Abstract
Bacterial denitrification is expressed in response to the concurrent exogenous signals of low-oxygen tension and nitrate or one of its reduction products. The mechanism by which nitrate-dependent gene activation is effected was investigated in the denitrifying bacterium Pseudomonas stutzeri ATCC 14405. We have identified and isolated from this organism the chromosomal region encoding the two-component sensor-regulator pair NarXL and found that it is linked with the narG operon for respiratory nitrate reductase. The same region encodes two putative nitrate or nitrite translocases, NarK and NarC (the latter shows the highest similarity to yeast [Pichia] and plant [Nicotiana] nitrate transporters), and the nitrate-regulated transcription factor, DnrE, of the FNR family. The roles of NarX and NarL in nitrate respiration were studied with deletion mutants. NarL activated the transcription of narG, narK, and dnrE but did not affect the denitrification regulons for the respiratory substrates nitrite, nitric oxide, and nitrous oxide. The promoters of narG, narK, and dnrE carry sequence motifs, TACYYMT, which correspond to the NarL recognition sequence established for Escherichia coli. The cellular response toward nitrate and nitrite was mediated by the sensor protein NarX, which discriminated weakly between these oxyanions. Our data show that the NarXL two-component regulatory system has been incorporated into the bacterial denitrification process of P. stutzeri for selective regulation of nitrate respiration.
Denitrification by prokaryotes is part of the global nitrogen cycle, where it is responsible for the balance of the nitrogen budget of the biosphere. In a pathway of four reaction steps, nitrate is successively reduced via nitrite, nitric oxide (NO), and nitrous oxide (N2O) to dinitrogen. Denitrification genes are usually expressed in response to nitrate or nitrite and a low oxygen level (although aerobic denitrification exists in specialized cases) (for a review, see reference 45). This requires the activation of sensory devices and signal transduction pathways by these respiratory substrates or their reduction products. We were interested in the mechanisms by which a denitrifying bacterium that is deprived of oxygen and shifted to N oxide utilization senses nitrate or nitrite and activates genes for anaerobic respiration.
In Escherichia coli the transcription factor NarL is an important regulator in cellular bioenergetics. NarL activates the operon for respiratory nitrate reductase, narGHJI, and other operons of ancillary systems required for nitrate respiration. At the same time, the factor acts as a repressor of operons for alternative modes of respiration. NarL is part of a two-component regulatory system, NarXL (reviewed in reference 13). The sensor-regulator pair is duplicated in NarQP, which exhibits a specificity toward target genes somewhat different from that of NarXL. Putative NarX and NarL homologs, requiring functional analysis, have surfaced as the result of projects to sequence the genomes of Haemophilus influenzae, Neisseria gonorrhoeae, Yersinia pestis, Bacillus subtilis, and Pseudomonas aeruginosa. Here we identify by a targeted approach the narXL genes of the denitrifying bacterium Pseudomonas stutzeri and study their phenotypic manifestations in deletion strains. The function of NarXL is to activate the operon encoding the initiator reaction for denitrification, i.e., respiratory nitrate reduction, but not to act as a global regulatory system for the overall denitrification process.
(Preliminary accounts of this work have been presented previously [21, 45].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type P. stutzeri (ATCC 14405), the mutant strain MK21, which is a spontaneously Smr mutant but otherwise represents wild-type traits, and the MK21 derivatives MRL118 (ΔnarL Kmr Smr) and MRX119 (ΔnarX Kmr Smr) were cultured at 30°C in synthetic medium with asparagine and citrate as major ingredients (10). The construction of the mutant strains MRL118 and MRX119 has been described previously (22). E. coli DH10B and XL1-Blue MR were grown at 37°C in Luria-Bertani medium. Where necessary, kanamycin, ampicillin, or streptomycin was added at a final concentration of 50, 100, or 200 μg ml−1, respectively. Cultures from which total RNA was prepared were grown in a 1-liter flask equipped with baffles and filled with 500 ml of medium. The optical density at 660 nm upon inoculation was about 0.3. The shaker speed of the gyratory incubator used was set at 240 rpm. Initial air saturation was estimated to be about 95% with a Clark-type electrode. Samples of oxygen-respiring cells were drawn after 3 h. For a shift to denitrifying conditions, cells were induced by nitrate (1 g/liter) for 1 h under O2 limitation by decreasing the shaker speed to 120 rpm, which lowered air saturation to about 0.5%. Full anaerobiosis is not required for the expression of the denitrification system of P. stutzeri provided that nitrate or nitrite is present (26). Samples for RNA extraction were drawn from cell suspensions that had reached an optical density of about 0.6 at 660 nm.
Purification of nitrate reductase and immunoblotting. Nitrate reductase from P. stutzeri was solubilized by heat and purified as described previously (6). The subunits were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the large subunit, NarG, was blotted onto a polyvinyl difluoride membrane. An N-terminal sequence, NRKQGEFADGHGETR, was obtained at the Protein Sequencing Facility, University of Konstanz.
For immunoblot analysis of enzymes, aerobically grown cells were transferred to fresh medium and induced for 8 h under O2 limitation with sodium nitrate (1 g/liter) or sodium nitrite (0.5 g/liter). Cells were harvested by centrifugation and washed twice with 25 mM Tris-HCl (pH 7.5)–10 mM MgCl2. They were suspended in the same buffer without MgCl2 and broken in the cold by sonication (Branson). The supernatant from centrifugation for 20 min at 39,000 × g was used as a cell extract for SDS-PAGE (27). Proteins were transferred to a nitrocellulose membrane by semidry electrotransfer. For Western blotting (38), polyclonal antisera were raised against the purified oxidoreductases. Quantitation was done by scanning laser densitometry with an ImageMaster scanner and software (Amersham Pharmacia Biotech). Protein concentration was determined by the Lowry procedure with bovine serum albumin as the standard.
Cloning of the narXL region.
A narL fragment was amplified from genomic DNA of strain MK21 with the primer pair 5′-CAAAGCTTSGACGACCACCCSMT-3′ and 5′-TCGAATTCARGTARCCGTCSGCRC-3′, designed from conserved NarL and NarP sequences and observing the codon preference of P. stutzeri genes. The restriction sites HindIII and EcoRI (boldfaced nucleotides in primer sequences) were added to allow the subsequent cloning of the PCR product. The PCR was carried out at an annealing temperature of 45°C. Amplification products were separated by electrophoresis, blotted onto a nylon membrane, and hybridized (16) with the narP probe of E. coli. A 285-bp fragment was isolated, cloned into pBluescript II SK(+) to give plasmid pBSnarL, and verified by sequencing as being homologous to narL from E. coli.
A genomic cosmid library of wild-type P. stutzeri was constructed with the SuperCos1 vector and E. coli XL1-Blue MR as the host (Stratagene). DNA was purified by a CsCl gradient and partially digested with Sau3A under conditions that yielded fragments of 30 to 50 kb. These fragments were cloned into the BamHI site of the vector by following the protocol of the supplier. Packaging was performed by using the GigapackIII XL packaging extract (Stratagene). For screening of the library by colony hybridization, we used an internal narL probe of 219 nucleotides, which was amplified from plasmid pBSnarL with the primers 5′-GCGTGACCTGCTGGATCTG-3′ and 5′-GCACATGGCTCTGCTCGTC-3′. For digoxigenin (DIG) labeling of probes, the PCR mixture was made up to contain 7 μM DIG-11-dUTP.
Cloning of a narG fragment, gene probes, and nucleic acid manipulations.
We translated the amino acid sequence GEFADGH, obtained from N-terminal sequencing of the NarG subunit, into the degenerate forward primer 5′-GGYGARTTCGCSGACGGYCAC-3′. The reverse primer 5′-TSGCCGGGATCGGSGAGAAGCC-3′ was designed from the conserved sequence GFSPIPAM, encoded at the 5′ ends of the narG genes of E. coli (positions 188 to 195) (5) and B. subtilis (positions 192 to 199) (24). A PCR fragment of 530 bp was amplified from genomic DNA of P. stutzeri MK21 at an annealing temperature of 55°C. The putative narG fragment was identified by Southern hybridization (16) with the E. coli narG gene. The ends were filled in with Klenow polymerase, and the fragment was cloned by blunt-end ligation into the EcoRV-cleaved plasmid pBluescript II SK(+). The insert in the resulting plasmid, pBSnarG, was sequenced with M13 universal and reverse primers to ensure its identity. An internal 356-bp PCR probe for Southern and Northern hybridizations was prepared from pBSnarG with the primer pair 5′-ATCCGCTCGCGCTGGCAGTA-3′ and 5′-CCATGCCGCGCTTGCTCTTG-3′ and was labeled with digoxigenin.
The 881-bp narG probe of E. coli was excised with PstI from plasmid pSL962 (33). A 410-bp fragment of narP was prepared by PCR with plasmid pVJS334 as the template (31) and the primers 5′-TCCTGGCTCTGAAGTGGTCG-3′ and 5′-CAAGCTGCAGAACATCC-3′. The digoxigenin-labeled probes for nirS and norB were amplified from cosmid c146 (41) with primer pairs located at positions 456 to 473 (5′-ACCGAAGCGGATGCAAG-3′) and 1127 to 1144 (5′-TGCGAAGCGACGATGGAC-3′) of the published sequence of nirS (25) and at positions 1192 to 1209 (5′-TCTGATCGGCCTGGCAGT-3′) and 1868 to 1885 (5′-ACCAGCAGCGGTGAGGAA-3′) of the published sequence of norB (46). The probe for the nosZ gene was a PstI digest from plasmid pNS220 (42).
DNA sequencing of cosmid g279 and PCR fragments was carried out by the dideoxy chain termination method with the Thermo Sequenase cycle-sequencing kit (Amersham Pharmacia Biotech) and 35S-labeled dATP (ICN). Unless specified otherwise, standard procedures were followed for DNA manipulations (32).
RNA isolation, Northern blotting, and primer extension analysis.
Total RNA was extracted from 12 ml of cell suspensions of MK21, MRL118, and MRX119 by the hot phenol method (1). Samples (10 μg) were separated electrophoretically in 1.2% agarose gels containing 0.4 M formaldehyde (2). RNA was transferred to a positively charged nylon membrane by downward capillary transfer (9). The dioxetane derivative CDP-Star was used as a chemiluminescent substrate for membrane-based detection of alkaline phosphatase conjugates.
In the primer extension analysis (2), 50 μg of RNA was used to map the 5′ end of the narG transcript. Reverse transcription was initiated from the γ-32P-end-labeled primer, 5′-TCCTGTTGAAGAAGCGCAGTTGATCGAG-3′, complementary to the 5′ end of the narG coding region. The sequencing reaction was performed with the same primer. The primer extension products and the sequencing reactions were analyzed on a 6% denaturing polyacrylamide gel. For mapping the narK transcript initiation sites, we used the primer 5′-AATCCAGAGGTTGCGATTGGCGATCC-3′ and the same conditions as for narG.
Nucleotide sequence accession number.
The narXL sequence data have been deposited with the DDBJ/EMBL/GenBank databases under accession no. AJ131854.
RESULTS
Isolation of the narXL region and linkage with the narG operon.
We based our strategy for the isolation of narXL from P. stutzeri on designing primers on the basis of a comparison of the amino acid sequences of NarL and NarP from E. coli (20, 31, 34) with those of the hypothetical NarP protein from H. influenzae, encoded by the open reading frame (ORF) HI0726 (19). In addition to the domains for DNA binding and phosphorylation, NarL and NarP proteins have the conserved sequences I(V)DDHPL(M) and GADGYL. These regions were translated into a degenerate primer pair to be used to amplify a narL fragment from genomic DNA of P. stutzeri (see Materials and Methods). A 285-bp fragment was detected with the narP probe of E. coli among the amplification products. This PCR fragment was isolated and served as a template for the preparation of a genuine narL probe of 219 nucleotides. The probe was used to locate the narL gene in a genomic library on cosmid g279. Genes encoding NarXL of E. coli are clustered with the narG operon and narK, encoding a putative nitrite transporter. To explore a possible linkage of narL with narG in P. stutzeri, we hybridized cosmid g279 with the narG probe; narG was found on this cosmid also.
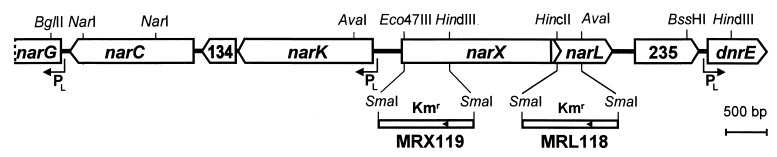
Having established a linkage of the two nar functions, we determined a double-stranded sequence of about 8 kb by using sequence-derived primers. The physical map of the narXL region is shown in Fig. 1. The narL-encoding ORF spans 657 bp. It is followed by ORF235, which has no noteworthy similarity with current sequence entries in data banks. The next ORF encodes the transcription factor DnrE, which belongs to the FNR family (41). In the 5′ direction narL overlaps 71 bp with an ORF that encodes a homolog of the nitrate sensor protein NarX. Upstream of narX two ORFs encode the hypothetical transporters, NarK and NarC, which show similarity to NarK of E. coli and fungal or plant nitrate transporters, respectively. Both belong to the family of major facilitator permeases (39). The highest similarity of NarC was found with putative nitrate transporters of the yeast Pichia angusta (26% identity in a 454-amino-acid overlap; accession no. Q92240) and the higher plant Nicotiana plumbaginifolia (29% identity in a 525-amino-acid overlap; accession no. O04431). A comparison with the products of two narK genes of P. aeruginosa, NarK1 and NarK2 (accession no. Y15252), shows that NarK2 is homologous to NarK of P. stutzeri, whereas NarK1 shows more similarity to the deduced nitrate or nitrite transporters NarK, NasA, and NarT of the gram-positive bacteria B. subtilis (11, 29) and Staphylococcus carnosus (17). In principle, the presence of two transporters would satisfy the requirement of movement of nitrate from the periplasm to the cytoplasm and the opposite translocation of nitrite. This makes the existence of NarC and NarK and their roles in denitrification and perhaps also nitrate assimilation an intriguing prospect. The third product of the narK region, encoded by ORF134, is weakly similar to the so-called conserved protein MTH153 of Methanobacterium thermoautotrophicum (accession no. AE000803) and the hypothetical protein ORF138 of Wolinella succinogenes (accession no. AJ000662), both of unknown function.
FIG. 1.
Organization of the narXL region of P. stutzeri and physical map of mutants. The map covers approximately 9.2 kb. narX overlaps narL by 71 bp. The maps for the narX and narL mutants are shown with the extent of deletions and orientation (arrowheads) of the kanamycin cassette. PL, NarL-regulated promoters. ORFs 235 and 134 are labeled according to the number of amino acids of their hypothetical gene products.
Properties of the derived NarL and NarX proteins.
NarL of P. stutzeri (NarLPs) consists of 218 amino acids, Mr 24,378; the protein has 51 and 47% positional identity with the E. coli proteins NarL and NarP, respectively. Because of the slightly higher amino acid identity with NarL of E. coli (NarLEc) and its function as regulator of the narG operon, we termed the newly isolated P. stutzeri gene narL. Figure 2 shows an alignment of NarLPs with homologous proteins. The crystal structure of NarLEc became known recently (3, 4). The high similarity of NarLPs with the E. coli protein allows predictions of secondary structure as shown in Fig. 2.
FIG. 2.
Structural features of NarL and NarX. Sequences were aligned with the CLUSTAL W program (37). Identical amino acids are marked by asterisks; similar amino acids are marked by colons. (A) Alignment of NarL and NarP proteins, identified in the bottom row. The structural predictions for NarLPs as deduced from the E. coli protein (4) are shown for the 10 α-helices and 5 β-strands. Helices 8 and 9 form the DNA-binding region. Boldfaced letters, E. coli residues important for phosphoryl transfer and the equivalent positions of NarLPs (aspartic acid residues 13, 14, and 59 and lysine 109) and homologous proteins. (B) Alignment of NarX and NarQ proteins, identified in the bottom row. The regions forming distinct structural elements are boxed and are discussed in the text. The predicted transmembrane helices for P. stutzeri and E. coli are boldfaced and are labeled TM1 and TM2.
The NarLPs residues Asp13, Asp14, Asp59, and Lys109 correspond to a set of conserved amino acids found in response regulator proteins. The aspartic acid residues form an acidic pocket which is part of the phosphoryl acceptor chemistry (30, 35). Mutation of Asp59 of NarLEc, the site of protein phosphorylation, has shown that this residue is necessary for the expression of formate dehydrogenase (the fdnG gene) or the repression of fumarate reductase (the frdA gene) in response to nitrate (15).
The derived NarX polypeptide of P. stutzeri (NarXPs) consists of 648 amino acids, Mr 71,791. NarXPs has 31% positional identity each with NarX and NarQ of E. coli. Hydropathy analysis and transmembrane prediction suggest two membrane-spanning regions (TM1 and TM2 [Fig. 2]) that delimit an internal periplasmic domain and a carboxy-terminal cytoplasmic domain. The latter exhibits the conserved regions, termed H, N, and D from the presence of key amino acids in these regions, which are characteristic for the histidine protein kinase family (36). The asparagine and histidine residues, which were identified by site-directed mutagenesis to be essential for kinase activity of NarXEc (8), are present in NarXPs. The conserved histidyl residue, which is subject to autophosphorylation, resides within the H region. In addition to the common characteristics of the members of the kinase family, the periplasmic P region, also known as the P-box, and the cytoplasmic C region, a stretch of conserved residues intercalated between the H and N regions, are conserved in nitrate- and nitrite-responsive sensory kinases. The P region is involved in binding of and distinguishing between nitrate and nitrite (7, 43), whereas the C region is a common feature of sensor proteins and is thought to be important in conferring specificity on sensor-response regulator interaction (30). NarXPs is C-terminally extended vis-à-vis NarX and NarQ from other sources. Certain sensor proteins with similarity to NarXPs, such as FixL, PhoR, EnvZ, and CpxA, also have extended C termini that show no sequence conservation.
NarX senses nitrate and nitrite, with some preference for nitrate.
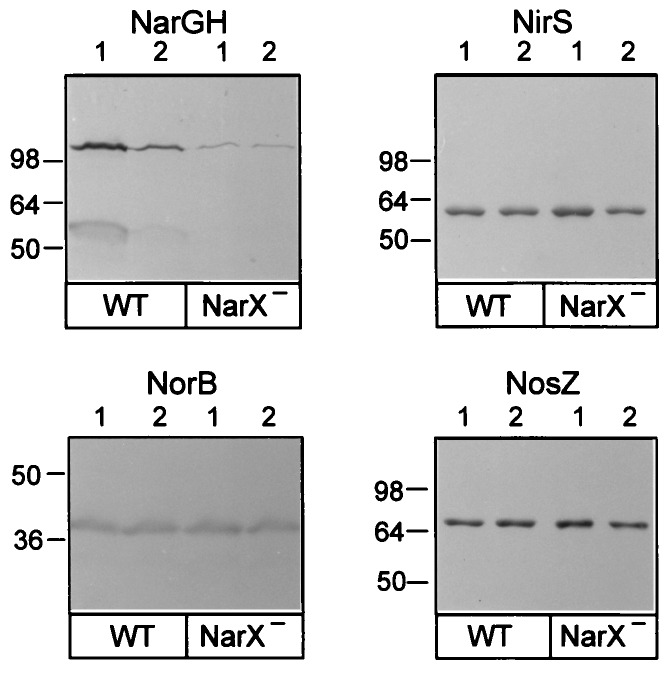
Nitrate and nitrite are the principal substrates for denitrification, and at a low oxygen tension, both induce the complete pathway of four consecutive enzymatic reactions. We were interested, therefore, in the specificity of NarXPs toward both substrates. We used the previously constructed narX deletion mutant MRX119, which has a 520-bp internal Eco47III-HindIII fragment replaced by a kanamycin resistance cassette (Fig. 1), to study by immunochemical means the expression of the structural genes for nitrate reductase, cytochrome cd1 nitrite reductase, NO reductase, and N2O reductase. MRX119 and the control strain MK21 were induced to denitrify nitrate or nitrite as described in Materials and Methods. The protein pattern of cell extracts was analyzed by SDS-PAGE, and the four denitrifying reductases were detected with polyclonal antisera (Fig. 3). Nitrate and nitrite both induced the synthesis of nitrate reductase in the wild type. Nitrite was about half as active an inducer as nitrate, as judged from the amount of NarG detected by Western blot analysis. The level of nitrate reductase was strongly reduced in MRX119 irrespective of which growth-supporting N oxide was present, with NarH falling below the detection limit. Lack of expression of nitrate reductase at wild-type levels in the narX mutant with nitrite as a substrate indicated that the nitrite signal is also processed by NarXPs. Induction of the other three denitrification enzymes elicited by either nitrate or nitrite was not affected by the disruption of narX, and they were present at wild-type levels (Fig. 3). The weak expression of nitrate reductase in MRX119 may depend on an alternative N oxide-responsive regulatory system, for which indirect evidence exists (22).
FIG. 3.
NarX functions as a sensory component with a preference for nitrate. Immunolabeling was done with polyclonal antisera raised against the purified nitrate reductase (NarGH; upper bands represent the NarG subunit, and lower bands represent the NarH subunit), cytochrome cd1 nitrite reductase (NirS), the cytochrome b subunit of NO reductase (NorB), and N2O reductase (NosZ). Lanes 1 and 2, cells cultured for 8 h with nitrate and nitrite, respectively (see Materials and Methods). Each panel shows the results obtained with strain MK21 (WT) and strain MRX119 (NarX−). Detection was carried out with a protein A-peroxidase conjugate and chloronaphthol (28). The NarG levels obtained by nitrate or nitrite induction differ in this experiment by a factor of 2.3. Amounts of cell extracts used were 48 μg each for NarGH and NorB and 6 μg each for NirS and NosZ. Mass references (in kilodaltons) were derived from the SeeBlue standard (Novex).
NarL acts selectively in denitrification as a transcriptional activator of the narG operon.
To study the role of NarL, we used the mutant MRL118 and monitored the expression of the four structural reductase genes for denitrification at the mRNA level. narL of MRL118 lacks a 358-bp HincII-AvaI fragment and carries a Kmr marker instead (Fig. 1). For comparative studies of gene expression, mutant MRX119 was included. If narXL is organized as an operon and NarL cannot be replaced by a homologous component, the phenotypes of narL and narX mutants should be indistinguishable.
We used an internal fragment from narG as a probe to detect transcription from the narG operon of P. stutzeri in Northern blot analysis. The operon from this bacterium has not yet been sequenced. However, nar sequences from P. aeruginosa (accession no. Y15252), Paracoccus denitrificans GB17 (subjective synonym, Thiosphaera pantotropha) (Q56356), Mycobacterium tuberculosis (O06559), Thermus thermophilus (Y10124), B. subtilis (X91819), S. carnosus (AF029225), and Streptomyces coelicolor (AL031515) show that without exception, nitrate-respiring and denitrifying bacteria, both gram negative and gram positive, have the nitrate reductase structural genes narG, narH, and narI, as well as a chaperone-like protein, encoded by narJ, in an invariant narGHJI gene cluster, probably in each case, as in E. coli (X16181), as an operon of four cistrons.
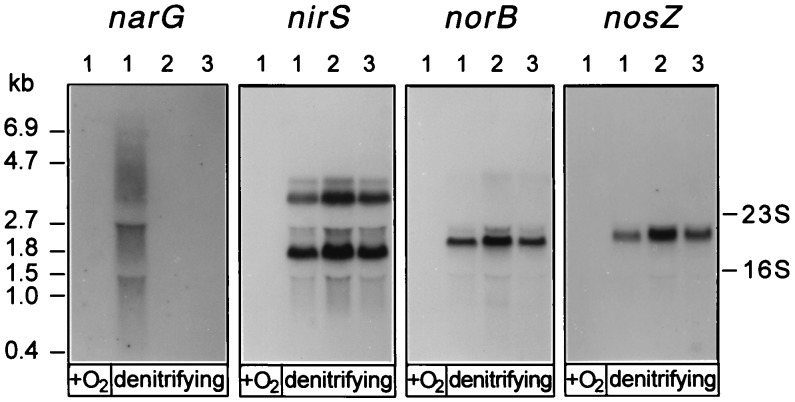
Specimens of total RNA from cells grown aerobically and from those shifted to denitrifying conditions were analyzed by RNA-DNA hybridization. The narG transcript was found only in denitrifying cells but not in O2-respiring wild-type P. stutzeri (Fig. 4). We noted mRNA instability on Northern blots in the form of considerable streaking, which was not observed with the transcripts of the other oxidoreductase genes. The size of the largest narG signal, 6.9 kb, corresponded to that expected from a narGHJI operon but would not be large enough to also include genes encoding the nitrate or nitrite transporters upstream, or other genes downstream of this operon. No narG transcripts were detected under denitrifying conditions in the narL and narX mutants MRL118 and MRX119, respectively. On the other hand, transcripts of the nirSTB operon (encoding cytochrome cd1 and two low-molecular-mass c-type cytochromes), the norCB operon (encoding the NO reductase complex), and the nosZ gene (encoding N2O reductase) were all present in both mutants (Fig. 4). No transcripts were detected in RNA isolated from aerobically grown cells, in agreement with the notion that induction of the denitrification apparatus occurs at a low oxygen tension. Our results show that NarL acts at the transcriptional level and activates the narG operon but not the other structural genes of denitrification oxidoreductases. We were unable to detect narXL transcripts, but the overlapping gene organization (Fig. 1) and the mutational results are indicative of a narXL operon.
FIG. 4.
NarL acts as a transcriptional activator for the narG operon. The DNA probe used for Northern blot analysis is given at the top of each panel. Total RNA was extracted from MK21 (lanes 1), MRL118 (ΔnarL) (lanes 2), and MRX119 (ΔnarX) (lanes 3). Cells were grown for 3 h with oxygen in the absence of nitrate (+O2) and then shifted to nitrate-denitrifying conditions and extracted 1 h after the shift (denitrifying). Transcripts from the nir operon are found as monocistronic nirS and polycistronic nirSTB messages (22). Size standards are the RNA molecular weight marker no. I (Boehringer GmbH, Mannheim, Germany) and the 16S and 23S rRNA species.
NarL-regulated promoters.
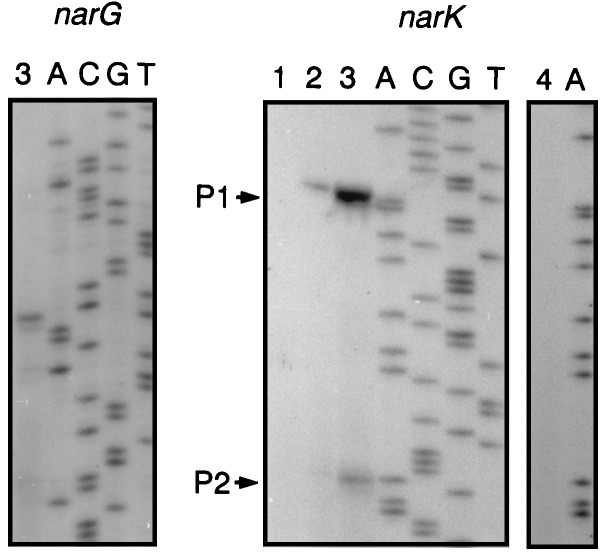
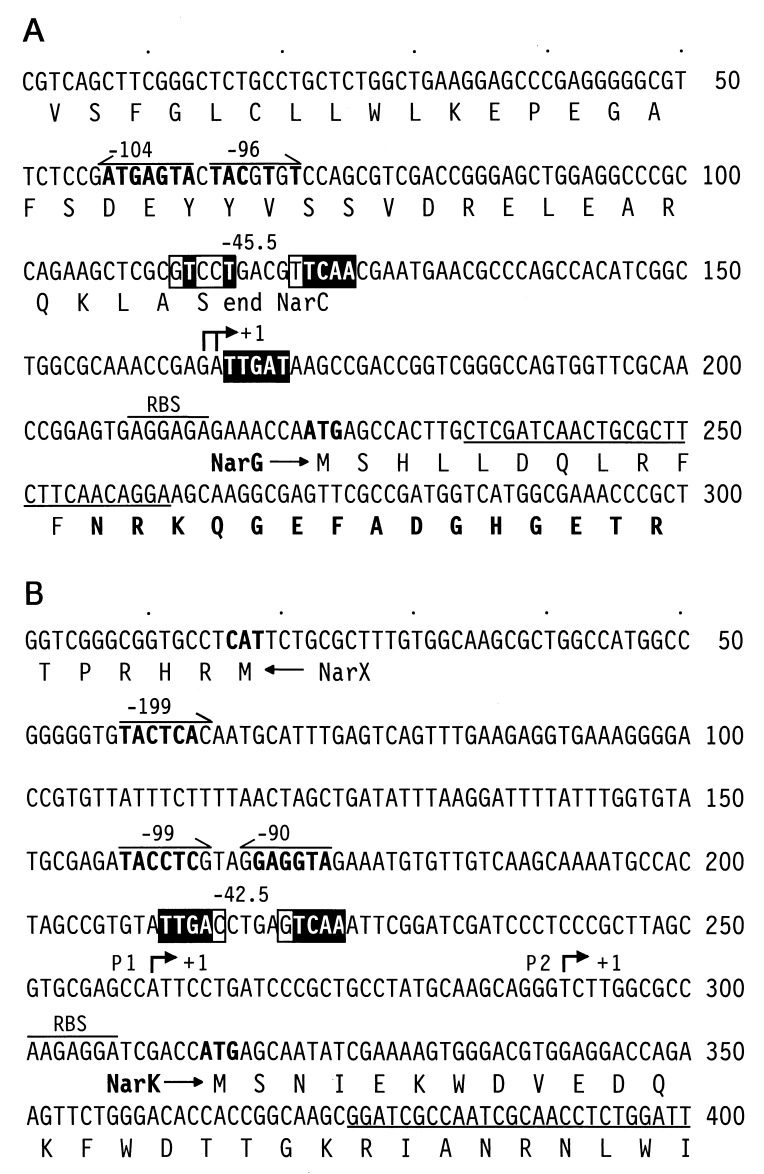
With the aim of identifying potential binding sites for the NarL regulator, we located the promoters of narG and narK by primer extension analysis (Fig. 5). The coding region of narG was independently identified from the N-terminal amino acid sequence obtained from the purified NarG subunit. The comparison of the nucleotide sequence-derived protein and the N terminus obtained from sequencing the isolated NarG subunit revealed that the first 11 amino acids were missing in the protein (Fig. 6A). The reason for this modification is unclear. We assume that the N terminus is processed by a protease, possibly activated during the heat treatment used for the isolation of nitrate reductase. The proteolytic activity, cleaving at the carbonyl site of phenylalanine, exhibits chymotrypsin specificity. The heterogeneity of the NarH subunit is a known phenomenon for heat-solubilized nitrate reductase (reference 6 and citations therein) but has not been reported so far for the NarG subunit. The 5′ end of narG was determined by primer extension (Fig. 5). Divergently oriented NarL sites centered around −100 nucleotides from the start site of transcription, as well as a degenerate FNR site, at a distance of −45.5 nucleotides, and an FNR half-site, TTGAT, downstream of position +1, form part of the promoter region (Fig. 6A). The FNR site of the P. stutzeri narG promoter may be involved in the anaerobic expression of this operon. Although an unprecedented multiplicity of four FNR factors has been identified in P. stutzeri, a candidate regulator to activate the narG operon in response to oxygen withdrawal is still missing (41).
FIG. 5.
Determination of the 5′ ends of the narG and narK transcripts by primer extension analysis. Total RNA was obtained from wild-type cells (MK21) grown aerobically (lane 1), under O2 limitation (lane 2), or under nitrate-denitrifying conditions (O2 limitation in the presence of nitrate) (lanes 3). The right panel for narK shows the lack of extension products of RNA from MRL118 (ΔnarL) (lane 4), which had been induced for denitrification identical to that of the wild type. Primer extension was performed with oligonucleotides complementary to the 5′ ends of the coding regions of narG and narK shown in Fig. 6. Lanes A, C, G, and T show the results of dideoxy sequencing reactions carried out with the same primers. For MRL118 only the dideoxyadenine reaction is shown.
FIG. 6.
NarL-dependent promoters of narG (A) and narK (B). The transcription start sites obtained from primer extension analysis are marked +1. Potential NarL sites are marked by half-arrows; nucleotides that correspond to the E. coli consensus are boldfaced. Putative FNR sites are boxed, and nucleotides within those sites that correspond to the E. coli consensus are highlighted. The oligonucleotides used for primer extension are underlined. RBS, ribosome binding site. The amino acid sequence obtained from the purified NarG subunit is shown in boldface in panel A.
Figure 5 shows that the determination of the transcription start site of narK revealed two putative promoters. Under aerobic conditions no transcripts were detected, but both promoters were weakly activated under O2-limited conditions. Transcription was enhanced by the addition of nitrate, and promoter P1 showed the stronger response. Since the pattern of appearance of P2 followed that of P1, we cannot rule out the possibility that the RNA species generating P2 is a processing or degradation product from the RNA giving rise to P1. In the narL mutant MRL118, no narK transcripts were detected (Fig. 5). The promoter P1 of narK shows sequence motifs corresponding to recognition sites for NarL and FNR at typical distances of these binding elements from the transcription initiation site (Fig. 6B). Again, this is in conformity with the observed induction pattern for denitrification.
DISCUSSION
We have argued that the denitrification process consists of three to four modules, i.e., partly independent respiratory systems utilizing nitrate, nitrite, nitric oxide, or N2O (44, 45). Only nitrite reduction is tightly coupled with the subsequent reaction, the reduction of NO to N2O, presumably to maintain NO at a low steady-state level and to limit the toxic effects of this radical. Our data show an autonomous element for regulating nitrate respiration in the form of the NarXL two-component system, distinct from regulators affecting nitrite denitrification, i.e., the reduction of nitrite to a gaseous product. The regulatory independence of denitrification in the strict sense from the initiator reaction supports our concept of a modular design.
A phylogenetic analysis by the CLUSTAL W program of the known NarL and NarP proteins, and putative homologs deduced from genomic sequencing projects, shows that the NarL proteins of the pseudomonads are most closely related to NarLEc (data not shown). NarL of E. coli binds to cognate promoters via heptameric sequences whose consensus is TACYYMT (12, 14, 40). The location of the NarL site is typically variable with respect to distance from and orientation toward the start of transcription (13). In anaerobically, nitrate-regulated promoters, the NarL site is usually found at greater distances from the transcription start site than the FNR site for binding the anaerobic regulator. We have shown so far that in P. stutzeri, transcription of narG, narK, and dnrE is activated by nitrate. DnrE is a transcription factor under the putative control of NarL. It belongs to the new DNR branch of regulators of the greater FNR family, which lack the cysteine motif for binding a [4Fe-4S] center as their major distinction (41). Those three genes show potential NarL sites which match or are highly similar to the consensus derived for E. coli (Table 1). DNA footprinting or mutational analysis is still required to attribute functionality to the heptameric motifs.
TABLE 1.
Heptameric sequence motifs in NarL-regulated promoters of P. stutzeri
| Gene | Recognition heptamera | Distanceb | Orientation of inverted repeat |
|---|---|---|---|
| narG | TACgTgT | −96 | Divergent from −104 site |
| TACTCAT | −104 | ||
| narK | TACCTCc | −90 | Convergent with −99 site |
| TACCTCg | −99 | ||
| TACTCAc | −199 | Isolated half-site | |
| dnrEc | TACCTCT | −39 | Convergent with −56 site |
| TACCTCT | −56 | ||
| TACTaCc | −140 | Isolated half-site | |
| narG of E. coli | TACYYMTd | −57 to −208 |
Nucleotides deviating from consensus are lowercased.
Number of nucleotides counted from −1 of the start of transcription to the middle position of the heptameric motif.
Data from reference 41.
Consensus sequence; Y represents T or C; M represents C or A.
The P and C regions are specific for NarX-type sensor proteins. They show a remarkable degree of conservation among the five proteins compared in Fig. 2. The P region was shown in an elegant mutational study to be responsible for the binding of nitrate and nitrite and to harbor elements essential for the discrimination of these ions. Whereas NarXEc is strongly biased towards nitrate (narG expression in a narQ null mutant is induced 100-fold by nitrate but only 4-fold by nitrite), no such bias is incorporated in NarQEc (43). The preference of NarXPs for nitrate is only about twofold. Extending a comparison of the P-box sequences of NarX and NarQ to include the sensor proteins of the pseudomonads lowers the identity score to 10 (from 15) of 18 consecutive amino acids. The amino acids Ser43, His45, and Lys49 of NarX (E. coli sequence numbering), supposedly of discriminatory quality compared with the positionally equivalent residues of NarQ proteins, Asp, Glu, and Ile, lose their differentiating value, but it is still to be noted that the P-box of NarXPs resembles that of NarQEc more closely than that of NarXEc. As suggested previously, elements outside of the P-box are also likely to participate in discriminating between nitrate and nitrite (43).
An important element of the P-box is the conserved Arg54, whose mutagenesis results in a ligand-unresponsive protein (7, 43). When nitrate or nitrite interact with a protein usually they require a transition metal for binding and their catalytic transformation. We have previously drawn attention to structures of protein nitrate complexes, even though they are unrelated to nitrate sensing, suggesting a mode of nitrate binding for the sensory domain of NarX (45). In Limulus polyphemus, hemocyanin nitrate occupies the site of the allosteric effector chloride (23), whereas in the tyrosine phosphatase of Yersinia enterocolitica, which takes part in the cellular regulation of pathogenicity, nitrate is bound by the phosphate-binding peptide loop (18). In each of these crystal structures, nitrate is hydrogen-bonded with two oxygens to the Nɛ and Nη atoms of an arginine residue. Further hydrogen bonds extend from the oxygen atoms of nitrate to the hydroxyl group of serine in hemocyanin or to amide nitrogens of the phosphate loop in tyrosine phosphatase. Both in hemocyanin and in the phosphatase, conformational changes distant from the binding site of nitrate are induced by anion binding. Thus, these models indicate the possibility that the binding of nitrate to the conserved arginine residue in the P region of NarX and transmembrane signaling may take place without a requirement for a transition metal in the nitrate sensor.
ACKNOWLEDGMENTS
We are indebted to John A. DeMoss and Valley Stewart for kindly providing plasmids and to the late I Rasched for protein sequencing. We thank H. Körner for a gift of NarG protein, B. Schreckenberger for technical assistance, and D. Jahn for sharing sequence information prior to publication.
The generous financial support of the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie is gratefully acknowledged.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus R P, Dickerson R E. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998;37:3665–3676. doi: 10.1021/bi972365a. [DOI] [PubMed] [Google Scholar]
- 4.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coliresponse regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 5.Blasco F, Iobbi C, Giordano G, Chippaux M, Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the naroperon and reassessment of the role of the α and β subunits in iron binding and electron transfer. Mol Gen Genet. 1989;218:249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- 6.Blümle S, Zumft W G. Respiratory nitrate reductase from denitrifying Pseudomonas stutzeri, purification, properties and target of proteolysis. Biochim Biophys Acta. 1991;1057:102–108. [Google Scholar]
- 7.Cavicchioli R, Chiang R C, Kalman L V, Gunsalus R P. Role of the periplasmic domain of the Escherichia coliNarX sensor-transmitter protein in nitrate-dependent signal transduction and gene regulation. Mol Microbiol. 1996;21:901–911. doi: 10.1046/j.1365-2958.1996.491422.x. [DOI] [PubMed] [Google Scholar]
- 8.Cavicchioli R, Schröder I, Constanti M, Gunsalus R P. The NarX and NarQ sensor-transmitter proteins of Escherichia colieach require two conserved histidines for nitrate-dependent signal transduction to NarL. J Bacteriol. 1995;177:2416–2424. doi: 10.1128/jb.177.9.2416-2424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Mackey K. One-hour downward capillary blotting of RNA at neutral pH. Anal Biochem. 1994;221:303–305. doi: 10.1006/abio.1994.1416. [DOI] [PubMed] [Google Scholar]
- 10.Coyle C L, Zumft W G, Kroneck P M H, Körner H, Jakob W. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina, purification and properties of a novel multicopper enzyme. Eur J Biochem. 1985;153:459–467. doi: 10.1111/j.1432-1033.1985.tb09324.x. [DOI] [PubMed] [Google Scholar]
- 11.Cruz Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin A J, Li J, Stewart V. Analysis of nitrate regulatory protein NarL-binding sites in the fdnG and narG operon control regions of Escherichia coliK-12. Mol Microbiol. 1996;20:621–632. doi: 10.1046/j.1365-2958.1996.5491074.x. [DOI] [PubMed] [Google Scholar]
- 13.Darwin A J, Stewart V. The NAR modulon systems: nitrate and nitrite regulation of anaerobic gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman & Hall; 1996. pp. 343–359. [Google Scholar]
- 14.Dong X-R, Li S F, DeMoss J A. Upstream sequence elements required for NarL-mediated activation of transcription from the narGHJI promoter of Escherichia coli. J Biol Chem. 1992;267:14122–14128. [PubMed] [Google Scholar]
- 15.Egan S M, Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coliK-12. J Bacteriol. 1991;173:4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engler-Blum G, Meier M, Frank J, Müller G A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 17.Fast B, Lindgren P-E, Götz F. Cloning, sequencing, and characterization of a gene (narT) encoding a transport protein involved in dissimilatory nitrate reduction in Staphylococcus carnosus. Arch Microbiol. 1996;166:361–367. doi: 10.1007/BF01682980. [DOI] [PubMed] [Google Scholar]
- 18.Fauman E B, Yuvaniyama C, Schubert H L, Stuckey J A, Saper M A. The X-ray crystal structures of Yersiniatyrosine phosphatase with bound tungstate and nitrate: mechanistic implications. J Biol Chem. 1996;271:18780–18788. doi: 10.1074/jbc.271.31.18780. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geohagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzaeRd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 20.Gunsalus R P, Kalman L V, Stewart R R. Nucleotide sequence of the narL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res. 1989;17:1965–1975. doi: 10.1093/nar/17.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Härtig E H, Zumft W G. Respiratory nitrate reductase of Pseudomonas stutzeriis regulated by a two-component system, NarXL, that is ineffective in nitrate control of denitrification sensu stricto. Biospektrum. 1998;4:51. [Google Scholar]
- 22.Härtig E, Zumft W G. Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeriduring the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J Bacteriol. 1999;181:161–166. doi: 10.1128/jb.181.1.161-166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazes B, Magnus K A, Kalk K H, Bonaventura C, Hol W G J. Nitrate binding to Limulus polyphemussubunit type II hemocyanin and its functional implications. J Mol Biol. 1996;262:532–542. doi: 10.1006/jmbi.1996.0533. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 25.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzericonsists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 26.Körner H, Zumft W G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl Environ Microbiol. 1989;55:1670–1676. doi: 10.1128/aem.55.7.1670-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Nakane P K. Simultaneous localization of multiple tissue antigens using the peroxidase-labeled antibody method: a study on pituitary glands of the rat. J Histochem Cytochem. 1968;16:557–560. doi: 10.1177/16.9.557. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa K-I, Akagawa E, Yamane K, Sun Z-W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 31.Rabin R S, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coliK-12. J Bacteriol. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sodergren E J, DeMoss J A. narI region of the Escherichia coli nitrate reductase (nar) operon contains two genes. J Bacteriol. 1988;170:1721–1729. doi: 10.1128/jb.170.4.1721-1729.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart V, Parales J, Jr, Merkel S M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coliK-12. J Bacteriol. 1989;171:2229–2234. doi: 10.1128/jb.171.4.2229-2234.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 37.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trueman L J, Onyeocha I, Forde B G. Recent advances in the molecular biology of a family of eukaryotic high affinity nitrate transporters. Plant Physiol Biochem. 1996;34:621–627. [Google Scholar]
- 40.Tyson K L, Cole J A, Busby S J W. Nitrite and nitrate regulation at the promoters of two Escherichia colioperons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol Microbiol. 1994;13:1045–1055. doi: 10.1111/j.1365-2958.1994.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 41.Vollack K-U, Härtig E, Körner H, Zumft W G. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses, and cognate metabolic proceses. Mol Microbiol. 1999;31:1681–1694. doi: 10.1046/j.1365-2958.1999.01302.x. [DOI] [PubMed] [Google Scholar]
- 42.Vollack K-U, Xie J, Härtig E, Römling U, Zumft W G. Localization of denitrification genes on the chromosomal map of Pseudomonas aeruginosa. Microbiology. 1998;144:441–448. doi: 10.1099/00221287-144-2-441. [DOI] [PubMed] [Google Scholar]
- 43.Williams S B, Stewart V. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coliK-12. Mol Microbiol. 1997;26:911–925. doi: 10.1046/j.1365-2958.1997.6262002.x. [DOI] [PubMed] [Google Scholar]
- 44.Zumft W G. The denitrifying prokaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 554–582. [Google Scholar]
- 45.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zumft W G, Braun C, Cuypers H. Nitric oxide reductase from Pseudomonas stutzeri: primary structure and gene organization of a novel bacterial cytochrome bccomplex. Eur J Biochem. 1994;219:481–490. doi: 10.1111/j.1432-1033.1994.tb19962.x. [DOI] [PubMed] [Google Scholar]