Abstract
Background
The rapid emergence of the Omicron variant and its large number of mutations led to its classification as a variant of concern (VOC) by the World Health Organization. Subsequently, Omicron evolved into distinct sublineages (eg, BA.1 and BA.2), which currently represent the majority of global infections. Initial studies of the neutralizing response toward BA.1 in convalescent and vaccinated individuals showed a substantial reduction.
Methods
We assessed antibody (immunoglobulin G [IgG]) binding, ACE2 (angiotensin-converting enzyme 2) binding inhibition, and IgG binding dynamics for the Omicron BA.1 and BA.2 variants compared to a panel of VOCs/variants of interest, in a large cohort (N = 352) of convalescent, vaccinated, and infected and subsequently vaccinated individuals.
Results
While Omicron was capable of efficiently binding to ACE2, antibodies elicited by infection or immunization showed reduced binding capacities and ACE2 binding inhibition compared to wild type. Whereas BA.1 exhibited less IgG binding compared to BA.2, BA.2 showed reduced inhibition of ACE2 binding. Among vaccinated samples, antibody binding to Omicron only improved after administration of a third dose.
Conclusions
Omicron BA.1 and BA.2 can still efficiently bind to ACE2, while vaccine/infection-derived antibodies can bind to Omicron. The extent of the mutations within both variants prevents a strong inhibitory binding response. As a result, both Omicron variants are able to evade control by preexisting antibodies.
Keywords: SARS-CoV-2, Omicron, antibody binding, multiplex, variants of concern
Using a large diverse cohort of vaccinated and convalescent individuals, we found that Omicron can bind to angiotensin-converting enzyme 2 (ACE2) more efficiently than inhibitory antibodies. Immunoglobulin G binding and inhibition of ACE2 binding were significantly reduced for both BA.1 and BA.2.
Since its initial outbreak in late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved into a global pandemic, characterized by multiple waves of infection within countries and regions. Following the initial global wave, subsequent waves were often triggered by the emergence of variants of concern (VOCs) [1] that outcompeted earlier variants. These emerging VOCs are presumed to have an increased rate of transmission or the ability to escape vaccine and infection-induced immunity [2–7]. Due to concerns that its numerous spike protein mutations would render it able to escape immune control and its rapid spread in South Africa, the World Health Organization classified Omicron as a VOC on 26 November 2021 [8]. Within days of this classification, Omicron had already been reported in multiple other countries and as of 26th April 2022, it is the dominant global variant [9]. The BA.2 lineage has now surpassed BA.1 [9], which is not only genetically distinct in key regions of the spike glycoprotein [10, 11], but has also evolved to give rise to BA.4, BA.5, and BA.2.12.1. Early studies at the end of 2021 of neutralization against BA.1 found a substantial reduction in neutralizing activity, particularly in individuals who did not receive 3 vaccine doses [12–17]. Because of the urgency of these studies to establish real-time data of whether there is an evasion toward vaccine-induced responses or therapeutics, they often included limited sample numbers, diversity of sample types (eg, only those vaccinated with Pfizer BNT162b2), or only directly compared Omicron to wild-type (WT) or a single other VOC. Complementary to these studies, we provide here a comprehensive analysis of the binding capacity, binding dynamics, and angiotensin-converting enzyme 2 (ACE2) binding inhibition of Omicron BA.1 and BA.2 against antibodies generated either by vaccination or natural infection compared to WT, currently recognized VOCs, and the Lambda and Mu variants of interest (VOIs). We also analyzed samples from a range of vaccines and dosing schemes currently available within the European Union, including booster vaccines, as well as convalescent samples from both adults and children from the first, second, and third waves of infection in Germany.
METHODS
Sample Cohort
Serum samples used in this study were originally collected for use in several other studies [18–21]. The sample set was designed to include a broad representation of samples from infected individuals (hereafter referred to as “convalescent”) from the different waves of SARS-CoV-2 within Germany, as well as vaccinated samples. We separated our cohort into 4 groups: vaccinated samples, convalescent samples, infected and later vaccinated samples, and prepandemic samples as controls.
Vaccinated study participants received either 2 homologous doses of AZD1222, BNT162b2, or mRNA-1273; 2 heterologous doses of AZD1222 and BNT162b2 or AZD1222 and mRNA-1273; or 3 doses of BNT162b2. To examine how antibody dynamics changed over time, we included samples from both 1–2 months post–second dose and 5–6 months post–second dose. We also included samples from vaccinated individuals who had a previous polymerase chain reaction (PCR)–confirmed infection and then received a single dose of BNT162b2 as per national guidelines. Samples from convalescent study participants were collected approximately 3 months after positive PCR. To assess differences between variants of SARS-CoV-2, we collected samples from those with a WT, Alpha, or Delta infection. WT samples came from both adults and children. To be considered a WT sample, the infection had to occur during the first wave of the SARS-CoV-2 pandemic in Germany (spring–summer 2020). Alpha samples were confirmed by PCR sequencing and were collected between January and May 2021. Delta samples were either confirmed by PCR sequencing, or where collected during a time period when all infections in Germany were considered to be Delta (September–November 2021). To represent naive samples, negative prepandemic samples were obtained from Central BioHub. An overview of the characteristics for each cohort subgroup can be found in Table 1.
Table 1.
Overview of Sample Characteristics for the Study Population
| Sample Type | Subgroup | No. of Samples | Median dT, Days (IQR) | No. (%) of Females | Median Age, Years (IQR) | History of Immunosuppressive Condition or Medication, No. (%) |
|---|---|---|---|---|---|---|
| Convalescent | WT (adults) | 30 | 104 (94–119) | 14 (47) | 62 (51–69) | 0 (0) |
| WT (children) | 20 | 124 (116–129) | 7 (35) | 11 (7–14) | 0 (0) | |
| Alpha | 30 | 88 (47–104) | 12 (40) | 56 (42–65) | 14 (47) | |
| Delta | 6 | 18 (10–23) | 5 (83) | 65 (56–73) | 4 (67) | |
| Infected and vaccinated | … | 25 | 54 (23–91) | 16 (64) | 55 (48–59) | 1 (4) |
| Vaccinated | A/A (1–2 mo) | 30 | 49 (48–52) | 20 (67) | 64 (60–66) | 2 (7) |
| A/A (4–6 mo) | 30 | 154 (146–158) | 23 (77) | 55 (48–60) | 0 (0) | |
| M/M (1–2 mo) | 30 | 51 (48–54) | 20 (67) | 59 (49–61) | 1 (3) | |
| M/M (4–6 mo) | 16 | 139 (131–145) | 9 (56) | 70 (51–83) | 1 (6) | |
| P/P (1–2 mo) | 30 | 51 (49–54) | 20 (67) | 58 (52–66) | 1 (3) | |
| P/P (4–6 mo) | 30 | 152 (141–160) | 25 (83) | 38 (30–53) | 0 (0) | |
| A/M | 20 | 153 (150–154) | 16 (80) | 41 (29–56) | 0 (0) | |
| A/P | 20 | 151 (144–157) | 19 (95) | 48 (42–56) | 0 (0) | |
| P/P/P | 20 | 14 (14–26.5) | 13 (65) | 33 (29–44) | 2 (12) | |
| Negative | … | 15 | … | 8 (53) | 37 (29–41) | 0 (0) |
Abbreviations: A/A, 2-dose AZD1222; A/M, first dose AZD1222, second dose mRNA-1273; A/P, first dose AZD1222, second dose BNT162b2; dT, time post-infection/last vaccination dose; IQR, interquartile range; M/M, 2-dose mRNA-1273; P/P, 2-dose BNT162b2; P/P/P, 3-dose BNT162b2; WT, wild-type (B.1 isolate).
Ethical Oversight
Informed written consent was obtained from all study participants. Ethical approval and oversight for the samples used in this study was provided by the following ethics committees: the Ethics Committee of the University Hospital Tuebingen (293/2020BO2, 764/2020/BO2 [amended 6 December 2021], B312/2020BO1 [amended 2 June 2021], 556/2021BO1), the Ethics Committee of the University of Tuebingen (179/2020/BO2, 188/2020A), and the Ethics Committee of Hannover Medical School (9086_BO_S_2020).
Mass Spectrometry of Omicron Receptor-Binding Domain
Receptor-binding domain (RBD) Omicron protein samples had their structure verified by liquid chromatography–mass spectrometry (LC-MS). In brief, samples were N-deglycosylated using PNGaseF reducing kit (New England Biolabs), diluted 1:3 with His-NaCl buffer and analyzed by liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. For full details, please consult the Supplementary Methods.
MULTICOV-AB Assay
MULTICOV-AB, a previously published multiplex immunoassay, was performed as described previously [22]. A full list of antigens included within the assay is shown in Table 2. Samples were randomly allocated to plates to ensure that at least 3 samples of every sample group were included on each plate. All samples were measured twice in 2 independent experiments. No sample failed quality control (QC). Raw median fluorescence intensity values were normalized to a QC sample for all antigens as per Becker et al [23]. Please consult the Supplementary Methods for full details.
Table 2.
Overview of Antigens Used in MULTICOV-AB and RBDCoV-ACE2 Assays
| Antigen | Manufacturer | Category Number | Mutations Covered |
|---|---|---|---|
| Spike WT (B.1) | NMI | … | … |
| RBD WT (B.1) | NMI | … | … |
| S1 domain WT (B.1) | NMI | … | … |
| S2 domain WT (B.1) | Sino Biological | 40590-V08B | … |
| Nucleocapsid WT (B.1) | Aalto Bioreagents | 6404-b | … |
| RBD Alpha (B.1.1.7) | NMI | … | N501Y |
| RBD Beta (B.1.351) | NMI | … | K417N, E484K, N501Y |
| RBD Gamma (P1) | NMI | … | K417T, E484K, N501Y |
| RBD Delta (B.1.617.2) | NMI | … | L452R, T478K |
| RBD Omicron (B.1.529/BA.1) | Sino Biological | 40592-V08H121 | G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H |
| Spike Omicron (B.1.1.529/BA.1) | Sino Biological | 40589-V08H26 | A67V, del HV69/70, T95I, G142D, del VYY 143-145, del N211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, H655Y, N679K, P681H, N764K, D796Y, F817P, N856K, A892P, A899P, A942P, Q954H, N969K, L981F, K986P, V987P |
| RBD Omicron (B.1.1.529/BA.2) | Sino Biological | 40592-V08H123 | G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H |
| Nucleocapsid Omicron (B.1.1.529/BA.2) | Sino Biological | 40588-V07E35 | P13L, del ERS 31-33, R203K, G204R, S413R |
| RBD Lambda (C.37) | NMI | … | L452Q, F490S |
| RBD Mu (B.1.621) | NMI | … | R346K, E484K, N501Y |
RBDCoV-ACE2 Assay
RBDCoV-ACE2, a previously published multiplex ACE2 inhibition assay [19], analyzes neutralizing antibody activity through ACE2 binding inhibition. A full list of antigens included in this assay can be found in Table 2. ACE2 binding inhibition was calculated as a percentage, with 100% indicating maximum ACE2 binding inhibition and 0% indicating no ACE2 binding inhibition. Samples with an ACE2 binding inhibition <20% are classified as nonresponders [19]. Please consult the Supplementary Methods for full details.
Biolayer Interferometry
Analysis of binding kinetics of RBD-specific antibodies in serum samples were performed using the Octet RED96e system (Sartorius) as per the manufacturer’s recommendations. Purified RBDs of WT, Delta, and Omicron were biotinylated with Sulfo-NHS-LC-LC-Biotin (Thermo Fisher Scientific) in 5 molar excess at ambient temperature for 30 minutes. Excess of biotin was removed by size exclusion chromatography using Zeba Spin Desalting Columns 7K MWCO 0.5 mL (Thermo Fisher Scientific) according to the manufacturer’s protocol. Data were analyzed using the Octet Data Analysis HT 12.0 software applying the 1:1 fitting model for the dissociation step. The binding profile response of each sample is illustrated as the mean wavelength shift in nm. For affinity determination, the 1:1 global fit of the Data Analysis HT 12.0 software was used. Please consult the Supplementary Methods for full details.
Statistical Analysis
Data were collated and matched to metadata in Excel 2016. Data visualization was done in RStudio (version 1.2.5001 running R version 3.6.1). Additional packages gplots and beeswarm were used for specific displays. The “lm” function of R’s stats library was used for linear regression analyzes. Correlation analyzes were performed using the “cor” function of R’s stats library. The “wilcox.test” function from R’s stats library was used to perform either Mann–Whitney U tests (2-sided) to estimate significance of observed differences between different groups, or Wilcoxon signed-rank tests (2-sided) to estimate significance of observed different between antigens. Graphs were exported from RStudio and further edited in Inkscape (version 0.92.4) to generate final figures. Biolayer inferometry graphical representation was prepared using GraphPad Prism Software (version 9.0.0).
RESULTS
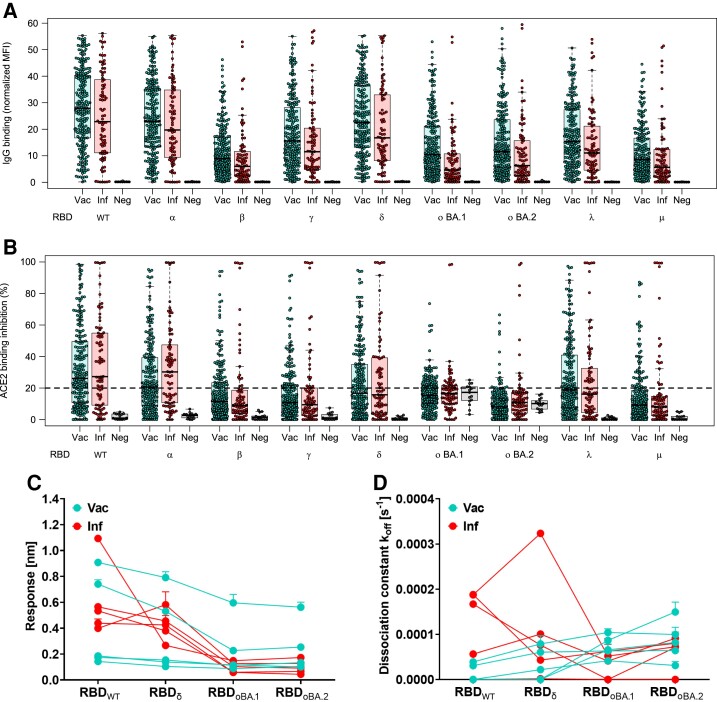
Initially, we examined immunoglobulin G (IgG) binding using MULTICOV-AB [22], a previously published SARS-CoV-2 multiplex immunoassay that was adapted to analyze binding toward RBDs from VOC/VOIs (a full list of antigens analyzed and their mutations contained within can be found in Table 2). For both vaccinated and convalescent samples, IgG binding toward both Omicron BA.1 and BA.2 for preexisting antibodies was significantly reduced compared to WT (BA.1: 2.3–4.1 median fold reduction, P < .001; BA.2: 2.1–3.0 median fold reduction, P < .001) (Figure 1, Supplementary Tables 1–4). This is similar to the statistically significant reduction in binding toward Beta (2.8–3.5 median fold reduction, P < .001) and Mu (3.0–3.6 median fold reduction, P < .001, Figure 1, Supplementary Tables 1–4). Within Omicron, BA.1 had a significantly increased reduction in IgG binding compared to BA.2 (1.1–1.2 median fold reduction, P < .001) (Figure 1, Supplementary Tables 2–4).
Figure 1.
Antibody binding response is significantly reduced for both BA.1 and BA.2. Binding response by preexisting antibodies generated through either infection or vaccination was measured with MULTICOV-AB (A) and RBDCoV-ACE2 (B) assays and Biolayer interferometry (C and D). A, Boxplot showing that immunoglobulin G binding is significantly reduced for both BA.1 and BA.2 as compared to other variants of concern (VOCs)/variants of interest (VOIs) for convalescent (n = 86) and vaccinated (n = 226) samples. Negative samples are included as controls (n = 15). B, Boxplot showing that ACE2 binding inhibition is significantly reduced for both BA.1 and BA.2 as compared to other VOCs/VOIs for both convalescent and vaccinated samples. Boxes represent the median with 25th and 75th percentiles; whiskers show the largest and smallest nonoutlier values. Outliers were determined by 1.5 interquartile range. C and D, Binding kinetics of receptor-binding domain (RBD)–specific antibodies from serum samples of convalescent and vaccinated individuals (both n = 5). Binding response (C) and dissociation constant (D) were determined by 1:1 fitting model of the individual serum samples between the different RBD variants. Median fold reductions for both (A) and (B) can be found as Supplementary Tables 1–3 and 5. Statistical differences between all variants was analyzed by Wilcoxon signed-rank test for both (A) and (B) and is available as Supplementary Tables 4 and 7. The response rate for (B) is available as Supplementary Table 6. Abbreviations: ACE2, angiotensin-converting enzyme 2; IgG, immunoglobulin G; Inf, infected; MFI, median fluorescence intensity; Neg, negative; RBD, receptor-binding domain; Vac, vaccinated; WT, wild-type.
Next, we analyzed ACE2 binding inhibition to determine how effective the antibodies were at blocking the RBD-ACE2 interaction, using RBDCoV-ACE2 [19], a multiplex ACE2-RBD inhibition assay. This assay mimics the interaction between ACE2 and the RBD, while the multiplex format allows for the simultaneous measurement of all VOCs/VOIs in a single well. To evaluate binding against other antigens of SARS-CoV-2, we also include the spike and S1 domain of WT and the spike and nucleocapsid of Omicron. In line with IgG binding, ACE2 binding inhibition against Omicron was significantly reduced compared to WT (Omicron BA.1: median, 15.2%–16.4% and BA.2: median, 8.0%–10.9%; WT: median, 26.1%–27.2%; both P < .001) (Figure 1, Supplementary Table 5), with less than half as many samples considered responsive (WT: 62.8%; BA.1: 30.1%–32.6%; BA.2: 8.4%–14.0%) (Figure 1B, Supplementary Table 6). Interestingly, although BA.2 revealed reduced ACE2 binding inhibition and sample response rate compared to BA.1, this was not significant (P = .08, Supplementary Table 7). While BA.2 had the lowest response rate of all variants examined, the median ACE2 binding inhibition and response rate for BA.1 was greater than for Beta (median, 8.9–11.4; response, 22.1%–31.0%), Gamma (median, 9.4–10.8; response, 25.5%–30.1%), and Mu (median, 8.1–9.1; response, 19.8%–22.6%).
To investigate the RBD binding of BA.1 and BA.2 further, we analyzed the binding kinetics of RBD-specific antibodies from vaccinated (2 doses of BNT162b2) and convalescent (WT) study participants by biolayer interferometry analysis (Figure 1C and 1D). Binding response and dissociation constant were measured for each sample as an indicator of amount and binding strength. Binding response toward BA.1 and BA.2 was reduced compared to both WT and Delta (Figure 1C), while most samples showed increased dissociation kinetics for Omicron (Figure 1D). Despite high variation observed for both the vaccinated and convalescent samples, Omicron always showed the lowest binding response (Supplementary Figure 1, Supplementary Table 8). When binding toward ACE2 itself was examined, Omicron BA.1 and BA.2 were still able to bind ACE2 with high affinity (Supplementary Figure 2).
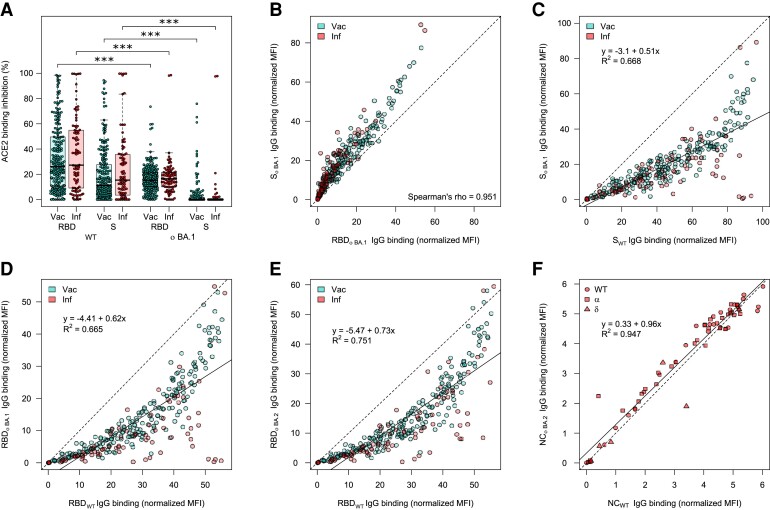
As mutations toward Omicron are not limited to the RBD, we also analyzed ACE2 binding toward the full-length spike protein. Omicron BA.1 ACE2 binding inhibition toward the spike protein was significantly reduced compared to WT (P < .001). Interestingly, the response rate underwent a similar 30% reduction from RBD to S for both WT (62.8% to 34.5%–43.0%) and Omicron (30.1%–32.6% to 3.5%–4.9%) (Figure 2A, Supplementary Table 9). IgG binding capacity toward spike appears to be conserved to a similar degree as the RBD (Figure 2B–D). However, this is substantially reduced compared to the nucleocapsid of BA.2, for which there was no change in binding capacity compared to WT for convalescent samples, regardless with which strain they had been previously infected (Figure 2E, samples highlighted according to strain).
Figure 2.
Angiotensin-converting enzyme 2 (ACE2) binding inhibition and correlations of binding capacity between BA.1, BA.2, and wild-type (WT) for different antigens. ACE2 binding inhibition (A) and immunoglobulin G (IgG) binding capacity (B–F) were compared for the Omicron BA.1 and BA.2 receptor-binding domain (RBD), and spike (S) to the WT RBD and S. A, Boxplot showing that ACE2 binding inhibition is significantly reduced toward BA.1 for both RBD and S for both vaccinated (n = 226) and convalescent (n = 86) samples. Boxes represent the median with 25th and 75th percentiles; whiskers show the largest and smallest nonoutlier values. Outliers were determined by 1.5 interquartile range. Statistical significance was calculated by Wilcoxon signed-rank test; ***P < .001. B, Correlation analysis of IgG binding capacity for the BA.1 spike compared to the BA.1 RBD. Spearman rank was calculated to assess ordinal association between the variables. C–F, Linear regressions of IgG binding capacity for the BA.1 S compared to WT S (C), BA.1 RBD compared to wild-type RBD (D), BA.2 RBD compared to WT RBD (E), and BA.2 nucleocapsid compared to WT nucleocapsid (F). R2 is included to indicate the correlation. Abbreviations: ACE2, angiotensin-converting enzyme 2; IgG, immunoglobulin G; Inf, infected; MFI, median fluorescence intensity; NC, nucleocapsid; RBD, receptor-binding domain; S, spike; Vac, vaccinated; WT, wild-type.
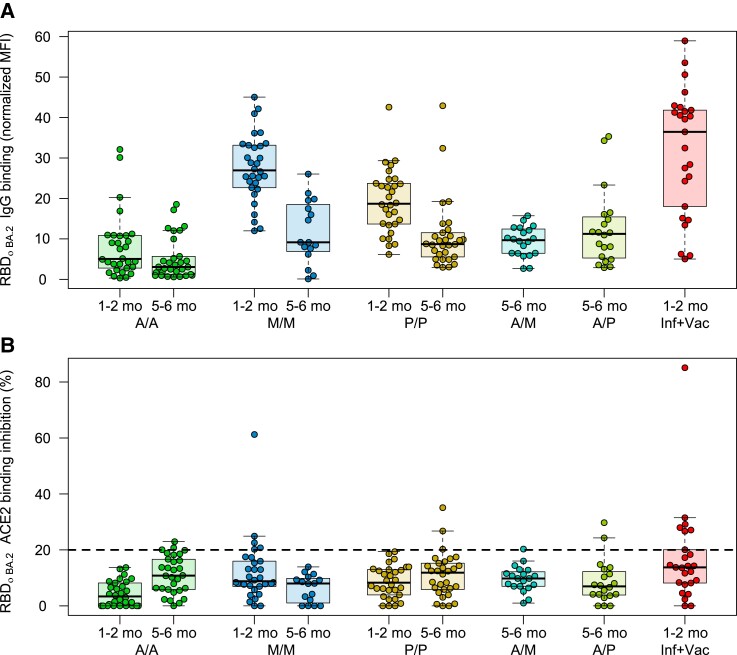
Next, we analyzed whether vaccine type (AZD1222, mRNA-1273, or BNT162b2) and number of doses (homologous or heterologous 2 dose, or homologous 3 dose) received resulted in differences with respect to Omicron binding response. To analyze how this response changed as time postvaccination increased, we also compared the responses at 1–2 months post–second dose and 5–6 months post–second dose for the homologous recipients. IgG binding capacity at 5–6 months was low for all recipients regardless of the administered vaccine, although homologous mRNA-based vaccination still had higher binding capacity at this timepoint than 1–2 months post–second dose for vector-based vaccination (median of 9.16, 8.7, and 5.04 for mRNA-1273, BNT-162b2, and AZD1222, respectively; Figure 3A). Among samples from 1–2 months postdosing, infected and then vaccinated had the greatest IgG binding capacity (median, 36.5), followed by 2-dose mRNA-1273 (median, 27.0), BNT162b2 (median, 18.7), and AZD1222 (median, 2.3). This pattern was consistent for both BA.1 (Supplementary Figure 3) and BA.2 (Figure 3). ACE2 binding inhibition was consistently low regardless of type received and timeframe postdose (Figure 3B).
Figure 3.
Differences in Omicron binding response among different populations of vaccinated samples. Binding response toward Omicron BA.2 was analyzed by either MULTICOV-AB (A) or RBDCoV-ACE2 (B) assays for samples from different vaccine schemes (n = 30 for all samples, except for mRNA-1273 at 5–6 months (n = 16), heterologous vaccine schemes (both n = 20), and infected and vaccinated (n = 25). To determine the effect of time postvaccination, samples from both 1–2 months and 5–6 months postvaccination were included. Boxes represent the median with 25th and 75th percentiles; whiskers show the largest and smallest nonoutlier values. Outliers were determined by 1.5 interquartile range. The 20% cutoff for nonresponders is indicated by the dashed line on (B). The equivalent data for BA.1 are provided as Supplementary Figure 3. Abbreviations: A/A, AZD1222; ACE2, angiotensin-converting enzyme 2; A/M, first dose AZD1222, second dose mRNA-1273; A/P, first dose AZD1222, second dose BNT162b2; IgG, immunoglobulin G; Inf, infected; MFI, median fluorescence intensity; M/M, mRNA-1273; P/P, BNT162b2; RBD, receptor-binding domain; Vac, vaccinated.
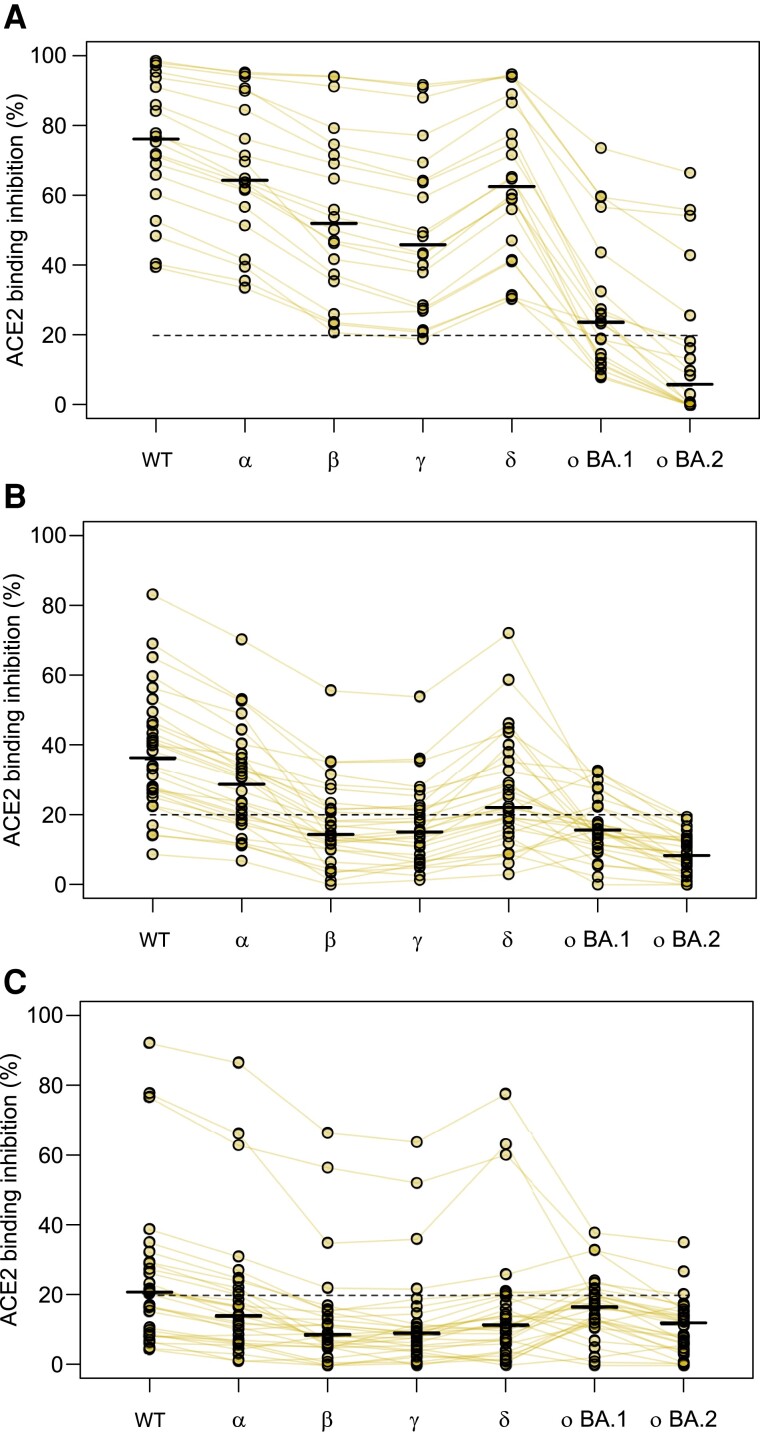
To determine whether a third dose results in increased binding responses against Omicron, we analyzed samples from individuals who had received a third dose of BNT162b2 and compared it to those who had received their second dose in a similar timeframe and individuals 5–6 months post–second dose, who would be eligible to receive a third dose (Figure 4). Further boosting was associated with higher Omicron ACE2 binding inhibition compared to 2 doses (27% and 0% of samples were responders toward BA.1 and BA.2 post–second dose, respectively; 55% and 25% were responders after boosting) suggesting that boosting offers increased protection against Omicron (Figure 4). However, this increase in protection was not limited to Omicron and was present for all VOCs (Figure 4). Compared to the second dose from a similar timepoint (30% and 10% responders to BA.1 and BA.2, respectively), boosting with the third dose increased ACE2 binding inhibition, substantially confirming this effect is generated by the third dose itself, and not by time postvaccination alone (Figure 4C).
Figure 4.
Angiotensin-converting enzyme 2 (ACE2) binding inhibition toward Omicron is boosted by a third vaccine dose. Changes in ACE2 binding response following the third dose of BNT162b2 for all variants within the study. Samples come from either boosted (n = 20, A), 1–2 months post–second dose of BNT162b2 (n = 20, B), or 5–6 months post–second dose of BNT162b2 (n = 20, C). Individual samples are highlighted by connected lines with bars representing medians. The 20% cutoff for nonresponders is indicated by the dashed line. Abbreviations: ACE2, angiotensin-converting enzyme 2; WT, wild-type.
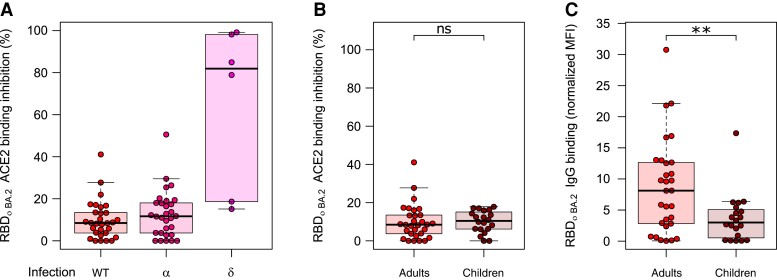
Last, we analyzed whether natural infection with different variants resulted in differences in binding responses. There was no difference in ACE2 binding inhibition between convalescent individuals infected with either WT or Alpha, with both having minimal inhibition against Omicron (Figure 5A, Supplementary Figure 3). While some samples with a previous Delta infection showed substantially more activity compared to WT or Alpha, they had been collected much sooner after the infection (median dT, 18 days) than WT (median dT, 104 days) or Alpha (median dT, 88 days). To evaluate whether children’s antibodies were more effective at binding toward Omicron than adults, we compared convalescent samples 3–4 months post-PCR from the first wave in children (n = 20), to convalescent samples 3 months post–positive PCR from the same wave in adults (n = 30) (Figure 5B and 5C, Supplementary Figure 4). There was no significant difference between adults and children in ACE2 binding inhibition (P = .48; Figure 5B), although children did have significantly reduced binding capacity toward Omicron (P = .01; Figure 5C).
Figure 5.
Differences in immunoglobulin G (IgG) binding response and angiotensin-converting enzyme 2 (ACE2) binding inhibition toward BA.2 among different populations of convalescent samples. Comparative ACE2 binding inhibition (A and B) and IgG binding capacity (C) between convalescent samples from different pandemic waves (A) and adults and children (B and C) for BA.2. A, There are no differences in ACE2 binding inhibition toward BA.2 for individuals infected with wild-type (WT) (n = 30), Alpha (n = 30), or Delta (n = 6). B, Children (n = 20) and adults (n = 30) have similar ACE2 binding inhibition toward BA.2 following WT infection, although they have significantly reduced IgG binding capacity (P = .01). C, Boxes represent the median with 25th and 75th percentiles; whiskers show the largest and smallest nonoutlier values. Outliers were determined by 1.5 interquartile range. Statistical significance was calculated by Mann–Whitney U test: **P < .01; ns, P < .05. The equivalent data for BA.1 are provided as Supplementary Figure 4. Abbreviations: ACE2, angiotensin-converting enzyme 2; IgG, immunoglobulin G; MFI, median fluorescence intensity; RBD, receptor-binding domain; WT, wild-type.
DISCUSSION
In this study, we provide an in-depth characterization of antibody binding to Omicron BA.1 and BA.2 compared to WT and all other VOCs and variants under investigation in a large, diverse sample cohort. The use of an ACE2 inhibition assay enabled the comparison of multiple variants of interest simultaneously, while also producing comparable results to classical virus neutralization assays [19]. Similar to others, we identified that antibody binding and ACE2 binding inhibition toward both Omicron BA.1 and BA.2 elicited by either immunization or previous infection were significantly reduced [14, 16, 24–28]. However, we provide here additional information on IgG binding capacity and ACE2 binding inhibition by the inclusion of a large variety of subcohorts representing the present diversity of immunity against SARS-CoV-2, the comparison of the Omicron variant to all other VOCs/VOIs, and the comparison between BA.1 and BA.2.
We found that both IgG binding capacity and ACE2 binding inhibition was significantly reduced for BA.1 and BA.2, similar to reductions for Beta and Mu, with the majority of samples being classified as nonresponsive toward Omicron for ACE2 binding inhibition. Like others researchers [15, 24, 28], we found that antibody binding responses toward Omicron were significantly increased upon administration of a booster dose. Boosting increases were not restricted to just Omicron IgG binding capacity and ACE2 binding inhibition, but were present against all VOCs/VOIs. No significant difference was present between BA.1 and BA.2. However, data regarding the longitudinal response post–third dose remains lacking, with some countries now offering a fourth dose for certain groups (eg, immunocompromised individuals). The increased responses following boosting also appear to apply for convalescent individuals, as seen by the increased IgG binding capacity and ACE2 binding inhibition for previously infected individuals who received a single dose, compared to those who had received any 2 vaccine doses. This increase in responses for individuals who have been both infected and vaccinated is in agreement with Pajon et al [29], who found that infections prior to vaccination resulted in a greater breadth of immune response, while Lechmere et al [26] found that breakthrough Delta infections among vaccinated individuals acted like a booster dose. Thus, both reinfection and a booster dose leads to appropriate affinity maturation of elicited antibodies. Among those who had received 2 doses, binding responses were consistent with other reports (eg, [16]) in identifying a significant decrease for those who received homologous 2-dose vector vaccination as opposed to homologous mRNA vaccination or heterologous vaccination.
We identified no significant difference in ACE2 binding inhibition toward Omicron for children compared to adults, although IgG binding capacity was significantly reduced. This is in contrast to previous research that has identified that children’s antibody titers are higher than adults following infection [21]; however, a limitation of our study is that the children and adult groups were not well-matched in terms of time post–positive PCR (median dT, 104 for adults and 124 for children) or disease severity (majority hospitalized adults vs asymptomatic/mildly symptomatic children). A larger investigation including vaccinated samples from children is needed to investigate any possible protective effect from previous infection and the antibody response toward Omicron itself in children in more detail.
Similar to Carreño et al [27], we identified that IgG binding capacity toward Omicron was not as severely reduced as ACE2 binding inhibition. Interestingly, while BA.2 had significantly increased IgG binding capacity to BA.1, it had reduced ACE2 binding inhibition. While mutations in BA.1 and BA.2 are not limited to the RBD, their reduction in activity was limited to the trimeric spike, with near-identical binding compared to WT for the nucleocapsid. Our analysis of both the spike- and RBD-derived ACE2 binding inhibition suggests that while both epitopes are sufficiently conserved to enable binding, their divergent mutated sequences affect their inhibitory response. Further investigation into this pattern and particularly the role of S1-derived antibodies in neutralization is required to understand the inhibitory protection offered against Omicron.
Overall, our results identify that while Omicron can efficiently bind ACE2 and vaccine/infection-induced antibodies can bind Omicron, the extent of the mutations within the RBD appear too divergent to enable RBD-directed antibodies to mount an inhibitory response. The dramatic reductions in both IgG binding and ACE2 binding inhibition toward Omicron, as opposed to other VOCs/VOIs, confirm that this variant remains capable of immune escape and requires careful sequence monitoring to identify any further sequence evolution. Importantly, booster doses elicit a significant increase in antibody response, which correlates with a significant increase in both IgG binding and ACE2 binding inhibition against Omicron. Our data add weight to a growing body of evidence that the continuous adaptation of vaccines toward novel highly contagious variants needs to be considered in order to control SARS-CoV-2.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. D. J., M. Be., A. D., and N. S.-M. conceived the study. D. J., M. Be., T. R. W., P. D. K., S. M., A. Z., U. R., A. D., and N. S. M. planned experiments. A. D. and N. S. M. supervised the study. D. J., M. Be., P. D. K., T. R. W., S. M., T. M. G., J. Gri., P. M., J. Gru., and B. T. performed experiments. C. H., Y. T. P., J. H., R. F., A. K., A. N., Y. M., S. S., J. S. W., K. A., G. U., M. M., T. B., K. S.-L., H. H., S. G., M. Bi., H. R., J. R., C. E., A. R. F., M. H., B. K., M. S., and G. K. collected samples or organized their collection. K. S.-L. and N. S. M. obtained funding. M. Be., T. R. W., and A. D. performed data analysis and generated the figures. A. D. wrote the first draft of the manuscript. All authors approved the manuscript prior to submission.
Acknowledgments. The authors thank other members of the Multiplex Immunoassays, Biological Development Center and Bioanalytics groups at the Natural and Medical Sciences Institute at the University of Tuebingen (NMI) for their support on this project. The authors also thank Joop van der Heuvel for his expertise in protein production; Ann Kathrin Horlacher and Mareike Walenta for assistance with sample processing and patient material storage; Ulrike Schmidt, Iris Schaefer, Richard Schaad, and Hannah Zug for technical support; Katharina Kienzle, Hartmut Mahrhofer, Hardy Richter, and Stefanie Döbele for help with sample collection; Andrea Evers-Bischoff, Andrea Bevot, and the Centre for Pediatric Clinical Studies at the University Hospital Tuebingen for organizational support in conducting the study; and all those involved in the organization of the MuSPAD and TuSeRe sample collection.
Disclaimer. The funders had no role in study design, data collection, and analysis; preparation of the manuscript; or the decision to submit the manuscription for publication.
Financial support. This work was supported by the State Ministry of Baden-Württemberg for Economic Affairs, Labour and Tourism (grant numbers FKZ 3-4332.62-NMI-67, FKZ 3-4332.62-NMI-68, and 7-4332.62-NMI/55); the Initiative and Networking Fund of the Helmholtz Association of German Research Centres (grant number SO-96); the European Union Horizon 2020 research and innovation program (grant agreement number 101003480, CORESMA), and the State Ministry of Lower Saxony for Science and Culture (grant agreement number 14-76103-1841, MWK HZI COVID-19).
Supplementary Material
Contributor Information
Daniel Junker, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Matthias Becker, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Teresa R Wagner, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany; Pharmaceutical Biotechnology, University of Tuebingen, Tuebingen, Germany.
Philipp D Kaiser, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Sandra Maier, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Tanja M Grimm, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Johanna Griesbaum, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Patrick Marsall, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Jens Gruber, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Bjoern Traenkle, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Constanze Heinzel, Institute of Tropical Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Yudi T Pinilla, Institute of Tropical Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Jana Held, Institute of Tropical Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Rolf Fendel, Institute of Tropical Medicine, University Hospital Tuebingen, Tuebingen, Germany; German Center for Infection Research, partner site Tuebingen, Tuebingen, Germany; Centre de Recherches Médicales de Lambaréné, Lambaréné, Gabon.
Andrea Kreidenweiss, Institute of Tropical Medicine, University Hospital Tuebingen, Tuebingen, Germany; German Center for Infection Research, partner site Tuebingen, Tuebingen, Germany; Centre de Recherches Médicales de Lambaréné, Lambaréné, Gabon.
Annika Nelde, Department of Peptide-Based Immunotherapy, University of Tuebingen and University Hospital Tuebingen, Tuebingen, Germany; Department of Internal Medicine, Clinical Collaboration Unit Translational Immunology, German Cancer Consortium, University Hospital Tuebingen, Tuebingen, Germany; Department of Immunology, Institute for Cell Biology, University of Tuebingen, Tuebingen, Germany; Cluster of Excellence iFIT (EXC2180) “Image-Guided and Functionally Instructed Tumor Therapies,” University of Tuebingen, Tuebingen, Germany.
Yacine Maringer, Department of Peptide-Based Immunotherapy, University of Tuebingen and University Hospital Tuebingen, Tuebingen, Germany; Department of Internal Medicine, Clinical Collaboration Unit Translational Immunology, German Cancer Consortium, University Hospital Tuebingen, Tuebingen, Germany; Department of Immunology, Institute for Cell Biology, University of Tuebingen, Tuebingen, Germany; Cluster of Excellence iFIT (EXC2180) “Image-Guided and Functionally Instructed Tumor Therapies,” University of Tuebingen, Tuebingen, Germany.
Sarah Schroeder, Department of Peptide-Based Immunotherapy, University of Tuebingen and University Hospital Tuebingen, Tuebingen, Germany; Department of Immunology, Institute for Cell Biology, University of Tuebingen, Tuebingen, Germany; Department of Otorhinolaryngology, Head and Neck Surgery, University of Tuebingen, Tuebingen, Germany.
Juliane S Walz, Department of Peptide-Based Immunotherapy, University of Tuebingen and University Hospital Tuebingen, Tuebingen, Germany; Department of Internal Medicine, Clinical Collaboration Unit Translational Immunology, German Cancer Consortium, University Hospital Tuebingen, Tuebingen, Germany; Department of Immunology, Institute for Cell Biology, University of Tuebingen, Tuebingen, Germany; Cluster of Excellence iFIT (EXC2180) “Image-Guided and Functionally Instructed Tumor Therapies,” University of Tuebingen, Tuebingen, Germany.
Karina Althaus, Center for Clinical Transfusion Medicine, Tuebingen, Germany; Institute of Clinical and Experimental Transfusion Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Gunalp Uzun, Center for Clinical Transfusion Medicine, Tuebingen, Germany.
Marco Mikus, Center for Clinical Transfusion Medicine, Tuebingen, Germany.
Tamam Bakchoul, Center for Clinical Transfusion Medicine, Tuebingen, Germany; Institute of Clinical and Experimental Transfusion Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Katja Schenke-Layland, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany; Department of Immunology, Institute for Cell Biology, University of Tuebingen, Tuebingen, Germany; Department for Medical Technologies and Regenerative Medicine, Institute of Biomedical Engineering, University of Tuebingen, Tuebingen, Germany; Division of Cardiology, Department of Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Stefanie Bunk, Infectious Diseases, Department of Internal Medicine I, University Hospital Tuebingen, Tuebingen, Germany.
Helene Haeberle, Department of Anaesthesiology and Intensive Care Medicine, University Hospital Tuebingen, Tuebingen, Germany.
Siri Göpel, German Center for Infection Research, partner site Tuebingen, Tuebingen, Germany; Infectious Diseases, Department of Internal Medicine I, University Hospital Tuebingen, Tuebingen, Germany.
Michael Bitzer, Infectious Diseases, Department of Internal Medicine I, University Hospital Tuebingen, Tuebingen, Germany; Center for Personalized Medicine, University of Tuebingen, Tuebingen, Germany.
Hanna Renk, University Children’s Hospital, Tuebingen, Germany.
Jonathan Remppis, University Children’s Hospital, Tuebingen, Germany.
Corinna Engel, University Children’s Hospital, Tuebingen, Germany; Center for Pediatric Clinical Studies, University Hospital Tuebingen, Tuebingen, Germany.
Axel R Franz, University Children’s Hospital, Tuebingen, Germany; Center for Pediatric Clinical Studies, University Hospital Tuebingen, Tuebingen, Germany.
Manuela Harries, Helmholtz Centre for Infection Research, Braunschweig, Germany.
Barbora Kessel, Helmholtz Centre for Infection Research, Braunschweig, Germany.
Berit Lange, Helmholtz Centre for Infection Research, Braunschweig, Germany.
Monika Strengert, Helmholtz Centre for Infection Research, Braunschweig, Germany; TWINCORE, Centre for Experimental and Clinical Infection Research, a joint venture of Hannover Medical School and the Helmholtz Centre for Infection Research, Hannover, Germany.
Gerard Krause, Helmholtz Centre for Infection Research, Braunschweig, Germany; TWINCORE, Centre for Experimental and Clinical Infection Research, a joint venture of Hannover Medical School and the Helmholtz Centre for Infection Research, Hannover, Germany.
Anne Zeck, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Ulrich Rothbauer, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany; Pharmaceutical Biotechnology, University of Tuebingen, Tuebingen, Germany.
Alex Dulovic, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
Nicole Schneiderhan-Marra, NMI Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany.
References
- 1. World Health Organization . Tracking SARS-CoV-2 variants. Available at:https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 1 May 2022. [PubMed]
- 2. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182:812–27.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Graham MS, Sudre CH, May A, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health 2021; 6:E335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021; 372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collier DA, De Marco A, Ferreira IATM, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021; 593:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 2021; 22:757–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pegu A, O'Connell SE, Schmidt SD, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 2021; 373:1372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization . Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern.2021. Available at:https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. Accessed 1 May 2022.
- 9. CoVariants . Shared mutations. Available at: https://covariants.org/shared-mutations/. Accessed 1 May 2022.
- 10. Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. cov-lineages.org . Lineage B.1.1.529.2022. Available at:https://cov-lineages.org/lineage.html?lineage=B.1.1.529. Accessed 1 May 2022.
- 12. Cele S, Jackson L, Khoury DS, et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv [Preprint]. Posted online 17 December 2021. doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 13. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022; 602:657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nemet I, Kliker L, Lustig Y, et al. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N Engl J Med 2022; 386:492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia-Beltran WF, StDenis KJ, Hoelzemer A, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022; 185:457–66.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med 2022; 386:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aggarwal A, Stella AO, Walker G, et al. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv [Preprint]. Posted online 15 December 2021. doi: 10.1101/2021.12.14.21267772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gornyk D, Harries M, Glöckner S, et al. SARS-CoV-2 seroprevalence in Germany. Dtsch Arztebl Int 2021; 118:824–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Junker D, Dulovic A, Becker M, et al. COVID-19 patient serum less potently inhibits ACE2-RBD binding for various SARS-CoV-2 RBD mutants. Sci Rep 2021; 12:7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heinzel C, Pinilla YT, Elsner K, et al. Non-invasive antibody assessment in saliva to determine SARS-CoV-2 exposure in young children. Front Immunol 2021; 12:753435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Renk H, Dulovic A, Seidel A, et al. Robust and durable serological response following pediatric SARS-CoV-2 infection. Na Commun 2022; 13:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Becker M, Strengert M, Junker D, et al. Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity. Nat Commun 2021; 12:1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Becker M, Dulovic A, Junker D, et al. Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat Commun 2021; 12:3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gruell H, Vanshylla K, Tober-Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med 2022; 28:477–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv [Preprint]. Posted online 8 December 2021. doi: 10.1101/2021.12.07.21267432. [DOI] [Google Scholar]
- 26. Lechmere T, Snell LB, Graham C, et al. Broad neutralization of SARS–CoV–2 variants, including Omicron, following breakthrough infection with Delta in COVID–19–vaccinated individuals. mBio 2022; 13:e0379821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carreño JM, Alshammary H, Tcheou J, et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2022; 602:682–8. [DOI] [PubMed] [Google Scholar]
- 28. Pajon R, Doria-Rose NA, Shen X, et al. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N Engl J Med 2022; 386:1088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goel RR, Apostolidis SA, Painter MM, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci Immunol 2021; 6:eabi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.