Abstract
Emerging viral diseases, such as Ebola, SARS, MERS, and the pathogen causing COVID-19, SARS-CoV-2, present a challenge for the development of therapeutics because of strict biosafety handling procedures and rapid mutation of their genomes. To facilitate the development of an adaptable and testable therapeutic model system, a colostrum exosome-based nanoparticle delivery system, EPM (exosome-PEI matrix), that overcomes stringent biosafety containment, was used to mimic the expression of viral proteins. This system would greatly expand the number of laboratories actively participating in the screening of potential therapeutics. EPM technology can deliver both plasmid DNA and siRNA to both simulate viral gene expression and screen potential antiviral siRNA therapeutics. Using this nanoplatform, three key SARS-CoV-2 proteins (the spike glycoprotein, nucleocapsid, and replicase) were expressed in vitro and in vivo. In vitro, several viral gene-targeting siRNAs were screened to determine knockdown efficiency, with some siRNA duplexes resulting in 80%–95% knockdown of corresponding protein expression. Moreover, in vivo experiments introducing the spike protein and nucleocapsid by EPM resulted in the production of antibodies against the viral antigen, measured up to 45 d after target delivery. Together, these findings support the efficacy of the EPM delivery system to establish a model for screening antiviral therapeutics-reduced biosafety level.
Keywords: delivery strategies, exosomal delivery, siRNA therapeutics, SARS-CoV-2, antibodies, BSL2 viral model system
Graphical abstract
An exosome and polyethyleneimine (EPM) nanoplex delivers nucleic acids, creating a model for the identification of antiviral therapeutics. Viral protein expression using EPM evokes the production of antigen-specific antibodies. EPM delivery requires less biosafety containment than live-virus assays with faster screening and is a promising platform for potential clinical delivery of siRNA.
Introduction
The emergence of SARS-CoV-2 (severe acute respiratory syndrome-coronavirus-2) and the resultant pandemic, in its third year, has resulted in the mobilization of a molecular toolbox now available in the genomics age, including gene-based vaccines. The need for therapeutics based on new technology is underscored by the lack of viable pharmaceuticals for treating SARS-CoV-2 infection. One obstacle in the discovery of therapeutics has been limited access to Biosafety Level 3 (BSL3) containment for studying live viruses such as SARS-CoV-2 and the lack of a model that can be used at available BSL2 facilities. Moreover, the rapid mutation rate of these highly infectious viruses, which often produces more dangerous variants that can evade vaccine and naturally acquired immunity, highlights the need for a new generation of rapidly adaptable antiviral therapeutics, such as targeted gene therapy.1 The decreased containment models in place, such as pseudoviral systems, while useful for studying cellular entry kinetics, are not useful for studying genome-based therapeutics as they typically include a single viral antigen of a single sequence. Thus, a strategy to explore genome-based viral therapeutics at a lower biosafety risk level for emerging pathogens such as SARS-CoV-2 is a high priority.
The concept of gene therapy is not new in the field of medicine. The discovery of small interfering RNA (siRNA) in 1999 has extended the reach of the gene therapy field to suppression of targeted protein expression. Several studies have used the delivery of siRNA to target an array of diseases in model systems, including Alzheimer’s and pancreatic cancer.2, 3, 4 Nearly 20 years ago, the use of antiviral siRNA was first reported to prevent and treat viral infections in a murine models.5 Despite the early promise of antiviral siRNA therapeutics, no antiviral siRNA has progressed from the lab bench to the clinic. A major hurdle for widespread clinical application of siRNA is a lack of suitable delivery systems to targeted locations that provide effective therapeutic interventions. Unprotected siRNA is not readily taken up by cells,6 while the presence of RNases in the body is sufficient to pose a significant threat to the integrity of the siRNA duplexes. To circumvent this issue, delivery systems have been developed to enhance cellular uptake and protect against extracellular nucleases.
Delivery modalities have typically been classified as either viral or non-viral. Viral-mediated siRNA delivery uses replication-deficient viral particles derived from viruses such as adenovirus. Delivery by adenoviruses has several reported drawbacks, including a lack of tissue tropism, transient effect of the delivery, dose limitation, and potential hepatotoxicity.7 Moreover, the delivery particles themselves can be immunogenic and the immune system may clear them before the payload has reached the target tissue.8 Non-viral transfection methods have also presented limitations. Polycations and cationic lipids have been used in vitro, although they offer little direct translatability to in vivo models.9,10 Like adenovirus-based delivery systems, liposomes and lipid nanoparticles (LNPs) lack tissue specificity and distribute equally in almost all organs, likely producing off-target effects of gene knockdown, while also suffering short blood circulation. The production of selective organ targeting (SORT) LNPs has achieved modest progress in increasing the delivery of nucleic acid-based therapeutics to certain organs, such as the liver and spleen,11 but fails to offer significant improvement in targeting to organs such as the lung, the target organ of SARS-CoV-2. Moreover, mammalian organs are complex systems containing multiple tissue types and multiple cell types, some of which are more susceptible to viral infection than others. Accumulation of the payload at the organ is not a guarantee of delivery to the tissues or cells that will receive most benefit from siRNA therapeutics.
Natural nanoparticles, such as exosomes, offer significant improvements over all other delivery systems for nucleic acid payloads targeted to specific tissue and cell types. Exosomes are ubiquitous in the human body, present in many bodily fluids, including plasma, urine, saliva, bronchial fluid, cerebral spinal fluid, breast milk, tears, and lymph,12, 13, 14, 15, 16 and are inherently capable of facilitating intercellular communication and delivery of biological molecules. These particles endogenously contain protein markers, such as CD47, that prolong circulation in the body and prevent phagocytosis by macrophages.17 Tissue targeting can be enhanced further by attaching a ligand to the surface-bound exosomal proteins.18 These ligands can be exploited to target both organs, such as the lung, and also cells within the lung that may be virally infected. As an example, lactoferrin receptor expression is increased in virally infected cells, likely due to the natural potent antiviral effects of lactoferrin (LF) both extracellularly and intracellularly.19
Like gene therapy, the use of exosomes as delivery vesicles is not new, but again limitations in mass production prohibit the clinical translatability. Traditionally, methods of bulk isolation of exosomes for drug delivery purposes have used human cell culture, specifically mesenchymal stem cells or fibroblasts.20 Bovine milk/colostrum exosomes used in the model system presented here offer advantages over human cell line-derived exosomes, with milk/colostrum containing a substantially higher abundance of exosomes that are well tolerated, lacking systemic and immunotoxicity.21,22 Milk/colostrum exosomes can also deliver small molecules21, 22, 23 and biologics.18,24 Recognized as overcoming many limitations of mass production from human cell lines, milk and colostrum exosomes have been loaded with a variety of molecules and used to study treatments in a variety of disease models, including cancer.25
Because exosomes are composed of a negatively charged lipid bilayer,26 the association between exosomes and nucleic acids must be facilitated by a modulator of these charge interactions. Polycations, such as polyethylenimine (PEI), have long been used for the transfection of eukaryotic cell lines;27,28 however, the toxicity of this molecule and limited cell uptake remains a concern in both animal models and clinical settings.29 By using PEI in the exosome and polyethyleneimine (EPM) nanoplex, the effective dose of PEI can be substantially reduced, and the cytotoxic effects minimized. In addition to successful loading of nucleic acids via interaction with PEI, the EPM can be functionalized with various targeting ligands. The versatility of the exosomal nanoplatform naturally lends itself to loading with targeting ligands. We have previously shown that folic acid can be attached to exosomes and enhance the delivery of payload to tumors, which have a higher expression of folate receptors than the surrounding tissue.18 Lactoferrin has been known for antimicrobial and antiviral properties, and many viruses make entry into the host cell via LF receptors.30 Therefore, the functionalizing of exosomes with LF could enhance the delivery of payload to virally infected cells.
Even with an optimized delivery system for siRNA therapeutics, the viral targets for those nucleic acid-based therapeutics also must be explored to find sequences that will provide maximum therapeutic effect. SARS-CoV-2 is nearly 30,000 bases long and it contains coding sequences for nearly 25 proteins.31 To increase the chance of success for meaningful siRNA therapeutic effect, viral targets must be chosen that, if decreased, would affect the viral titer in the host. The spike glycoprotein mediates the cellular entry of all coronaviruses via its interaction with the angiotensin-converting enzyme 2 (ACE2) receptor.32 This protein has two subunits, S1 and S2; of those, it is the S1 containing the specific receptor binding domain (RBD) that interacts with the ACE2 receptor. The RBD of the S1 interacts with the highest affinity antibodies, indicating it is the most highly immunogenic antigen of SARS-CoV-2.33 The viral nucleocapsid (N) is important in viral genome packaging and viral particle release34 and also induces antibody production.35 Both of these structural proteins have been suggested as promising targets for siRNA-mediated inhibition of SARS-CoV-2 as they tend to be highly conserved, with fewer mutations than other viral coding sequences.36 Although the viral replicase (RdRp) is not a structural protein and therefore is not highly antigenic, it is essential to viral replication and often the target of other viral therapeutics, such as remdesivir.
Many different siRNA duplexes can be designed that could target degradation of the mRNA of the S1, N, or RdRp. To screen each individual duplex against the live virus would be expensive, time-consuming, and limited to the labs authorized to handle organisms requiring BSL3 containment. The ability of exosomes to carry and deliver nucleic acid cargoes that can introduce the production of a target protein or reduce the production of an endogenously expressed protein, make the exosomal delivery system an ideal model for developing therapeutic approaches for dangerous pathogens such as SARS-CoV-2 at lower biosafety containment. Here, we demonstrate the utility of exosomes to develop a testable model of human disease at a lower biosafety level by the introduction of mRNA transcribed from expression plasmids for viral antigens and the subsequent development of siRNA targeted against the sequences using colostrum exosome-based delivery system. This approach can be more broadly applied to develop additional disease models to facilitate the development of therapeutics at lower biosafety levels that are more widely accessible to the research community. The versatility of the exosomal delivery system also lends itself to functionalization with targeting ligands, such as LF, which can help to direct the payload to virally infected respiratory cells.
Results
Entrapment efficiency of plasmid DNA (pDNA)
The ability of the EPM technology to entrap siRNA has been well characterized in our previous study.18 With either 2 or 10 μg of pDNA loaded, exosomes alone were not able to entrap pDNA, and PEI alone resulted in 40% entrapment, with detectable pDNA remaining in the supernatant of the reaction (Figure 1A). However, both pDNA loads were entrapped >90% using EPM technology (Figures 1A and 1B). With typical 75 μg exosomes and 2 and 10 μg pDNA, entrapment was >90%.
Figure 1.
Characterization of the EPM-pDNA
(A) Entrapment efficiency of the EPM was compared to polyethyleneimine (PEI) alone using 32P-labeled pDNA as a tracer. Exosomes were incubated with 40 μg PEI and 2–10 μg non-radioactive pEMP and 5′-32P-labeled pEMP as tracer in 150 μL PBS, in triplicate. After 15 min incubation, the reaction mixture was precipitated to recover EPM-5′-32P-pEMP and suspended in PBS. To determine the entrapment efficiency, relative distribution of the radioactivity was determined by spotting aliquots of EPM-5-32P-pEMP pellet (P) and the supernatant (S) on a piece of PEI-cellulose thin layer (dot blot analysis). Due to variable volumes, one-third of the total pellet and one-tenth of the supernatant were spotted; appropriate dilution factors were taken into consideration when determining entrapment. The amount of radioactivity was measured by Packard InstantImager. (B) Quantification of entrapment efficiency from (A). Raw data are shown in Table S1. (C) Lack of cytotoxicity of EPM formulations. When treated with either exosomes alone, EPM, or EPM loaded with nucleic acids as used in this study, HEK293T cells had similar survival as compared to untreated cells, indicating the lack of detectable cytotoxicity. PEI 60K alone had some associated cytotoxicity, but this effect was mitigated by the presence of exosomes in the formulation. The data shown represent means and SDs of 3 biological replicates. Analysis of cell survival was done using a 1-way ANOVA with Dunnett’s post hoc analysis compared to the untreated control (∗∗p < .01).
EPM and EPM loaded with nucleic acid does not show cytotoxicity in vitro
Human embryonic kidney cells (HEK293T) were incubated with representative formulations and formulation components for detection of cytotoxicity. These formulations included exosomes alone, EPM alone, and the EPM loaded with both types of nucleic acids used for this study. Cells were incubated with the formulations or exosomes alone for the same period of time as used in expression studies (48 h) and then viability was assessed by mixing with trypan blue and counting live cells using a Countess 3 Cell Counter (Thermo Fisher Scientific, Waltham, MA). As compared to untreated cells cultured under the same conditions, none of the formulations or the exosomes alone resulted in any detectable cytotoxicity (Figure 1C). PEI used alone as a transfection agent did produce some cytotoxicity, but this effect was mitigated by the presence of exosomes in the formulation (Figure 1C).
Establishment of viral antigen expression model for SARS-CoV-2 in vitro
Exosome nanoparticles were used to entrap and deliver expression constructs for SARS-CoV-2 proteins; the S1 subunit of the S, the N, and the RdRp. Exosomal formulations composed of exosomes, PEI, and the plasmid encoding one of the aforementioned viral proteins were applied to HEK293T cells. After a 48-h incubation, protein expression of S1 protein of approximately 90 kD (Figure 2A), the N protein of approximately 50 kD (Figure 2B) and the RdRp of approximately 106 kD (Figure 2C) was detected via western blot.
Figure 2.
Expression of viral antigens in vitro
Expression constructs were delivered to HEK293T cells via EPM nanoplatform. Antigens introduced included the S1 subunit of the spike glycoprotein (A), the nucleocapsid (N) (B), and the RNA-dependent RNA polymerase (RdRp) (C). Expression of antigens was detected by western blot, and β-actin was used as a loading control. (D) Dose response of S1 expression, varying exosomal load, and pDNA payload after 48 h incubation. Fold change is indicated as compared to 75 μg exosomes, 40 μg PEI, and 1.5 μg pDNA, the selected standard reaction. The samples shown are pooled biological triplicates undergoing single replicate analysis. (E) Time course of N expression using the standard reaction. The blot shown is representative of 3 biological replicates analyzed individually. Fold change shown with respect to expression level after 12 h. (F) Time course of S1 expression using the standard reaction. The blot shown is representative of 3 biological replicates. Fold change shown with respect to expression level after 12 h. Data expression as means ± SDs for at least 3 biological replicates and significance determined by 1-way ANOVA with post-hoc comparison of all means to EPM-pDNA for target antigen alone (∗p < 0.05, ∗∗∗p < 0.001).
To determine the optimal loading conditions, the amounts of exosomes and pDNA were varied and viral protein expression assessed. Typically, the exosomal protein used was either 75 or 150 μg. pDNA payload ranged from 0.5 to 4.5 μg. The expression of S1 could be detected following treatment with all of the formulations (Figure 2D). The expression of RdRp and N followed a similar pattern (Figures S1 and S2). In vitro, the balance is highly skewed in favor of the delivery system, with many more exosomes than cells, as the goal is testing the nucleic acid being delivered. That said, the exogenous expression limitation is likely reached using smaller amounts of exosomal protein and pDNA and increasing either does not significantly increase expression in the cell. As there was no significant increase in expression by using more exosomal protein or pDNA, we chose 75 μg exosomes and 1.5 μg pDNA as our standard formulation.
Expression of the viral proteins as introduced by EPM was time dependent. Using the standard formulation (75 μg exosomes, 40 μg PEI, and 1.5 μg pDNA), expression of the N could be detected as early as 12 h after treatment, although maximal expression occurred after 48 h (Figures 2E and S3). Expression of the S1 glycoprotein was barely detectable until 24 h post-treatment and this too was highest after 48 h incubation (Figures 2F and S4). As such, we chose 48 h as the optimal treatment time for all of the subsequent studies. As the EPM-mediated delivery of pDNA is a multi-step process, it stands to reason that there should be a time-dependent response. The EPM-pDNA complex must first be taken into the cell, then released from the endosome, and the DNA travel to the nucleus from transcription.
Screen of antiviral siRNA
Multiple siRNA duplexes were designed based on the plasmid sequence of each key viral antigen. The design algorithm could not predict the efficacy of gene knockdown, so the optimal siRNA sequence was determined by simultaneously delivering two EPM formulations: (1) loaded with the corresponding expression plasmid and (2) loaded with the test siRNA duplex. To eliminate the potential cytotoxicity of increased PEI concentration as a result of two exosomal formulations, the media volume was doubled to maintain a similar concentration of PEI in the transfection media. As a positive control for the expression level of the target protein, the EPM formulation bearing the corresponding expression plasmid was delivered simultaneously with an “empty” sham EPM formulation. Again, each EPM-pDNA formulation contained 75 μg exosomes, 40 μg PEI, and 1.5 μg of the respective plasmid (pDNA-S1, pDNA-N, or pDNA-R). The efficacy of each siRNA duplex was screened with formulations containing 40 pmol of each duplex, 75 μg exosomes, and 40 μg PEI.
For the S1 expression model, a total of six siRNA candidates were tested, for the initial screen in single sample analysis (Figure S5; Table S3). Three candidates (siRNA-S1-2, siRNA-S1-3, and siRNA-S1-6) were analyzed in biological triplicates. Because a scrambled siRNA sequence was not included in this analysis, we included two (siRNA-S1-2 and siRNA-S1-3) that had shown a high degree of expression knockdown in the initial screen and one (sirRNA-S1-6) that had shown lesser knockdown, to ensure that any modulation of gene expression was specific to the siRNA sequence and not an artifact. The degree of knockdown detected by western blot analysis varied from >80% using siRNA-S1-3 (p < 0.05) to <75% with siRNA-S1-6 (p < 0.05) relative to EPM-pDNA-S1 and sham EPM treatment (Figures 3A and S6). Using PEI alone with siRNA-S1-3 demonstrated a statistically insignificant reduction of approximately 40% protein expression (Figure 3A).
Figure 3.
Analysis of siRNA duplexes efficacy in mediation reduction in expression of antigen levels
EPM was used to deliver siRNA duplexes concurrently with expression constructs encoding viral antigens to HEK293T cells. (A) Three siRNA duplexes were screened for efficiency in knocking down S1 expression; PEI-delivered siRNA was included for comparison. (B) Three siRNA duplexes were screened for efficiency in knocking down N expression. (C) Three siRNA duplexes were screened for efficiency in knocking down RdRp expression. (D) Dose response of S1 siRNA duplex (siR-S1-2) was tested, and a concentration-dependent response was detected. All western blots shown are representative blots of the 3 biological replicates. Data expression as means ± SDs for 3 biological replicates and significance determined by 1-way ANOVA with post hoc comparison of all means to EPM-pDNA for target antigen alone (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). All siRNA inhibition (fold change) is relative to the expression level of the target antigen when delivered by EPM along with the “empty” sham treatment.
For gene knockdown of the N mRNA, three (siRNA-N1, siRNA-N4, and siRNA-N5) of the six siRNA candidates tested demonstrated significant gene knockdown (Figure S7; Table S4). Two siRNA candidates (siRNA-N1 and siRNA-N5) showed >80% knockdown (p < 0.001), while siRNA-N4 demonstrated an approximately 75% gene expression decrease (p < 0.001) (Figures 3B and S8).
Following a similar protocol for viral replicase RdRp, five duplexes were tested for knockdown efficacy (Figure S9; Table S5), with three candidates (siRNA-R1, siRNA-R2, siRNA-R5) significantly reducing protein expression. Two (siRNA-R1 and siRNA-R2) showed nearly 95% knockdown of protein expression (p < 0.001), while the siRNA-5 showed about 85% inhibition (p < 0.001) (Figures 3C and S10).
Dose response of antiviral siRNA in cell culture
A dose response with lower concentrations was used to more completely assess the efficacy of siRNA to modulate target protein expression. Using S1 siRNA as a model, siRNA-S1-2 duplex was tested at 5, 10, and 40 pmol loaded on an EPM formulation of 75 μg exosomes and 40 μg PEI. These doses were co-delivered with the standard S1 expression construct-loaded EPM, 75 μg exosomes, 40 μg PEI, and 1.5 μg pDNA-S1. The highest siRNA dose, 40 pmol, showed the greatest knockdown of protein expression (>80%) but as little as 5 pmol still resulted in nearly 60% knockdown (p < 0.01), indicating that lower doses may be sufficient for physiologically meaningful impairment of viral antigen expression (Figures 3D and S11).
Two additional duplexes, siRNA-S1-3 and siRNA-S1-4, were analyzed at lower concentrations and compared to siRNA-S1-2 for knockdown efficiency. siRNA-S1-4 showed >90% knockdown at doses as low as 5 pmol (Figure S13) and nearly 70% knockdown at a dose as low as 2 pmol (Figure S13). Consequently, this sequence, siRNA-S1-4, was chosen for use in the siRNA cocktail (see below).
Delivery of antiviral antigens simultaneously in vitro
In a viral infection, multiple viral proteins are simultaneously expressed inside the host cell as viral particles are assembled. Thus, a single antigen expression does not accurately model an infection scenario. To reflect infection, plasmids bearing the sequences more accurately for all three viral antigens were delivered simultaneously via EPM formulation (EPM-pDNA-S1NR) to determine whether this model system could be used to express multiple antigens. To optimize this delivery strategy, two alternative methods were tested: (1) loading all three plasmids on the same EPM formulation EPM-pDNA-S1NR or (2) loading each plasmid on an individual formulation prepared separately and treating with one-third of each formulation (3-EPM-pDNA). For EPM-pDNA-S1NR, 75 μg of exosomal protein was incubated with 40 μg of PEI and 1.5 μg of each plasmid. For 3-EPM-pDNA, the standard formulation was prepared for each plasmid, with 75 μg exosomal protein and 1.5 μg pDNA and then one-third of the final formulation of each plasmid mixed together. Each treatment was incubated with HEK293T cells for 48 h and protein expression assessed by western blot. With the exception of RdRp, the protein expression of the viral antigens decreased when multiple plasmids were delivered; however, delivery of the three plasmids on EPM-pDNA-S1NR resulted in a higher expression than plasmids delivered by individual EPM formulations (Figures 4A–4C). For the remainder of the cocktail pDNA studies, the three viral antigen expression plasmids were delivered as a single formulation.
Figure 4.
Simultaneous delivery of multiple expression constructs to produce complex protein combinations
All three plasmids (pDNA-S1, pDNA-N, and pDNA-R), each containing an individual coding sequence for S1, N, or RdRp, respectively, were loaded onto the same EPM formulation. Expression was achieved by transfection with this formulation and to transfection for each viral antigen as an individual plasmid for S1 (A), N (B), and RdRp (C). siRNA was also loaded as a cocktail targeting the most efficacious siRNA duplexes for each individual antigen. EPM loaded with plasmids for all 3 antigens (EPM-pDNA-S1NR) was delivered to HEK293T cells. To test the cross-reactivity of siRNA duplexes, each siRNA duplex was delivered individually. Primary antibodies for each target were used to probe western blots of the same samples for S1 (D), N (E), and RdRp (F). Western blots shown in (D–F) are representative blots from biological triplicates (along with Figures S12–S14), all analyzed for the expression of 3 protein targets, S1, N, and RdRp; (D) and (E) are from the same biological replicate analyzed on the same western blot, probed for both S1 and N, so the same β-actin is shown for both. (F) A second blot analyzed for the same set of samples as (D) and (E) using RdRp primary antibody. Data expression as means ± SDs for at least 3 biological replicates and significance determined by 1-way ANOVA with post hoc comparison of all means to EPM-pDNA for target antigen alone. (∗p < 0.05, ∗∗p < 0.01). All of the siRNA inhibition (fold change) is relative to the expression level of the target antigen when delivered by EPM along with the “empty” sham treatment.
Delivery of antiviral siRNA cocktail in vitro
Using the simultaneously delivery of multiple viral antigens via EPM as the model system, a cocktail of siRNA was also delivered to determine whether multiple siRNA sequences could be effectively delivered using the EPM nanoplatform for successful gene knockdown.
For this experiment, EPM-pDNA formulations delivered all three viral antigens to HEK293T cells. To remove the effect of any non-specific knockdown, EPM-siRNA with only a single duplex was tested against all three antigens. For the single-sequence siRNA formulation, 40 pmol of the specific siRNA duplex was loaded. For the siRNA cocktail, 40 pmol of each test candidate was loaded, for a total of 120 pmol. Each antigen was analyzed individually via western blot.
All three siRNA duplexes were able to demonstrate gene knockdown of the target antigen when delivered as a siRNA cocktail. The decrease in expression was slightly less than when the siRNA candidates were delivered individually, but not statistically significant. While siRNA-S1-4 delivered alone resulted in an 80% knockdown of S1 expression (p < 0.01), when delivered as part of the siRNA cocktail, gene expression decreased by almost 70% (p < 0.05) (Figures 4D and S14). The siRNA-N1 delivered alone by EPM decreased N expression by >95% (p < 0.05), with a slight decrease in efficiency when siRNA-N1 was delivered as part of the cocktail, resulting in almost 80% knockdown of N expression (p < 0.05) (Figures 4E and S15). Finally, siRNA-R1 alone knocked down RdRp expression by nearly 90% (p < 0.05) and nearly 60% when delivered as part of the siRNA cocktail (p < 0.05) (Figures 4F and S16). Of note, no off-target gene knockdown was observed across the three viral antigens. For example, co-incubation of the EPM-pDNA-S1NR with siRNA-S1-4 did not result in the decrease of either N or RdRp expression (Figures 4D–4F). A similar lack of cross-reactivity was observed with both siRNA-N1 and siRNA-R1.
Target antigens can be delivered in vivo
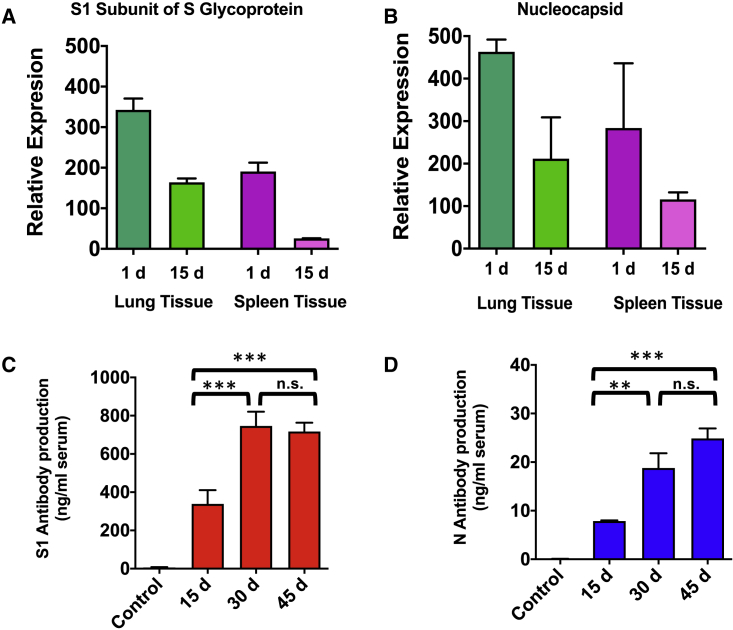
To meet the goal of establishing a model for testing antiviral therapeutics in BSL2 containment, production of viral antigens using the EPM delivery system needed to be demonstrated in an animal model. For application in vivo, 10 μg of each plasmid was loaded onto 75 μg of exosomal protein with 40 μg of PEI. For qRT-PCR experiments, total RNA was isolated from approximately 50 mg of lung and spleen tissues. After Trizol isolation of RNA, quality and concentration were assessed (Tables S6 and S7). EPM introduction of the expression constructs, the mRNA of two antigens, S1, and N could be detected by qRT-PCR in both the lung and spleen 1 d after the last dose. Higher expression of the S1 mRNA was found in the lung as compared to the spleen (Figure 5A), as was the case for N mRNA (Figure 5B). Fifteen days after the last dose, mRNA for both S1 and N were still detectable in both the lung and spleen (Figures 5A and 5B). A similar protocol was followed for the introduction of the viral replicase, RdRp, in the murine model. Expression of this protein could also be detected in the lung and spleen of treated mice (Figure S17). The viral replicase is an important target for therapeutics; however, as it is not highly antigenic, additional studies were not pursued with this protein.
Figure 5.
Expression of viral antigens in a mouse model
Female BALB/C wild-type mice were treated with EPM-pDNA (10 μg/dose) in 3 groups: EPM-pDNA-S1 (n = 4), EPM-pDNA-N (n = 4), and EPM alone (n = 3). Mice were dosed intravenously for 5 consecutive days and then euthanized at either 1 or 15 d following the last dose. (A) S1 expression could be detected in both the lung and spleen at both time points, although higher expression was seen 1 d after final dose. (B) N expression followed a similar pattern, with higher expression in the lung compared to the spleen. Data represent averages ± SDs (n = 4). Relative expression determined using the Pfaffl method. (C) Antibody production following introduction of viral antigens via EPM-pDNA. S1-specific antibodies were detected 15 d after the last dose, increased after 30 d, and remained at the same level after 45 d. (D) N-specific antibodies were detected 15 d post-inoculation and increased up to 45 d. Antibody production is compared to the 15-d detection in both cases. Data for both S1 and N antibody detection represent averages ± SDs (n = 3).
EPM delivery of SARS-CoV-2 S1 glycoprotein and the N protein results in production of antigen-specific antibodies in a murine model
As an indirect means to determine whether antigens produced in the mouse were immunogenically identical to the antigens produced by the live viral particles, BALB/C wild-type mice were injected with pDNA for expression of the most immunogenic proteins, S1 and N, using the EPM model system every other day for three doses (10 μg pDNA per protein per dose), and blood samples were taken every 15 d for up to 45 d to detect S1- and N-specific antibodies using enzyme-linked immunosorbent assays (ELISAs). Both the S1- and N-specific immunoglobulin G (IgG) antibodies were detected as early as 15 d after the last EPM-pDNA injection. The S1-specific antibody response increased from 15 to 30 d (p < 0.001) but remained the same at 45 d post-inoculation (Figure 5C). However, the N-specific antibody response increased by >2-fold from 15 to 30 d (p < 0.001), but after that, the increase was only modest (Figure 5D).
Exosomes can be functionalized with LF
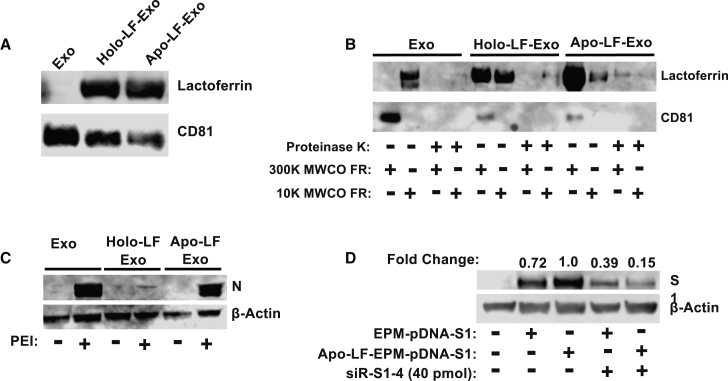
Here, we demonstrate that LF can be loaded onto colostrum exosomes and does not interfere with the delivery of nucleic acid payload in vitro. LF exists in two different conformations as a result of interaction with iron ions, the iron-rich hololactoferrin (holo-LF), and the iron-depleted apolactoferrin (apo-LF). After incubation and subsequent molecular weight cutoff (MWCO) filtration, both apo-LF and holo-LF were successfully loaded onto exosomes (Figure 6A). Exosomal marker CD81 detection was used to confirm that the LF was associated with the exosomal particles, not free in solution. To determine whether LF was surface associated with exosomes, we treated LF-Exo formulations, along with unloaded exosomes, with proteinase K. Proteinase K treatment resulted in almost complete loss of LF and CD81 (surface bound) from both the LF-Exo preparations and the control exosomes (Figure 6B). The loaded LF appeared to be more stably associated when apo-LF was used, compared to holo-LF, as LF appeared in the flow through of the 300-K MWCO filter, even in the absence of proteinase treatment (Figure 6B).
Figure 6.
Functionalization of exosomes with lactoferrin (LF)
(A) LF, both iron-saturated (holo) and iron-depleted (apo) was loaded onto exosomes following brief co-incubation as detected by western blot analysis. (B) The association of LF with exosomes is surface bound, as evidenced by the removal of LF with proteinase K treatment. (C) apo-LF exosomes complex with PEI serves as a nanoplatform to delivery pDNA-N to HEK293T cells; the holo-LF exosomes delivered pDNA-N show significantly lower antigen expression. (D) Delivery of siRNA by apo-LF-EPM in vitro, resulting in comparable knockdown of target gene expression as non-functionalized EPM.
Functionalization of exosomes with LF does not interfere in nucleic acid delivery in vitro
To determine whether LF-functionalized EPM can deliver a coding sequence, LF-EPM and EPM were ionically complexed with pDNA encoding the N. Because LF is a positively charged moiety, formulations were tested with and without PEI to determine whether LF alone could associate nucleic acids with exosomes. Transfection of HEK293T showed that EPM and apo-LF-EPM resulted in a similar expression level of the N protein; however, expression was markedly lower from the holo-LF-EPM (Figure 6C). Consequently, apo-LF was used for the remainder of the LF studies. Although LF is positively charged, LF-Exo was not capable of entrapping and delivering nucleic acid, and therefore PEI is necessary to establish the ionic complex (Figure 6C).
Apo-LF-EPM could also successfully deliver siRNA for the purpose of gene knockdown. As proof of principle, siRNA-S1-4 was loaded onto both EPM and apo-LF-EPM and co-incubated with EPM-pDNA-S1. Apo-LF-EPM-delivered siRNA-S1-4 showed nearly 85% knockdown of the S1 glycoprotein, compared to just over 60% knockdown of the EPM-delivered siRNA-S1-4 compared to the same control (Figure 6D). These data indicate that LF-functionalized exosomes could deliver nucleic acids as part of the EPM complex and enhance efficacy presumably due to higher cell uptake. The addition of LF as a targeting ligand could therefore enhance the application of this model system by increasing trafficking to cells more likely to be easy access points for viral particles.
Discussion
Despite efforts to prevent and contain infectious diseases, several emerging viral diseases, such as Ebola, H1N1 swine flu, Middle East respiratory syndrome (MERS), SARS, Zika, and the current pandemic, SARS-CoV-2, have plagued humanity. Mitigating the outbreaks of these viral diseases has been accompanied by the added complexity of handling these pathogens exclusively in BSL3 containment. Compared to lower biosafety laboratories (BSL2), the number of BSL3 facilities is significantly fewer, and therefore dramatically curtails research efforts to develop vaccines and therapeutics. The rate of growth and development in the field of genetics and bioinformatics has greatly enhanced the speed and accessibility to sequence information about viral pathogens. The first genome of the novel coronavirus was published on January 10, 2020,37 <1 month after the first case was reported in Wuhan, China. With the toolbox available for modern scientists, this sequence information from a virus requiring BSL3 containment can be converted to innocuous nucleic acids that can be used in a lower safety level viral model system. Moreover, as soon as genomic sequences become available for new emerging variants, the model can be modified to encompass those new variants by changing the nucleic acid sequence of the expression plasmids and/or siRNA. Here, we present an alternative model for developing antiviral therapeutics using BSL2 level containment by producing select viral antigens and screening viral therapeutics via EPM technology in both in vitro and in vivo systems. Although this system may not apply to the screening of all of the potential viral therapeutics, it will expand the breadth of model systems available at lower biosafety containment.
Using SARS-CoV-2 as a model virus, we expressed three different viral proteins, the S1 subunit of the spike glycoprotein, the N, and the replicase both in vitro and in vivo, using plasmids delivered by EPM technology. The expression of these viral proteins in vivo was sufficient to evoke the production of antibodies specific to viral antigens produced for the purpose of antibody detection (i.e., not proteins expressed from the same plasmid constructs), indicating that this model system could produce viral proteins with the same antigenic potential. In vitro, several different siRNA duplexes against all three targets were screened in a BSL2 facility, without the need for BSL3 containment.
The EPM technology is premised on the isolation of nanoparticles from bovine colostrum, a readily available, biocompatible, and scalable source of exosomes.18 The use of exosomes as delivery vesicles for nucleic acids for the purpose of gene therapy is not new, but the sources of these exosomes may hinder the progress of the technology. Bovine colostrum contains exosomal particles several orders of magnitude higher abundance compared to those sourced from mammalian cell culture.20 Exosomal formulations can then be made using PEI and nucleic acid, producing the EPM platform.18 These formulations do not demonstrate detectable cytotoxicity, and we have previously shown the lack of systemic toxicity of functionalized EPM.38 While PEI can produce toxicity when used alone as a transfection reagent, that cytotoxicity is mitigated in the presence of exosomes, potentially because of the decreased concentration of PEI in the EPM complex necessary to achieve the efficient transfection. In this study, we demonstrated that coding sequences for viral antigens can be delivered either individually or in combination for the expression of these proteins in vitro and in vivo. While the expression of these antigens is innocuous as it is in absence of the complete viral particle, the proteins themselves are still immunogenic, as demonstrated by the production of specific antibodies to the introduced viral targets. This finding supports the use of the EPM technology as a delivery platform for nucleic acid-based vaccines.
The delivery of these antigens in our model system is via pDNA, and the SARS-CoV-2 virus itself is an RNA virus. The expression of the viral antigens in this manner allows the exploration of therapeutics at both the levels of RNA and protein. This model system could be used for the expression and targeting of any viral antigen, whether RNA or DNA virus, as the intermediates produced in infected cells are biologically similar. The pDNA is transported into the cell via the EPM, released in the cytoplasm, and then travels to the nucleus for transcription. The resulting mRNAs are transferred back to the cytoplasm, where RNA from viruses would also be present in a virally infected host. The presence of these antigen-specific mRNA in the cytoplasm gives us the opportunity to test therapeutic agents such as siRNA. We screened several siRNA sequences for each antigen independently and then demonstrated the capacity of the EPM nanoplatform to deliver these nucleic acid duplexes either individually or together in vitro. As the efficacy of an siRNA duplex against a specific mRNA target is based on the sequence of each, this interaction and the efficacy of gene knockdown by any particular siRNA can be assessed using EPM delivery in vitro before determination of the siRNA efficiency at viral replication inhibition. The in vitro model with EPM-delivered nucleic acids can allow for high-throughput screening of many siRNA sequences before testing the most effective sequences against the live virus. As such, the majority of the work could be done in BSL2 level containment.
In addition, the evidence of production of antibodies in the murine model specific to the SARS-CoV-2 antigens indicates the efficacy of this particular model system for testing therapeutics in the absence of the viral particles, as well as the development of a new strategy for vaccine delivery. As was found in the SARS-CoV-2 pandemic, many of the individuals most susceptible to the virus had underlying health conditions that potentially weakened their immune systems. As such, some of the other delivery platforms for nucleic acid vaccines, such as viral vectors, may not be effective in these individuals. The EPM nanopolyplex provides a biocompatible, targetable modality for delivering nucleic acid-like siRNA and DNA for the purpose of disease intervention.
Importantly, the plasticity of the EPM delivery system allows for high-throughput screening of siRNA-based antiviral therapeutics, even as new viral variants emerge. By simply changing the coding sequence of the expression plasmid based on new genomic information, putative siRNA duplexes can be screened in a few days, decreasing the time necessary to identify therapeutic siRNA candidates. Certainly, the live virus is the true test of the efficacy of any therapeutic but much of the initial screening to narrow down the pool of therapeutics that are tested in animal and live virus models could be completed at lower containment at a faster pace.
Finally, the versatility of the EPM technology allows for the addition of targeting ligands to the platform that can enhance delivery of antiviral therapeutics such as siRNA to the affected tissues. LF has long been known to be antiviral and antibacterial, acting as a modulator of the immune system and sequestering iron.30 New evidence suggests that cell surface-associated LF receptors, specifically heparin sulfate proteoglycans (HSPGs) are the entry points for viruses such as SARS-CoV-2,39 as well as a host of others, including human papillomavirus (HPV),40 and HIV.41 Functionalizing the EPM platform with LF increasing the selective delivery of the antiviral therapeutics, such as siRNA, to virally infected respiratory cells, for the virus in the current model, SARS-CoV-2 and other viruses of public health interest that extort LF receptors to make entry into host cells. As with the overall model developed, targeted delivery of functionalized exosomes could be examined in a BSL2 facility and optimized, before the use of live virus models.
The use of siRNA to treat respiratory diseases is not novel, but it has had trouble making it from the bench into the clinic. Despite many attempts to deliver either naked siRNA or siRNA delivered by liposomes or liponanoparticles, no antiviral siRNA has as yet made it to market.36 Clinical trials have been started and abruptly ended as a result of the lack of efficacy.36 However, the ability of mutants, such as the Omicron variant of SARS-CoV-2, to overcome both vaccine-acquired and naturally acquired immunity, indicate that available vaccines may not signal the end of the pandemic. The delivery of siRNA to a respiratory pathogen does present hurdles that require an effective delivery modality. The lung provides a challenging environment for the update of delivery particles, including the presence of alveolar macrophages that clear the airways of any foreign particles. In addition to its versatility in a model system, the EPM-based siRNA delivery system holds much promise as a therapeutic because of its ability to avoid engulfment by cells of the immune system.
In short, the use of the EPM nanoplatform as a delivery vehicle for nucleic acids broadens the approach for the study of antiviral therapeutics. The EPM technology itself has shown no toxicity in wild-type mice over 28 d of treatment.18 The matrix has the ability to deliver multiple nucleic acid sequences both in vitro and in vivo, facilitating the study of viral antigens as individual proteins and mRNA sequences, rather than as part of the infectious viral particle. Ultimately, each therapeutic will have to be tested against the live virus; however, the use of the EPM delivery platform for nucleic acids will facilitate the increased speed of early efficacy screening at a lower biosafety level, permitting the faster and more efficient development of antiviral treatments and therapies. The EPM platform, based on naturally trafficking vesicles, also lends itself to use as a therapeutic delivery system for siRNA because of its versatility, extended time in circulation, and ability to be modified to target specific organs.
Materials and methods
Preparation of plasmids for viral antigen expression
Plasmids containing the coding sequences for the spike glycoprotein subunit S1, N, and RdRp were obtained from Sino Biological (Wayne, PA). Coding sequences for each of the listed antigens were amplified by standard PCR. In a typical reaction, 0.5 μg plasmid was used as a template for DreamTaq polymerase (Thermo Fisher Scientific). The production of amplicons was confirmed by gel electrophoresis. Upon confirmation, 4 μL of the PCR reaction was added to the linearized mammalian expression vector pcDNA 3.3 TOPO-TA (Thermo Fisher Scientific). Inserted fragments were sequenced to ensure proper orientation. If cloned into the vector correctly, then viral antigen expression was under the control of the cytomegalovirus (CMV) promoter to achieve constitutive expression. Plasmids were maintained in bacterial culture and isolated in bulk for transfection studies. A similar procedure was followed to create an expression plasmid for the full-length spike glycoprotein; however, due to lower expression level, likely because of the larger size of the protein and the concentration of epitope determinants in the S1 subunit of this protein containing the receptor binding domain, we chose to concentrate on the smaller glycoprotein.
Design of siRNA
Putative siRNA duplexes targeting viral antigens encoded in the above plasmids were designed using the Custom Dicer-Substrate siRNA design tool from Integrated DNA Technologies (Coralville, IA). The complete coding sequence from each antigen was input into the design tool, and based on the algorithm of the tool, several duplexes were designed. Duplexes complementary to a variety of locations in the coding sequence were selected (some close to the 5′ end of the mRNA, others in the middle, and some close to the 3′ end) for testing. For this model design, siRNA duplexes with unmodified phosphate backbones were used as they have shown the same efficacy of gene knockdown as demonstrated by siRNA duplexes with backbone modifications in our previous work.18
Loading of nucleic acid in the EPM
Bovine exosomes were harvested from colostrum powder by differential centrifugation as described for milk exosomes,21 with modifications.18 Briefly, colostrum powder was rehydrated in deionized water to a final concentration of 5% w/v. Exosomes were isolated by three centrifugations (13,000 × g, 30 min; 65,000 × g, 60 min; and 135,000 × g, 2 h). The resultant pellet was resuspended in PBS and protein concentration determined using Pierce BCA Protein Assay (Thermo Fisher Scientific). The exosomal suspension was stored at a concentration of 8 mg/mL at −80°C until use. All of the components of the EPM formulation were passed through a 0.22-μm syringe filter for sterilization before mixing. Formation of the exosomal complex with plasmid DNA (EPM-pDNA) or with siRNA duplexes (EPM-siRNA) was accomplished by incubating the nucleic acids with exosomes and PEI of mol wt 60,000 (PEI-60K) (Ethylenimine Technologies, Lake Wylie, SC). Reaction components typically consisted of 75 μg exosomes and 0.025% PEI in 150 μL PBS. After the incubation of exosomes with PEI at ambient temperatures, 1.5–10 μg pDNA or 5 to 40 pmol siRNA was added and incubation continued. Precipitation of the complex and removal of free PEI was achieved by adding polyethylene glycol (Sigma-Aldrich, St. Louis, MO), mol wt 2,000 (PEG-2K), and incubating at 4°C for 1 h. The EPM-pDNA or EPM-siRNA was harvested by centrifugation at 13,000 × g for 15 min. The resulting pellet was suspended in PBS (pH 7.4).
Preparation of 32P-labeled pDNA
A control plasmid (pEMP) lacking a sequence to be expressed was used for entrapment studies. pEMP was digested using OliI (AleI) enzyme (Thermo Fisher Scientific) following the manufacturer’s instructions. The digested plasmid was 5′-32P labeled with T4 polynucleotide kinase and [γ-32P] ATP (>6,000 Ci/mmol) using a molar excess of the pDNA. Any residual ATP was removed using Illustra ProbeQuant G-50 Micro Column (GE Healthcare, Chicago, IL), and the labeled pDNA was purified by a plasmid isolation kit (New England Biolabs, Ipswich, MA). The purity of the labeled pDNA was determined by polyacrylamide gel electrophoresis.
Determining loading efficiency of pDNA onto EPM
To determine the entrapment and loading of pDNA in the EPM, 32P-labeled pDNA was used as a tracer along with non-radioactive pDNA. Loading/entrapment reaction of the pDNA in EPM was performed as described for siRNA and pDNA-entrapped exosomes were collected by ExoQuick (SBI Biosciences, Palo Alto, CA) precipitation followed by high-speed centrifugation. The total formulation volume, including exosomes, PEI, pDNA, and precipitating reagent was 180 μL. After precipitation, each formulation was centrifuged at 13,000 × g for 15 min. The supernatant (180 uL) was collected, and the resultant pellet resuspended in 15 μL PBS. One-third (5 μL) of the pellet and 1/10 (18 μL) of the supernatant were spotted on PEI cellulose for analysis. Entrapment efficiency was calculated by the distribution of the radioactivity present in the EPM-pDNA recovered by precipitation and in the supernatant and the radioactivity quantitation using ImageJ (NIH) software. The total volume of the percentage of entrapment and loading efficiencies of pDNA were calculated as follows:
Cell culture
Human embryonic kidney cells HEK293T (American Type Culture Collection [ATCC], Manassas, VA) were cultured in DMEM high glucose (Gibco, Waltham, MA) supplemented with 10% FBS and 2 mM l-glutamine and grown at 37°C in 5% CO2.
In vitro measurement of cytotoxicity
HEK293T cells were plated at a density of 7.5 × 104 cells per well in a 24-well plate and allowed to attach. Formulations (prepared as described above), 40 μg PEI 60K, or 75 μg exosomes were added directly to the culture media. Incubation was done in 1.5 mL culture media for 48 h, and then cells were harvested for the detection of viability. Cells were detached and washed with PBS before resuspension in culture media. Equal volumes of trypan blue and cell suspension were mixed briefly by pipetting and cell viability assessed using a Countess 3 Cell Counter (Thermo Fisher Scientific). Cell survival was calculated as a percentage of treated cells alive compared to untreated cells. Treatments were performed in biological triplicate. Three biological replicates were analyzed using a one-way ANOVA followed by Dunnett’s post hoc analysis, compared to the untreated control.
In vitro transfection of EPM-siRNA and EPM-pDNA
In vitro, exogenous gene expression experiments were performed in HEK293T cells. Cells were seeded in 12-well plates at a density of 1.5 × 105 cells per well, allowed to attach, and treated with EPM-pDNA alone for expression experiments or EPM-pDNA and EPM-siRNA for knockdown experiments for 48 h. Nucleic acid loads varied by application, including 0.5–4.5 μg pDNA or 10–40 pmol siRNA duplex. For the preparation of some formulations, multiple pDNA or multiple siRNA complexes were loaded at similar doses. Additional controls, such as EPM (without nucleic acid), PEI-siRNA, and exosomes alone, were also included. Protein lysates were prepared from transfected cells and protein expression analyzed by western blot to determine the expression level of introduced viral antigens and knockdown of targets.
Western blot analysis
Whole-cell lysates were analyzed for the expression of the viral antigen of interest as described previously.24 Briefly, treated cells were detached from culture dishes using 0.25% trypsin-EDTA (Thermo Fisher Scientific) and lysed using radioimmunoprecipitation assay (RIPA) buffer containing phosphatase/protease inhibitors (Thermo Fisher Scientific). Total protein, 20–30 μg, was loaded in polyacrylamide gels under reducing conditions. Two different primary antibodies for the spike glycoprotein were obtained from Sino Biological, one polyclonal and one monoclonal antibody. The polyclonal antibody produced some nonspecific detection in the untreated samples that was ameliorated with the use of the monoclonal antibody. A monoclonal antibody for the N was also obtained from Sino Biological. A polyclonal antibody for the RdRp was purchased from GeneTex (Irvine, CA). β-Actin was used as a loading control and the antibody monoclonal (Cell Signaling Technology, Danvers, MA). Lactoferrin and CD81 antibodies were purchased from Santa Cruz Biotechnologies (Dallas, TX). Secondary antibodies were conjugated with IRDye 800CW and 680RD. Blots were imaged using Image Studio for LI-COR Odyssey CLx (LI-COR Biotechnologies, Lincoln, NE) and quantified with Empiria Studio software (LI-COR Biotechnologies).
Application of EPM-pDNA in vivo for detection of antigens
All animals were maintained according to the Institutional Animal Care and Use Committee (IACUC) guidelines. For introduction of viral antigenic proteins, following acclimation, 5- to 6-week-old female BALB/C wild-type mice (Charles River Laboratories, Indianapolis, IN) were randomized into 3 groups. Three groups of animals received EPM-pDNA (10 μg/dose) formulations bearing the coding sequence for a single viral protein, EPM-pDNA-S1 (n = 4) and EPM-pDNA-N (n = 4); one group (n = 3) received vehicle (EPM) alone intravenously daily for 5 d. Seven days following the last dose, animals were euthanized and select organs (liver, lung, kidney, and spleen) were harvested for analysis of viral antigen expression by qRT-PCR. Harvested organs were snap frozen in liquid nitrogen and then the total RNA was isolated.
qRT-PCR for target gene expression
Total RNA was isolated from tissues using the Trizol (Invitrogen, Waltham, MA) method. Approximately 50 mg of each tissue of interest was homogenized in Trizol and chloroform phase separation was used to separate RNA from DNA. Quality and concentration of total RNA was assessed using Cytation C5 Confocal & Widefield Microscopy with Multi-mode Plate Reading (BioTek, Winooski, VT) before qRT-PCR.
One-Step SYBR green qRT-PCR kit (Quanta Biosciences, Gaithersburg, MD) was used to perform cDNA synthesis and PCR amplification from 50 ng total RNA according to the manufacturer’s instructions. The kit includes Moloney murine leukemia virus (MMLV) reverse transcriptase for cDNA synthesis and AccuStart Taq DNA polymerase for PCR amplification. Primer stocks were maintained at 10 μM and all of the RNA samples were diluted to 25 ng/μL. A typical reaction included 12.5 μL SYBR Green master mix, 8 μL RNase-free water, 0.5 μL 50× MMLV reverse transcriptase, 1 μL of each primer stock, and 2 μL diluted RNA. qRT-PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems, Bedford, MA) and Ct measured using 7500 Software. Following a 10-min incubation at 45°C for cDNA synthesis, initial denaturation was at 95°C for 10 min, followed by 40 amplification cycles (95°C for 15 s, 60°C for 1 min). The melt curve proceeded from 60°C to 95°C with a 1% increase every 30 s.
Transcript detection of introduced viral sequences was normalized to endogenous β-actin expression (primer sequences in Table S2) and analyzed using the ΔΔCT method. Reactions were performed in technical triplicate for biological triplicate samples.
Application of EPM-pDNA in vivo for production of antigen-specific antibodies
For the detection of antibody production to the S1 antigen, female BALB/C mice (5- to 6-week-old) were randomized into 2 groups (n = 4) and received EPM alone (group 1) and EPM-pDNA-S1 + EPM-pDNA-N (group 2). All of the regimens were administered intravenously three times per week (10 μg pDNA/antigen/dose). Animals were provided purified AIN-93M diet and water ad libitum. Body weight gains, diet intake, and overall animal physical health were monitored weekly. Fifteen days after the final dose, blood was collected via jugular vein and blood collection was repeated every 15 d for a total of 45 d post-inoculation. At the end of the study, animals were euthanized by CO2 asphyxiation. Blood and select organs (lung, liver, spleen, and kidneys) were collected and stored at −80°C for further analysis.
Detection of antibodies from murine serum
Murine serum was separated from blood samples collected at different time points. Since the commercial kits for the detection of antibody in rodents are not available, we developed an ELISA assay for the detection of antibodies. Briefly, S1 (NR-52366) and N (NR-48761) Antigens (100 ng/well in PBS) were used to coat the high-sorb plate (Thermo Fisher Scientific), and allowed to absorb overnight at 4°C. The next day, the plate was washed four times with wash buffer (PBS containing 1% Tween 20 [PBST]), and coated wells were blocked with blocking buffer (3% bovine serum albumin [BSA]) at room temperature for 1 h on a slow-speed platform orbital. Following blocking, plates were washed 4 times with PBST and 100 μL of mouse sera diluted with 3% BSA at 2 different dilutions (100X and 250X) was added to each well and incubated for 1 h at room temperature. The dilution of sera was based on the commercial antibody standard curve, and no effect of the dilution factor was noted at the used dilutions. Commercial antibodies for S1 and N were serially diluted in blocking buffer, and different concentrations were used to generate the standard curve. After four washes, HRP-conjugated anti-mouse IgG antibody (Cell Signaling Technology), at a dilution of 1:5,000 in blocking buffer was added to each well and incubated again for 1 h at room temperature. After washing the plate 4 times, 100 μL KPL SureBlue TMB substrate (Sera Care Life Sciences, Milford, MA) was added and incubated for 10 min. The reaction was stopped by adding 100 μL 2 M H2SO4 and the plate was read at 450 nm using a SpectraMax spectrophotometer.
Functionalizing exosomes with LF
To load bovine LF (Professional Whey, Erina, Australia), exosomes were briefly incubated with bovine LF (2:1, w/w). Both holo-LF (iron containing) and apo-LF (iron depleted) were tested; apo-LF was prepared by acidifying holo-LF to pH 4.0 in the presence of ethanol (2.5%).42 The unbound LF was removed by ultrafiltration using 300 K MWCO filter. To confirm exosomal loading of LF, the 300 K MWCO filter-retained (FR) material was analyzed by western blot. To determine whether LF-functionalized EPM can deliver a coding sequence, LF-EPM and EPM were ionically complexed with pDNA in the same manner as described for exosomes previously. LF-EPM were also complexed with siRNA to determine the efficiency of gene knockdown with siRNA delivered by this platform. For siRNA intervention with viral antigen expression, the “empty” LF-EPM was used as a sham treatment to determine the basal level of protein expression.
Characterization of LF-loaded exosomes
To determine whether LF was surface bound or internalized during loading, LF-loaded exosomes were treated with proteinase K (Roche, Basel, Switzerland). Briefly, colostrum exosomes, apo-LF-loaded exosomes, and holo-LF-loaded exosomes were diluted to 0.5 mg/mL, incubated with proteinase K (10 μg/mL) for 45 min at room temperature. After incubation, the reaction was passed through a 300-K MWCO filter twice to separate digested peptides from exosome particles. Retentate (FR) samples were analyzed by western blot. Flowthrough was concentrated using 10 K MWCO filters and the FR was used for analysis.
Statistical analysis
Statistical analysis was performed using Graphpad Prism version 8.0 (La Jolla, CA). Data are presented as means ± SDs of three to four replicates. One-way ANOVA followed by post hoc analysis compared to control was used to compare treatment groups to control. Details of specific statistical analyses appear in each figure legend. All western blot data were quantified by Empiria Studio software. Signal from the target protein as normalized to β-actin expression and treatment compared to control. In each assay, a p-value of <0.05 was considered significant.
Acknowledgments
This work was supported by funds from 3P Biotechnologies, Louisville, KY and, in part, from NCI SBIR grants (R44-CA-221487 and R41-OD-031942) and the Agnes Brown Duggan Endowment (to R.C.G.). The authors would also like to thank Sarah Wilcher for her assistance with the animal studies. The viral antigen NR-52366 was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein Receptor Binding Domain (RBD) from SARS-related coronavirus-2, Wuhan-Hu-1 with C-terminal histidine tag, recombinant from HEK293F cells. The viral antigen NR-48761 was obtained through BEI Resources, NIAID, NIH: SARS-CoV nucleocapsid (N) protein, recombinant from Escherichia coli, NR-48761. The graphical abstract was created with Biorender.com.
Author contributions
M.W. created plasmids, designed siRNA, conducted cell culture experiments, and wrote the manuscript. F.A. designed and supervised in vivo studies and contributed to data analysis. R.K. and N.T. conducted in vivo studies. J.J. isolated and characterized exosomes for formulation preparation. S.A. assisted with in vitro experiments. D.J.S., W.S., and R.C.G. contributed to the conceptualization of experiments, data analysis, and critique of manuscript.
Declaration of interests
R.C.G. holds positions both at the University of Louisville and 3P Biotechnologies. The authors have filed an international patent application (PCT) based on part of the results reported in this paper.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.08.011.
Supplemental information
Document S2. Article plus supplemental information
Data availability
The authors confirm that data supporting the findings of this research are available within the body of the article or in supplemental material.
References
- 1.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 3.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S., Takanashi M., Sudo K., Ueda S., Ishikawa A., Matsuyama N., Fujita K., Mizutani T., Ohgi T., Ochiya T., et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitko V., Musiyenko A., Shulyayeva O., Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Lu Z., Wientjes M.G., Au J.L. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shayakhmetov D.M., Li Z.Y., Ni S., Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J. Virol. 2004;78:5368–5381. doi: 10.1128/jvi.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muruve D.A. The innate immune response to adenovirus vectors. Hum. Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 9.Simberg D., Weisman S., Talmon Y., Barenholz Y. DOTAP (and other cationic lipids): chemistry, biophysics, and transfection. Crit. Rev. Ther. Drug Carrier Syst. 2004;21:257–317. doi: 10.1615/critrevtherdrugcarriersyst.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- 10.Zuhorn I.S., Visser W.H., Bakowsky U., Engberts J.B., Hoekstra D. Interference of serum with lipoplex-cell interaction: modulation of intracellular processing. Biochim. Biophys. Acta. 2002;1560:25–36. doi: 10.1016/s0005-2736(01)00448-5. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 13.Grigor'eva A.E., Tamkovich S.N., Eremina A.V., Tupikin A.E., Kabilov M.R., Chernykh V.V., Vlassov V.V., Laktionov P.P., Ryabchikova E.I. [Characteristics of exosomes andmicroparticles discovered in human tears] Biomed. Khim. 2016;62:99–106. doi: 10.18097/PBMC20166201099. [DOI] [PubMed] [Google Scholar]
- 14.Milasan A., Tessandier N., Tan S., Brisson A., Boilard E., Martel C. Extracellular vesicles are present in mouse lymph and their level differs in atherosclerosis. J. Extracell. Vesicles. 2016;5:31427. doi: 10.3402/jev.v5.31427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlotogorski-Hurvitz A., Dayan D., Chaushu G., Korvala J., Salo T., Sormunen R., Vered M. Human saliva-derived exosomes: comparing methods of isolation. J. Histochem. Cytochem. 2015;63:181–189. doi: 10.1369/0022155414564219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akers J.C., Ramakrishnan V., Kim R., Skog J., Nakano I., Pingle S., Kalinina J., Hua W., Kesari S., Mao Y., et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8:e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oldenborg P.A. CD47: a cell surface glycoprotein which regulates multiple functions of hematopoietic cells in health and disease. ISRN Hematol. 2013;2013:614619. doi: 10.1155/2013/614619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munagala R., Aqil F., Jeyabalan J., Kandimalla R., Wallen M., Tyagi N., Wilcher S., Yan J., Schultz D.J., Spencer W., et al. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. 2021;505:58–72. doi: 10.1016/j.canlet.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berlutti F., Pantanella F., Natalizi T., Frioni A., Paesano R., Polimeni A., Valenti P. Antiviral properties of lactoferrin--a natural immunity molecule. Molecules. 2011;16:6992–7018. doi: 10.3390/molecules16086992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendt M., Kamerkar S., Sugimoto H., McAndrews K.M., Wu C.C., Gagea M., Yang S., Blanko E.V.R., Peng Q., Ma X., et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munagala R., Aqil F., Jeyabalan J., Gupta R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371:48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal A.K., Aqil F., Jeyabalan J., Spencer W.A., Beck J., Gachuki B.W., Alhakeem S.S., Oben K., Munagala R., Bondada S., et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine. 2017;13:1627–1636. doi: 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Munagala R., Aqil F., Jeyabalan J., Agrawal A.K., Mudd A.M., Kyakulaga A.H., Singh I.P., Vadhanam M.V., Gupta R.C. Exosomal formulation of anthocyanidins against multiple cancer types. Cancer Lett. 2017;393:94–102. doi: 10.1016/j.canlet.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aqil F., Munagala R., Jeyabalan J., Agrawal A.K., Kyakulaga A.H., Wilcher S.A., Gupta R.C. Milk exosomes - natural nanoparticles for siRNA delivery. Cancer Lett. 2019;449:186–195. doi: 10.1016/j.canlet.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Feng X., Chen X., Zheng X., Zhu H., Qi Q., Liu S., Zhang H., Che J. Latest trend of milk derived exosomes: cargos, functions, and applications. Front. Nutr. 2021;8:747294. doi: 10.3389/fnut.2021.747294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hata T., Murakami K., Nakatani H., Yamamoto Y., Matsuda T., Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem. Biophys. Res. Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 27.Longo P.A., Kavran J.M., Kim M.S., Leahy D.J. Transient mammalian cell transfection with polyethylenimine (PEI) Methods Enzymol. 2013;529:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C., Tang T., Uludag H., Cuervo J.E. Molecular dynamics simulations of DNA/PEI complexes: effect of PEI branching and protonation state. Biophys. J. 2011;100:2754–2763. doi: 10.1016/j.bpj.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kafil V., Omidi Y. Cytotoxic impacts of linear and branched polyethylenimine nanostructures in a431 cells. Bioimpacts. 2011;1:23–30. doi: 10.5681/bi.2011.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kell D.B., Heyden E.L., Pretorius E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front. Immunol. 2020;11:1221. doi: 10.3389/fimmu.2020.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravichandran S., Coyle E.M., Klenow L., Tang J., Grubbs G., Liu S., Wang T., Golding H., Khurana S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta A., Michler T., Merkel O.M. siRNA therapeutics against respiratory viral infections-what have we learned for potential COVID-19 therapies? Adv. Healthc. Mater. 2021;10:e2001650. doi: 10.1002/adhm.202001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandimalla R., Aqil F., Tyagi N., Gupta R. Milk exosomes: a biogenic nanocarrier for small molecules and macromolecules to combat cancer. Am. J. Reprod. Immunol. 2021;85:e13349. doi: 10.1111/aji.13349. [DOI] [PubMed] [Google Scholar]
- 39.Chang R., Ng T.B., Sun W.Z. Lactoferrin as potential preventative and adjunct treatment for COVID-19. Int. J. Antimicrob. Agents. 2020;56:106118. doi: 10.1016/j.ijantimicag.2020.106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buck C.B., Day P.M., Trus B.L. The papillomavirus major capsid protein L1. Virology. 2013;445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazurier J., Spik G. Comparative study of the iron-binding properties of human transferrins. Biochem. Biophys. Acta. 1980:399–408. doi: 10.1016/0304-4165(80)90112-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S2. Article plus supplemental information
Data Availability Statement
The authors confirm that data supporting the findings of this research are available within the body of the article or in supplemental material.