Abstract
Background
Limited data exist on antibody responses to mixed vaccination strategies that involve inactivated coronavirus disease 2019 (COVID-19) vaccines, particularly in the context of emerging variants.
Methods
We conducted an open-label trial of a third vaccine dose of a messenger RNA (mRNA) vaccine (BNT162b2, Fosun Pharma/BioNTech) in adults aged ≥30 years who had previously received 2 doses of inactivated COVID-19 vaccine. We collected blood samples before administering the third dose and 28 days later and tested for antibodies to the ancestral virus using a binding assay (enzyme-linked immunosorbent assay [ELISA]), a surrogate virus neutralization test (sVNT), and a live virus plaque reduction neutralization test (PRNT). We also tested for antibodies against the Omicron variant using live-virus PRNT.
Results
In 315 participants, a third dose of BNT162b2 substantially increased antibody titers on each assay. Mean ELISA levels increased from an optical density of 0.3 to 2.2 (P < .001), and mean sVNT levels increased from an inhibition of 17% to 96% (P < .001). In a random subset of 20 participants, the geometric mean PRNT50 titers rose substantially, by 45-fold from day 0 to day 28 against the ancestral virus (P < .001) and by 11-fold against the Omicron variant (P < .001). In daily monitoring, post-vaccination reactions subsided within 7 days for more than 99% of participants.
Conclusions
A third dose of COVID-19 vaccine with an mRNA vaccine substantially improved antibody levels against the ancestral virus and the Omicron variant with a well-tolerated safety profile in adults who had received 2 doses of inactivated vaccine 6 months earlier.
Clinical Trials Registration
Keywords: COVID vaccine, immunogenicity, booster, BNT162b2, CoronaVac
In this open-label trial of Chinese adults aged ≥30 years who received 2 doses of inactivated coronavirus disease 2019 vaccine 6 months earlier, third-dose messenger RNA vaccine substantially improved antibody levels against the ancestral virus and Omicron variant with a well-tolerated safety profile.
The accrual of population immunity to coronavirus disease 2019 (COVID-19), acquired through natural infection or vaccination, will eventually bring an end to the pandemic and allow life to return to normal. Major vaccine technologies being used for COVID-19 include inactivated-virus, viral-vectored, recombinant protein-based, and messenger RNA (mRNA) vaccines [1]. Previously, we showed that 2 doses of the mRNA vaccine BNT162b2 (Fosun Pharma/BioNTech) conferred approximately 10-fold higher post-vaccination neutralizing antibody titers than 2 doses of the aluminum hydroxide–adjuvanted inactivated virus vaccine CoronaVac (Sinovac) [2, 3], while T-cell responses to the 2 vaccines were similar [3]. The emergence of variants of concern and decreases in vaccine effectiveness within a few months after the second vaccine dose have resulted in recommendations for third doses [4].
Various studies have reported immunogenicity and safety of third doses using the same vaccine, that is, homologous booster vaccination [5, 6]. However, few studies have evaluated a heterologous booster, such as the use of a third dose of an mRNA vaccine in individuals who previously received 2 doses of inactivated vaccine [7–11]. Costa Clemens et al conducted a noninferiority, randomized trial of a heterologous third dose of BNT162b2 or other vaccines against a homologous third dose of CoronaVac in adults who had received 2 doses of CoronaVac [8]. They showed that all heterologous booster groups had a substantial rise in neutralizing antibody titers against the Omicron and Delta variants [8]. They also reported more local reactions but fewer systemic reactions to BNT162b2 compared with adenoviral-vectored vaccines given as third doses.
Here, we report a trial of the immunogenicity and reactogenicity to a third dose of BNT162b2 in Chinese adults who had previously received 2 doses of inactivated COVID-19 vaccine.
METHODS
Study Design
The “mRNA vaccination to boost antibodies against SARS-CoV-2 in recipients of inactivated vaccines” (mBoost) study is an open-label, single-arm, clinical trial to measure the antibody responses and reactogenicity of an mRNA vaccine (BNT162b2) given as a third dose in adults aged ≥30 years who previously received 2 doses of an inactivated COVID-19 vaccine. CoronaVac and BNT162b2 have been available to adults in Hong Kong since March 2021. Some Hong Kong residents could have received 2 doses of inactivated vaccine BIBP (Sinopharm) instead from mainland China or overseas. The BNT162b2 vaccine used in Hong Kong, known as the Pfizer/BioNTech vaccine elsewhere, is distributed solely by Fosun Pharma in Greater China.
Enrollment invitations were extended to community-dwelling adults in Hong Kong through mass promotion efforts including advertisements in newspapers and social media. Interested adults were invited to visit the study website and complete an online screening form for initial assessment of enrollment eligibility and confirmed again shortly before vaccination. Individuals were eligible if they were aged ≥30 years and had previously received 2 doses of an inactivated COVID-19 vaccine, with the most recent dose ≥90 days prior to enrollment. We excluded individuals who reported a history of COVID-19 infection, received any dose of COVID-19 vaccine other than an inactivated vaccine, or were unsuitable to receive an mRNA vaccine including but not limited to allergies to the active substance or other vaccine ingredients. We also excluded individuals with diagnosed medical conditions related to their immune system, those who used medication that impairs the immune system in the last 6 months except for topical steroids and short-term oral steroids (course lasting ≤14 days), those who had used immunoglobulins or any blood products within 90 days prior to enrollment, and any females who were pregnant or intending to become pregnant in the coming 3 months.
We collected 20-mL clotted blood specimens on the day of enrollment and vaccination (day 0) and again after 28 days, with additional blood draws planned after 182 and 365 days. This was a single-arm study with no need for randomization, and the participants and the study staff were aware of the type of vaccination administered (no blinding). After vaccination, participants were observed for 30 minutes to record any immediate events. We then asked participants to report the presence of local or systematic events/reactions daily (post-vaccination) for 7 days using an online e-diary. If the participant was still reporting any events/reactions on day 7, additional daily monitoring continued for up to 3 additional weeks until symptoms resolved. On each day, participants were also asked to grade whether the reported symptoms overall had interfered with their usual activities (mild, moderate, and severe). Participants are interviewed at 28, 182, and 365 days after vaccination to collect information on underlying medical conditions and any medical encounters including hospitalizations during the study. Participants were provided with a free tympanic thermometer at enrollment and a gift voucher of $13.00 at the follow-up blood draws. Study data were collected and managed using REDCap electronic data capture tools [12, 13].
Ethics
Written informed consent was obtained from all participants. The University of Hong Kong Institutional Review Board approved the study protocol.
Outcome Measures
The primary outcome measure is the vaccine immunogenicity at 28 days after receipt of the third dose of BNT162b2, measured as geometric mean titer (GMT) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serum neutralizing antibodies against the vaccine strain (ie, the ancestral virus) using the plaque reduction neutralization test (PRNT). As the Omicron strain emerged in late 2021, we also evaluated vaccine immunogenicity against the Omicron strain. The secondary outcome measures include the incidence of solicited reactions or medical encounters following vaccination and the GMT at other post-vaccination time points (days 182 and 365) and their corresponding geometric mean fold-rise (GMFR) from baseline, for which data collection is continuing and the results will be reported in due course.
Sample Size Justification
For the primary outcome of vaccine immunogenicity, that is, the GMT of SARS-CoV-2 serum neutralizing antibodies against the vaccine strain (ancestral virus) at 28 days after vaccination, a sample size of 300 individuals was chosen based on feasibility [14].
Laboratory Methods
Blood samples were delivered to our study laboratory at the University of Hong Kong as soon as possible, with the optimal delivery time within 24 hours. Sera were extracted from the clotted blood within 48 hours and stored at −80°C until subsequent testing. We tested heat-inactivated sera at 56°C for 30 minutes with 3 assays: our in-house enzyme-linked immunosorbent assay (ELISA) for the receptor-binding domain (RBD) of the spike protein, a surrogate virus neutralization test (sVNT; GenScript), and PRNT as previously described [10, 15, 16]. We aimed to test sera collected at baseline and day 28 with the ELISA and sVNT in all participants and with PRNT in a randomly selected subset of 20 participants. We demonstrated a good correlation (r = 0.77) between PRNT50 and sVNT neutralization percentage for the ancestral virus in our earlier studies [17]. The ELISA was not designed as a quantitative assay, although it has a dynamic range of between 0 and 5. We have demonstrated a good correlation (r = 0.83) between ELISA optical density (OD)450 and sVNT neutralization percentage for the ancestral virus [17]. For ELISA, a single 1:100 serum dilution was tested; for sVNT, a single 1:10 serum dilution was tested. For PRNT, serial 2-fold serum dilutions from 1:10 to 1:320 were tested. PRNT assays were carried out using ancestral SARS-CoV-2 BetaCoV/Hong Kong/VM20001061/2020 isolated in Hong Kong in January 2020 in Vero-E6 cells (American Type Culture Collection CRL-1586) and the Pango lineage B.1.1.529 Omicron variant designated hCoV-19/Hong Kong/VM21044713_WHP5047-S5/2021 isolated in Vero-E6 TMRSS2 cells (Vero E6 cells overexpressing TMPRSS2, kindly provided by Dr S Matsuyama and colleagues). The passage level 3 virus aliquots were used. Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin–streptomycin (all from ThermoFisher Scientific, Waltham, MA). Virus sequences are available from the Global Initiative on Sharing All Influenza Data as EPI_ISL_412028 and EPI_ISL_6716902. The PRNT50 and PRNT90 titers were the highest serum dilutions neutralizing ≥50% and ≥90% of input plaques, respectively. The World Health Organization control serum provided by the National Institute for Biological Standards and Control 20/136 gave PRNT50 titers of 320 and 320 against the ancestral virus and 20 and 40 against the Omicron variant in 2 titrations [10].
Statistical Analyses
We assessed the GMT of SARS-CoV-2 (PRNT) neutralizing antibody titers, surrogate virus neutralization percentages, and the mean concentrations of SARS-CoV-2 spike RBD immunoglobulin (Ig) G (proxy by OD450) at day 28. For sVNT, measured negative neutralization percentages were transformed to zero. PRNT titers were taken as the reciprocal of the serum dilution and were interval-censored, for example, a sample that was able to neutralize virus at a 1:20 dilution but not at a 1:40 dilution was reported as 20 to indicate that the titer was ≥20 and <40. We imputed titers <10 with the value 5 and titers ≥320 with the value 640 for estimation of GMTs. We compared antibody levels at day 28 after receipt of BNT162b2 to baseline using Wilcoxon signed rank tests. Correlation of PRNT90 titers measured at day 28 against the ancestral strain and the Omicron variant was estimated using the Spearman rank correlation coefficient. Reactogenicity end points were described as frequency (%) for local and systemic reactions or events among participants who reported health status for at least 1 day in the week following receipt of the third dose of BNT162b2. Statistical analyses were conducted using R version 4.1.2.
RESULTS
From 13 October 2021 through 18 January 2022, we screened 436 individuals, of whom 366 (84%) were eligible and 315 (86%) were enrolled and administered the BNT162b2 vaccine. We collected paired day 0 and day 28 blood samples in 312 (99%) vaccinated participants. Most participants were older Chinese adults (median age, 54 years; interquartile range, 47–62; 35% were obese, and about one-third reported at least 1 chronic medical condition including hypertension (18%), hypercholesterolemia (13%), and diabetes (7%); and almost all (98%) reported receiving 2 doses of CoronaVac rather than another inactivated vaccines (Table 1). Although adults who had received 2 doses of inactivated virus vaccine at least 90 days earlier were eligible to enroll in our study (Supplementary Figure 1), 75% of participants reported receiving the second dose typically around 6–7 months earlier (Table 1). A detailed flow chart of participant enrollment is provided in Supplementary Figure 1. The study is ongoing and day 182 and 365 samples will be collected in spring 2022 and autumn 2022, respectively.
Table 1.
Characteristics of Study Participants at Baseline
| Characteristic | Participants Tested Using ELISA and sVNT | Random Subset of 20 Participants Tested Using ELISA, sVNT, and PRNT |
|---|---|---|
| (N = 312) | (N = 20) | |
| n (%) | n (%) | |
| Female | 120 (38) | 9 (45) |
| Age (median, IQR), years | 54 (47–62) | 55 (47–64) |
| Ethnicity | ||
| ȃChinese | 306 (98) | 20 (100) |
| Obesity (for Asian populations), body mass index | ||
| ȃUnderweight (<18.5) | 6 (2) | 0 (0) |
| ȃNormal (18.5–22.9) | 114 (37) | 5 (25) |
| ȃOverweight (23.0–24.9) | 84 (27) | 7 (35) |
| ȃObese (≥25.0) | 108 (35) | 8 (40) |
| Chronic medical conditions | ||
| ȃAny | 98 (32) | 6 (30) |
| ȃLung disease, including chronic obstructive pulmonary disease and asthma | 1 (0) | 0 (0) |
| ȃHeart disease | 7 (2) | 0 (0) |
| ȃHypertension | 57 (18) | 5 (25) |
| ȃDiabetes | 22 (7) | 1 (5) |
| ȃHypercholesterolemia | 42 (13) | 3 (15) |
| ȃKidney disease | 4 (1) | 0 (0) |
| ȃLiver disease | 4 (1) | 1 (5) |
| ȃCancer | 5 (2) | 1 (5) |
| Prior COVID-19 vaccination | ||
| ȃ2-dose CoronaVac (Sinovac) | 305 (98) | 20 (100) |
| ȃ2-dose BIBP (Sinopharm) | 7 (2) | 0 (0) |
| Days between first and second dose of COVID-19 vaccination, median (IQR) | 28 (28–29) | 28 (28–29) |
| Days between second and third (study) dose of COVID-19 vaccination, median (IQR) | 206 (192–217) | 197 (172–208) |
| Smoking | ||
| ȃEver | 33 (11) | 5 (25) |
| ȃCurrent | 22 (7) | 3 (15) |
We enrolled 315 adults who previously received 2 doses of inactivated COVID-19 vaccine and administered the BNT162b2 vaccine as a third vaccine dose. Among them, ELISA and sVNT against ancestral severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus were performed in paired day 0 and day 28 sera available from 312 vaccinated participants, and we randomly selected 20 participants and also performed PRNT against the ancestral SARS-CoV-2 virus and the Omicron variant.
Abbreviations: COVID-19, coronavirus disease 2019; ELISA, enzyme-linked immunosorbent assay; IQR, interquartile range; PRNT, plaque reduction neutralization test; sVNT, surrogate virus neutralization test.
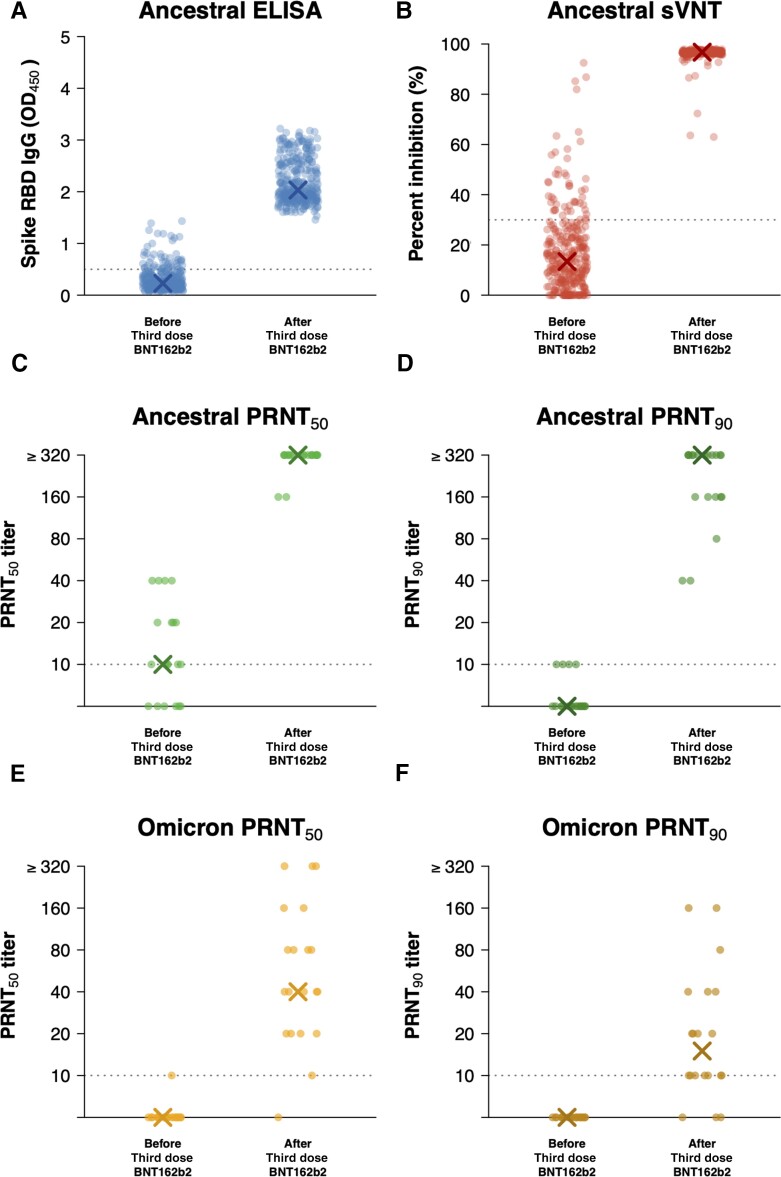
The third dose of BNT162b2 substantially increased antibody titers measured using the various assays (Figure 1). Mean ELISA levels increased from an OD of 0.3 to 2.2 (P < .001), and mean sVNT levels increased from an inhibition of 17% to 96% (P < .001; Table 2). We randomly selected a subset of 20 participants for further PRNT testing against the ancestral virus and the Omicron strain (Table 1, Supplementary Table 1). The ELISA and sVNT levels in these 20 participants were similar to levels in the 312 vaccinated participants (Supplementary Figure 2). In these 20 participants, the geometric mean PRNT50 titer (GMTPRNT50) against the ancestral strain at day 0 was 12. Assuming a titer value of 640 for each of the 18 of 20 (90%) day 28 sera samples that were positive at the highest dilution of 1:320, we estimated the GMTPRNT50 at day 28 to be 557. The corresponding GMFRPRNT50 from day 0 to day 28 was estimated to be 45 (P < .001). The geometric mean PRNT90 titer (GMTPRNT90) against the ancestral strain at day 0 was 6. At day 28, 12 of 20 samples were positive at the highest dilution of 1:320, and similarly we estimated the GMTPRNT90 at day 28 to be 309, with the corresponding GMFRPRNT90 to be 54 (P < .001). Furthermore, in these same 20 participants, the GMTPRNT50 against the Omicron strain at day 0 was 5 and rose to 59 at day 28, a GMFRPRNT50 of 11 given that the day 0 titers were almost all assigned values of 5 corresponding to the floor of the assay at <10 (P < .001). The GMTPRNT90 against the Omicron strain at day 0 was also 5 and rose to 19 at day 28, giving a GMFRPRNT90 of 4 (P < .001). We did not see any evidence to suggest a correlation between the PRNT90 titers against the Omicron variant and that against the ancestral virus (r = 0.30, P = .21; Supplementary Figure 3).
Figure 1.
Third dose of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) BNT162b2 vaccine boosts serum antibodies against the ancestral strain and Omicron variant in adults who previously received 2 doses of inactivated vaccines. A, ELISA for serum IgG against SARS-CoV-2 spike RBD of the ancestral strain. B, sVNT against the ancestral strain. Live virus PRNT against ancestral strain with end points at (C) 50% (PRNT50) or (D) 90% (PRNT90). Live virus PRNT against the Omicron variant with end points at (E) 50% (PRNT50) or (F) 90% (PRNT90). X in each panel indicates the median level. Data for ELISA and sVNT were available from 312 vaccinated participants for whom paired day 0 and day 28 sera were collected, while data for PRNT were available from a random sample of 20 participants. Abbreviations: ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; OD, optical density; PRNT, plaque reduction neutralization test; RBD, receptor-binding domain; sVNT, surrogate virus neutralization test.
Table 2.
Antibody Response After Third-Dose BNT162b2 Vaccine in Adults Who Previously Received 2 Doses of Inactivated Coronavirus Disease 2019 Vaccine
| Summary Measure | Participants Tested Using ELISA and sVNT | Participants Tested Using ELISA, sVNT, and PRNT | ||||
|---|---|---|---|---|---|---|
| (N = 312) | (N = 20) | |||||
| Day 0 | Day 28 | P Value | Day 0 | Day 28 | P Value | |
| Ancestral ELISA | ||||||
| ȃMean (SD) | 0.3 (0.2) | 2.2 (0.4) | <.001 | 0.3 (0.3) | 2.0 (0.3) | <.001 |
| ȃMedian (IQR) | 0.2 (0.2–0.4) | 2.0 (1.9–2.5) | 0.2 (0.2–0.3) | 1.9 (1.8–2.0) | ||
| Ancestral sVNT | ||||||
| ȃMean (SD) | 17% (16) | 96% (3) | <.001 | 21% (16) | 97% (1) | <.001 |
| ȃMedian (IQR) | 13% (5–23) | 97% (96–97) | 21% (6–32) | 97% (97–97) | ||
| Ancestral PRNT50 | ||||||
| ȃGMT (SD) | N/A | 12 (2) | 557 (2) | <.001 | ||
| ȃMedian (IQR) | 10 (5–20) | ≥320 (≥320–≥320) | ||||
| ȃGMFR | 45 | |||||
| Ancestral PRNT90 | ||||||
| ȃGMT (SD) | N/A | 6 (1) | 309 (3) | <.001 | ||
| ȃMedian (IQR) | 5 (5–5) | ≥320 (160–≥320) | ||||
| ȃGMFR | 54 | |||||
| Omicron PRNT50 | ||||||
| ȃGMT (SD) | N/A | 5 (1) | 59 (4) | <.001 | ||
| ȃMedian (IQR) | 5 (5–5) | 40 (20–100) | ||||
| ȃGMFR | 11 | |||||
| Omicron PRNT90 | ||||||
| ȃGMT (SD) | N/A | 5 (1) | 19 (3) | <.001 | ||
| ȃMedian (IQR) | 5 (5–5) | 15 (10–40) | ||||
| ȃGMFR | 4 | |||||
Abbreviations: COVID-19, coronavirus disease 2019; ELISA, enzyme-linked immunosorbent assay; GMFR: geometric mean fold rise in PRNT titer from day 0 to day 28; GMT, geometric mean titer; IQR, interquartile range; PRNT, plaque reduction neutralization test; SD, standard deviation; sVNT, surrogate virus neutralization test.
ELISA and sVNT against ancestral severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus were performed in paired day 0 and day 28 sera available from 312 vaccinated participants, and we randomly selected 20 participants and also performed PRNT against ancestral SARS-CoV-2 virus and the Omicron variant. Significant differences between antibody levels at day 0 and day 28 were evaluated using Wilcoxon signed rank tests. For the estimation of GMT for PRNT50 and PRNT90 titers, titers <10 were imputed as 5 and titers ≥320 were imputed as 640.
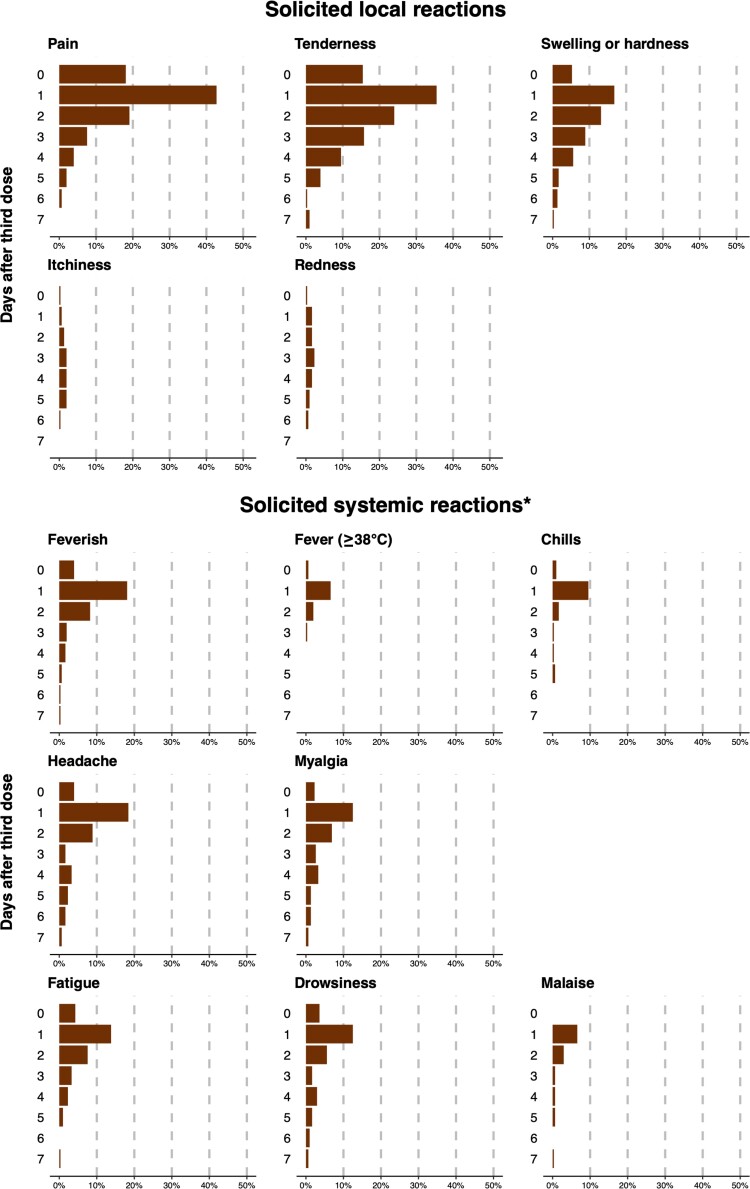
Among the 315 participants who received the BNT162b2 vaccine, 304 (97%) reported health status for at least 1 day in the week following receipt of BNT162b2, including 234 (77%) who reported every day and another 38 (13%) who reported for ≥7 days. There were 193 of 304 (63%) participants who reported feeling unwell for an average of 2.9 days (standard deviation, 1.8 days), and only 3 (<1%) participants reported feeling unwell beyond 7 days after the third dose of BNT162b2 vaccine. Within the 7 days after receipt of BNT162b2, the most commonly reported local reactions were pain (46%) and tenderness (44%) at the injection site, while systemic reactions were reported by a minority of participants (Table 3). These symptoms usually subsided within 7 days after vaccination (Figure 2). Among 312 participants whose information is available, 6 (2%) reported having sought medical consultation within 1 month of the third dose, but none were hospitalized. Among these 6 participants, 4 reported seeking medical consultation possibly for discomfort associated with vaccination within the week after vaccination, and the remaining 2 participants sought medical consultation due to back pain or stress outside the 7-day window.
Table 3.
Solicited Local and Systemic Reactions on the Day After and Anytime Within 7 Days After a Third-Dose BNT162b2 Vaccine in Adults Who Previously Received 2 Doses of Inactivated Coronavirus Disease 2019 Vaccine
| Post-Vaccination Reactions | Anytime Within 7 Days After Third-Dose BNT162b2 Vaccine | Day 1 After Third-Dose BNT162b2 Vaccine |
|---|---|---|
| (N = 304) | (N = 304) | |
| n (%) | n (%) | |
| Local reactions at the injection site | ||
| ȃPain | 141 (46) | 130 (43) |
| ȃTenderness | 133 (44) | 108 (36) |
| ȃSwelling or hardness | 71 (23) | 51 (17) |
| ȃItchiness | 20 (7) | 2 (1) |
| ȃRedness | 15 (5) | 5 (2) |
| Systemic reactions and general symptoms | ||
| ȃFeverish | 72 (24) | 55 (18) |
| ȃFever ≥38.0°C | 25 (8) | 20 (7) |
| ȃChills | 32 (11) | 29 (10) |
| ȃHeadache | 75 (25) | 56 (18) |
| ȃMyalgia | 54 (18) | 38 (12) |
| ȃFatigue | 56 (18) | 42 (14) |
| ȃDrowsiness | 51 (17) | 38 (12) |
| ȃMalaise | 31 (10) | 20 (7) |
| ȃPain at the injection site | 25 (8) | 14 (5) |
| ȃLoss of appetite | 21 (7) | 12 (4) |
| ȃDizziness | 21 (7) | 11 (4) |
| ȃRunny nose | 20 (7) | 12 (4) |
| ȃNasal congestion | 19 (6) | 9 (3) |
| ȃArthralgia | 17 (6) | 10 (3) |
| ȃSneezing | 17 (6) | 8 (3) |
| ȃSore throat | 17 (6) | 10 (3) |
| ȃChest discomfort | 13 (4) | 9 (3) |
| ȃDiarrhea | 13 (4) | 4 (1) |
| ȃAbdominal distention | 11 (4) | 6 (2) |
| ȃPalpitations | 11 (4) | 5 (2) |
| ȃInsomnia | 11 (4) | 5 (2) |
| ȃPhlegm | 10 (3) | 5 (2) |
| ȃNausea | 9 (3) | 5 (2) |
| ȃCough | 8 (3) | 4 (1) |
| ȃAbdominal pain | 7 (2) | 4 (1) |
| ȃConstipation | 6 (2) | 4 (1) |
| ȃBody itching | 5 (2) | 2 (1) |
| ȃFlushing of the face | 4 (1) | 3 (1) |
| ȃSkin rash | 3 (1) | 1 (0) |
| ȃChest pain | 3 (1) | 0 (0) |
| ȃEnlarged lymph nodes | 3 (1) | 0 (0) |
| ȃFace swelling | 2 (1) | 0 (0) |
| ȃArm or leg swelling | 1 (0) | 1 (0) |
| ȃMuscle spasms | 1 (0) | 1 (0) |
| ȃConfusion | 1 (0) | 1 (0) |
| ȃDyspnea | 1 (0) | 0 (0) |
| ȃNosebleeds | 0 (0) | 0 (0) |
| ȃAgeusia | 0 (0) | 0 (0) |
| ȃAnosmia | 0 (0) | 0 (0) |
| ȃConjunctivitis | 0 (0) | 0 (0) |
| ȃVomiting | 0 (0) | 0 (0) |
| ȃFacial drooping/weakness | 0 (0) | 0 (0) |
| ȃLoss of consciousness | 0 (0) | 0 (0) |
| ȃSeizure | 0 (0) | 0 (0) |
Daily frequencies within 7 days after third-dose BNT162b2 vaccine for selected reactions are shown in Figure 2.
Figure 2.
Solicited local and systemic reactions during the 7 days after a third dose of BNT162b2 vaccine in adults who previously received 2 doses of inactivated coronavirus disease 2019 vaccine. *For solicited systemic reactions, only feverish/fever ≥38.0°C/chills and those reported in at least 10% of participants are shown.
DISCUSSION
Work by our research group and others indicates that 2 doses of inactivated COVID-19 vaccine confer moderate increases in antibody levels at 1 month after the second dose [2, 3]. Antibody levels then gradually decline but can be boosted by receipt of a third dose. Zeng et al reported that a homologous third dose of inactivated vaccine given to healthy young adults at 8 months after the second dose boosted neutralizing antibodies against the ancestral strain by about 21-fold [6]. Here, we show that a third dose of mRNA vaccine (BNT162b2) after 2 doses of inactivated virus vaccine (in most cases, CoronaVac), all originally formulated against the ancestral virus, boosted PRNT50 titers against the ancestral strain by 45-fold. Similarly, Costa Clemens et al showed that a third dose of mRNA or adenoviral-vectored vaccines induced greater rises in anti-spike binding antibodies compared with a third dose of inactivated vaccine among CoronaVac recipients [8]. Together, these data suggest a substantial improvement in heterologous boosting using mRNA vaccine compared with homologous boosting using inactivated vaccine against the ancestral virus.
Most participants in the present study were older adults, and their antibody levels against Omicron were at a very low level at day 0 prior to the third dose, consistent with other recent studies [8, 9]. Almost one third of participants had underlying medical conditions (Table 1). We show that a third dose of BNT162b2 conferred substantial rises in GMTPRNT50 against the Omicron variant to 59, a factor of 11-fold. In our separate initial investigation that included sera from individuals with various intervals (51–234 days) between second and third doses [10] and some of whom were selected for third-dose vaccination due to low (<60%) post-second dose serum surrogate neutralizing antibodies [18], administration rates of BNT162b2 about 3 months after 2 doses of CoronaVac showed a GMT of 59 against Omicron (compared with 305 against the ancestral virus after the boost) [10]. In comparison, Costa Clemens et al reported an increase in mean neutralizing antibodies (FRNT50) against Omicron from a baseline titer of 10 to 223 four weeks after a heterologous third-dose of BNT162b2 vaccination, and heterologous boosting with a third dose of mRNA or adenoviral-vectored vaccines induced a greater rise in neutralizing antibodies against both Omicron and Delta variants than homologous boosting with inactivated vaccines [8]. A GMTPRNT50 of 59 against Omicron at day 28 in the present study is higher than the GMTPRNT50 of 27 against ancestral virus after 2 doses of CoronaVac reported in our earlier study [2]. While there is still a lack of consensus on the antibody threshold for protection [19–22], given that 2 doses of inactivated vaccine were estimated to provide at least 50% protection against infection with the ancestral virus [23–25], the antibody levels against Omicron estimated in the present study could correspond to a moderate degree of effectiveness in protection against Omicron. In summary, these studies suggest that a heterologous third dose of mRNA vaccine after 2 doses of inactivated vaccine substantially boosts neutralizing antibody titers against both the ancestral virus and Omicron variant and will clearly provide improved protection against the Omicron variant.
Earlier studies have investigated post-vaccination reactions after 2 or 3 doses of inactivated [6, 25, 26] or mRNA vaccination [27, 28]. These studies suggest that fatigue, myalgia, and chills were common after receipt of both inactivated [25] and mRNA vaccines [27]; that nausea was common after inactivated vaccination [25]; and that fever and headache were common after mRNA vaccination [27]. Similar to reactions after mRNA vaccination, feverishness, and headache were commonly reported in our study, while gastrointestinal symptoms such as nausea and diarrhea were less commonly reported. No hospitalizations were reported within the month after vaccination in our study, although our sample size would not have been large enough to detect rare events.
Our study has several limitations. We have only performed PRNT against the ancestral strain and Omicron variant up to a dilution of 1:320. However, more than half of our study participants were positive for antibodies to the ancestral strain at this dilution, indicating antibody titers ≥320 at day 28, for which we assumed a value of 640 in the estimation of GMTs. Conversely, day 0 samples were typically negative even at the starting dilution of 1:10. Therefore, our results may underestimate the true values for the GMTPRNT50 and GMTRPRNT50 against the ancestral strain after a heterologous third-dose of BNT162b2 vaccination because of the ceiling effects. Nevertheless, we were able to conclude that a third dose of mRNA vaccine would provide substantial benefits against both ancestral and Omicron strains. Our results on immunogenicity may be subject to selection bias, including volunteer bias, since in Hong Kong older adults are more inclined to receive the inactivated vaccines and younger adults to received mRNA vaccines [29]. Our results on reactogenicity may be subject to information bias as this was an open-label trial and it is recognized that there could be more post-vaccination reactions after mRNA vaccination compared with inactivated vaccination [30].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Each author’s contributions to the article are listed here according to the CRediT model. Conceptualization: N. H. L. L., G. M. L., B. J. C.; methodology: N. H. L. L., S. M. S. C.; formal analysis: N. H. L. L., M. M.-S.; investigation: N. Y. M. A., Y. Y. N., L. L. H. L., K. C. K. C., J. K. C. L., Y. W. Y. L., L. C. H. T., S. C., K. K. H. K.; funding acquisition: B. J. C.; project administration: N. H. L. L., S. M. S. C., J. S. M. P., B. J. C.; supervision: N. H. L. L., S. M. S. C., D. K. M. I., L. L. M. P., G. M. L., J. S. M. P., B. J. C.; writing the original draft: N. H. L. L., M. M.-S., J. S. M. P., B. J. C.; reviewing and editing the manuscript: N. H. L. L., S. M. S. C., M. M.-S., N. Y. M. A., Y. Y. N., L. L. H. L., K. C. K. C., J. K. C. L., Y. W. Y. L., L. C. H. T., S. C., K. K. H. K., D. K. M. I., L. L. M. P., G. M. L., J. S. M. P., B. J. C.
Acknowledgments. The authors gratefully acknowledge colleagues including Eileen Yu, Teresa So, and Zacary Chai for technical support in preparing and conducting this study; Anson Ho for setting up the database; Julie Au and Lilly Wang for administrative support; Hetti Cheung, Victoria Wong, and Bobo Yeung at Hong Kong University (HKU) Health System; Cindy Man and other colleagues at the HKU Community Vaccination Centres at Gleneagles Hospital; and all the study participants for facilitating the study.
Disclaimer. The funding bodies had no role in the design of the study; the collection, analysis, and interpretation of data; or writing of the manuscript.
Financial support. This project was supported by the Theme-based Research Scheme T11-712/19-N of the Research Grants Council (RGC) of the Hong Kong Special Administrative Region, China (B. J. C.). B. J. C. is supported by a RGC Senior Research Fellow Scheme grant (HKU SRFS2021-7S03) from the Research Grants Council of the Hong Kong Special Administrative Region, China.
Supplementary Material
Contributor Information
Nancy H L Leung, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park, New Territories, Hong Kong Special Administrative Region, China.
Samuel M S Cheng, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Mario Martín-Sánchez, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Niki Y M Au, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Yvonne Y Ng, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Leo L H Luk, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Karl C K Chan, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
John K C Li, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Yonna W Y Leung, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Leo C H Tsang, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Sara Chaothai, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Kelvin K H Kwan, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Dennis K M Ip, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China.
Leo L M Poon, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Hong Kong University (HKU)-Pasteur Research Pole, School of Public Health, Li Ka Shing Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Centre for Immunology and Infection, Hong Kong Science and Technology Park, New Territories, Hong Kong Special Administrative Region, China.
Gabriel M Leung, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park, New Territories, Hong Kong Special Administrative Region, China.
J S Malik Peiris, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Hong Kong University (HKU)-Pasteur Research Pole, School of Public Health, Li Ka Shing Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Centre for Immunology and Infection, Hong Kong Science and Technology Park, New Territories, Hong Kong Special Administrative Region, China.
Benjamin J Cowling, World Health Organization Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing (LKS) Faculty of Medicine, University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region, China; Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park, New Territories, Hong Kong Special Administrative Region, China.
References
- 1. Krammer F. SARS-CoV-2 vaccines in development. Nature 2020; 586:516–27. [DOI] [PubMed] [Google Scholar]
- 2. Lim WW, Mak L, Leung GM, Cowling BJ, Peiris M. Comparative immunogenicity of mRNA and inactivated vaccines against COVID-19. Lancet Microbe 2021; 2:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mok CKP, Cohen CA, Cheng SMS, et al. Comparison of the immunogenicity of BNT162b2 and CoronaVac COVID-19 vaccines in Hong Kong. Respirology 2022; 27:301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bar-On YM, Goldberg Y, Mandel M, et al. Protection of BNT162b2 vaccine booster against Covid-19 in Israel. N Engl J Med 2021; 385:1393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falsey AR, Frenck RW Jr, Walsh EE, et al. SARS-CoV-2 neutralization with BNT162b2 vaccine dose 3. N Engl J Med 2021; 385:1627–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zeng G, Wu Q, Pan H, et al. Immunogenicity and safety of a third dose of CoronaVac, and immune persistence of a two-dose schedule, in healthy adults: interim results from two single-centre, double-blind, randomised, placebo-controlled phase 2 clinical trials. Lancet Infect Dis 2022; 22:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Intapiboon P, Seepathomnarong P, Ongarj J, et al. Immunogenicity and safety of an intradermal BNT162b2 mRNA vaccine booster after two doses of inactivated SARS-CoV-2 vaccine in healthy population. Vaccines (Basel) 2021; 9:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costa Clemens SA, Weckx L, Clemens R, et al. Heterologous versus homologous COVID-19 booster vaccination in previous recipients of two doses of CoronaVac COVID-19 vaccine in Brazil (RHH-001): a phase 4, non-inferiority, single blind, randomised study. Lancet 2022; 399:521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez-Then E, Lucas C, Monteiro VS, et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med 2022; 28:481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng SMS, Mok CKP, Leung YWY, et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med 2022; 28:486–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zuo F, Abolhassani H, Du L, et al. Heterologous immunization with inactivated vaccine followed by mRNA-booster elicits strong immunity against SARS-CoV-2 Omicron variant. Nat Commun 2022; 13:2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. World Health Organization . Guidelines on clinical evaluation of vaccines: regulatory expectations. Geneva, Switzerland: WHO, 2017. [Google Scholar]
- 15. Perera RA, Mok CK, Tsang OT, et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill 2020; 25:2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perera R, Ko R, Tsang OTY, et al. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat, and hamster sera. J Clin Microbiol 2021; 59:e02504-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau EH, Hui DS, Tsang OT, et al. Long-term persistence of SARS-CoV-2 neutralizing antibody responses after infection and estimates of the duration of protection. EClinicalMedicine 2021; 41:101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mok CKP, Chen C, Yiu K, et al. A randomized clinical trial using CoronaVac or BNT162b2 vaccine as a third dose in adults vaccinated with two doses of CoronaVac. Am J Respir Crit Care Med 2022; 205:844–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Earle KA, Ambrosino DM, Fiore-Gartland A, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 2021; 39:4423–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med 2021; 27:2032–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021; 27:1205–11. [DOI] [PubMed] [Google Scholar]
- 22. Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022; 375:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al Kaabi N, Zhang Y, Xia S, et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA 2021; 326:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim JH, Marks F, Clemens JD. Looking beyond COVID-19 vaccine phase 3 trials. Nat Med 2021; 27:205–11. [DOI] [PubMed] [Google Scholar]
- 25. Tanriover MD, Doganay HL, Akova M, et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 2021; 398:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao Y, Hao X, Wang X, et al. Humoral immunogenicity and reactogenicity of CoronaVac or ZF2001 booster after 2 doses of inactivated vaccine. Cell Res 2022; 32:107–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi A, Koch M, Wu K, et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med 2021; 27:2025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McMenamin ME, Nealon J, Lin Y, et al. Vaccine effectiveness of two and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong. [Preprint]. doi: 10.1101/2022.03.22.22272769. [DOI] [PMC free article] [PubMed]
- 30. McDonald I, Murray SM, Reynolds CJ, Altmann DM, Boyton RJ. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. NPJ Vaccines 2021; 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.