Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants continue to emerge, and effective tracking requires rapid return of results. Surveillance of variants is typically performed by whole genome sequencing (WGS), which can be financially prohibitive and requires specialized equipment and bioinformatic expertise. Genotyping approaches are rapid methods for monitoring SARS-CoV-2 variants but require continuous adaptation. Fragment analysis may represent an approach for improved SARS-CoV-2 variant detection.
Methods
A multiplex fragment analysis approach (CoVarScan) was validated using PCR targeting variants by size and fluorescent color. Eight SARS-CoV-2 mutational hot spots in variants of concern (VOCs) were targeted. Three primer pairs (recurrently deleted region [RDR] 1, RDR2, and RDR3–4) flank RDRs in the S-gene. Three allele-specific primers target recurrent spike receptor binding domain mutants. Lastly, 2 primer pairs target recurrent deletions or insertions in ORF1A and ORF8. Fragments were resolved and analyzed by capillary electrophoresis (ABI 3730XL), and mutational signatures were compared to WGS results.
Results
We validated CoVarScan using 3544 clinical respiratory specimens. The assay exhibited 96% sensitivity and 99% specificity compared to WGS. The limit of detection for the core targets (RDR1, RDR2, and ORF1A) was 5 copies/reaction. Variants were identified in 95% of samples with cycle threshold (CT) <30 and 75% of samples with a CT 34 to 35. Assay design was frozen April 2021, but all subsequent VOCs have been detected including Delta (n = 2820), Mu, (n = 6), Lambda (n = 6), and Omicron (n = 309). Genotyping results are available in as little as 4 h.
Conclusions
Multiplex fragment analysis is adaptable and rapid and has similar accuracy to WGS to classify SARS-CoV-2 variants.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) have emerged harboring epidemiologically significant genetic mutations impacting transmission kinetics (1), vaccine responses (2, 3), mortality (4, 5), and monoclonal antibody therapy (3, 6–9). Rapid identification of SARS-CoV-2 VOCs is therefore of exigent need in clinical and public health arenas. Whole-genome sequencing (WGS) remains the current gold standard for SARS-CoV-2 variant identification; however, broad adoption is challenging due to requirements for specialized equipment and bioinformatics expertise (10). While these limitations may be mitigated through centralized high-volume testing, consequent increased turnaround time for batching, data deconvolution, and sequence analysis often exceed an actionable timeframe for contact tracing or patient assessment.
Genotyping has arisen as a method for variant screening or determination. Single-nucleotide polymorphism (SNP)-based genotyping approaches have been applied in several platforms including high-resolution melt curve analysis, molecular beacons, TaqMan probes, CRISPR-Cas, and digital droplet PCR. Most of these approaches rely on thermocycler platforms that can detect 4 to 5 colors maximum. Thus, the number of targets is limited and required frequent adjustment when new variants arose. Furthermore, the limited number of targets meant these assays could screen for variants but could not provide definitive identification. Alternatively, Sanger sequencing of the spike gene receptor-binding domain (RBD) has proven useful for identifying variants without the need for assay adaptation. Although Sanger sequencing is more comprehensive, it is more labor intensive to analyze results compared to targeted genotyping methods.
In this work, we describe the development and assessment of a rapid, cost-effective, high-throughput multiplex fragment analysis assay named CoVarScan to detect SARS-CoV-2 VOCs in a routine clinical setting. Fluorescently labeled RT-PCR amplicons are analyzed by capillary electrophoresis, and SARS-CoV-2 VOCs are identified by unique mutation signatures. Our goal was to assess whether fragment analysis can be configured to accurately detect multiple deletions, insertions, and SNPs with alternately labeled primers, and whether it can be scaled for use in a rapid, high-throughput environment. Such use could be beneficial since the method features modest reagent requirements, utilizing instrumentation already found in many routine clinical and public health laboratories.
Methods
This work was reviewed and approved by the UT Southwestern Institutional Review Board and deemed not human subject research due to an exception for disease surveillance.
clinical specimens
Positive nasopharyngeal or nasal specimens in viral transport media (Remel, Thermo Scientific) or Abbott Universal Collection Kit media from the pathology service line at the University of Texas Southwestern Medical Center were utilized for assay verification. SARS-CoV-2 positivity was determined by either RT-PCR using the Alinity m SARS-CoV-2 assay (Abbott Molecular), Xpert® Xpress SARS-CoV-2 assay or Xpert® Xpress SARS-CoV-2 Flu/RSV assay (Cepheid), or isothermal amplification (IDNOW, Abbott Diagnostics). A total of 3544 (April 4, 2021–February 7, 2022) SARS-CoV-2–positive prospective specimens collected from asymptomatic, symptomatic, and confirmed COVID-19 patients were utilized. Included specimens had RT-qPCR cycle threshold (CT) values of <35 (Abbott Alinity M SARS-Cov-2 assay) or a positive qualitative result from either Cepheid or IDNOW COVID-19 assays.
covarscan assay
RT-PCR was performed with fluorescently labeled primers to create amplicons of different size and color, which were separated by capillary electrophoresis (ABI 3730XL, Applied Biosystems) (11). Details regarding this method, the WGS method and bioinformatic pipeline are present in the online Supplemental Methods.
While fragment analysis could theoretically accommodate over 20 targets using different combinations of florescent dyes and amplicon sizes, we found that targeting 8 defined hotspot regions is sufficient to differentiate all currently described SARS-CoV-2 variants of interest and VOCs (Supplemental Tables 1 and 2). SARS-CoV-2 variants display convergent evolution with certain regions recurrently mutated. We leveraged this insight to target 5 recurrently deleted regions (RDRs; S:RDR1, S:RDR2, S:RDR3–4, ORF1A, and ORF8) (Supplemental Table 2) and 3 SNPs (S:N501Y, S:L452R, and S:E484K), permitting differentiation of all VOCs (Fig. 1, A). Since its initial inception, CoVarScan has been optimized for parameters including primer melting temperatures, PCR annealing temperature, SNP primer specificity, and balanced primer concentrations.
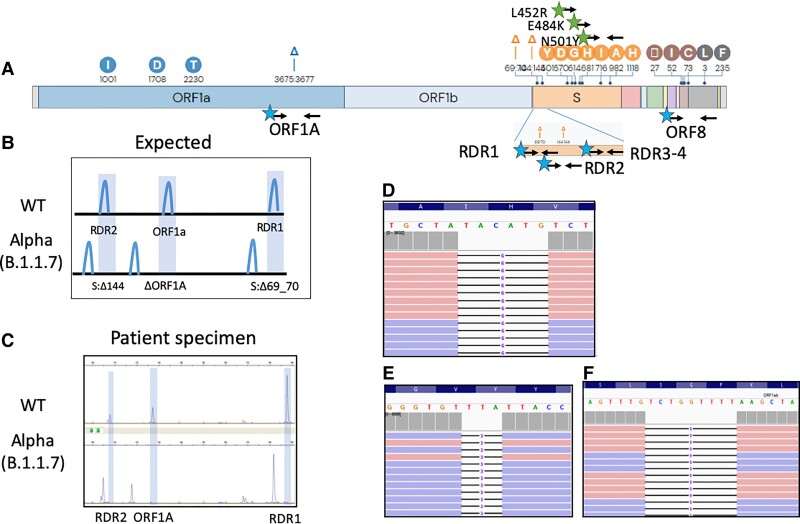
Fig. 1.
Fragment analysis identifies the SARS-CoV-2 Alpha VOC. (A), Genetic variations of the SARS-CoV-2 Alpha VOC. Black arrows indicate primer binding sites. Stars indicating fluorescent dye (blue, FAM; green, HEX) primer labels; (B), Expected deletions present in the SARS-CoV-2 Alpha variant compared to WT as visualized by fragment analysis; (C), Electrophoretogram representative of 2 patients with WT and Alpha SARS-CoV-2 infections; (D–F), Integrated genome viewer visualization of the WGS reads confirming the presence of the ORF1A:3675_3677del (D), S:69_70del (E), and S:144del (F) initially identified by capillary electrophoresis.
The core assay detects 2 S-gene deletion mutations (S:Δ69_70- RDR1 and S: Δ144-RDR2) initially described in the Alpha VOC, and a deletion in ORF1A (ORF1A:Δ3675_3677) present in Alpha (4), Beta (South Africa, B.1.351) (12), Gamma (Brazil, P.1) (13), Iota (New York, B.1.526), Lambda (Peru, C.37), and Omicron (B.1.1.529) VOCs (Supplemental Tables 3 and 4). CoVarScan was expanded to include additional allele-specific primers to detect recurrent RBD mutations (S:N501Y, S:E484K, and L452R), conferring either antibody resistance or increased transmission by higher ACER2 binding affinity (3, 6). Lastly, primers flanking ORF8-N gene junction and RDR3–4 were added to differentiate Beta (S:Δ242_244) and Gamma VOCs (Ins28269-28273) (Supplementary Tables 1 and 2).
Sensitivity, specificity, and accuracy were established by comparing results to WGS of the same specimens. Detailed performance instructions are present in the Supplementary Materials and Methods.
interpretive criteria
Samples were considered positive if capillary electrophoresis peaks were (a) the expected size (x-axis) (Supplemental Table 5), (b) >50 fluorescence units (double background fluorescence), and (c) the HEX:FAM or FAM:HEX signal was >10:1 to call an allele as mutant or wild-type (WT). A small number of recurrent artifacts were observed (Supplemental Table 6). Considering all targets together, one can identify WT, Alpha, Beta, Gamma, Iota, Eta, Epsilon, Delta, Lambda, Mu, Omicron, and other SARS-CoV-2 variants (Supplemental Table 3).
limit of detection
The limit of detection for each assay target was estimated using spiked Alpha, Beta, and Gamma in vitro transcribed RNA (Twist Biosciences). A 2-fold dilution series ranged from 2000 down to 1 copy per reaction. Specimens were analyzed in triplicate, and then full limit of detection was performed with 20 replicates at 100 and 5 copies per reaction.
Results
core assay: fragment length polymorphism detection in spike rdr1, rdr2, and orf1a
We initially designed primers flanking the mutation (S:Δ69_70) responsible for the S-gene target failure (ThermoFisher TaqPath assay) found in the Alpha variant (B.1.1.7, U.K. origin). This mutation is in the RDR1 of the N-terminal side of the S-gene (1, 14). As the S: Δ69_70 mutation was not unique to the Alpha variant, we targeted 2 additional deletions: S: Δ144 in the RDR2 gene and ORF1A: Δ3565_3567 (9 bp deletion). These 3 targets produced aberrant PCR products specific to the Alpha variant. (Fig. 1, A–F).
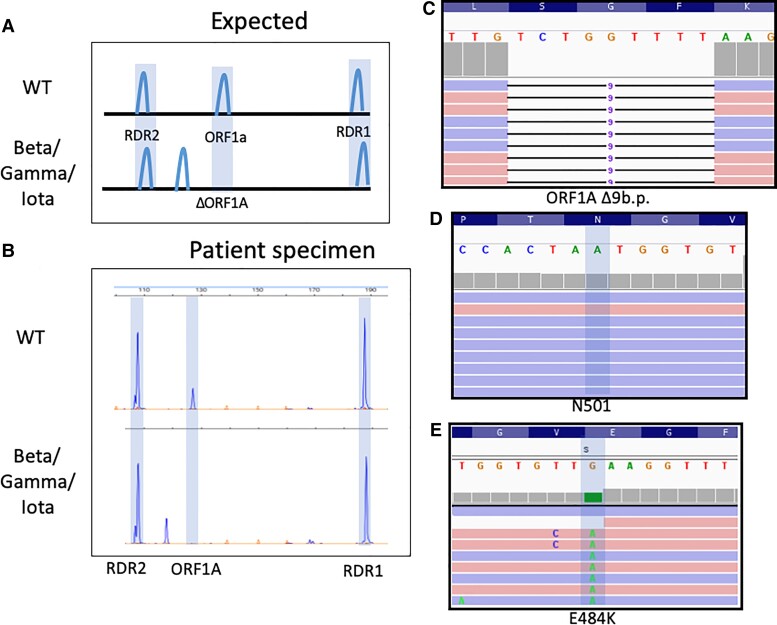
The 3 targets, RDR1, RDR2, and ORF1A, form the core assay. However, allele-specific targets were required to differentiate Beta, Gamma, and Iota VOCs. When initial SARS-CoV-2 samples were tested, some contained an isolated ORF1A 9 bp deletion (Fig. 2, A and B). This was originally ascribed to the Beta (B.1.351, South Africa origin) (15, 16) and Gamma (P.1, Brazil origin) (17) variants, which have 2 RBD mutations (S:N501Y and S:E484K). However, sequencing revealed a variant with only S:E484K, known as Iota (B.1.526, New York origin) (Fig. 2, C–E) (18).
Fig. 2.
Detection of Iota (B.1.526, New York) SARS-CoV-2 variant by fragment analysis. (A), Expected deletions present in Gamma, (P.1, Brazil), Beta (B.1.351, South Africa), and Iota (B.1.526, New York) VOCs compared to WT; (B), Electrophoretograms representative of clinical WT SARS-CoV-2 infection and one with SARS-CoV-2 exhibiting a 9-bp deletion in ORF1A characteristic of Beta/Gamma/Iota. Integrated genome viewer (IGV) visualization of the WGS reads confirming the presence of the ORF1A:del3675_3677 (C), which is found in Beta/Gamma/Iota. Lack of the N501Y (D) and presence of the E484K (E) mutation along with other mutations led to classification of this SARS-CoV-2 strain as Iota.
Allele-specific primers, S:N501Y. In additional to detecting amplicons of different sizes, fragment analysis can detect SNPs by either restriction digestion or allele-specific primers. We chose allele-specific primers to avoid additional handling and off-target effects of restriction enzymes.
WT specific (FAM) and mutant specific (HEX) fluorescently labeled primers were designed to contain terminal 3′ nucleotides specific to the target allele (Fig. 1, A). To reduce cross-reactivity, mutations were added to the second to last (2′) and third to last (3′) nucleotide to destabilize improper primer binding or extension (Fig. 3, A). S:N501Y was targeted as it increases angiotensin-converting enzyme 2 receptor binding affinity 7-fold and was present in the first 3 VOCs described (Alpha, Beta, and Gamma), representing convergent evolution (15, 17, 19).
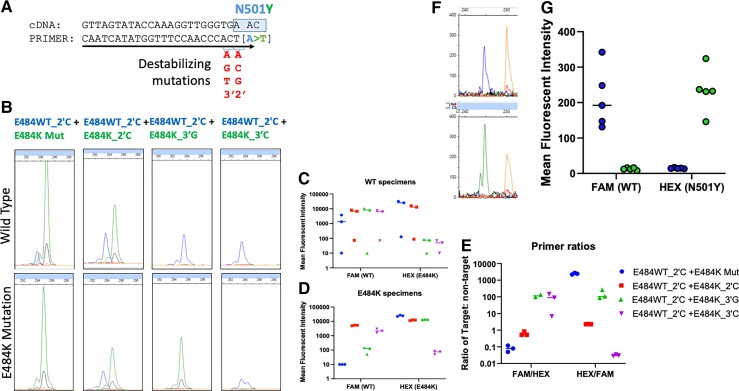
Fig. 3.
Allele-specific primers for S:N501Y and S:E484K. (A), Scheme for designing allele-specific primers with destabilizing mutations; (B), Electrophoretograms of 4 combinations of E484WT- and E484K-labeled primers with (C) graphs quantifying fluorescence intensity in WT specimens, (D) E484K mutated specimens, and (E) the ratio of the 2. (F), Electrophoretogram of signal from N501 and N501Y allele-specific primers; and (G), graphical representation of FAM and HEX signal in response to WT and N501Y specimens.
RT-qPCR with SYBR green amplification detection was performed to screen primers (Supplemental Table 7) for on-target vs off-target amplification. UTSW_N501Y_3′T was labeled with HEX dye as it was the lead candidate with the lowest CT for mutant sequence, a single melt curve peak, and least WT amplification across multiple annealing temperatures (Supplemental Fig. 1). The N501_WT, unmutated primer, was selected for FAM dye labeling. For fragment analysis, the combination of UTSW_N501Y_3′T primer was labeled with HEX, and UTSW_N501_WT was labeled with FAM; this combination properly discriminated WT from mutant sequence (Fig. 3, F and G).
Allele-specific primer, S:E484K. In the Beta and Gamma SARS-CoV-2 variants, the S:E484K mutation in the RBD aids in evasion of specific immune responses (20). As with N501Y targeting, mismatch allele-specific primers were selected for E484K (Supplemental Table 8). These primers used the same common reverse primer as N501Y due to close genomic proximity. UTSW_E484_WT_2′C had the lowest CT value against WT sequence (WT_2′C-ON) (Supplemental Fig. 2). The WT_2′TC-OFF cross-reactive CT value was higher at 60°C (CT = 31) compared to the mutant-specific signal at 60°C, so primer UTSW_E484_WT_2′C was labeled with FAM. UTSW_E484K_3′TA primer amplified its intended target at a very low CT value (4.0–4.5) across annealing temperatures with single melt curve peaks. Additionally, amplification was undetectable for WT sequence (Supplemental Fig. 2). Therefore, primer UTSW_E484K_3′A was selected and labeled with HEX.
However, when this primer combination was tested by capillary electrophoresis, HEX signal was absent when the S:E484K mutation was present, but cross-reactive FAM signal remained. Therefore, 4 alternative primers (E484K_Mut, E484K_2′C, E484K_3′G, and E484K_3′C) were labeled with HEX and tested against WT (n = 3) and E484K specimens (n = 3; Iota, New York variants) (Fig. 3, B). When tested against these specimens, primers were eliminated for (a) amplifying WT sequence (E484_Mut, E484K_2′C) (Fig. 3, B and C) or (b) not amplifying mutant sequence (E484K_3′C) (Fig. 3, B and D). The combination of E484WT_2′C+E484K_3′G produced a specific signal for both WT and E484K by capillary electrophoresis (Fig. 3, E); therefore, the combination of E484WT_2′C+E484K_3′G was selected for the assay.
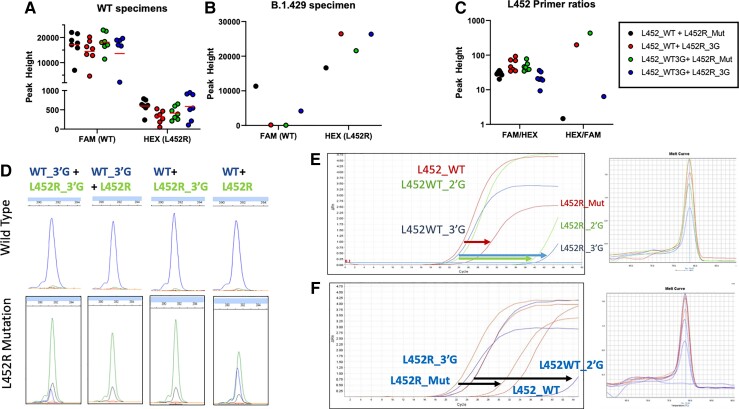
Allele-specific primer S:L452R. In March 2021, the Epsilon variant (B.1.427/B.1.429) from California emerged with a mutation (S:L154R) resistant to therapeutic antibodies (bamlanivimab) (21). The site is a hotspot for variation as it is also present all 3 Indian variant lineages (B.1.617.1, B.1.617.2, B.1.617.3), and an alternative mutation, S:L452Q, is present in the Lambda variant (C.37).
As in silico prediction models used for S:E484K allele-specific primers did not produce optimal results, we created all possible mutations in the terminal second and third nucleotides of primers (Supplemental Table 9). All primers were combined with the common reverse primer used for N501Y and E484K reactions (Common_230) and cDNA for (a) the WT or (b) S:L452R allele to compare amplification by qPCR CT value.
The WT_3′G primer had the lowest CT value (closest to the unmutated WT primer on-target binding) (Fig. 4, E; Supplemental Fig. 3). When presented with the L452R mutant, the WT_3′G primer did not amplify mutant sequence (Fig. 4, F; Supplemental Fig. 4). The L452R_3′G mutant-specific primer amplified mutant sequence as well as the unmutated L452R_Mut-specific primer. All mutated L452R primers (including L452R_3′G) did not amplify WT sequence (Supplemental Figs. 3–4). Melt curve analysis confirmed all PCR reactions amplified a single PCR product (Fig. 4, E and F).
Fig. 4.
Allele-specific primers for S:L452R. (A), Electrophoretograms of 4 combinations of L452WT- and L452R-labeled primers; (B, C), RT-qPCR curves of various L452WT- and L452R-specific primers binding to either WT or mutant sequences. Arrows demonstrate how the same primer is less reactive to WT sequence (C) or how mutant primers are less reactive to WT sequence (B). Melting curves are shown to the right to confirm production of a single PCR product for each case. (D), Graphs quantifying fluorescence intensity in WT specimens and (E) a B.1.429 mutated specimen positive for S:L452R, and (F) the ratio of the 2.
Selected mutated primers (WT_3′G, L452R_3′G) and nonmutated (WT, L452R_Mut) primers were tested by capillary electrophoresis (Fig. 4, A–D). Four combinations were created and reacted against WT (n = 7) and S:L452R sequence (n = 1 Epsilon) from patient specimens. The combination of WT+L452R_3′G and WT_3′G+L452R_Mut created a strong, specific WT signal (Fig. 4, A). However, WT+L452R and WT_3′G+L452R_3′G were eliminated for off-target WT signal against mutant sequence (Fig. 4, E). The WT_3′G+L452R_Mut combination was selected for the assay because it had higher HEX/FAM ratio than WT+L452R_3′G (Fig. 4, F).
Discriminating Gamma (P.1) vs Beta (B.1.351) SARS-CoV-2 variants. The targets described so far produce an identical mutational signature in Beta and Gamma VOCs (ORF1A 9 bp deletion, N501YMUT, E484KMUT). Thus, additional targets are required to discriminate these variants. An RDR4 spike deletion (S:Δ242_244) is present in Beta but absent in Gamma variants. Conversely, a 4 bp insertion between ORF8 and N genes is present in Gamma but absent from Beta SARS-CoV-2 VOCs. Thus, we created fluorescently labeled primers spanning the RDR3–4 and ORF8/N regions to create fragments 284 and 226 bp in size, respectively, that would not interfere with other targets. Adding these ORF8 and RDR3–4 fragment analysis targets completed assay design, providing a unique mutational signature for these major VOCs.
multiplex optimization
CoVarScan was optimized to balance PCR product size by annealing temperature and primer input levels. The shortest amplicons often oversaturated the fluorescent detectors while several longer PCR products had much lower intensity. To optimize primer concentrations, the highest peak height PCR product (RDR2 and ORF1A) primers were reduced to 50 nM. Additionally, RDR3–4, ORF8, N501Y, and E484K PCR product detection was much improved at 200 nM primer concentrations (Supplemental Table 1). Annealing temperatures from 60°C to 65°C were tested, and the optimal performance was set at 61°C (Supplemental Fig. 5).
clinical verification
To validate CoVarScan for clinical use, we used SARS-CoV-2 WGS as a comparative method to confirm the variants detected were accurate. Specificity was established using 34 SARS-CoV-2–negative specimens, 60 SARS-CoV-2–positive WT specimens, and 20 nontemplate control samples. WGS experienced contamination in the nontemplate control samples (n = 9). WGS contamination results from multiple rounds of PCR and may require multiple negatives to performed in a single run. The lack of contamination in CoVarScan by comparison demonstrates the robustness of the assay. Fragment analysis unexpectedly detected 3 L452R and 2 N501Y mutations in the WT sample batch. Upon review, all mutations were analytically accurate. As the N501Y mutation is not variant defining, samples with N501Y were included, and 2 of the L452R mutated cases were called Epsilon variants by updated (more accurate) bioinformatic software, so were excluded. Thus, only one of the L452R mutated cases was called a false positive, and the remainder of the samples were negative for signal (n = 34 negative samples, n = 20 nontemplate control samples) or negative for a variant (n = 57 WT), resulting in a negative percent agreement of 99.1% (111/112, Supplemental Table 10).
In total, 3544 specimens were sent for WGS, but 166 (99 + 67) (Supplemental Table 11) were excluded for not returning a lineage, leaving 3378 total results. Positive percent agreement was 95.0% (3210/3378). Variants sampled included Alpha (n = 125), Beta (n = 3), Gamma (n = 21), Delta (n = 2820), Lambda (n = 5), Mu (n = 6), Iota (n = 6), and Omicron (n = 309). The discrepancy rate was 1.0% (32/3242), which was most likely caused by variability in sensitivity of the two assays. Overall sensitivity of CoVarScan was 96.0% (3242/3378) (Supplemental Table 12).
Limit of detection. Limit of detection studies used Alpha variant control RNA in triplicate diluted serially from 1000 copies to a single copy/reaction. Triplicate testing detected all targets at 63 copies/reaction, and the three core targets were detected at 4 copies/reaction (Supplemental Fig. 6). Sensitivity was confirmed at 100 copies/reaction (full 8-plex panel) and 5 copies/reaction (core panel targets: RDR1, RDR2, ORF1A) with 20 replicates showing 95% detection for the full and core panel, respectively (Supplemental Table 13, Supplemental Fig. 6).
Method comparison of 8-plex PCR to WGS. PCR is often more sensitive compared to massively parallel sequencing methods, and we compared the ability to classify variants by 8-plex fragment analysis vs WGS at various CT values. Percent positivity for both methods was >90% for CT < 32, dropping to 74% for the PCR method at a CT value of 34 and 35 (Supplemental Table 14, Supplemental Fig. 7).
emerging variants
A common issue facing variant genotyping approaches is that they must be adaptable to novel emerging variants. We hypothesized that targeting mutational hotspots with fragment analysis was sufficient to evaluate mutational signatures specific to any SARS-CoV-2 variant (Fig. 5, B; Supplemental Table 4). Thus, the final validated assay method was used from April 2021 onward without modification (Fig. 5, A).
Fig. 5.
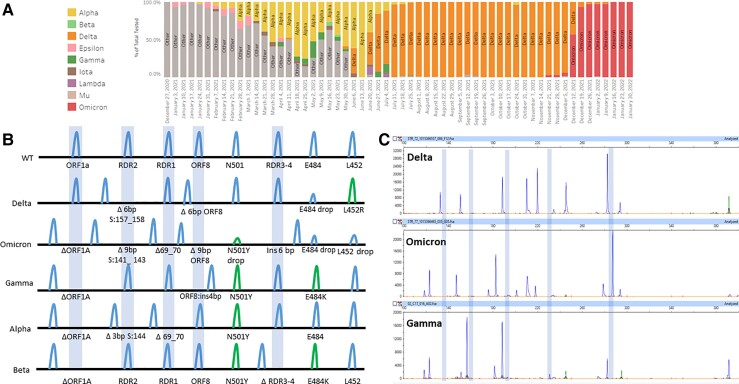
Epidemiologic data derived from mutational signatures. (A), Epidemiologic data produced from the fragment analysis and WGS; (B), Mutational signatures of the major variants of concern as detected by fragment analysis; (C), Electrophoretograms of Gamma, Delta, and Omicron variants.
In April 2021, B.1.617.2 was the first variant to be encountered that was not accounted for in 8-plex assay design. However, we found a characteristic mutational signature: RDR2Δ6 bp + ORF8Δ6 bp + S:L452RMut + S:E484drop out. S:E484 target dropout was attributed to the S:478K mutation overlapping the E484 primer binding site. The RDR2 deletion matches the S:Del157_158 mutation (Fig. 5, C), and the ORF8 deletion matches the ORF8:Del119_120 mutation characteristic of the Delta VOC.
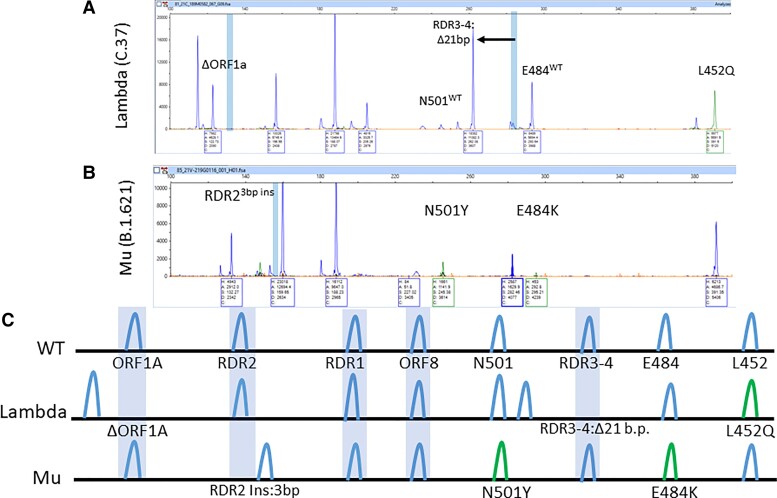
Two months later, in June 2021, the Lambda variant from Peru (C.37) was found in 5 specimens and was defined by the signature of ORF1AΔ9 bp + RDR3_4Δ21 bp + L452RMut. Although Lambda has an L452Q mutation present, the mutant-specific primer cross-reacts and is able to contribute to the unique mutation signature (Fig. 6, A and C).
Fig. 6.
Expected and actual mutational signatures of Lambda and Mu variants. (A), Electrophoretogram of Lambda variant (C.37) characterized by ORF1A Δ9 bp, RDR3–4 Δ21 bp, and L452Mut; (B), Electrophoretogram of Mu variant (B.1.621) characterized by RDR2 3 bp insertion, N501Y, and E484K; (C), Expected mutational signature of Lambda and Mu variants.
The Mu variant (B.1.621) from Colombia was concerning due to similar RBD mutations as Beta and Gamma VOCs when found in July but could be distinguished by its unique mutational signature: RDR2Ins3 bp + N501YMut + E484KMut (Fig. 6, B and C). This insertion at RDR2 indicates the RDR regions are prone not only to deletions but insertions too. This further demonstrates a key feature of the fragment analysis assay to detect PCR amplicons of different sizes in hotspot regions.
Most recently, the Omicron variant emerged and was phenotypically linked with increased spread and immune resistance (22). The key mutations of this variant created a unique signature that was predicted (Fig. 5, B), observed (Fig. 5, C), and WGS confirmed. The mutational signature is ORF1AΔ9 bp, RDR2Δ9 bp, RDR1Δ6 bp, RDR3–4ins6 bp, and N501YMut. The characteristic deletion pattern was detected in 3 cases of Omicron and allowed early reporting and tracking by the public health department.
In total, positive percent agreement is 95.0% concordant with WGS. Thus, the 8-plex CoVarScan assay has identified SARS-CoV-2 variants with accuracy comparable to that of the WGS reference method (Supplemental Table 11).
Discussion
Here we have validated a novel fragment analysis-based approach to analyze SARS-CoV-2–positive respiratory specimens for VOCs in a clinical setting. Among the advantages are decreased time to result, adaptability to new variants without modification, and use of noncomplex, routine instrumentation and analysis.
Prospectively, the application of a reliable method for VOC determination provides economic advantages. In the United States, the Delta VOC became dominant in July 2021. In practice, if a specimen was identified as Delta (or any other dominant VOC) by this assay, it would not require sequencing as WGS is primarily used for the detection of emerging variants. Thus, screening for epidemiologically dominant VOCs using a cost-effective method allows conservation of WGS resources for investigation of nondominant VOC lineages, enabling more targeted variant monitoring in SARS-CoV-2–positive samples. Based upon our estimates, when the prevalence of a VOC exceeds 40% to 50%, it may be more cost-effective to screen samples routinely by fragment analysis with reflex to WGS for nonpredominant lineages compared to prospective WGS on all specimens.
Fragment analysis differs from RT-PCR–based screening methods for VOCs in several ways. While various nonsequencing approaches have been described to screen for SARS-CoV-2 VOCs, they lack the flexibility and scalability of fragment analysis. Some RT-PCR approaches rely upon allele-specific molecular beacon probes that discriminate between WT and mutant alleles by identifying shifts in high-resolution melt curves (23–25) with S:N501Y/S:E484K alleles or dot plot populations in digital droplet PCR (26). While some commercial assays have used Taqman probe methods to successfully multiplex 4 targets (27, 28) in a single reaction, fluorescent channels are limited to 4 to 5 channels in most RT-PCR platforms (27). Similarly, multiplex RT-qPCR has been described for detection of VOCs by targeting the ORF1AΔ9 bp and RDR2Δ3 bp deletion (18, 29). A CRISPR-Cas13 method can also discriminate variant alleles but can only do so one at a time (30). A drawback to these approaches is that assay throughput may be lower (and more expensive) if multiple wells are required to test enough targets for classification. While these tests are useful for screening, our method can classify a variant with similar accuracy as sequencing without target modification. Novel VOCs have continued to emerge, but despite the diversity of these lineages, CoVarScan has not required modification since April 2021, differentiating all named VOCs (31, 32). This adaptability is an important characteristic as new variants emerge.
Additionally, some RT-qPCR assays detect variants by target loss such as with S-gene target failure in the ThermoFisher TaqPath assay (14). This method has been used in the United Kingdom to track the prevalence of B.1.1.7 and now Omicron variants. However, there are S-gene target failure cases that arise sporadically (33). In contrast, this assay gives improved specificity by measuring the size of multiple deletions. Mutational signatures of the 8 targets can be as epidemiologically specific as WGS. This feature differentiates fragment analysis from other approaches that merely screen for variants. For example, the epidemiologic specificity of ORF1AΔ among the most prevalent variants with ORF1AΔ mutations is 88% (567 563/643 640); S: Δ144, 98% (553 578/563 775); and S: Δ69_70, 96% (572 995/594 454). Considered together, these mutations are 98% specific for Alpha, differentiating it from other SARS-CoV-2 strains that may have one of the other deletions (34).
CoVarScan has high-throughput capacity as it can be performed in a single well with 96 tests run at a single time by capillary electrophoresis with autoinjection (ABI 3730XL). Result interpretation requires approximately 1 h for 96 samples but can be automated using PCR fragment size-specific “bins” in the GeneMapper Software (Applied Biosystems). This is an important feature as genotyping variant assays have emerging clinical utility (35).
Some limitations of this assay exist. It is not as sensitive as some RT-PCR methods. However, the assay has sensitivity similar to or higher than most WGS platforms (CT <28–30). With an increasing number of targets, primer dimers may form; however, the length of these dimers should all be <70 bp, so they should not interfere with classifying the current targets of this assay. Some recurrent artifacts have been found, but they do not overlap with any targeted PCR amplicons (Supplemental Table 6). This assay cannot identify all possible SNPs that may occur. CoVarScan thus cannot differentiate Delta from “Delta Plus.” Some mutations such as the S:K417N mutation in “Delta Plus” (AY.1/AY.2) were concerning for antibody resistance but remained <0.1% of total Delta lineages.
In summary, we have applied fragment analysis to SARS-CoV-2 VOC detection for the first time. CoVarScan can detect characteristic viral genomic changes found in all variants at single-nucleotide resolution. The assay is cost-effective, rapid, and scalable. Extensive testing in a routine clinical setting has revealed this fragment analysis–based approach performs as accurately as WGS, supporting its use for both rapid variant surveillance and clinical decision-making if significant resistance to an antibody therapy is found.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical assistance of Vanessa Torres and Ginger Koetting. Additionally, we thank Prithvi Raj and Vanessa Schmid for sequencing assistance, and all members of the Once Upon a Time Human Genomics laboratory, UTSW COVID diagnostic laboratory, the UTSW clinical microbiology laboratory, and Texas Health Resources laboratories for archiving and retrieving specimens.
Contributor Information
Andrew E Clark, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Zhaohui Wang, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Emily Ostman, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Hui Zheng, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Huiyu Yao, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Brandi Cantarel, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA; Bioinformatics Core Facility, University of Texas Southwestern Medical Center, Dallas, TX, USA; Department of Bioinformatics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Mohammed Kanchwala, McDermott Center for Human Growth and Development, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Chao Xing, McDermott Center for Human Growth and Development, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Li Chen, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Pei Irwin, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Yan Xu, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Dwight Oliver, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Francesca M Lee, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Jeffrey R Gagan, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Laura Filkins, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA; Department of Pathology and Laboratory Medicine, Children’s Health System of Texas, Dallas, TX, USA.
Alagarraju Muthukumar, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Jason Y Park, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA; McDermott Center for Human Growth and Development, University of Texas Southwestern Medical Center, Dallas, TX, USA; Department of Pathology and Laboratory Medicine, Children’s Health System of Texas, Dallas, TX, USA.
Ravi Sarode, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Jeffrey A SoRelle, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Nonstandard Abbreviations
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VOC, variants of concern; WGS, whole-genome sequencing; SNP, single nucleotide polymorphism; RBD, receptor-binding domain; CT, cycle threshold; WT, wild-type; RDR, recurrently deleted region.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
J.Y. Park, Clinical Chemistry, AACC.
Consultant or Advisory Role
J.Y. Park, scientific advisory board member of HU Group (Miraca Holdings); Baylor Genetics, Fujirebio and SRL Labs are subsidiaries. L. Filkins, Scientific Advisory Board member for Avsana Labs.
Stock Ownership
J. SoRelle, Myriad Genetics; L. Filkins, restricted stock, Avsana Labs.
Honoraria
None declared.
Research Funding
J. SoRelle, residual COVID-19 positive samples, paying for epidemiologic surveillance with the method, sequencing costs from UTSW; L. Filkins, Biofire Diagnostics, Biomerieux.
Expert Testimony
None declared.
Patents
J. SoRelle is listed as an inventor for a pending patent application based on this work.
Other Remuneration
L. Filkins, support for attending meetings and/or travel from ASM Clin Micro Open 2019, 12/5-12/7/2019.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Previous Publication
A version of this article was previously posted as a preprint on medRxiv as https://www.medrxiv.org/content/10.1101/2021.04.15.21253747v1.
References
- 1. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021;593:266–9. [DOI] [PubMed] [Google Scholar]
- 2. Muik A, Wallisch A-K, Sänger B, Swanson KA, Mühl J, Chen W, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 2021;371:1152–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021;593:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Public Health England . Investigation of novel SARS-CoV-2 variant of concern 202012/10—technical briefing 5 London SE1 8UG. London (UK): Public Health England; 2021. [Google Scholar]
- 5. Iacobucci G. Covid-19: new UK variant may be linked to increased death rate, early data indicate. BMJ 2021;372:n230. [DOI] [PubMed] [Google Scholar]
- 6. Cele S, Gazy I, Jackson L, Hwa S-H, Tegally H, Lustig G, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 2021;593:142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021;29:463–76.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021;184:2384–93.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022;602:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filkins L, SoRelle JA, Schoggins J, Park JY. Laboratory action plan for emerging SARS-CoV-2 variants. Clin Chem 2021;67:720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark AE, Wang Z, Cantarel B, Kanchwala M, Xing C, Chen L, et al. Multiplex fragment analysis identifies SARS-CoV-2 variants. Preprint at https://www.medrxiv.org/content/10.1101/2021.04.15.21253747 (2021). [DOI] [PMC free article] [PubMed]
- 12. Tegally H, Wilkinson E, Lessells RJ, Giandhari J, Pillay S, Msomi N, et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat Med 2021;27:440–6. [DOI] [PubMed] [Google Scholar]
- 13. Nonaka CKV, Franco MM, Gräf T, de Lorenzo Barcia CA, de Ávila Mendonça RN, de Sousa KAF, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis 2021;27:1522–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. ThermoFisher. Solutions for surveillance of the S gene mutation in the B.1.1.7 (501Y.V1) SARS-CoV-2 strain lineage. https://www.thermofisher.com/blog/behindthebench/solutions-for-surveillance-of-the-s-genemutationin-the-b117-501yv1-sars-cov-2-strain-lineage/(Accessed April 16, 2021).
- 15. COVID-19 Genomics UK Consortium. COG-UK report on SARS-CoV-2 Spike mutations of interest in the UK. COVID-19 Genomics UK Consortium. January 15, 2021. https://minhalexander.files.wordpress.com/2021/02/cog-uk-report-on-sars-cov-2-spike-mutations-of-interest-in-the-uk-15-thjanuary-2021-report-2_cog-uk_sars-cov2-mutations.pdf(Accessed December 16, 2021).
- 16. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Preprint at https://www.medrxiv.org/content/10.1101/2020.12.21.20248640(2020).
- 17. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021;372:815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Annavajhala MK, Mohri H, Wang P, Nair M, Zucker JE, Sheng Z, et al. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature 2021;597:703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, et al. Emergence and rapid transmission of SARS-CoV;2, B.1.1.7 in the United States. Cell 2021;184:2587–94.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wise J. Covid-19: the E484K mutation and the risks it poses. BMJ 2021;372:n359. [DOI] [PubMed] [Google Scholar]
- 21. Eli Lilly and Company. Fact Sheet for Healthcare Providers: Emergency Use Authorization (EUA) of Bamlanivimab and Etsevimab. https://pi.lilly.com/eua/bam-and-ete-eua-factsheet-hcp.pdf(Accessed December 16, 2021).
- 22. Madhi SA, Kwatra G, Myers JE, Jassat W, Dhar N, Mukendi CK, et al. Population immunity and COVID-19 severity with omicron variant in South Africa. N Engl J Med 2022;386:1314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banada P, Green R, Banik S, Chopoorian A, Streck D, Jones R, et al. A simple reverse transcriptase PCR melting-temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. J Clin Microbiol 2021;59:e0084521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Y, Lee A, Composto K, Cunningham MH, Mediavilla JR, Fennessey S, et al. A novel diagnostic test to screen SARS-CoV-2 variants containing E484K and N501Y mutations. Emerg Microbes Infect 2021;10:994–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matic N, Lowe CF, Ritchie G, Stefanovic A, Lawson T, Jang W, et al. rapid detection of SARS-CoV-2 variants of concern, including B.1.1.28/P.1, British Columbia, Canada. Emerg Infect Dis 2021;27:1673–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perchetti GA, Zhu H, Mills MG, Shrestha L, Wagner C, Bakhash SM, et al. Specific allelic discrimination of N501Y and other SARS-CoV-2 mutations by ddPCR detects B.1.1.7 lineage in Washington State. J Med Virol 2021;93:5931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seegene Inc . Seegene’s latest COVID-19 test can simultaneously target 4 genes of SARS-CoV-2 and recognize multiple virus variants. https://seegenetech.com/seegenes-latest-covid-19-test-can-simultaneously-target-4-genes-of-sars-cov-2-and-recognize-multiple-virus-variants/ (Accessed December 16, 2021).
- 28. PerkinElmer . PKamp™ VariantDetect™ SARS-CoV-2 RT-PCR assay. https://perkinelmer-appliedgenomics.com/home/sars-cov-2-testing-solutions/pkamp-variantdetect-sars-cov-2-rt-pcr-assay/(Accessed December 16, 2021).
- 29. Vogels CBF, Breban MI, Ott IM, Alpert T, Petrone ME, Watkins AE, et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol 2021;19:e3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hd P, Lee RA, Najjar D, Tan X, Soenksen LR, Angenent-Mari NM, et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv 2021;7:eabh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero PE, Dávila-Barclay A, Gonzáles L, Salvatierra G, Cuicapuza D, Solis L, et al. C.37: Novel sublineage within B.1.1.1 currently expanding in Peru and Chile, with a convergent deletion in the ORF1a gene (Δ3675-3677) and a novel deletion in the Spike gene (Δ246-252, G75V, T76I, L452Q, F490S, T859N). April 2021. https://virological.org/t/novel-sublineage-within-b-1-1-1-currently-expanding-in-peru-and-chile-with-a-convergent-deletion-in-the-orf1a-gene-3675-3677-and-a-novel-deletion-in-the-spike-gene-246-252-g75v-t76i-l452q-f490s-t859n/685(Accessed December 16, 2021).
- 32. McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, Duprex WP. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021;371:1139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. UK Health Security Agency . UKHSA: SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 35. January 28, 2022. https://www.gov.uk/government/publications/ukhsa-sars-cov-2-variants-of-concern-and-variants-under-investigation-in-england-technical-briefing-35-28-january-2022(Accessed December 16, 2021).
- 34. Outbreak.info: a standardized open-source database of COVID-19 resources and epidemiology data. S:del144/145, S:del69/70, ORF1a:del3675/3677 Variant Report 2021. https://outbreak.info/situation-reports?pango&muts=S%3ADEL69%2F70&muts=ORF1A%3ADEL3675%2F3677&muts=S%3ADEL144%2F145(Accessed May 10 2022).
- 35. Greninger AL, Bard JD, Colgrove RC, Graf EH, Hanson KE, Hayden MK, et al. Clinical and infection prevention applications of severe acute respiratory syndrome coronavirus 2 genotyping: an infectious diseases Society of America/American Society for Microbiology consensus review document. J Clin Microbiol 2022;60:e0165921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.