Abstract
A search for homologs of the Bacillus subtilis PhoP response regulator in the group A streptococcus (GAS) genome revealed three good candidates. Inactivation of one of these, recently identified as csrR (J. C. Levin and M. R. Wessels, Mol. Microbiol. 30:209–219, 1998), caused the strain to produce mucoid colonies and to increase transcription of hasA, the first gene in the operon for capsule synthesis. We report here that a nonpolar insertion in this gene also increased transcription of ska (encoding streptokinase), sagA (streptolysin S), and speMF (mitogenic factor) but did not affect transcription of slo (streptolysin O), mga (multiple gene regulator of GAS), emm (M protein), scpA (complement C5a peptidase), or speB or speC (pyrogenic exotoxins B and C). The amounts of streptokinase, streptolysin S, and capsule paralleled the levels of transcription of their genes in all cases. Because CsrR represses genes unrelated to those for capsule synthesis, and because CsrA-CsrB is a global regulatory system in Escherichia coli whose mechanism is unrelated to that of these genes in GAS, the locus has been renamed covR, for “control of virulence genes” in GAS. Transcription of the covR operon was also increased in the nonpolar insertion mutant, indicating that CovR represses its own synthesis as well. All phenotypes of the covR nonpolar insertion mutant were complemented by the covR gene on a plasmid. CovR acts on operons expressed both in exponential and in stationary phase, demonstrating that the CovR-CovS pathway is separate from growth phase-dependent regulation in GAS. Therefore, CovR is the first multiple-gene repressor of virulence factors described for this important human pathogen.
The group A streptococcus (GAS) (Streptococcus pyogenes) is an important bacterial pathogen, restricted to growth in humans, that produces various diseases. These range in severity from mild suppurative infections of the throat and skin, such as pharyngitis and pyoderma, to more serious and life-threatening invasive diseases, such as fasciitis, myositis, and streptococcal toxic shock syndrome (4, 5). In many cases, it appears that single GAS strains can cause all or most of these syndromes.
Most strains of GAS can produce many factors that are known or suspected to be involved in the survival, spread, and persistence of the organism within the human host. M protein, long considered the major GAS virulence factor (24), plays several roles in pathogenesis. It increases resistance of the GAS cell to complement-mediated killing by polymorphonuclear leukocytes (39), it is important for attachment to keratinocytes in skin infections (37), and it promotes bacterium-bacterium interaction following attachment to tonsillar epithelial cells during throat infections (7). In addition to M protein, GAS is enveloped in a hyaluronate capsule that also retards phagocytosis by immune cells and has been shown in several animal models to be critically important for virulence of several GAS strains (1, 20, 32, 50). The cell-associated protein streptolysin S (SLS) is a cytolysin produced by GAS that can lyse eukaryotic cells, including polymorphonuclear leukocytes and platelets, and GAS mutants defective in SLS production were shown to be less virulent in a mouse model (3).
Although other GAS proteins have not been tested directly in animal models, some of the following have activities strongly suggesting important roles in virulence. The secreted protein streptolysin O (SLO), a cytolysin similar to SLS, also functions to modulate proinflammatory responses of keratinocytes to which GAS cells have attached (44). Streptokinase is an enzyme that catalyzes the conversion of plasminogen to plasmin, which is a serine protease with a broad activity spectrum (2). The streptococcal cysteine protease encoded by the speB gene has been shown to be a potent protease capable of cleaving both human and streptococcal proteins (9, 33, 35). Thus, streptokinase and the cysteine protease may help GAS to spread through tissue and to invade deeper layers by promoting dissolution of the connective matrix. Streptococcal toxins include SpeA, SpeC, and SpeMF, which are superantigens capable of stimulating proliferation of T lymphocytes and release of cytokines (21, 45, 53). It has been hypothesized that toxin expression is important in the development of streptococcal toxic shock syndrome.
Since GAS encounters a series of different niches in the human host during the various stages of the many diseases it causes, expression of a changing repertoire of virulence determinants is of great advantage to the bacterium. Thus, it seems likely that GAS strains are able to respond to environmental changes by regulating the expression of their virulence factors. At this time, the only regulatory protein known to control expression of more than one virulence factor in response to different environments is the multiple gene regulator of GAS, Mga. Mga activates transcription of several genes, including those encoding M protein (emm) (47), C5a peptidase (scpA) (10), and itself (mga) (36). Mga is also required for expression of a secreted inhibitor of complement (encoded by sic) (23) and, when their genes are present in a GAS strain, of M-like proteins and of serum opacity factor (30, 41). Since Mga is positively autoregulated, a negative regulator of its expression awaits discovery.
Many potential virulence genes of GAS are expressed only in specific phases of the growth cycle. In addition to genes regulated by Mga, which are expressed in the exponential but not in the stationary phase of growth, some Mga-independent genes are differentially expressed at different growth stages (3, 11, 28). Although no mechanism or pathway has yet been identified for these regulatory processes, there is at least one global regulatory system affecting virulence gene expression in GAS that is independent of Mga.
Pathogenic bacteria commonly use global networks of regulation to control expression of different virulence factors in response to changing environmental cues throughout the infection process. Often this regulation is accomplished through a signal transduction system comprised of two components. The first is a surface-located sensor kinase that recognizes a specific environmental signal, such as osmotic pressure or pH, and transduces this into a phosphorylation event. This phosphate group is then passed to the second component, a response regulator protein that binds DNA at a specific site or sites to alter the frequency of initiation of transcription from the operons it regulates. The genes encoding the sensor and regulator components are often adjacent on the chromosome and expressed as a polycistronic message.
To identify genes involved in environmental regulation, possibly including regulation of the Mga regulon, the GAS M1 genome sequence (42) was searched for potential two-component regulatory gene pairs. Here we report that immediately upstream of isp (a gene encoding an immunogenic secreted protein), located directly upstream of mga in the GAS chromosome (29), are two open reading frames (ORFs) exhibiting significant homology to the PhoP-PhoS two-component signal transduction pathway in Bacillus subtilis (19). A further search of the entire GAS M1 genome sequence revealed two other sets of unlinked ORFs encoding homologs of the B. subtilis phoPS genes. During the course of this work, Levin and Wessels described one of these loci as being involved in repression of the capsule synthesis operon, and they called it csr, for “capsule synthesis regulator” (25). Because we demonstrate here that CsrR regulates genes in addition to those for capsule synthesis, and because CsrA-CsrB is a global regulatory system in Escherichia coli (43) and other bacteria whose mechanism differs from that of these GAS genes, the CsrR repressor of GAS was renamed CovR, for “control of virulence genes” in GAS. Mutations were isolated in two of these three apparent sensor-regulator pairs, and their roles in control of transcription of the Mga regulon and other virulence factors of GAS are described below.
MATERIALS AND METHODS
Bacterial strains and media.
All GAS strains used are derivatives of the streptomycin-resistant serotype M6 strain JRS4 (48). Cultures of GAS were grown at 37°C without agitation in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY). E. coli DH5α was used as the host for plasmid constructions and was grown at 37°C with shaking in Luria broth. Concentrations of drugs used were as follows: chloramphenicol, 2 μg/ml for GAS and 25 μg/ml for E. coli; kanamycin, 200 μg/ml for GAS and 50 μg/ml for E. coli; spectinomycin, 50 μg/ml for GAS and 100 μg/ml for E. coli.
DNA manipulations.
Plasmid DNA was isolated from E. coli with the Wizard Maxiprep and Miniprep systems (Promega). Genomic DNA was isolated from GAS by the method of Chassy (8). DNA fragments were isolated from agarose gels by the QIAquick gel extraction kit (Qiagen). T4 DNA polymerase (NEB) was used to blunt restriction nuclease-digested DNA ends. Restriction enzyme digestions and T4 DNA ligations were performed according to the manufacturer’s recommended protocol (NEB). PCR was performed with Taq DNA polymerase (Sigma) unless otherwise stated. PCR-generated fragments were purified with the QIAquick PCR purification system (Qiagen).
Construction of plasmids for insertional inactivation of irr, covR, and sycF.
Plasmids for inactivation of the genes encoding three different potential response regulators were constructed as follows. Fragments internal to the coding regions of the genes irr, covR, and sycF were amplified by PCR with high-fidelity Pfu polymerase (Stratagene) with the following primers: irr, GGTGACGTTTTGCTAAATAA and AAAGCGAATAACTATGATCC; covR, TAGTGAGAGAAATCTCATCG and TATGAAGTCATTGTTGAGGT; sycF, TGACAAAAGAAGGTTATGAC and GCTAGATGATGCAATAGTTC. The PCR-derived fragments were blunt-end ligated into SmaI-digested pUC-Spec (20), a suicide vector unable to replicate in GAS, to produce pJRS550 (irr), pJRS551 (covR [Fig. 1A]), and pJRS552 (sycF). These plasmids were introduced into JRS4 by electroporation, and transformants were grown on THY agar with selection for spectinomycin. Spectinomycin-resistant transformants, resulting from insertion of plasmids into the GAS chromosome and inactivation of the target gene, were named JRS550 (irr1) and JRS551 (covR1 [Fig. 1A]). JRS551 was verified by PCR analysis across the plasmid-chromosome junctions with the following primer pairs: covR1 5′ junction, cgcggatccAGAGGATAAGGGTTGGTATA (COVR-L [arrowhead 1, Fig. 1A]) and gcgtgatcaAGAAGCCAATGAAATCTATA (SPEC-R [arrowhead 2, Fig. 1A]); covR1 3′ junction, gcgtgatcaCAATTAGAATGAATATTTCCC (SPEC-L [arrowhead 3, Fig. 1A]) and ccggaattcATGACTTATTTCTCACGAAT (COVR-R [arrowhead 4, Fig. 1A]). JRS550 was verified by PCR analysis across the irr1 plasmid-chromosome junctions with the following primer pairs: irr1 5′ junction, gcgggatccAATCTAGAAAGGGAAGTTAT (IRR-L) and SPEC-R; irr1 3′ junction, SPEC-L and gcggaattcTGATGACCAAAAAGGTTTTT (IRR-R) (lowercase letters represent nonhomologous sequences). All primer sequences are given left to right in 5′-to-3′ order.
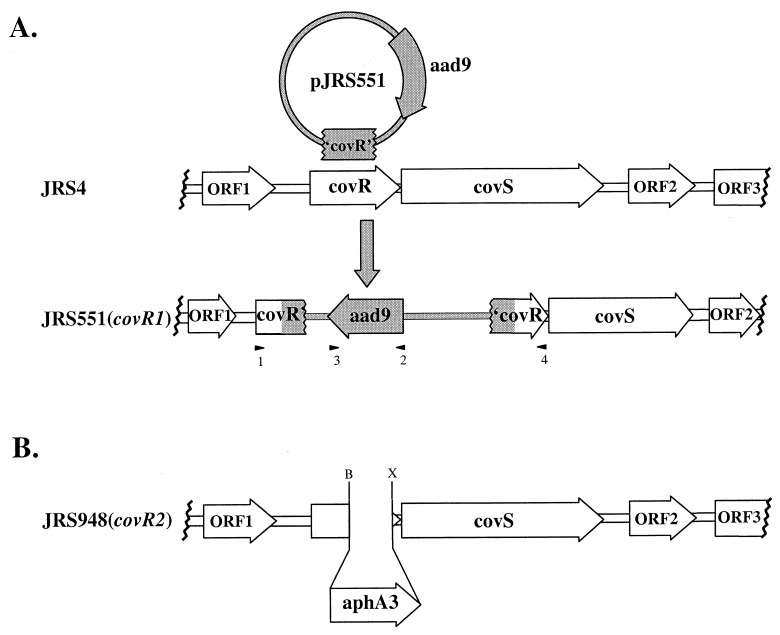
FIG. 1.
Inactivation of covR in the serotype M6 GAS strain JRS4. (A) Region surrounding covR in the chromosome of GAS. Coding regions (open arrows) are as predicted from the M1 genome sequence database (42). Plasmid pJRS551 (shaded circle) contains a fragment internal to covR (′covR′) and confers spectinomycin resistance (aad9). JRS551 was produced by homologous recombination, which inserted pJRS551 into the wild-type covR gene in the JRS4 chromosome. Arrowheads represent primers used to confirm plasmid insertion into the chromosome: 1, COVR-L; 2, SPEC-R; 3, SPEC-L; 4, COVR-R (see Materials and Methods). (B) Region surrounding covR2 in JRS948, with all adjacent ORFs labeled as in panel A. The covR2 allele consists of a nonpolar aphA3 gene replacing 347 bp from the 3′ end of the covR gene, located between sites for BsgI (B) and XcmI (X).
Construction of the nonpolar covR2 strain JRS948.
A fragment containing covR, most of covS, and the upstream ORF ORF1 (Fig. 1A) was amplified by PCR from the JRS4 chromosome by using the primers cggggtaccATTAGGAGAAGATGATGTTAGC and cggggtaccCGCGAATCATGTCTAACATA and introducing unique KpnI sites at both ends (indicated by lowercase boldface type). The resulting 2.5-kb PCR fragment was digested with KpnI and cloned into KpnI-digested pBluescriptII KS− (Stratagene) to produce pJRS943. pJRS943 was digested with BsgI-XcmI to remove a 347-bp fragment from the 3′ end of covR, which contains the predicted DNA binding domain. The deletion was replaced with an 850-bp SmaI fragment of pUC18K (31) containing a nonpolar aphA3 cassette to produce the covR2 allele in pJRS947.
Plasmid pJRS9160 is a derivative of the gene replacement vector pJRS233 (40), which contains the temperature-sensitive pWV01 origin (38). A 3.0-kb KpnI fragment harboring the covR2 allele from pJRS947 was cloned into KpnI-digested pJRS9160 to produce plasmid pJRS948. This plasmid was introduced into JRS4 by electroporation, and erythromycin-resistant transformants were selected at 30°C, a temperature permitting replication of the plasmid. The covR2 allele was exchanged for wild-type covR sequences following growth at the nonpermissive temperature (37°C), with selection for resistance to kanamycin (covR2 marker) to produce JRS948 (Fig. 1B). The presence of the covR2 allele in the chromosome of JRS948 was confirmed by PCR analysis across covR with primers COVR-L and COVR-R (see above and arrowheads 1 and 4, Fig. 1A). As expected, the mutant produced a PCR fragment 491 bp larger than the wild type.
Construction of the covR2-complementing plasmid pJRS951.
A PCR fragment containing all of covR and 287 bp upstream of its start of translation was amplified from the JRS4 chromosome by using primers ccggaattcCAAGGGTTGTTTGATGAATA and ccggaattcATGACTTATTTCTCA, which introduce unique EcoRI sites at both ends (indicated by lowercase boldface type). The resulting 997-bp fragment was digested with EcoRI and ligated into EcoRI-digested pLZ12 (13), a chloramphenicol-resistant vector able to replicate in GAS, to produce pJRS951.
RNA hybridization analysis.
Total RNA was harvested from GAS strains with the FastPrep system (Bio 101), followed by treatment with DNaseI for 1 h at room temperature. PCR analysis was used to detect possible DNA contamination. RNA hybridization experiments were performed as previously described (28), except that RNA was cross-linked to the membrane by baking at 80°C for 2 h. PCR-derived DNA probes were amplified from the JRS4 chromosome with some primers pairs described elsewhere (28): mga, OYR4 and OYL13; emm, OM6-30 and OM6-19; scpA, SCPA-1 and SCPA-10; slo, SLO-L1 and SLO-R1; ska, SKA-L2 and SKA-REV; speB, SPEB-1 and SPEB-2; speC, SPEC-L1 and SPEC-R1; speMF, SPEMF-L2 and SPEMF-R1. Primer pairs synthesized for this study include the following: sagA, ATTTTAGCTACTAGTGTAGCTG and TTTACCTGGCGTATAACTTC; covS, AATGCCTTAAGCTACTCTAA and GTTGTAGATGTCTATATTCG; aphA3, gcgtgatcaGAAAAGAGGAAGGAAATAATA and gcgggatccTAAAAAGCTTGTAGTTAAAG; rpsL, gccgaattcGAATGTAGATGCCTACAATTAACCA and cccaagcttTTTACGACTCATTTCTCTTTATCCC; hasA, ATCTATTTGGAACATCAACTGTAGG and HASA-R (28). The covR primers GATGACTAATATGAATCGTGTC and COVR-R amplified the 204-bp fragment from the 3′ end of the gene. DNA probes were labeled with [α32P]dATP incorporated by the Klenow fragment of DNA polymerase (NEB). Hybridization experiments were repeated at least twice with RNA isolated independently.
Protein activity assays.
Hyaluronic acid production was quantitated as described elsewhere (46). Briefly, GAS cultures were grown to late exponential phase; then, the hyaluronic acid from a 1.0-ml sample was reacted with 1-ethyl-2-[3-(1-ethylnaphtho-[1,2-d]thiazolin-2-ylidene)-2-methylpropenyl]naptho-[1,2-d]thiazolium bromide (Sigma) and the absorbance was measured at 640 nm. Values obtained for each strain were compared to standards of known concentrations of hyaluronic acid (Sigma). Values are reported as grams of hyaluronic acid per CFU.
Streptolysin S activity was demonstrated by the ability of GAS to lyse bovine erythrocytes. GAS cells were grown to early stationary phase, washed, and resuspended in phosphate-buffered saline, pH 7.4. Streptolysin S was released from the GAS surface (34), and serial dilutions of the supernatant were incubated with bovine erythrocytes at a final concentration of ca. 0.35% for 30 min at 37°C. Lysis of erythrocytes, determined by the amount of hemoglobin released into the supernatant, was detected spectrophotometrically by monitoring the absorbance of the reaction at an optical density of 541 nm. Hemolytic units correspond to the reciprocal of the dilution that produces 50% lysis of erythrocytes compared to that produced by the same volume of water (44). Trypan blue completely inhibited the reaction, demonstrating that streptolysin O made no significant contribution to the activity measured.
Streptokinase activity in GAS supernatants was determined by ability of GAS to convert human plasminogen to the active form, plasmin. The amount of para-nitroaniline released from H-d-valyl-leucyl-lysin p-nitroanaline (Sigma) was measured as absorbance at 405 nm, as described previously (49). Units of streptokinase are defined by comparison with purified enzyme (Sigma).
RESULTS
B. subtilis PhoP-PhoS homologs are present in GAS.
In many two-component signal-transducing systems, the gene for the sensor kinase is adjacent to that for the cognate response regulator. In the case of mga, on the other hand, emm, which encodes the M protein, lies immediately downstream and isp, which encodes an immunogenic secreted protein, is upstream. However, immediately upstream of isp are two ORFs encoding proteins with homology to the sensor-transducer system PhoP-PhoS of B. subtilis (19). Based on their chromosomal locations, these genes are termed irr (for “isp-adjacent response regulator”) and ihk (for “isp-adjacent histidine kinase”), respectively.
When these genes were identified, additional homologs of the response regulator protein PhoP were sought by searching the University of Oklahoma streptococcal genome sequencing database (42). Two additional distinct ORFs encoding proteins homologous to PhoP were identified on different sequenced “contigs.” Directly downstream of each of these homologs lies a second ORF whose predicted product appears to be a classical sensor kinase with homology to the B. subtilis PhoS protein. One of these sets of homologs, which we call covRS, corresponds to the recently identified csrRS, a locus in GAS involved in repressing transcription of the hasABC capsule synthesis operon in a serotype M3 GAS strain (25) (Fig. 1). The last of the three PhoP-PhoS homolog pairs has considerable homology with the genes yycF (70%) and yycG (45%), which encode a two-component signal-transducing system in B. subtilis whose function has not yet been elucidated. In this work, these genes are called sycF (for “streptococcal yycF”) and sycG (for “streptococcal yycG”).
Chromosomal inactivation of PhoP homologs in GAS.
To investigate the functions of the three sets of potential two-component system genes in GAS virulence, chromosomal insertions within the respective loci were constructed in the serotype M6 strain JRS4 (48). A PCR-amplified fragment internal to the coding sequence of the PhoP response regulator homolog from each pair was cloned into the suicide vector pUC-Spec (20). The resulting plasmids were introduced into JRS4 by electroporation and integrated into the GAS chromosome via homologous recombination, resulting in inactivation of the targeted gene (see Fig. 1A for JRS551). Mutant alleles were verified by PCR analysis of the chromosome region containing the plasmid insertion.
The mutant GAS strain JRS550, containing an insertionally inactivated irr gene, was constructed by integration of plasmid pJRS550 into the JRS4 chromosome. In a similar fashion, plasmid pJRS551 was used to inactivate the covR gene in JRS4 to produce JRS551 (Fig. 1A). Inactivation of covR in JRS551 resulted in a mucoid colony phenotype on agar plates compared to the normally small, round colonies of the parental strain JRS4. Repeated attempts to mutagenize the third PhoP homolog (sycF) in JRS4 by the method described above were unsuccessful. Recently, yycF in B. subtilis was shown to be essential for growth of that bacterium, since it could not be inactivated (15). Therefore, construction of a mutation in the GAS sycF homolog was not pursued.
covR represses transcription of several virulence operons.
To determine if either covR or irr is involved in the pathogenesis of GAS, the amounts of transcripts for several different genes with proven or potential roles in GAS virulence were compared in mutant and wild-type strains. Mga, the multiple gene regulator of GAS, activates expression of several genes involved in pathogenesis. Transcription of Mga regulon genes responds to environmental signals, including carbon dioxide level and temperature, and may rely on a signal transduction system to react to environmental change. Therefore, three genes activated by Mga were studied: emm (encoding M protein) (47), scpA (encoding the complement C5a peptidase) (52), and mga itself (6). The first gene in the operon required for capsule synthesis, hasA (12, 14, 51), was selected because colonies of JRS551 were much more mucoid than those of the parental strain JRS4, because capsule production has been found in several animal models to be an important virulence factor (1, 20, 32), and because of the recent identification of csrR (covR) as a repressor of this gene in an M3 GAS strain (25). The three other genes chosen for study, slo (encoding SLO) (22), sagA (required for SLS production) (3), and ska (encoding streptokinase) (18), are predicted to have roles in the pathogenesis of GAS and may also be dependent on environmental stimuli for maximal expression.
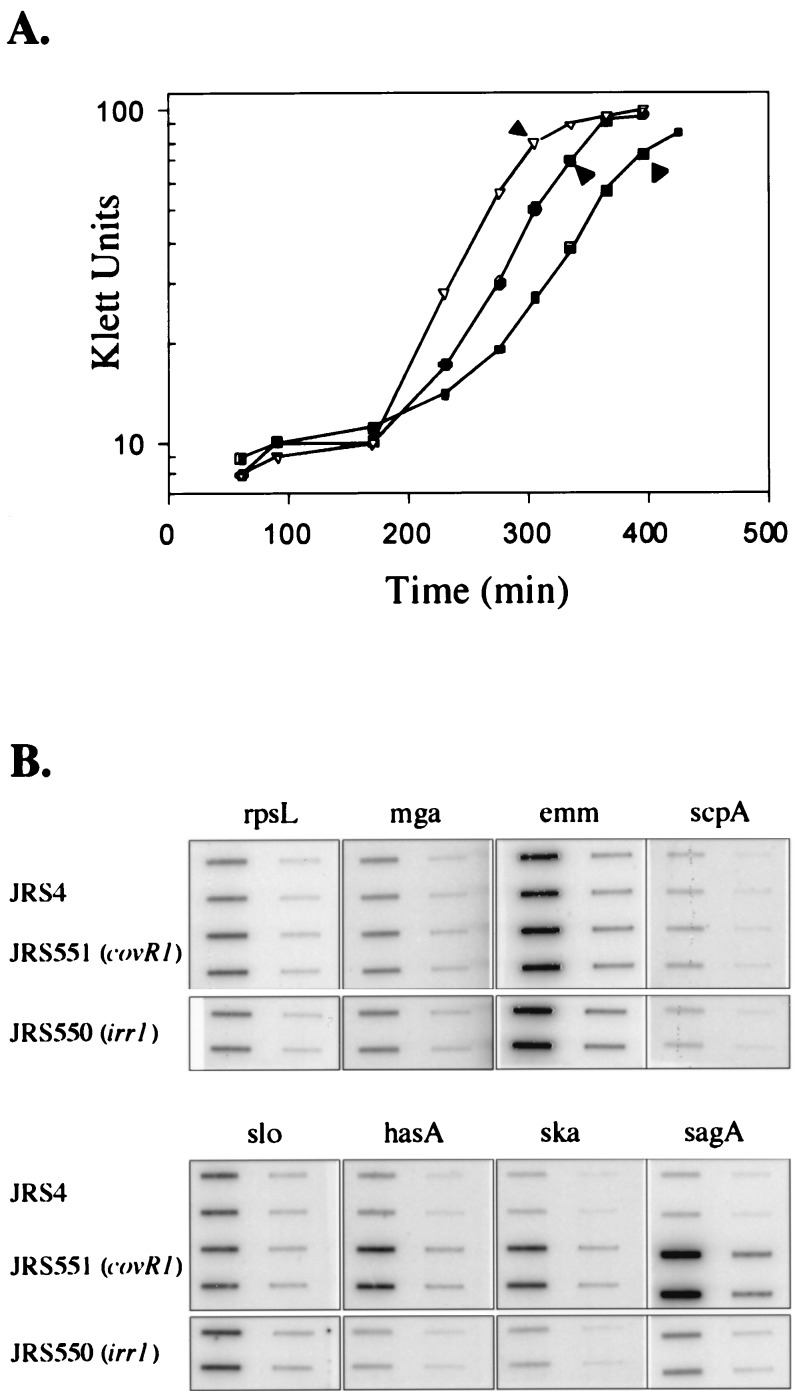
RNA was isolated in late exponential phase (Fig. 2A) from the irr mutant strain JRS550 and its wild-type parent JRS4 and assayed by hybridization to specific PCR-derived probes for each of the seven genes listed above. To assure that equal amounts of mRNA from each strain were loaded on the blot, all were probed with rpsL (which encodes a ribosomal protein). When JRS550 was compared to JRS4, no significant differences in transcription of any gene probed were seen (Fig. 2B). Since there was no detectable effect on the regulation of these virulence genes in the absence of Irr, JRS550 was not investigated further in this study.
FIG. 2.
Transcription of virulence genes in wild-type and mutant GAS strains. (A) Growth curves showing times of isolation of RNA (arrowheads). Open triangles represent JRS4 (wild type), filled circles represent JRS550 (irr1), and filled squares represent JRS551 (covR1). (B) Hybridization of specific DNA probes to RNA harvested at the times indicated in panel A. On each filter, the left column contains 4 μg of RNA and the right column contains 0.4 μg of RNA. Duplicates are arranged vertically. Membranes were reacted with PCR-derived DNA probes internal to the coding region of the GAS virulence genes mga, emm, scpA, slo, hasA, ska, and sagA, as shown (Materials and Methods). A probe from rpsL was used to determine that equal amounts of RNA were loaded. Filters with all RNA samples were hybridized together. Results reported are representative of hybridizations from at least two independent RNA isolations.
When transcript levels in the covR1 mutant JRS551 were analyzed, no differences from the mRNA levels in JRS4 were detected for mga, emm, scpA, or slo (Fig. 2B). However, transcript levels of the three other genes—hasA, ska, and sagA—were all found to be much higher in the mutant strain than in the parent (Fig. 2B). Therefore, covR appears to repress not only the transcription of hasA but also the transcription of ska and sagA.
covR-mediated repression is not growth phase regulated.
In JRS4 and in the M1 genome sequence, the ORFs downstream of covS read in the same direction as covR and covS and may be cotranscribed with them (Fig. 1). It was reported that in an M3 strain of GAS, covR and covS (csrR and csrS) are cotranscribed as a single message (25). In Fig. 1 of that report, the direction of transcription of the ORF immediately downstream of covS was shown incorrectly (26); it actually matches that in the other strains investigated. Although no transcriptional linkage was found between covR and covS and adjacent ORFs in the M3 GAS strain (25), these genes may be cotranscribed in JRS4. Thus, since insertion of the plasmid that creates the covR1 mutation in strain JRS551 would be expected to cause premature termination, it might exert a polar effect on covS and the downstream ORFs. To determine whether covR alone is responsible for repression of hasA, ska, and sagA, a nonpolar mutation in covR was constructed (Fig. 1B). A 347-bp fragment from the 3′ end of covR was deleted and replaced with a nonpolar kanamycin resistance cassette containing the aphA3 gene followed by a ribosomal binding site (31). This produced the covR2 allele in plasmid pJRS948. Allelic exchange was used to replace the wild-type covR allele in the chromosome of strain JRS4 with this covR2 mutation to produce the GAS strain JRS948 (Fig. 1B). As observed for the covR1 strain JRS551, the JRS948 colonies appeared to be highly mucoid on agar plates. Transcription studies (see “CovR represses its own synthesis,” below) confirmed the nonpolar nature of covR2.
Since expression of hasA, sagA, speC, and speMF has been shown to change during the cell cycle (3, 11, 28), we compared the transcript levels of hasA, ska, sagA, speC, and speMF in the nonpolar mutant strain JRS948 with those in its parent, JRS4, at two different times: in late exponential phase and 150 min into stationary phase (Fig. 3A). We included speB in our analysis as a gene whose expression was not expected to be influenced by phase of growth (28) but might be expressed differently in the covR mutant. In agreement with a previous report (11), hasA was transcribed at both times but showed maximal transcript levels during late-exponential growth (compare Fig. 3B and C). Similarly, the amount of ska transcript appeared to be greater at the earlier time point. On the other hand, as previously demonstrated (3, 28), the levels of sagA and speMF expression were higher in stationary phase than in late exponential phase. The levels of transcript for speB and speC remained very low, and regulation by CovR was not evident (data not shown).
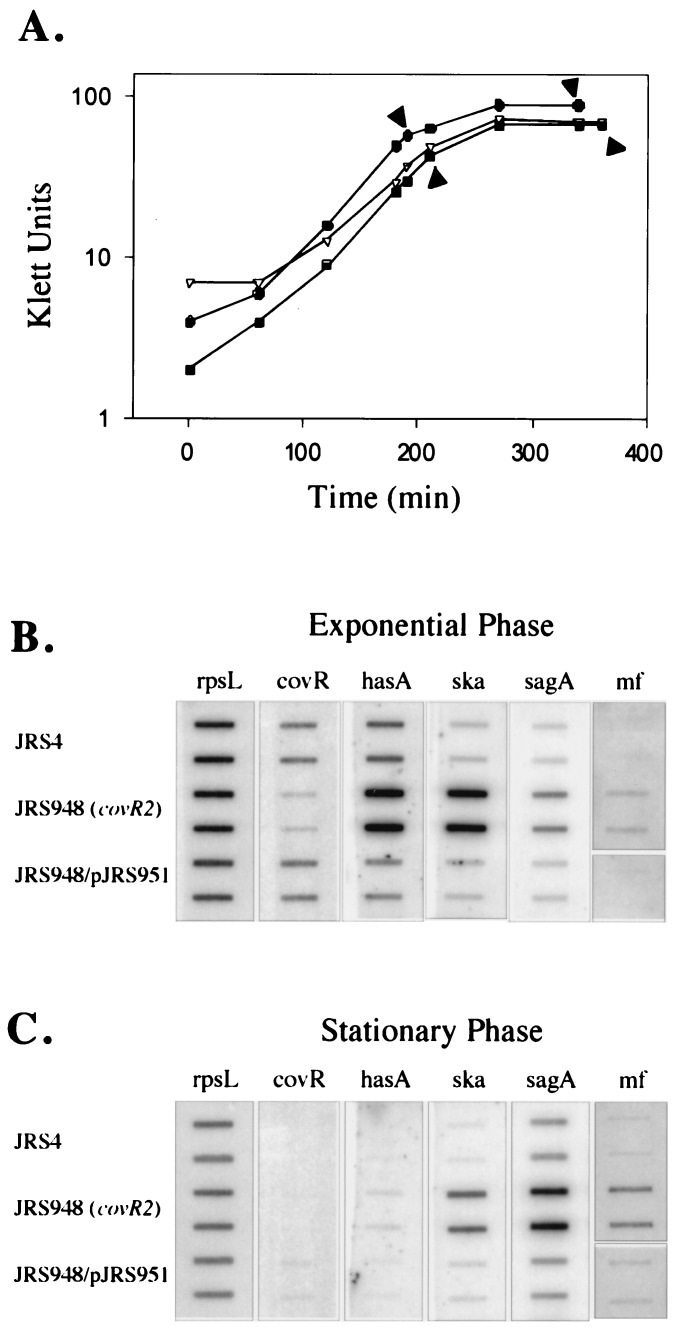
FIG. 3.
Complementation of transcription of covR2. (A) Growth curves showing times of isolation of RNA (arrowheads). Filled circles represent JRS4, open triangles represent JRS948 (covR2), and filled squares represent JRS948/pJRS951 (covR2/covR+). (B and C) Hybridization of specific DNA probes to RNA was as described in the legend to Fig. 2B, except that 4 μg of RNA was used throughout and the DNA probe for speMF was internal to the coding region (Materials and Methods). The covR probe is restricted to the 204 bp at the 3′ end of the gene.
Although their maximal expressions occurred at different times in the growth cycle, the levels of message for hasA, ska, sagA, and speMF in the nonpolar covR2 mutant JRS948 were higher than in JRS4 at both times (Fig. 3B and C). This confirms the results presented above (Fig. 2B) for the polar covR1 mutant JRS551 in late exponential phase. Although this indicates that CovR-mediated repression occurs both in log phase and in stationary phase, the level of covR transcript appears to be maximal during exponential growth (Fig. 3A and B).
The covR2 mutation can be complemented in trans.
To verify that covR alone was responsible for repression of hasA, ska, sagA, and speMF expression, complementation of the covR2 defect in JRS948 was attempted. Since the location of the covR promoter has not been determined, the DNA fragment cloned with the covR gene was designed to include a potential promoter that has homology to the consensus −10 and −35 sequences. This fragment was cloned into pLZ12 to generate pJRS951 (see Materials and Methods). Strain JRS948/pJRS951 produced covR transcript at a level comparable to the wild-type parent, JRS4 (Fig. 3B). This demonstrates that the native promoter for covR is within the 287 bases upstream of the coding region. In the complemented strain, the levels of transcript for hasA, ska, sagA, and speMF were comparable to those in JRS4 (Fig. 3B and C). Therefore, covR expressed from its native promoter can fully complement the phenotypic defects of the covR2 allele, as measured by transcription.
Protein activity correlates with transcript level.
To determine whether the amount of protein produced from each gene correlates with transcriptional expression of ska, sagA and the has operon, the product of each was assayed in the covR2 mutant and its complemented derivative (see Materials and Methods). As previously mentioned, both JRS551 (which contains the covR1 mutation) and JRS948 (with the nonpolar covR2 mutation) produce highly mucoid colonies on agar plates compared to the small, round colony phenotype of the JRS4 parent strain. When the covR2 mutation was complemented by pJRS951, the colonies appeared to be similar to the parental strain. Quantitation of hyaluronic acid produced by each strain in liquid culture showed that the wild-type strain JRS4 and the complemented strain JRS948/pJRS951 produced negligible amounts (0.02 and 0.05 pg per CFU, respectively), while there were about 1.4 pg of hyaluronic acid per CFU from the mutant strain JRS948. Thus, the amount of hyaluronic acid capsule reflects the amount of hasA transcript in these strains.
Production of streptokinase was assayed by the ability of the organism to activate plasminogen, as described in Materials and Methods. No activity was detected (<0.01 units/ml) from either JRS4 or the complemented mutant (JRS948/pJRS951), while the covR2 mutant strain JRS948 produced a significant amount of enzyme (2.2 units/ml).
SLS activity was measured by lysis of bovine erythrocytes. SLS was released from the GAS cell surface with magnesium (see Materials and Methods), and activity in cell-free supernatants was analyzed. Activity of SLO, which is not inhibited by trypan blue, did not account for any hemolysis seen. Less than 1 hemolytic unit was detected in supernatants of JRS4 and the complementing strain JRS948/pJRS951, whereas approximately 275 hemolytic units were found in the supernatant from JRS948, the covR2 mutant. Therefore, the relative amounts of SLS in the mutant and wild-type parallel the relative amounts of message for sagA.
CovR represses its own synthesis.
Response regulator proteins often regulate their own expression as well as that of their cotranscribed cognate sensor kinase genes. To determine whether covR is autoregulated, the effect of the mutation on transcription of the covRS operon was investigated. The inserted aphA3 gene in JRS948 (Fig. 1B) has no promoter and should be transcribed from the covR promoter. Furthermore, both the covR and covS genes were found to be cotranscribed in an M3 strain of GAS, suggesting that transcription of covS is driven from the promoter located upstream of covR (25). Because the structure of the covRS locus in the M6 strain used in our study appears to be similar to both the reported M3 strain and the sequenced M1 strain, the amount of covS transcript should reflect that of covR.
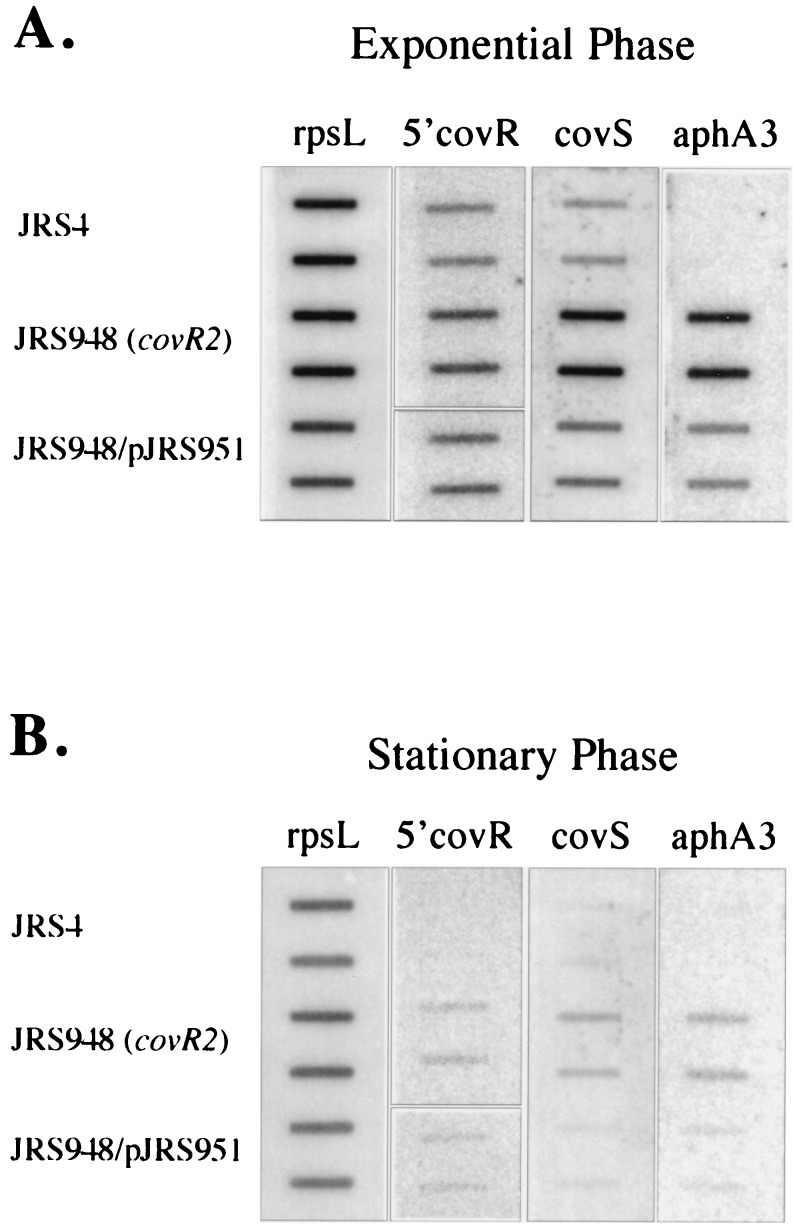
Using slot blot hybridization analysis, the amounts of message of covS and aphA3 in the covR2 mutant JRS948 were compared to those in the parent JRS4 strain. To control for the amount of mRNA loaded on the gel from each strain, the transcript of rpsL was used as before. In JRS948 there is substantially more transcript from covR and covS than in JRS4 (Fig. 4). Therefore, CovR appears to repress transcription from its own promoter. In support of this, the low level of covR and covS transcript in the complementing strain JRS948/pJRS951 is similar to that in the wild-type. Furthermore, the aphA3 transcript reflects the same repression by CovR, since it too is lower when the covR2 mutant strain JRS948 is complemented by pJRS951 (Fig. 4). From all these strains, a comparison of the amounts of covR and covS (and, when present, aphA3) message at different growth phases (Fig. 4) indicates that there is more transcript for this operon in exponential than in stationary phase.
FIG. 4.
Effects of CovR on expression of the covRS operon. RNA was the same as that in Fig. 3 and was harvested at the times indicated in Fig. 3A. Hybridization was as described in the legend to Fig. 2B, except that 4 μg of RNA was used throughout. The covR probe is restricted to the 305 bp at the 5′ end of the gene.
DISCUSSION
A new sensor-regulator gene pair that regulates GAS virulence factors.
For GAS to adapt and survive at various locations within the human host during an infection, it must be able to sense the changing surroundings and regulate its many virulence genes in response to available signals. Mga activates expression of several virulence genes in response to different growth conditions, and yet many other virulence genes are not subject to control by Mga (28). To help identify new pathways for environmental regulation of virulence factors, three different loci in the GAS genome that are predicted to encode homologs of bacterial two-component signal-transducing systems were identified and two of them were characterized.
We report here that one of these sensor-regulator gene pairs, encoding CovR-CovS, represents a new regulatory pathway affecting expression of several GAS virulence genes that are not regulated by Mga. In addition, this pathway appears to be completely independent of the established Mga regulon. It seems likely that CovR-CovS is present in most strains of GAS, since this locus from a serotype M3 GAS strain had homology with all 28 GAS strains tested (25). In this work, we have demonstrated that CovR-CovS also represses expression of three additional virulence factors, streptokinase, SLS, and mitogenic factor. Since we investigated a limited set of genes, it remains possible that additional virulence genes of GAS are regulated by CovR.
We were not able to assign a function to the other two sensor-regulator gene pairs investigated in this study. Inactivation of irr, the gene encoding the response element located directly upstream of isp and mga in the GAS genome, showed no effect on expression of the specific virulence genes studied here. However, it is possible that irr functions only under environmental growth conditions not used in these studies or that irr acts on genes not investigated in this work.
Our inability to inactivate the sycF response regulator gene, encoding a homolog of B. subtilis yycF, precluded further study of it. However, since Fabret and Hoch reported a similar inability to inactivate yycF in B. subtilis, this locus may be essential for GAS growth, as it appears to be for B. subtilis (15). It is unlikely that this locus is virulence specific, because it is apparently essential under laboratory growth conditions and because the gene in the nonpathogenic B. subtilis is highly homologous to the gene in GAS.
CovR is the first repressor of multiple genes described for GAS.
Transcriptional regulators previously described for GAS activate transcription of the genes they control. These include Rof, the regulator of expression of protein F (a surface-located adhesin), and RopB, which regulates expression of SpeB (a cysteine protease). Currently, there is no evidence that RopB and Rof act on any additional genes (17, 27). In contrast, Mga activates transcription of several genes that are probably important for virulence and, in addition, it positively autoregulates its own expression (6, 10, 36). Positive autoregulation allows for a rapid increase in production of activated gene transcripts, although some form of negative regulation is required to limit the response. No such negative regulator has been identified yet for the Mga regulon, and it is not clear why rapid and extensive expression of these genes would be required in response to an environmental signal.
CovR regulation, on the other hand, involves repression of transcription and represents the first multiple gene repressor described in the GAS. Inactivation of covR in the serotype M6 GAS strain JRS4 resulted in a significant increase in transcript levels for the genes encoding streptokinase (ska), SLS (sagA), mitogenic factor (speMF), and the first enzyme of capsule synthesis (hasA). In addition, CovR negatively regulates transcription of its own operon. Because a decrease in the amount of CovR derepresses the covR promoter, there is always an adequate amount of CovR available to respond to an environmental signal. This facilitates a rapid response to an environmental signal. Thus, the negative feedback loop produced by autorepression of CovR allows GAS to turn off the activated CovR regulatory circuit rapidly. By limiting expression of the sensor and regulator components, the length of the response can be restricted. Such a system provides an exquisitely sensitive ability to respond rapidly to potentially short-term environmental changes. In keeping with this idea, the genes identified as being CovR-repressed encode products that may be relatively short lived. Capsule is broken down by hyaluronidase, streptokinase and mitogenic factor most likely diffuse away from the cell, and SLS may be removed from the GAS surface by proteases.
Does CovR act directly?
The response regulator elements of two-component systems are usually DNA binding proteins that act directly at the promoters of their target genes in response to their signal. CovR possesses conserved sequences homologous to those required for a response regulator and is required for repression of ska, sagA, hasA, and covR transcription. CovR may act directly on the promoters of some or all of these genes, or it may act indirectly through a cascade involving another regulatory circuit. If it acts directly on all four promoters, one might expect to find a consensus site within each promoter at which CovR binds. However, because we were not able to identify a conserved sequence within the region upstream of the start of translation for each of the CovR-regulated genes, we can make no predictions about the mode of action of CovR.
A regulatory network controls expression of GAS virulence genes.
Some bacterial pathogens use a global regulatory network to control their many virulence genes in a coordinated fashion. Although both Mga and CovR regulate expression of more than one gene, the genes they regulate do not appear to overlap. Levin and Wessels found that loss of CovR in an M type 3 GAS strain has no obvious effect on the amount of Mga-dependent M protein or hemolytic activity in the culture supernatant (SLO) (25). We previously found that transcription of the CovR-regulated genes hasA, ska, speMF (28), and sagA (16) is not affected by Mga, and we report here that transcription of the Mga-regulated genes emm, scpA, and mga is not affected by CovR. Therefore, each regulatory circuit seems to target a separate subset of virulence genes. Whether GAS has a global regulator that controls both the Mga and CovR pathways remains to be determined, but it is apparent that growth phase affects expression of both sets of genes.
Many different GAS virulence genes exhibit growth phase-dependent regulation. Some genes, such as emm, scpA, mga, and hasA, show maximal expression during exponential growth, while others, including sagA and speMF, are expressed at higher levels in stationary phase (3, 28). We have shown here that even in a strain lacking CovR, expression of CovR-regulated genes is growth phase dependent. Furthermore, CovR represses these genes in both exponential and stationary phase. Thus, the CovR pathway is independent of growth phase and represents a separate regulatory pathway. This means that the genes encoding SLS, capsule synthesis, streptokinase, and mitogenic factor are regulated by at least two distinct signals, one for each pathway.
The phase of growth also correlates with expression of genes in the Mga regulon, since these genes are shut off as cells enter stationary phase. However, since CovR does not repress Mga regulon genes and Mga does not affect CovR regulon genes, both must be independently controlled by a pathway or pathways subject to growth phase-related signals (Fig. 5).
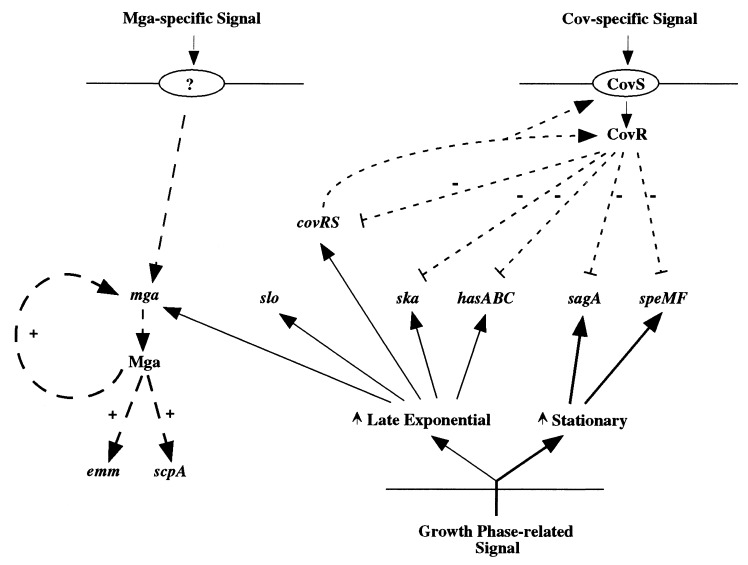
FIG. 5.
Model of regulatory networks in GAS. Positive regulation is indicated by +, and negative regulation is indicated by −. Lines with short dashes represent CovR regulation, and lines with long dashes represent regulation by Mga. Solid lines indicate regulation correlated with growth phase either in exponential phase (light lines) or stationary phase (heavy lines).
With three distinct pathways of regulation now described for GAS, the complexity of the ability of this organism to respond to various niches within the human host begins to emerge. The Mga-specific and CovR-specific pathways appear to act independently of each other and most likely respond to different environmental cues. Growth phase-dependent regulation, whose mechanism(s) has not yet been studied, affects genes in both pathways without the need for either Mga or CovR. For genes that are regulated by two pathways, the amount of transcript produced is affected by both. As a result, it is difficult to predict the degree to which such a gene is expressed at any location in the host in which the GAS finds itself. It is presumably the interaction of these regulatory networks (Fig. 5), and possibly others not yet discovered, that endow this organism with the ability to produce so many different types of disease syndromes. A better understanding of these mechanisms of GAS gene control might enable us to intervene in clinical situations to prevent growth and dissemination of this serious human pathogen.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R37-AI20723, and K.S.M. was supported in part by National Research Service award AI09460 from the National Institute of Allergy and Infectious Diseases.
We thank Carla Easter and Michael Caparon for providing the protocol for the SLS assay.
REFERENCES
- 1.Ashbaugh C D, Warren H B, Carey V J, Wessels M R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Investig. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajaj A P, Castellino F J. Activation of human plasminogen by equimolar levels of streptokinase. J Biol Chem. 1977;252:492–498. [PubMed] [Google Scholar]
- 3.Betschel S D, Borgia S M, Barg N L, Low D E, De Azavedo J C. Reduced virulence of group A streptococcal Tn916mutants that do not produce streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno A L. Group A streptococcal infections and acute rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 5.Bronze M S, Dale J B. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am J Med Sci. 1996;311:41–54. doi: 10.1097/00000441-199601000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caparon M G, Stephens D S, Olsen A, Scott J R. Role of M protein in adherence of group A streptococci. Infect Immun. 1991;59:1811–1817. doi: 10.1128/iai.59.5.1811-1817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chassy B M. A gentle method for the lysis of oral streptococci. Biochem Biophys Res Commun. 1976;68:603–608. doi: 10.1016/0006-291x(76)91188-8. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee M S, Gerlach D, Yu C E, Ferretti J J. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect Immun. 1993;61:3719–3723. doi: 10.1128/iai.61.9.3719-3723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 11.Crater D L, van de Rijn I. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J Biol Chem. 1995;270:18452–18458. doi: 10.1074/jbc.270.31.18452. [DOI] [PubMed] [Google Scholar]
- 12.DeAngelis P L, Papaconstantinou J, Weigel P H. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993;268:19181–19184. [PubMed] [Google Scholar]
- 13.de Vos W M. Genetic improvement of starter streptococci by the cloning and expression of the gene coding for a non-bitter proteinase. Biomolecular engineering programme—final report. In: Magnien E, editor. Biomolecular engineering in the european community: achievements of the research programme (1982–1986)—final report. Lancaster, England: Martinus Nijhoff; 1986. pp. 465–472. [Google Scholar]
- 14.Dougherty B A, van de Rijn I. Molecular characterization of a locus required for hyaluronic acid capsule production in group A streptococci. J Exp Med. 1992;175:1291–1299. doi: 10.1084/jem.175.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federle, M. J., J. G. Smith, and J. R. Scott. 1998. Unpublished data.
- 17.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang T T, Malke H, Ferretti J J. The streptokinase gene of group A streptococci: cloning, expression in Escherichia coli, and sequence analysis. Mol Microbiol. 1989;3:197–205. doi: 10.1111/j.1365-2958.1989.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 19.Hulett F M. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:933–939. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- 20.Husmann L K, Yung D L, Hollingshead S K, Scott J R. Role of putative virulence factors of Streptococcus pyogenesin mouse models of long-term throat colonization and pneumonia. Infect Immun. 1997;65:1422–1430. doi: 10.1128/iai.65.4.1422-1430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imanishi K, Igarashi H, Uchiyama T. Activation of murine T cells by streptococcal pyrogenic exotoxin type A. Requirement for MHC class II molecules on accessory cells and identification of V beta elements in T cell receptor of toxin-reactive T cells. J Immunol. 1990;145:3170–3176. [PubMed] [Google Scholar]
- 22.Kehoe M, Timmis K N. Cloning and expression in Escherichia coli of the streptolysin O determinant from Streptococcus pyogenes: characterization of the cloned streptolysin O determinant and demonstration of the absence of substantial homology with determinants of other thiol-activated toxins. Infect Immun. 1984;43:804–810. doi: 10.1128/iai.43.3.804-810.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kihlberg B M, Cooney J, Caparon M G, Olsen A, Bjork L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 24.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 25.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 26.Levin, J. C., and M. R. Wessels. 1999. Personal communication.
- 27.Lyon W R, Gibson C M, Caparon M G. A role for Trigger Factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIver K S, Scott J R. Role of mgain growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIver K S, Subbarao S, Kellner E M, Heath A S, Scott J R. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect Immun. 1996;64:2548–2555. doi: 10.1128/iai.64.7.2548-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenesM49 demonstrates that VirR functions as a positive regulator of streptococcal C5a peptidase and M protein in OF+ strains. Dev Biol Stand. 1995;85:149–152. [PubMed] [Google Scholar]
- 31.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexnerientry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moses A E, Wessels M R, Zalcman K, Alberti S, Natanson-Yaron S, Menes T, Hanski E. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A streptococcus. Infect Immun. 1997;65:64–71. doi: 10.1128/iai.65.1.64-71.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musser J M, Stockbauer K, Kapur V, Rudgers G W. Substitution of cysteine 192 in a highly conserved Streptococcus pyogenesextracellular cysteine protease (interleukin 1β convertase) alters proteolytic activity and ablates zymogen processing. Infect Immun. 1996;64:1913–1917. doi: 10.1128/iai.64.6.1913-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofek I, Bergner-Rabinowitz S, Ginsburg I. Oxygen-stable hemolysins of group A streptococci. 8. Leukotoxic and antiphagocytic effects of streptolysins S and O. Infect Immun. 1972;6:459–464. doi: 10.1128/iai.6.4.459-464.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohara-Nemoto Y, Sasaki M, Kaneko M, Nemoto T, Ota M. Cysteine protease activity of streptococcal pyrogenic exotoxin B. Can J Microbiol. 1994;40:930–936. doi: 10.1139/m94-149. [DOI] [PubMed] [Google Scholar]
- 36.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mryregulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 37.Okada N, Pentland A P, Falk P, Caparon M G. M protein and protein F act as important determinants of cell-specific tropism of Streptococcus pyogenesin skin tissue. J Clin Investig. 1994;94:965–977. doi: 10.1172/JCI117463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otto R, de Vos W M, Gavrieli J. Plasmid DNA in Streptococcus cremoris: influence of pH on selection in chemostats of a variant lacking a protease plasmid. Appl Environ Microbiol. 1982;43:1272–1277. doi: 10.1128/aem.43.6.1272-1277.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Casal J, Caparon M, Scott J. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenesrestores its ability to resist phagocytosis. Res Microbiol. 1992;143:549–558. doi: 10.1016/0923-2508(92)90112-2. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Casal J, Price J A, Maguin E, Scott J R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 41.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural virregulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, B. A., S. P. Linn, L. Song, X. Yuan, S. Clifton, M. McShan, and J. Ferretti. 1 April 1999, revision date. Streptococcal genome sequencing project. [Online.] University of Oklahoma, Norman. http://www.genome.ou.edu/strep.html. [26 April 1999, last date accessed.]
- 43.Romeo T. Global regulation by the small RNA-binding protein CsrA and the noncoding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz N, Wang B, Pentland A, Caparon M. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol Microbiol. 1998;27:337–346. doi: 10.1046/j.1365-2958.1998.00681.x. [DOI] [PubMed] [Google Scholar]
- 45.Schlievert P M. Role of superantigens in human disease. J Infect Dis. 1993;167:997–1002. doi: 10.1093/infdis/167.5.997. [DOI] [PubMed] [Google Scholar]
- 46.Schrager H M, Rheinwald J G, Wessels M R. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Investig. 1996;98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott J R, Fischetti V A. Expression of streptococcal M protein in Escherichia coli. Science. 1983;221:758–760. doi: 10.1126/science.6192499. [DOI] [PubMed] [Google Scholar]
- 48.Scott J R, Guenthner P C, Malone L M, Fischetti V A. Conversion of an M− group A streptococcus to M+by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tewodros W, Norgren M, Kronvall G. Streptokinase activity among group A streptococci in relation to streptokinase genotype, plasminogen binding, and disease manifestations. Microb Pathog. 1995;18:53–65. doi: 10.1016/s0882-4010(05)80012-9. [DOI] [PubMed] [Google Scholar]
- 50.Wessels M R, Bronze M S. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proc Natl Acad Sci USA. 1994;91:12238–12242. doi: 10.1073/pnas.91.25.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wessels M R, Goldberg J B, Moses A E, DiCesare T J. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect Immun. 1994;62:433–441. doi: 10.1128/iai.62.2.433-441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wexler D E, Chenoweth D E, Cleary P P. Mechanism of action of the group A streptococcal C5a inactivator. Proc Natl Acad Sci USA. 1985;82:8144–8148. doi: 10.1073/pnas.82.23.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yutsudo T, Murai H, Gonzalez J, Takao T, Shimonishi Y, Takeda Y, Igarashi H, Hinuma Y. A new type of mitogenic factor produced by Streptococcus pyogenes. FEBS Lett. 1992;308:30–34. doi: 10.1016/0014-5793(92)81043-l. [DOI] [PubMed] [Google Scholar]