was utilized to transform E. coli, with selection for Apr. The transformants were analyzed, and the majority were found to carry a plasmid, named pRSZ02, with a 3.4-kb genomic insert. A 1.3-kb EcoRI fragment from pRSZ02 was cloned into the same site of transcriptional fusion vector pJM783 (31), generating plasmid pRSZ03. In this plasmid, a 0.9-kb EcoRI-to-NdeI fragment found downstream of the tasA gene was artificially fused to the 5′ region of tasA. Thus, a 2.5-kb NaeI (adjacent to NdeI)-to-SstI (on the lacZ gene of pJM783) fragment was isolated from pRSZ03 and recloned into the 5.5-kb-long backbone of pJM783 that had been digested with SmaI and SstI. This step created the integrational plasmid pRSZ04, which carries 0.4 kb of DNA from the tasA 5′ region (as well as the first 51 codons of the gene) fused to the lacZ gene (Fig. 1A). To create a tasA-lacZ transcriptional fusion in an amyE integrational vector, oligomers N12 (5′-GTTCGCGCGCAATACAC-3′) and N13 (5′-CAAGCGTACCTGATGC-3′) were used to generate by high-fidelity PCR a 400-bp fragment encompassing the 5′ region and the first 42 codons of tasA. Digestion of the PCR product with EcoRI and RsaI produced a DNA fragment that could be cloned between the EcoRI and SmaI sites of the amyE integrational vector pSN32 (provided by I. Sá-Nogueira), thereby creating the tasA-lacZ transcriptional fusion plasmid pRSZ05 (Fig. 1A).
Oligonucleotides P5 (5′-GCACGAATTCCAAACCCGGCATTTATGC-3′) and P3 (5′-GCGTGGATCCTCTCCCCCGGATGAACGT-3′) generated by high-fidelity PCR a DNA fragment containing 377 bp of DNA upstream of the sipW start codon, as well as its initial 32 codons. The PCR product was purified, digested with EcoRI and BamHI, and cloned between the same sites of plasmid pAC5 (24), creating the amyE integrational plasmid pRSZ06, which carries a sipW-lacZ fusion. A 2.5-kb EcoRI-to-SstI (on lacZ) fragment isolated from pRSZ06 was cloned into EcoRI- and SstI-cut pJM783, originating the integrational plasmid pRSZ07 (Fig. 1A).
Lastly, fusions of yqxM to lacZ were generated as follows. First, a 1,543-bp PCR fragment encompassing the yqxM regulatory region was amplified with oligonucleotides OM127 (5′-TAAGAGTGTCGACGGATTCGGGAACAG-3′), and OM128 (5′-CGCATTTTGCTAGCCTCATAGGCTCCG-3′). The PCR fragment, flanked by engineered SalI and NheI sites, was cloned between the SalI and SpeI sites of pMLK83 (18), creating pOZ5. Second, a 1,084-bp EcoRI fragment was obtained from pOZ5, which contained 520 bp of DNA upstream of yqxM and the first 188 codons of its coding sequence. The fragment encompassing the 5′ region of yqxM was inserted at the EcoRI site of both pSN32 and pJM783, thereby creating the yqhM-lacZ plasmids pRSZ08 and pRSZ09, respectively (the first of which can be used to transfer the fusion to the amyE locus) (Fig. 1A).
Construction of strains carrying tasA-, sipW-, and yqxM-lacZ fusions at the tasA region or ectopically inserted at the amyE locus.
Plasmids pRSZ04 and pRSZ09 were used to transfer the tasA- and yqxM-lacZ fusions to the corresponding regions of homology in the chromosome of the wild-type strain PY79. The corresponding strains, AZ404 and AZ409, were the result of Campbell-type recombination at the corresponding region of homology with the chromosome as verified by PCR analysis of total genomic DNA. No transformants were ever obtained in similar crosses involving pRSZ07, a possible explanation for which is given in the Results section. Chromosomal DNA from strain AZ404 (tasA-lacZ) was prepared and used to transfer the fusion by transformation to the sporulation mutants KS450 (gerE36), AZ393 (spo0H::sp), BK395 (spoIIID83), BK556 (spoIVCB23), SC500 (spoIIIGΔ1), SC1159 (spoIIAC1), and SC1163 (spoIIGB55), generating the Cmr strains AZ412, AZ413, AZ414, AZ415, AZ416, AZ417, and AZ418 (Table 1). The antibiotic resistance marker in strain AZ404 was changed from Cmr to Specr by transformation with pCm::Sp (40). Strain AZ410 (Cms Specr) was then transformed with chromosomal DNA from strain AH679 (Cmr [Table 1]), yielding the Pspac-spo0H tasA-lacZ strain AZ411 (Table 1).
Plasmids pRSZ05, pRSZ06, and pRSZ08 (see above) were incubated with ScaI, and the linearized DNA was used to transform the Emr strain AH131 (ΔamyE::erm) to Cmr (Table 1). Transformants were the results of a double crossover (marker replacement) recombination event that introduced a single copy of the tasA-, sipW-, or yqhM-lacZ fusions at the amyE locus. The resulting strains were named AZ405, AZ406, and AZ408, respectively (Table 1). These were also shown to be deficient in α-amylase production, as assayed by growth on 1% starch plates followed by staining of the agar with Gram’s iodine stain (8).
Isolation of spore coat proteins and N-terminal sequence analysis.
Spores were harvested by centrifugation of DSM cultures 24 and 48 h after the onset of sporulation. The spore suspension was washed, and the spores were purified on a step gradient of Renocal-76 (Squibb Diagnostics), as described elsewhere (14, 16). Coat proteins were extracted from about 2 U of optical density at 580 nm (OD580) of purified spores by boiling the suspension for 8 min in the presence of 125 mM Tris–4% SDS–10% (vol/vol) 2-mercaptoethanol–1 mM DTT–0.05% bromophenol blue–10% glycerol at pH 6.8 (14, 16). Alternatively, an alkali treatment was employed. The spores were incubated in the presence of 0.1 M sodium hydroxide at 0°C for 30 min. After centrifugation, the supernatants with the solubilized coat proteins were subjected to electrophoretic fractionation in 10 or 12.5% polyacrylamide gels containing SDS (SDS-polyacrylamide gel electrophoresis [PAGE]). For the N-terminal sequence analysis, the electrophoretically resolved proteins were electrotransferred to polyvinylidene difluoride (PVDF) membranes and subjected to several cycles of the Edman degradation reaction.
Purification of TasA and generation of a polyclonal antiserum.
The portion on the tasA gene encoding the mature form of the TasA protein was amplified by PCR with Pfu polymerase and oligonucleotides OM252 (5′-GGAGGACCATGGGGGCAGCATTTAACGACA-3′) and OM253 (5′-GCTGTTAAATATTTTTATCCTCGCTATGC-3′). The 726-bp-long PCR product was cut with NcoI and inserted between the NcoI and EcoRV sites in plasmid pET-30a(+) (Novagen). This created pMS2, consisting of an in-frame fusion (verified by sequence analysis) between a six-His–S.Tag-encoding sequence and tasA. Plasmid pMS2 was introduced into the E. coli strain BL21(DE3)pLysS, generating a strain in which the TasA fusion protein could be produced under the control of the T7lac promoter. The fusion protein, found predominantly in the soluble fraction, was purified over a HisTrap column (Pharmacia Biotech), as described by the manufacturer. The purified fusion protein was transferred to a PVDF membrane and subjected to on-membrane cleavage with Lys-C protease. Several of the resulting peptides were purified by high-pressure liquid chromatography, and their masses were determined by MALDI mass spectrometry at the Emory Microchemical Facility and compared to the TasA-deduced peptide map. Two peptides were sequenced by Edman degradation, yielding sequences DFQFENNGSLAIK and ANGGSNTSPEDFLSQFEVTLLTVGK (Fig. 1B). The results of this analysis suggested that the complete TasA protein had been produced in E. coli. Gel-purified TasA antigen was then sent to Eurogentec (Seraing, Belgium) for the immunization of rabbits.
Immunoblotting.
Samples (15 ml) of DSM cultures of various strains were collected at 1-h intervals throughout growth and sporulation. The cells were harvested by centrifugation and resuspended in 1 ml of lysis buffer, and the suspension was passed twice through a French pressure cell at 19,000 lb/in2 (39). Proteins in 30-ml samples of culture supernatants were precipitated with an equal volume of ice-cold 10% trichloroacetic acid, incubated on ice for 20 min. The precipitated proteins were recovered by centrifugation, and the pellet was washed with ice-cold ethanol and resuspended in 1/100 of the original buffer in SDS loading dye (125 mM Tris, 4% SDS, 10% [vol/vol] 2-mercaptoethanol, 1 mM DTT, 0.05% bromophenol blue, 10% glycerol at pH 6.8). Samples of 30 μg of total protein were electrophoretically resolved on SDS–12.5% polyacrylamide gels (SDS-PAGE), and the resolved proteins were electrotransferred to nitrocellulose membranes. The membranes were incubated overnight in phosphate-buffered saline–Tween (8 mM sodium phosphate [pH 7.5], 150 mM NaCl, 0.1% Tween 20), containing 5% low-fat milk. The membranes were then incubated for 1 h at room temperature with an anti-TasA antiserum (at a dilution of 1:1,000), in phosphate-buffered saline–Tween 20 containing 0.5% milk. Incubation with a secondary antibody conjugated to horseradish peroxidase (Amersham Biotech) was for 20 min at a 1:3,000 dilution. The immunoblots were washed and developed with enhanced chemiluminescence reagents, as described by the manufacturer (Amersham Biotech).
Electron microscopy.
Spores were purified as described above from DSM cultures of strain MB24 (wild type) and of its congenic derivative AH1700 (tasA), approximately 24 h after the onset of sporulation. The spores were fixed and embedded essentially as described before (17). Electron microscopy analysis and photography were conducted on a Philips EM301 microscope, operated at 80 keV.
Germination efficiency and spore resistance properties.
Purified spores were heat activated and diluted in 10 mM Tris-HCl (pH 8.0) buffer containing 1 mM glucose, 1 mM fructose, and 10 mM KCl (GFK). After 15 min at 37°C, germination was induced by addition of 10 mM l-alanine or 10 mM l-asparagine. Germination was monitored at 5-min intervals, by monitoring the decrease in the OD580 of the suspension, until a constant reading was reached (27). Spore viability was assessed as CFU per milliliter on 2× YT plates before and after heat and lysozyme treatment (30).
β-Galactosidase assays.
β-Galactosidase activity was measured during sporulation and exponential growth. Samples (1 ml) were taken at appropriate times, and the specific activity of β-galactosidase was determined with the substrate o-nitrophenol-β-d-galactoside as described previously (27, 36).
RESULTS
TasA is the predominant polypeptide extracted from gerE mutant spores.
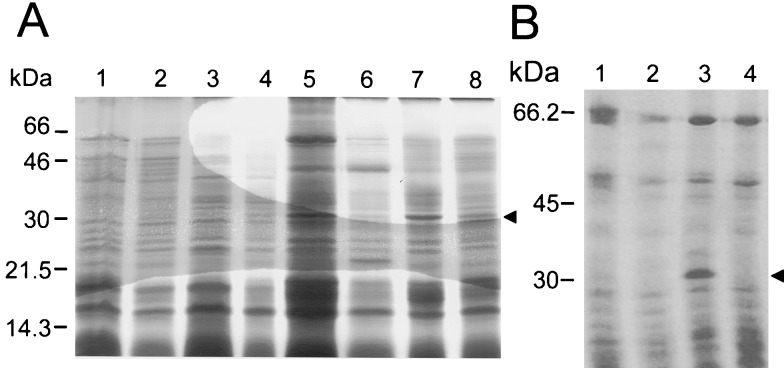
We have been determining the N-terminal sequences of several of the proteins that are extractable from the coats of wild-type spores of B. subtilis, with the goal of identifying the complete collection of polypeptides that compose it. However, there are two main problems with this approach. One results from the complexity of the protein sample that can be solubilized from wild-type coats (15). Another problem results because of extensive cross-linking; about 30% of the total coat protein in wild-type spores is refractory to the extraction procedures normally employed and therefore not amenable to electrophoretic analysis (17, 49). The gerE mutant of B. subtilis is pleiotropically impaired in the transcription of several of the cot genes (36, 44, 48) and forms spores with altered coat layers which lack several of the proteins that can be extracted from wild-type coats (25). Most bands found in SDS-DTT extracts from gerE mutant spores are also found in extracts made from wild-type spores (see, for example, reference 17), and thus it is unlikely that these bands represent proteins that artifactually bind to gerE mutant coats. We reasoned that by using gerE mutant spores certain proteins would be more readily extractable. We purified gerE mutant spores 24 h after the onset of sporulation and analyzed their coat composition by extracting the coat proteins, either by NaOH or by SDS-DTT treatments (see Materials and Methods). The solubilized proteins were then fractionated on 12.5% polyacrylamide gels containing SDS (Fig. 2). The results in Fig. 2B indicate that a polypeptide of about 30 kDa was the predominant species extracted from gerE mutant spores by treatment with NaOH (Fig. 2B, lane 3). The 30-kDa polypeptide could also be extracted from gerE mutant spores by treatment with SDS-DTT (Fig. 2A, lane 5). A prominent band of about 66 kDa was also found in extracts from gerE mutant spores. This could correspond to CotA, which has the same apparent mobility (11) and is encoded by the GerE-repressed gene (38). The 30-kDa species was not noticeably extracted from similarly aged wild-type spores (Fig. 2, lanes 1). Increased extractability of the polypeptide directly correlated with the presence of the gerE mutation. In contrast, extractability was not significantly enhanced by the cotE allele (Fig. 2A, lanes 4 and 8), known to prevent outer coat assembly (52). To determine the identity of the 30-kDa polypeptide, the electrophoretically resolved coat proteins were transferred to PVDF membranes and the N-terminal amino acid sequence of the polypeptide was determined by the Edman degradation reaction. The sequence obtained (AFNDIKSKDATFA [Fig. 1B]) revealed that the 30-kDa component corresponded to the deduced product of a gene in the comGG-sinR intergenic region (22), after removal of a 27-residue amino-terminal extension resembling a signal peptide (see below). In a different study (42), the same gene had been named tasA, for translocation-dependent antimicrobial spore component. This designation, which emphasizes the gene’s multiple functions, was herein retained. We note that TasA shows sequence similarity, albeit weak, to the C-terminal half (residues 353 to 569) of the ActA protein of Listeria monocytogenes (Fig. 1B), involved in the reorganization of the host’s cell actin cytoskeleton (10, 21, 32). In addition, we note that TasA has regions of sequence identity with the Saccharomyces cerevisiae USO1 protein, a cytoskeletal component required for intracellular protein transport (29). Finally, TasA also has regions similar to parts of the Streptococcus pyogenes FcrA protein (accession no. S35760), and to a human toll-like receptor (accession no. U88879). These regions of the TasA protein, as well as the percentages of identity to the ActA, USO1, FcrA, and toll-like sequences are indicated in Fig. 1C.
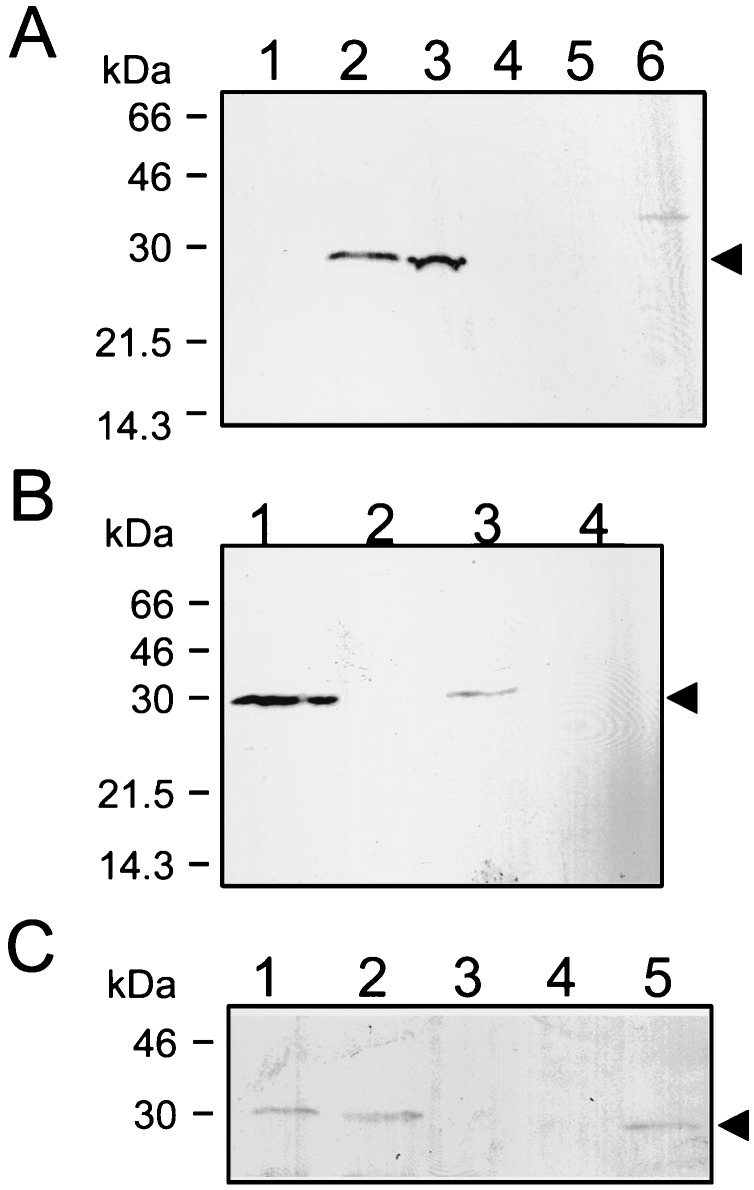
FIG. 2.
Extraction of TasA from gerE mutant spores. Spores of a wild-type strain and various mutant strains were purified, and the coat proteins were extracted by treatment with a buffer containing SDS-DTT (A) or with alkali (B). The extracted proteins were resolved in 12.5% polyacrylamide gels containing SDS, and the gels were stained with Coomassie brilliant blue. The spores used for the extraction of the coat proteins were as follows: (A) lane 1, wild type; lane 2, tasA mutant; lane 3, cotE mutant; lane 4, cotE tasA mutant; lane 5, gerE mutant; lane 6, gerE tasA mutant; lane 7, cotE gerE mutant; lane 8, gerE cotE tasA mutant; (B) lane 1, wild type; 2, tasA mutant; lane 3, gerE mutant; lane 4, gerE tasA mutant. The arrowheads indicate the position of the 30-kDa TasA polypeptide which is readily extracted from gerE mutant spores. Also indicated are the positions of the molecular mass markers (in kilodaltons).
Properties of a tasA insertional mutant.
To investigate a possible role of the tasA locus in coat assembly, we generated a tasA insertional mutant. Competent cells of a wild-type strain, MB24, were transformed with plasmid pRSZ01, which carries a 516-bp EcoRI-HindIII DNA fragment internal to the tasA coding region (Fig. 1). A Cmr integrant, the result of a single Campbell-like recombination event between the plasmid and the region of homology in the host genome, was named AZ402 and chosen for further study. Plasmid pRSZ01 was also introduced in a series of congenic strains bearing mutations in either the gerE or the cotE locus or both. We then used treatments for the extraction of coat proteins (see Materials and Methods), to release proteins from purified spores of different strains prepared at 24 h of sporulation. The extracted proteins were then resolved by SDS-PAGE. As shown in Fig. 2, disruption of tasA strongly reduced the amount of the 30-kDa component extracted from gerE or gerE cotE mutant spores by the SDS-DTT treatment (Fig. 2A, lanes 6 and 8). Moreover, the tasA mutation completely prevented the alkali extraction of the prominent 30-kDa component from spores carrying the gerE mutation (Fig. 2B, lanes 3 and 4). The material seen in the 30-kDa region of the gel after SDS-DTT extraction of gerE tasA or gerE cotE tasA mutant spores is likely to correspond to a tasA-independent component with the same mobility as TasA. The component of about 30 kDa that can be released from wild-type coats by treatment with SDS-DTT (Fig. 2A, lane 1) is not changed by the tasA mutation. Thus, its solubilization is tasA independent. This contaminating component is evidently not solubilized by the alkali treatment from either the wild type or a gerE tasA double mutant (Fig. 2B, lanes 1 and 4). Inactivation of tasA did not noticeably alter the pattern of proteins released from gerE or cotE mutant spores (Fig. 2A, lanes 4 and 6, and 2B, lane 4). We conclude that extraction of the 30-kDa component from the coats of gerE mutant spores requires a functional tasA locus. TasA either is a minor component or is refractory to solubilization in wild-type spores prepared at 24 h of sporulation (see also below).
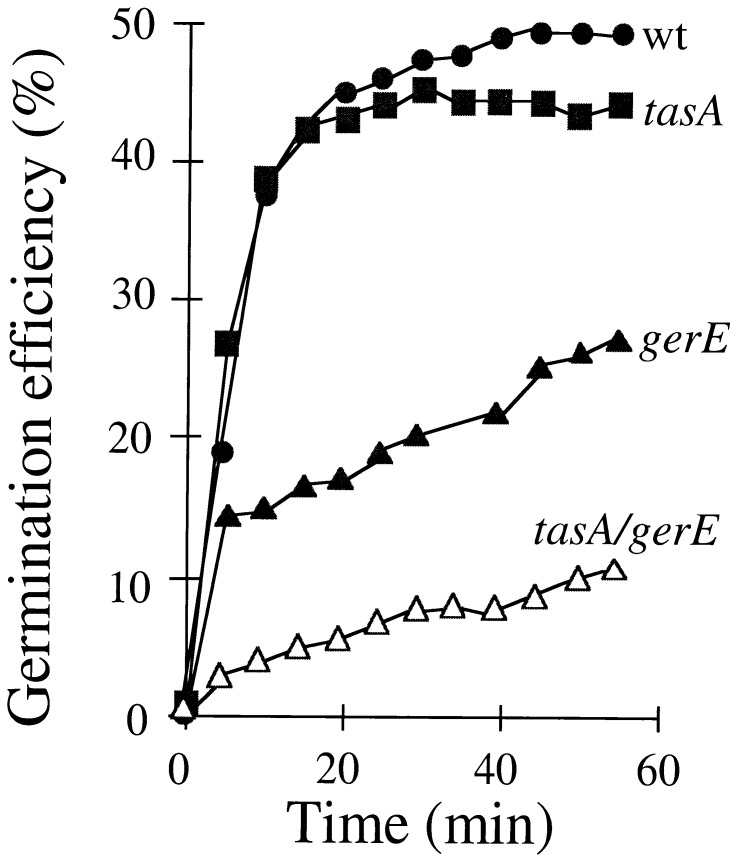
Spores from the congenic strains PY17 (wild type), AZ402 (tasA), KS450 (gerE36), and AZ403 (gerE36 tasA) were prepared and used to measure lysozyme resistance and the efficiency of germination. These are spore properties known to be determined in part by the coat layers (25, 52). We also measured heat resistance of the spores, a property that is determined by the status of the cortex layer (see, for example, reference 33). The tasA mutation did not alter the heat resistance properties of wild-type or gerE mutant spores (data not shown). In addition, the tasA mutation did not confer lysozyme sensitivity upon wild-type spores, nor did it accentuate the lysozyme-sensitive phenotype of gerE mutant spores (data not shown). We also found that disruption of tasA did not impair germination in response to l-alanine (Fig. 3) or to l-asparagine (data not shown) in GFK. However, the efficiency of spore germination (the percentage of spores that germinate) in response to l-alanine in GFK appeared reduced in spores doubly mutant for tasA and gerE, compared to gerE single-mutant spores (Fig. 3). Thus, gerE and tasA act synergistically in germination. Because disruption of tasA did not affect heat resistance, the mutation is unlikely to cause any gross alteration of the cortex layer. Rather, TasA either is involved in the response to germinants or is needed for accurate coat formation, which, in turn, is required for efficient germination.
FIG. 3.
Germination efficiency of wild-type and mutant spores. Spores produced by a wild type (wt), as well as by tasA, gerE, and gerE tasA mutants, were purified, and the kinetics of germination were examined. Germination was induced by l-alanine in GFK, as previously described (27), and monitored by the decrease in OD600 of the spore suspension. The efficiency of germination is defined as the ratio between the optical density of the culture at a given time after exposure to the germinant mixture and the original optical density of the suspension.
Disruption of tasA renders the spores asymmetric.
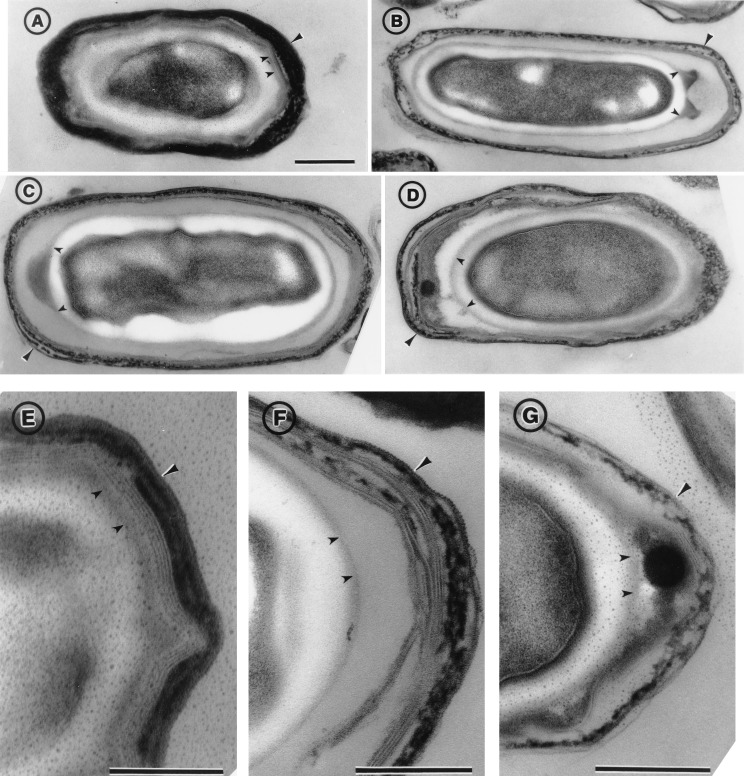
Because our results implicated tasA in both the assembly and the function of the coat structure (see above), we wanted to examine the impact of the same tasA insertional mutation on the normal morphological pattern of the coat layers. Cultures of a wild-type strain and a congenic tasA derivative (see Materials and Methods) were incubated until 48 h after the onset of sporulation in DSM. At that time, the spores were collected, washed, purified by equilibrium sedimentation in step gradients of Renocal-76, and processed for electron microscopy analysis (17). In wild-type spores, the coat layers consist of a diffuse or amorphous undercoat, no more than 10 to 20 nm wide, which separates the electron-translucent cortex from the inner coat (3, 15). The undercoat adheres tightly to the interior surface of the inner coat (see smaller arrowheads in Fig. 4A and E). The inner coat consists of three to five lightly staining laminae and is typically 20 to 40 nm wide (Fig. 4A and E). Closely apposed to the inner coat is a wider (40 to 90 nm) outer coat, which typically consists of three to five electron-dense striations (Fig. 4, larger arrowheads in panels A and E) (15).
FIG. 4.
Electron microscopy of wild-type and tasA mutant spores. Spores were collected from DSM cultures of a wild type (MB24 [A and E]) and the tasA mutant (AH1700 [B to D, F, and G]), 48 h after the initiation of sporulation. The spores were purified by centrifugation through Renocal gradients and processed for electron microscopy analysis as described in Materials and Methods. The large arrowheads point to the outer coat structure. The smaller arrowheads indicate the boundary between the cortex and the undercoat, which is probably defined by the forespore outer membrane. Darkly staining material (between the two small arrowheads) accumulates in the undercoat region of the tasA mutant but not in wild-type spores. Scale bar, 0.2 μm.
Examination of the tasA mutant revealed several distinctive features, the most striking of which is that the mutant spores tend to accumulate electron-dense material (between the two small arrowheads in Fig. 4B, C, D, and G) on one pole of the spore. This asymmetry was found for all the spore longitudinal sections showing abnormal accumulation of electron-dense material (about 20% of the total number of longitudinal sections examined). This material accumulates in the region between the cortex and the inner coat and in some cases is in close proximity to the inner coat (Fig. 4D and G). In other cases, the electron-dense material accumulates on the outside of a thin layer (indicated by the two smaller arrowheads) that seems to define the outer boundary of the cortex region and may correspond to the forespore outer membrane (see longitudinal sections in Fig. 4B and C; see also reference 37). In this case, there is virtually no accumulation of electron-dense undercoat material apposed to the interior surface of the inner coat (see in particular Fig. 4B and C), as occurs in the wild type (Fig. 4A and E). Instead, the space between the presumptive forespore outer membrane and the inner coat is greatly expanded, whether accumulation of electron-dense material occurs at the cortex-undercoat boundary (for example, Fig. 4F). Note that the relative volume of the cortex region remains essentially unchanged (compare the cortex in the wild-type spore in Fig. 4A with the cortex in the mutant in Fig. 4B, C, or D, or the wild-type spore in Fig. 4E with the mutant in Fig. 4G). The expanded undercoat region in the tasA mutant is filled with a lightly staining material that presumably fails to accumulate against the inner coat layer (for example, Fig. 4B, C, and F). However, this region may have essentially the same polypeptide composition as that in wild-type spores (Fig. 2). We also note that both coat layers are reduced and misassembled in the mutant (Fig. 4B, D, and F) and that the outer coat has a diffuse appearance (longer arrowhead in Fig. 4G). Sporadically, we also noted a pattern of indentations on the outermost layer of the outer coat (longer arrowhead in Fig. 4F) that is reminiscent of a similar feature of a mutant thought to have a specific deficiency in outer coat formation (16).
The observation of a morphological phenotype mainly associated with the coat layers is in agreement with our inference that TasA is required for proper coat assembly and spore germination (see above). Moreover, the asymmetric phenotype of the tasA mutant suggests that, at an early stage of sporulation, the TasA protein localizes to the polar septum and that after engulfment TasA may influence coat assembly from an asymmetric localization. In addition, TasA appears to be required for correct formation of the outer coat.
Assembly of TasA into wild-type and mutant spores.
To investigate whether TasA could associate with the spore coats, we raised a polyclonal antibody against the TasA protein produced in E. coli as an N-terminal His-tag fusion (see Materials and Methods). We used the anti-TasA antiserum in immunoblots of material extracted from wild-type and various mutant spores. Spores of 24- and 48-h cultures were purified by centrifugation through gradients of Renocal and subjected to the SDS-DTT procedure described in Materials and Methods, known to solubilize most of the coat proteins. The TasA protein was detected in material extracted from gerE mutant spores prepared 24 h after the onset of sporulation, suggesting its association with the coat components (Fig. 5A, lane 3). The TasA antigen was not detected in material extracted from spores doubly mutant for gerE and tasA (Fig. 5A, lane 6). TasA antigen was also detectable in material purified from cotE mutant spores of the same age (Fig. 5A, lane 2). In both cases, the component detected was TasA dependent, as it was eliminated by the insertional inactivation of tasA (lanes 4, 5, and 6). In confirmation of earlier results (Fig. 2), we failed to detect TasA among the sample of proteins released from wild-type spores prepared 24 h after the onset of sporulation (Fig. 5A, lane 1). Surprisingly, immunoblots of material prepared from wild-type spores purified 48 h after the commencement of sporulation revealed that TasA was present among the polypeptides solubilized by the SDS-DTT treatment (Fig. 5B, lane 1). In contrast, the level of TasA extracted from gerE mutant spores prepared at 48 h of sporulation was reduced (Fig. 5B, lane 3), compared to wild-type spores of the same age or to gerE mutant spores of 24 h (Fig. 5B, lane 1, and 5A, lane 3). To determine whether TasA was loosely associated with old (48-h) wild-type spores, the coat proteins were extracted before or after washing of the spores with a 1 M KCl solution (45). The results from these experiments are shown in Fig. 5C. Even though in this particular experiment the amount of antigen released was lower than that in the experiment in Fig. 5B, treatment with KCl did not change the amount of TasA antigen released from the coats of wild-type spores of 48 h (lanes 1 and 2, respectively). In comparison, the same treatment completely washed TasA off gerE mutant spores (Fig. 5C, lanes 4 and 5). In all cases, proteins were extracted from an equal number of spores (see Materials and Methods).
FIG. 5.
Detection of TasA in wild-type and mutant spores. Immunoblot analysis of material extracted from purified spores produced by a wild-type strain and various mutant strains. (A) Spores of a wild-type strain (MB24, lane 1) or of the following mutant strains were purified 24 h after the onset of sporulation: cotE mutant (AH1823, lane 2), gerE mutant (AH94, lane 3), tasA mutant (AH1822, lane 4), cotE tasA mutant (AH1824, lane 5), and gerE tasA mutant (AH1827, lane 6). (B) Spores used were purified 48 h after the onset of sporulation from cultures of a wild-type strain (MB24, lane 1), a tasA mutant (AH1822, lane 2), a gerE mutant (AH94, lane 3), or a gerE tasA double mutant (AH1827, lane 4). (C) Coat proteins were extracted from purified spores before or after washing of the spore suspension with 1 M KCl (45). Lane 1, wild type, washed; lane 2, wild type, before washing; lane 3, tasA mutant, not washed; lane 4, gerE mutant, washed; lane 5, gerE mutant, before washing. Proteins were resolved on SDS-containing 12.5% polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were then probed with an anti-TasA antiserum. The arrowheads indicate the positions of TasA antigen. The positions of molecular mass markers (in kilodaltons) are also indicated.
One possibility is that TasA is present (but refractory to extraction) in wild-type spores prepared at 24 h of sporulation. The gerE mutation greatly increases TasA extractability, presumably because TasA has a diminished capacity to interact with or to be retained by the altered coat layers. As the result, as gerE mutant spores age, TasA is lost from the coat layers (Fig. 5B, lane 3). In contrast, over time TasA becomes extractable from wild-type spores by the SDS-DTT treatment, although it resists solubilization by a high-salt solution (Fig. 5C, lane 1).
ςH-dependent tasA transcription is enhanced in the predivisional cell at the onset of sporulation.
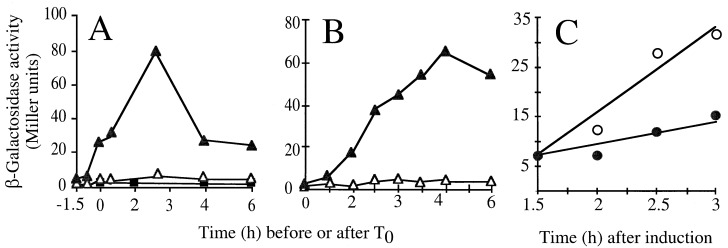
The results in the section above indicated that TasA is required for the normal assembly of the spore coat. The expression of all genes directly involved in coat assembly occurs in the mother cell compartment of the sporangium (2, 7, 14, 16, 20, 27, 36, 44, 48, 50). However, no cot mutations were known to result in asymmetry of the coat layers. Moreover, the morphology of spores mutant for tasA suggested that the locus acted in part at septation, which is earlier than all other cot genes (Fig. 4). We therefore decided to examine its transcriptional regulation. Plasmid pRSZ04 (Fig. 1A) carries an NaeI-EcoRI segment encompassing the upstream region and the first 51 codons of the tasA coding region joined to the E. coli lacZ gene. The plasmid was transferred to a wild-type recipient by Campbell-type recombination, involving cloned DNA and homologous sequences in the genome. This cross created strain AZ404, which carries a tasA-lacZ fusion integrated at the tasA locus. Production of β-galactosidase was then monitored throughout growth and sporulation of AZ404 in DSM. Transcription of tasA-lacZ, which was detected during growth, was enhanced early in sporulation, reaching maximum levels some 2 h after the onset of the process (Fig. 6A). Expression of tasA was also detected during stationary phase in 2× YT medium (Fig. 6B). Consistent with the results of Fig. 6A, indicating an early expression of tasA during sporulation, tasA-directed β-galactosidase production was not diminished in strains mutated in the spoIIAC, spoIIGB, spoIIID, spoIIIG, spoIVCB, or gerE gene (data not shown). These loci encode transcriptional factors that control intermediate to late gene expression during sporulation and include the regulators responsible for the transcription of all cot genes so far characterized (15). In contrast, tasA-directed β-galactosidase production was strongly impaired in DSM (Fig. 6A) or 2× YT medium (data not shown), by a mutation in the spo0H gene, encoding ςH. To confirm that tasA expression was under ςH control, we isolated a strain containing a tasA-lacZ fusion and the spo0H gene under the control of the IPTG-inducible Pspac promoter (47). To do this, strain AZ410 (tasA-lacZ spc) was transformed with chromosomal DNA isolated from strain AH679 (Pspac-spo0H), with selection for Cmr Specr cells. The results in Fig. 6C show that addition of IPTG to a culture of the resulting strain (AZ411) in 2× YT medium triggers synthesis of β-galactosidase, confirming that tasA is under ςH control. However, the requirement for ςH for expression of tasA may be indirect. ςH is required at the onset of sporulation for the activation of the transcription factor Spo0A, which then acts at several promoters utilized by both ςA- and ςH-containing RNA polymerase (26, 43). In any case, other genes regulated by either ςA or ςH during the early stages of sporulation have the same temporal pattern of expression of tasA and are transcribed prior to septation (43). Their products are thought to partition between both compartments issued from the asymmetric division of sporulation (43). The finding that tasA is expressed during vegetative growth and early sporulation in a ςH-dependent manner suggests that TasA is present in both cell chambers of the sporangium.
FIG. 6.
tasA is controlled by ςH. The figure illustrates the time course of β-galactosidase production by various strains bearing a tasA-lacZ transcriptional fusion integrated at the tasA locus. Different growth conditions were used. (A) Strains were induced to sporulate in DSM, and T0 defines the onset of sporulation. (B) YT medium (2×) was used (T0 corresponds to an OD600 of about 0.3, whereas maximum enzyme activity was reached about 3 to 4 h later). Enzyme production was measured in strain AZ404 (tasA-lacZ) (dark triangles) and its congenic sigH mutant (AZ393, dark squares). (C) A Pspac-spo0H strain carrying the tasA-lacZ fusion was grown in 2× YT medium to a low OD600 value (about 0.2), at which point the culture was divided in half. IPTG was added to one flask (open circles) but not to the other (closed circles). Samples were collected at the indicated times, and the specific activity of β-galactosidase was determined with the substrate o-nitrophenol-β-d-galactopyranoside (ONPG). Background levels of β-galactosidase synthesis were estimated for the wild-type strain PY79 (open triangles in panels A and B).
In support of our suggestion (based on the ultrastructural analysis of tasA spores; see above) that tasA acts early in sporulation, the results in this section indicate that tasA represents a new, very early class of coat genes, transcribed prior to creation of the mother cell compartment.
tasA is the third gene in an operon.
The tasA gene maps at 218° on the B. subtilis chromosome in the comGG-sinR intergenic region and is preceded by two open reading frames, yqxM and sipW (22, 42, 46) (Fig. 1A). A region of dyad symmetry, possibly a factor-independent transcription terminator, separates tasA from sinR (Fig. 1A). Because the tasA-lacZ fusion in plasmid pRSZ04 was Campbell integrated at the tasA locus, the possibility existed that in strain AZ404 (see above) expression of the transcriptional fusion was driven by a promoter located upstream of the locus. We checked for the presence of a promoter in the tasA region by fusion of 400 bp of DNA derived from the region just upstream of tasA to lacZ in an amyE integrational vector. The resulting plasmid, pRSZ05, was used to replace the ΔamyE::erm marker in strain AH131 (Table 1) with the tasA-lacZ fusion linked to a Cmr marker. The resulting strain was named AZ405 (Table 1). In contrast to the corresponding Campbell-integrated tasA-lacZ fusion (strain AZ404; see above), no β-galactosidase activity was detected in AZ405 (data not shown). Moreover, TasA was no longer seen among the proteins extracted from spores of a gerE mutant strain containing the Campbell-integrated tasA-lacZ fusion (strain AZ412 [data not shown]). These results indicate that the region 5′ of tasA cloned in pRSZ05 (Fig. 1) does not carry sequences able to promote transcription initiation and strongly suggest that tasA is part of a larger transcriptional unit.
To define the boundaries of the tasA-containing operon, we tried to inactivate the yqxM and sipW genes, which precede tasA on the chromosome (Fig. 1A). However, several attempts to inactivate these genes, by either a single or a double recombination event, were unsuccessful. These observations suggested that YqxM and/or SipW (but not TasA) could be essential for growth and viability of B. subtilis, at least in the background of our reference strain, MB24 (Table 1). We note that other investigators were able to insertionally inactivate the sipW gene (42, 46). As an alternative strategy, we created fusions of yqxM and sipW to lacZ in amyE integrational plasmids (pRSZ08 and pRSZ06), as well as equivalent fusions for Campbell-type integration at the corresponding chromosomal loci (pRSZ09 and pRSZ07). The results of these studies are summarized in Fig. 1A. No promoter activity was detected upstream of sipW in strain AZ406 (sipW-lacZ at the amyE locus), but several attempts to integrate the sipW-lacZ fusion-bearing plasmid pRSZ07 at the sipW locus were never successful. In keeping with the notion that sipW may be essential, this result suggested that the promoter for sipW was upstream of the region cloned in the plasmid (Fig. 1A). In contrast to the situation with sipW, pRSZ09 (carrying a yqxM-lacZ fusion) could be easily integrated by a Campbell-type mechanism at the yqxM locus, generating strain AZ409. We infer that the yqxM region cloned in plasmid pRSZ09 carries all the 5′ sequences required for efficient expression of the gene. In support of this interpretation, the pattern of β-galactosidase production in strain AZ409 was similar to that of AZ408 (data not shown), which carries an equivalent yqxM-lacZ fusion ectopically integrated at the amyE locus (Fig. 1A). Moreover, the pattern of β-galactosidase formation and the genetic dependency on spo0H in strain AZ409 or AZ408 (data not shown) were indistinguishable from those observed for strain AZ404 (tasA-lacZ at the tasA locus [see above]). We conclude that yqxM is the first, sipW is the middle, and tasA is the third and last gene of an operon controlled by ςH.
TasA is a secreted protein.
The primary structure of TasA, deduced from the B. subtilis genome sequence (22), includes 27 amino-terminal residues that were not found when the 30-kDa coat-associated polypeptide was subjected to Edman degradation (Fig. 1B) (see above). The N-terminal extension of TasA resembles signal peptides from B. subtilis (28): it has three positively charged residues (KKK) near its N terminus, followed by a central hydrophobic region flanked by a G residue, five positions prior to the deduced cleavage site, between two alanines (vertical arrow in Fig. 1B). The coat-associated TasA thus results from the removal of a signal peptide-like sequence from the preprotein. The presence of a signal peptide and the cotranscription of tasA with sipW, which encodes a type I SPase (46), strongly suggested that maturation of pre-TasA could involve SipW (9, 46). We used the anti-TasA antibody to investigate the pattern of TasA accumulation throughout growth and sporulation of a gerE mutant (Fig. 7A). We found that the antiserum identified a cross-reactive component of about 30 kDa, which in agreement with the transcriptional analysis (Fig. 6A) was first detected at 2 h of sporulation and persisted in whole-cell extracts until at least 6 h of sporulation (Fig. 7A, lanes 1 to 5). TasA was not detected in whole-cell extracts prepared from a ςH mutant or from a gerE tasA double mutant at 2 h of sporulation (Fig. 7A, lanes 7 and 8). We infer that the antiserum raised against purified TasA recognizes the TasA protein in whole-cell extracts, as it does in purified coat material. In addition, we found that a component with the same electrophoretic mobility as the species detected in whole-cell extracts and purified coat material could be found in culture supernatants from the tasA+ strain MB24 (but not from a tasA mutant), at 2 h of sporulation (Fig. 7B). TasA antigen was more difficult to detect in the supernatants of gerE mutant cultures, raising the possibility that gerE controls the level of TasA secretion (data not shown). Together with the result of the N-terminal sequence analysis, these observations suggest that the 30-kDa antigen is the mature form of TasA (see also below). Thus, TasA is found extracellularly concomitantly with its accumulation in sporulating cells.
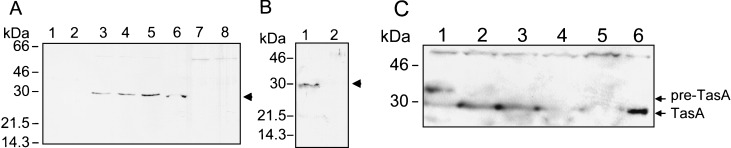
FIG. 7.
TasA production and pre-TasA processing during growth and sporulation. (A) Samples of DSM cultures of strain AH94 (gerE36) were harvested during the logarithmic phase of growth (lane 1), at T0 (lane 2), T2 (lane 3), T4 (lane 4), and T6 (lane 5), and whole-cell extracts were prepared. The cultures were allowed to sporulate, and coat material was then isolated from spores purified 24 h after T0 (lane 6). DSM cultures of a ςH mutant (strain AH17; lane 7) or of a gerE tasA insertional mutant (strain AH1827, lane 8) were also collected at T2. (B) Samples of supernatants of a culture of a tasA+ strain (AH94, lane 1) or of a tasA mutant (AH1700, lane 2) were collected at T2, and the proteins were concentrated as described in Materials and Methods. (C) The inducer IPTG was added to a DSM culture of the Pspac-tasA strain AH1802 at T0. Samples were collected at 30 (lane 1), 60 (lane 2), 90 (lane 3), and 120 min (lane 4) after the addition of IPTG, and whole-cell lysates were prepared. Samples of DSM cultures of a tasA mutant (lane 5) and of a tasA+ strain (lane 6) at T2 were also analyzed. Proteins in all samples were resolved on 12.5% polyacrylamide gels containing SDS and transferred to nitrocellulose membranes. The membranes were probed with an anti-TasA antiserum. The arrows indicate the positions of the TasA antigen. Molecular mass markers (in kilodaltons) are also indicated.
SipW is involved in pre-TasA processing at the onset of sporulation.
Because the tasA operon was transcribed at low levels during the vegetative phase of growth (Fig. 6), we reasoned that induction of tasA expression during growth could lead to accumulation of the precursor protein. Plasmid pMS3, carrying a Pspac-tasA fusion, was integrated into the chromosome by a single crossover (Fig. 1A). In the resulting strain AH1802, tasA is separated from the first cistrons in the operon, whose expression remains under the control of the native promoter. In addition, as the result of integration of pMS3, expression of a full-length copy of tasA was placed under the control of the IPTG-inducible Pspac promoter. However, after addition of IPTG to mid-log-phase cultures of AH1802, we failed to detect TasA in whole-cell extracts, a fact that could indicate instability of TasA or its efficient secretion to the growth medium (data not shown). When IPTG was added at the onset of sporulation, a species with an apparent molecular mass of about 35 kDa formed about 70% of the TasA antigen detected 30 min after induction (Fig. 7C, lane 1). No other species is detected by the antibody in the 30- to 46-kDa range (for example, Fig. 7A). Therefore, the 35-kDa band is likely to be a TasA precursor. One hour after induction, as expression of the truncated yqxM sipW operon started to peak (Fig. 6A), the 35-kDa precursor was completely converted to the processed 30-kDa form (Fig. 7C, lanes 2 to 4). We suggest that, at least at the onset of sporulation, processing of pre-TasA requires sipW expression and that SipW uses pre-TasA as a substrate. We further suggest that the imbalance created between TasA production and processing at the initiation of sporulation was possible because, at the onset of sporulation, SipW may be the only SPase capable of processing pre-TasA. These results do not exclude, however, the possibility that during the exponential phase of growth other type I SPases can contribute to its maturation.
The processed form of TasA that is detected in whole-cell extracts prepared from sporulating cells may be translocated across the cell membrane while remaining associated with the cell envelope or, as suggested by the ultrastructural analysis (see above), may be secreted to a membrane-delimited cellular compartment, such as that formed by the asymmetric sporulation septum.
DISCUSSION
The 30-kDa product of the tasA gene (22, 42) is one of the proteins that can be more efficiently extracted from gerE mutant spores by treatment with NaOH. The TasA polypeptide can also be extracted from wild-type spores prepared 48 h after the onset of sporulation by an SDS-DTT treatment. Because both processes are known to solubilize spore coat polypeptides (11, 14, 16, 27, 36), we investigated the role of TasA in spore coat assembly. Our results suggest that TasA is involved in the assembly of the coat layers. Unlike all other genes involved in coat assembly, the expression of which is confined to the mother cell compartment of the sporangium (15), tasA is transcribed in the predivisional cell under ςH control. No other regulators of the sporulation process known to act in either the forespore or the mother cell were found to influence tasA expression. Second, TasA is made as a secretory preprotein, and it is the processed form that is found in mature spores, implicating protein secretion in coat assembly.
tasA is the third cistron of an operon that also encodes the type I SPase SipW (46). From the time of septation onward, the mature form of TasA is found in culture supernatants, as well as in whole-cell extracts, and pre-TasA processing appears to be entirely dependent upon sipW expression. Mature TasA is detected in whole-cell extracts prepared during sporulation of a tasA+ strain, presumably because the protein is secreted to and accumulates in the cellular compartment defined by the sporulation septum. In support of this model, the ultrastructural analysis of the tasA mutant revealed that the spores formed were asymmetric, accumulating misassembled material at one spore pole. Moreover, this observation suggests that, after engulfment, TasA retains an asymmetric position. Presumably, this location corresponds to the spore pole proximal to the initial site of septation. Translocation of TasA to the septum depends upon SipW function, but SipW is unlikely to localize exclusively in the septal membranes, since TasA is also detected in the supernatant of sporulating cultures. SipW belongs to the endoplasmic reticulum (ER)-type subfamily of type I SPases, making B. subtilis the only known eubacterium that contains SPases of both the prokaryotic and ER types (9, 46). We propose that SipW has properties that allow it to efficiently translocate proteins into the septal compartment formed during sporulation, which in that respect may be an analog of the eukaryotic ER organelle. However, SipW contributes to protein secretion during the vegetative growth phase (reference 46 and this work), and the tasA sipW operon is also induced postexponentially in 2× YT medium, although the physiological significance of this, if any, is uncertain (this work).
One important inference from the present work is that intrasporangial protein secretion is used as a mechanism to direct components to specific cellular locations during spore differentiation and that this process is somehow required for proper coat assembly and normal germination properties. TasA has sequence similarities with proteins known to promote interactions between cytoskeletal elements (USO1 and ActA), to promote adhesion between cells (FcrA), or to act as pattern recognition receptors (the extracytoplasmic domain of the human TLR) (10, 21, 29, 32). In spores of a tasA mutant, the region comprising the undercoat, defined by the forespore outer membrane (see also Fig. 1 in reference 37) and the inner coat, is considerably expanded. In wild-type spores, this region appears compacted and completely apposed to the inner coat structure. Accumulation of misassembled coat-like material in the tasA mutant seems to occur on the outside of the forespore outer membrane. In light of its similarity to proteins implicated in adhesion, cytoskeletal organization, or pattern recognition, it is tempting to speculate that TasA acts from the septal compartment to promote adhesion between the cortex and undercoat structures (see also below). Albeit weak, the sequence similarity of TasA to the C-terminal half of the ActA protein of L. monocytogenes, a bacterial surface protein involved in the reorganization of the cytoskeletal elements of the host cell (10, 21, 32), is particularly interesting. Like TasA, ActA is also made as a secretory preprotein, but it associates with the cell via a C-terminal membrane anchor which is absent in TasA. When the actA gene is expressed in eukaryotic cells, the ActA polypeptide is targeted to the mitochondrial membrane via its C-terminal membrane anchor. The ActA protein is then able to induce actin accumulation (as well as other cytoskeletal elements) around mitochondria, mimicking its function in the bacterial cells (32). Thus, in a parallel with our model for TasA function, ActA can act as a nucleator of the polymerization of actin filaments around the subcellular structure on which it localizes (32). In tasA mutant spores, the misassembled material that accumulates at the cortex-undercoat boundary (sometimes in close contact with the inner coat structure) appears to be asymmetrically located. Whether TasA acts from the septal compartment (or cortical region), this observation suggests that TasA may function from only one pole of the spore, to nucleate the organization of the undercoat, which then can proceed all around the spore without further TasA participation (Fig. 8). Thus, the material that accumulates in the mutant could correspond to a scar resulting from the absence of TasA function during the initial stages of the process. In contrast, the rest of the undercoat region, although lacking the compaction seen in the wild type, is not involved in the initial nucleation stages and therefore does not display the same phenotype.
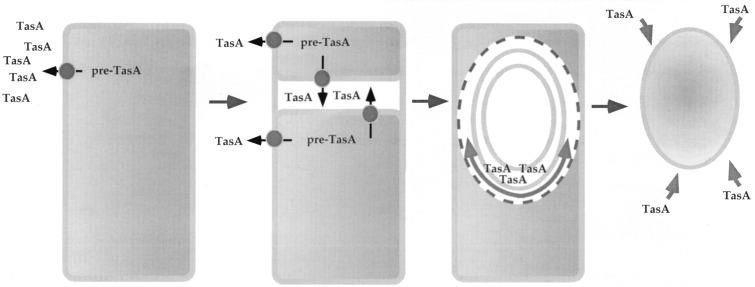
FIG. 8.
Model for TasA function in coat assembly. The figure illustrates the various morphological stages of development in relation to TasA production. TasA production is initiated under ςH command early in sporulation, before the asymmetric division that marks morphological stage II. In this early period, TasA is secreted to the culture medium, where its mature form accumulates. Processing is thought to rely on the SipW type I signal peptidase. After formation of the sporulation septum, the mature form of TasA accumulates in the space defined by the two septal membranes. After engulfment, TasA may localize preferentially on the septum-proximal side of the spore. From this position, TasA may nucleate the organization of the undercoat material, which then proceeds around the entire forespore. In the absence of TasA, a scar is left at the nucleation position. Finally, after release of the spore upon lysis of the mother cell, TasA in the culture medium can associate with the spore coats. Alternatively (not represented), in old spores, the spore-associated TasA becomes more extractable.
Our results indicate that TasA has a role in coat assembly. Our results do not imply an association of TasA with the coat layers, nor do they exclude an association of TasA with other spore structures. Because the localization of TasA within the sporangium is presently unknown, we cannot decide with certainty whether TasA is a true coat component. At least at an early stage in spore maturation, most of the mature form of TasA in sporulating cells is presumably confined to the septal lumen. Thus, TasA could in principle become associated with the cortex, which is formed in this region, or even with more internal spore structures (42). Its involvement in coat assembly does not requires TasA to be a coat component. The effects herein described on the assembly of the coat layers can be explained by assuming that TasA somehow (perhaps via other as yet unknown proteins) acts from the cortical region. In the light of the available evidence, this explanation is favored. Thus, TasA may be not a coat component but rather a protein required for the normal assembly of the coat structure.
Whether or not TasA is a true coat protein, it may be cross-linked in a GerE-dependent manner. The gerE locus has already been implicated in the cross-linking of the CotJC protein, a component of the inner coat layers (14, 39), and gerE is required for the production of a coat-associated transglutaminase (19, 20). The weak association of TasA with gerE mutant spores is not likely to be caused by a profound defect in the cortex structure (gerE mutant spores are heat resistant and appear to have a normal cortex) but may indicate absence of TasA cross-linking. In contrast, our failure to extract TasA from wild-type spores within 24 h suggests that in these spores TasA may be in a form that resists extraction. In support of this idea, we note that most of the CotJC antigen can be released from wild-type spores (39) (see also above). tasA mutant spores also show alterations of the outer coat structure. Because TasA can be detected in wild-type spores from 48-h cultures, one possibility is that TasA in the culture medium can associate with the coat layers (Fig. 8). In contrast to certain extracellular proteins that can loosely associate with the spores (45), TasA may adhere tightly to wild-type coats. Alternatively, spore-associated TasA could become more extractable as wild-type spores age (Fig. 8). In either case, coat maturation appears to continue long after lysis of the mother cell and concomitant release of the spore.
ACKNOWLEDGMENTS
We are grateful to Richard Losick for the gift of several stains and for critical reading of the manuscript. We are also grateful to Carla Caruso (Università della Tuscia, Viterbo, Italy) and J. Pohl (Emory Microchemical Facility) for the N-terminal sequence analysis, to Adam Driks for sharing unpublished information, and to Paulo Tavares for helpful discussions.
This work was supported by grants CNR (Progetti Finalizzati “Biotecnologie”) to E. Ricca, Praxis XXI/PCNA/C/BIA/129/96 to R. Zilhão, Convénio de Cooperação Científica JNICT/CNR (Proc. 423/CNR/SCIAE) to R. Zilhão and E. Ricca, and by GM54395 from the National Institutes of Health to C. P. Moran, Jr. M.S. and A.O.H. were the recipients of pre- and postdoctoral fellowships from Junta Nacional de Investigação Científica e Tecnológica (J.N.I.C.T.), respectively.
REFERENCES
- 1.Aronson A I, Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson A I, Song H-Y, Bourne N. Gene structure and precursor processing of a novel Bacillus subtilisspore coat protein. Mol Microbiol. 1988;3:437–444. doi: 10.1111/j.1365-2958.1989.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 3.Aronson A I, Ekanayake L, Fitz-James P C. Protein filaments may initiate the assembly of the Bacillus subtilisspore coat. Biochimie. 1992;74:661–667. doi: 10.1016/0300-9084(92)90138-5. [DOI] [PubMed] [Google Scholar]
- 4.Beall B, Driks A, Losick R, Moran C P., Jr Cloning and characterization of a gene required for assembly of the Bacillus subtilisspore coat. J Bacteriol. 1993;175:1705–1716. doi: 10.1128/jb.175.6.1705-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne N, Fitz-James P C, Aronson A I. Structural and germination defects of Bacillus subtilisspores with altered contents of a spore coat protein. J Bacteriol. 1991;173:6618–6625. doi: 10.1128/jb.173.20.6618-6625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutting S, Zheng L, Losick R. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J Bacteriol. 1991;173:2915–2919. doi: 10.1128/jb.173.9.2915-2919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biology methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons, Ltd.; 1990. pp. 27–74. [Google Scholar]
- 9.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wächter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenesrequired for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan W, Zheng L, Sandman K, Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987;196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 12.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 13.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 13a.Henriques, A. O. Unpublished data.
- 14.Henriques A O, Beall B W, Roland K, Moran C P., Jr Characterization of cotJ, a ςE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilisspores. J Bacteriol. 1995;177:3394–3406. doi: 10.1128/jb.177.12.3394-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriques, A. O., and C. P. Moran, Jr. Structure and assembly of the bacterial endospore coat. Submitted for publication. [DOI] [PubMed]
- 16.Henriques A O, Beall B W, Roland K, Moran C P., Jr CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–1897. doi: 10.1128/jb.179.6.1887-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques A O, Melsen L R, Moran C P., Jr Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J Bacteriol. 1998;180:2285–2291. doi: 10.1128/jb.180.9.2285-2291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karow M L, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilisand their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi K, Suzuki S, Hashiguchi K, Yamanaka S. 9th International Conference on Bacilli, Lausanne, Switzerland, 15 to 19 July 1997. 1997. Transglutaminase expressed in sporulating cells of Bacillus subtilis, abstr. 39. [Google Scholar]
- 20.Kobayashi K, Kumazawa Y, Miwa K, Yamanaka S. ɛ-(γ-Glutamyl)lysine cross-links of spore coat proteins and transglutaminase activity in Bacillus subtilis. FEMS Microbiol Lett. 1996;144:157–160. [Google Scholar]
- 21.Kochs C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. Listeria monocytogenes-induced actin assembly requires the actAgene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 22.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 23.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis“-12, -24” promoter of the levanase operon and evidence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 25.Moir A. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J Bacteriol. 1981;146:1106–1116. doi: 10.1128/jb.146.3.1106-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 27.Naclerio G, Baccigalupi L, Zilhão R, de Felice M, Ricca E. Bacillus subtilis spore coat assembly requires cotHgene expression. J Bacteriol. 1996;178:4375–4380. doi: 10.1128/jb.178.15.4375-4380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagarajan V. Protein secretion. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 713–726. [Google Scholar]
- 29.Nakajima H, Hirata A, Ogawa Y, Yonehara T, Yoda K, Yamasaki M. A cytoskeleton-related gene, USO1, is required for intracellular protein transport in Saccharomyces cerevisiae. J Cell Biol. 1991;113:245–260. doi: 10.1083/jcb.113.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biology methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons, Ltd.; 1990. pp. 391–450. [Google Scholar]
- 31.Perego M, Hoch J A. Isolation and sequence of the spo0E gene: its role in initiation of sporulation in Bacillus subtilis. Mol Microbiol. 1987;1:125–132. doi: 10.1111/j.1365-2958.1987.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 32.Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham D L, Illades-Aguiar B, Setlow P. The Bacillus subtilis dacBgene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J Bacteriol. 1995;177:4721–4729. doi: 10.1128/jb.177.16.4721-4729.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sacco M, Ricca E, Losick R, Cutting S. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995;177:372–377. doi: 10.1128/jb.177.2.372-377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakae Y, Yasuda Y, Tochikubo K. Immunoelectron microscopic localization of one of the spore germination proteins, GerAB, in Bacillus subtilisspores. J Bacteriol. 1995;177:6294–6296. doi: 10.1128/jb.177.21.6294-6296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of a promoter for a spore coat protein gene in Bacillus subtilisand studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 39.Seyler R, Henriques A O, Ozin A, Moran C P., Jr Interactions and assembly of cotJ-encoded products, constituents of the inner layers of the Bacillus subtilisspore coat. Mol Microbiol. 1997;25:955–966. doi: 10.1111/j.1365-2958.1997.mmi532.x. [DOI] [PubMed] [Google Scholar]
- 40.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 41.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stöver A G, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilisspore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 44.Takamatsu H, Chikahiro Y, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. A spore coat protein, CotS, of Bacillus subtilisis synthesized under the regulation of ςK and GerE during development and is located in the inner coat layer of spores. J Bacteriol. 1998;180:2968–2974. doi: 10.1128/jb.180.11.2968-2974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tesone C, Torriani A. Protease associated with spores of Bacillus cereus. J Bacteriol. 1975;124:593–594. doi: 10.1128/jb.124.1.593-594.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W J, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor machinery of Bacillus subtilis: identification of a eubacterial homolog of archael and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson A I. Regulation of the transcription of a cluster of Bacillus subtilisspore coat genes. J Mol Biol. 1994;240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Fitz-James P C, Aronson A I. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J Bacteriol. 1993;175:3757–3766. doi: 10.1128/jb.175.12.3757-3766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]
- 51.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. Sporulation regulatory protein GerE from Bacillus subtilisbinds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol. 1992;226:1037–1050. doi: 10.1016/0022-2836(92)91051-p. [DOI] [PubMed] [Google Scholar]
- 52.Zheng L, Donovan W, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilisendospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]