Abstract
Escherichia coli mutants lacking activities of all known cytosolic ATP-dependent proteases (Lon, ClpAP, ClpXP, and HslVU), due to double deletions [ΔhslVU and Δ(clpPX-lon)], cannot grow at low (30°C) or very high (45°C) temperatures, unlike those carrying either of the deletions. Such growth defects were particularly marked when the deletions were introduced into strain MG1655 or W3110. To examine the functions of HslVU and other proteases further, revertants that can grow at 30°C were isolated from the multiple-protease mutant and characterized. The revertants were found to carry a suppressor affecting either ftsZ (encoding a key cell division protein) or sulA (encoding the SulA inhibitor, which binds and inhibits FtsZ). Whereas the ftsZ mutations were identical to a mutation known to produce a protein refractory to SulA inhibition, the sulA mutations affected the promoter-operator region, reducing synthesis of SulA. These results suggested that the growth defect of the parental double-deletion mutant at a low temperature was due to the accumulation of excess SulA without DNA-damaging treatment. Consistent with these results, SulA in the double-deletion mutant was much more stable than that in the Δ(clpPX-lon) mutant, suggesting that SulA can be degraded by HslVU. As expected, purified HslVU protease degraded SulA (fused to the maltose-binding protein) efficiently in an ATP-dependent manner. These results suggest that HslVU as well as Lon participates in the in vivo turnover of SulA and that HslVU becomes essential for growth when the Lon (and Clp) protease level is reduced below a critical threshold.
Protein turnover in Escherichia coli mostly depends on several ATP-dependent proteases that are present in the cytosol (Lon [also called La], ClpAP [Ti], ClpXP, and HslVU) or associated with the inner membrane (FtsH [HflB]) (12). Some of them (Lon and FtsH) form homo- or hetero-oligomers, whereas others (ClpAP, ClpXP, and HslVU) are two-component proteases that consist of a catalytic subunit (ClpP and HslV) and an ATPase subunit, which presumably confers substrate specificity (ClpA, ClpX, and HslU). These enzymes can degrade not only misfolded or abnormal proteins but also some physiologically important proteins that are normally unstable. Among the naturally unstable proteins whose stability is modulated by these proteases are heat shock ς factor (ς32), cell division inhibitor SulA, and transcription activator RcsA.
ς32 (encoded by rpoH) is specifically required for the transcription of heat shock genes and is found at a very low level under nonstress conditions due to both its extreme instability (half-life, 1 min) and restricted translation of rpoH mRNA; the ς32 level is rapidly and transiently enhanced during the heat shock response by both stabilization and translational induction (15, 47). Interestingly, most of the ATP-dependent proteases or their subunits are heat shock proteins, and their synthesis is coordinately regulated with that of other heat shock proteins through transcriptional activation mediated by ς32. Conversely, the stability of ς32 is tightly modulated by the DnaK/DnaJ chaperones and proteases, particularly membrane-bound FtsH (17, 40); however, an active role of cytosolic proteases, including HslVU, has been suggested by both in vivo (25) and in vitro (24, 43) analyses.
SulA and RcsA, on the other hand, are well-known substrates for the Lon protease (12). SulA (encoded by sulA/sfiA) is a member of the SOS regulon, and its synthesis is induced by DNA-damaging agents, such as mitomycin C (42); when induced, it inhibits septation by binding to FtsZ, a key cell division protein. SulA is normally unstable in the wild type (half-life, 1 to 2 min) but is greatly stabilized in lon mutants (half-life, 20 to 30 min) (32), leading to an excess accumulation that renders the cell hypersensitive to DNA damage. RcsA (half-life, 5 min) specifically activates the transcription of cps genes involved in colanic acid synthesis. The stabilization of RcsA in lon mutants (half-life, 20 min) results in the overproduction of capsular polysaccharide and the formation of mucoid colonies (41).
We recently identified HslVU as a protease that can participate in the in vivo turnover of ς32 as well as of heterologous proteins, such as human prourokinase (25). HslVU consists of two rings of six catalytic subunits (HslV) flanked by rings of six or seven ATPase subunits (HslU) on both sides (5, 26, 31, 33, 34, 46). In the course of characterizing mutants lacking all known cytosolic proteases, we found mutants (derived from strain MG1655 or W3110) that exhibit clear growth defects at both low (30°C) and very high (45°C) temperatures. These findings prompted us to further dissect the function of HslVU by isolating and characterizing revertants that can grow at 30°C. We found that HslVU plays at least an auxilliary role in the degradation of SulA.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli K-12 strains and plasmids used are listed in Table 1. Each deletion of the chromosomal protease genes was transduced into wild-type strain MG1655 (or W3110) from derivatives of FS1576 (C600 thy recD1009) carrying the deletion (25). To construct an ftsZ2691 mutant with the wild-type (protease-positive) background, the leu::Tn5 mutation (kanamycin resistance) of CBK012 (44) was first transduced into KY2691, and the closely linked leu::Tn5 and ftsZ2691 mutations were then transduced into MG1655 by selection for kanamycin resistance; the resulting ftsZ2691 transductants (temperature sensitive) were confirmed by nucleotide sequencing. To construct the multiple-protease mutant lacking SulA (KY3052), the sulA::Tn5 mutation of GC2597 (9) was transduced into KY2350 at 42°C, and one of the kanamycin-resistant transductants lacking SulA (confirmed by immunoblotting) was established as KY3052. Phage P1 or T4GT7 was used for all transduction experiments.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| MG1655 | Prototrophic E. coli K-12 | B. Bachmann |
| KY2966 | MG1655 ΔhslVU1172::tet | This study |
| KY2347 | MG1655 Δ(clpPX-lon) 1196::cat | This study |
| KY2350 | KY2347 ΔhslVU1172::tet | This study |
| KY2691 | KY2350 ftsZ2691 | This study |
| KY2981 | KY2350 sulA2981 | This study |
| KY3052 | KY2350 sulA::Tn5 | This study |
| CBK012 | thyA leu::Tn5 | 44 |
| GC2597 | sulA::Tn5 pyrD thr leu his lac gal malB srl::Tn10 sfiC str | 9 |
| Plasmids | ||
| pMW118 | ori(pSC101) bla lacZα | |
| pKV1238 | pMW118 Δlacp | This study |
| pTrc99A | ori(pMB1) bla lacIqtrcp | |
| pKV1025 | pTrc99A hslV | This study |
| pKV1022 | pTrc99A hslU | This study |
Plasmids pMW118 and pTrc99A were obtained from Nippon Gene, Tokyo, Japan, and Amersham Pharmacia Biotech, respectively. To delete the lac promoter from pMW118, the plasmid was cut with HindIII, partially digested with ApaLI, blunted with T4 DNA polymerase, and ligated to obtain pKV1238. The hslVU operon was cut out from pKV1004 (25) and ligated to pKV1238 to obtain pKV1238-hslVU. pKV1238-lon carrying a 2.9-kb NcoI-MunI fragment that contains the entire lon operon or pKV1238-clpPX carrying a 2.8-kb SplI-HindIII fragment that contains the entire clpPX operon was constructed by excising the respective DNA fragments from Kohara’s λ clone 148 (28). The promoterless hslV or hslU was inserted under the control of the trc promoter on pTrc99A lacking the initiation codon (ATG) within the NcoI site (pKV1025 or pKV1022, respectively).
Media and chemicals.
L broth was described elsewhere (39); ampicillin (50 μg/ml), kanamycin (10 μg/ml), chloramphenicol (10 μg/ml), or tetracycline (10 μg/ml) was added when necessary. Mitomycin C (final concentration, 2.5 μg/ml) was used to induce the synthesis of SulA. Chemicals were obtained from Nacalai Tesque, Kyoto, Japan, or Wako Pure Chemicals, Osaka, Japan.
Protein purification.
Cells of strain KY2691 harboring pKV1025 or pKV1022 were grown to the mid-log phase in L broth, and the production of HslV or HslU was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 1 mM). After 1.5 h of incubation at 37°C, cells were harvested by centrifugation, suspended in 50 mM Tris-HCl (pH 7.5)–1 mM EDTA–1 mM dithiothreitol–10% (vol/vol) glycerol (buffer B) (26), and sonicated. All purification steps were carried out at 4°C. After precipitation with polyethyleneimine as described previously (26), the supernatant was loaded onto a HiLoad Q-Sepharose column (Amersham Pharmacia Biotech), and proteins were eluted with buffer B containing a linear gradient of KCl.
To purify HslV, fractions from the ion-exchange chromatography were loaded onto a HiTrap Blue column (Amersham Pharmacia Biotech), and proteins were similarly eluted with a linear KCl gradient. Ammonium sulfate (final concentration, 1 M) was added to the fractions containing HslV, which were then applied to a HiTrap Phenyl-Sepharose HP column (Amersham Pharmacia Biotech) equilibrated with buffer B containing 1 M ammonium sulfate. The flowthrough fraction was concentrated and applied to a HiPrep Sephacryl S-300 column (Amersham Pharmacia Biotech) equilibrated with buffer B plus 0.2 M KCl, and the HslV fractions were concentrated and stored at −70°C. The purity was estimated to be >95% by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie brilliant blue. HslU was purified by directly loading the fractions from the ion-exchange chromatography (HiLoad Q-Sepharose column) onto a HiPrep Sephacryl S-300 column. The resulting HslU fractions of about 90% purity were similarly stored at −70°C. Protein concentrations were determined by Bradford protein assays (Bio-Rad) (2).
Enzyme assays.
The reaction mixture (60 μl) for enzyme assays contained 50 mM Tris-HCl (pH 8.0), 0.1 M KCl, 1 mM dithiothreitol, 0.02% Triton X-100, 25 mM MgCl2, 4 mM ATP, 0.96 μg of HslV, 2.4 μg of HslU, and 2.4 μg of substrate. The mixture was incubated at 37°C, and the reaction was terminated by mixing with an equal volume of 2× SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. MBP-SulA (SulA fused to maltose-binding protein [MBP]) and MBP-LacZα (α fragment of LacZ fused to MBP) were kindly donated by Y. Ishii and Y. Kato (Kyushu Institute of Technology, Fukuoka, Japan).
Other procedures.
Nucleic acid manipulation (35), SDS-PAGE (39), and immunoblotting (25) were performed as described previously. Antisera against SulA and Lon were generously supplied by M. Maurizi and S. Gottesman (National Cancer Institute, Bethesda, Md.).
RESULTS
Isolation of suppressors from the multiple-protease mutant.
The E. coli double-deletion mutant [ΔhslVU Δ(clpPX-lon)] lacking all cytosolic ATP-dependent proteases (Lon, ClpAP, ClpXP, and HslVU) was isolated from wild-type strain MG1655 as described in Materials and Methods and designated KY2350. This mutant exhibited clear growth defects at or below 37°C or at a very high temperature (45°C); the efficiency of plating on L agar was markedly reduced compared to those of MG1655 or isogenic single-deletion mutants lacking Lon, ClpAP, and ClpXP (KY2347) or HslVU (KY2966) (Table 2). [Note that the Δ(clpPX-lon) deletion eliminated the activities of ClpAP, ClpXP, and Lon.] Such growth defects presumably result from the excessive accumulation of one or more protein substrates that are normally degraded by these proteases. When a moderately low-copy-number plasmid (pKV1238) carrying lon, clpPX, or hslVU was introduced into this mutant, the resulting strains all grew at 30°C (data not shown), suggesting that a certain common substrate(s) for these proteases accumulates in the original mutant (KY2350) and inhibits growth at the restrictive temperatures. In order to identify such a potential substrate(s), pseudorevertants that can grow at 30°C due to extragenic suppressors were isolated and characterized.
TABLE 2.
Relative efficiency of platinga
| Strain | No. of colonies formed at
temp (°C) of:
|
|||||
|---|---|---|---|---|---|---|
| 20 | 30 | 37 | 42 | 44 | 45 | |
| MG1655 (wild type) | 1.5 | 1.5 | 1.4 | 1 | 0.88 | 1.4 |
| KY2966 (ΔhslVU) | 0.8 | 1.3 | 0.88 | 1 | ND | 0.97 |
| KY2347 [Δ(clpPX-lon)] | 0.72b | 0.22b | 0.41 | 1 | 1.5 | 1.6 |
| KY2350 [ΔhslVU Δ(clpPX-lon)] | 1.4 × 10−5b | 3.3 × 10−5b | 1.0 × 10−2 | 1 | 1.5 | 1.6 × 10−4 |
| Revertants | ||||||
| KY2691 (class I) | 0.67b | 0.62b | 1.2 | 1 | 0.34c | <10−4 |
| KY2981 (class II) | 0.82b | 1.2b | 1.1 | 1 | 0.91 | 0.56c |
Cells were grown in L broth at 42°C, diluted, and plated on L agar. Plates were incubated at the indicated temperatures for 24 h (30 to 45°C) or for 72 h (20°C) to score the numbers of colonies. The numbers obtained were normalized to that at 42°C. ND, not determined.
Mucoid colonies.
Small colonies.
When mutant cells grown in L broth at 42°C were plated at 30°C, spontaneous revertants were obtained at fairly high frequencies (10−5 to 10−4) after 24 h. Among the 60 independent fast-growing revertants tested, 3 showed defects in growth at 44 to 45°C on L agar plates, unlike the others. Quantitative analyses revealed that the former revertants (class I) exhibited markedly reduced efficiencies of plating at 45°C, whereas the rest (class II) showed almost normal efficiencies of plating (Table 2). Both the class I and the class II revertants formed mucoid colonies at 30°C, as did the Δ(clpPX-lon) mutant (KY2347), suggesting that the RcsA activator which is supposedly stabilized by the Δ(clpPX-lon) mutation remained stable in the revertants. It thus appeared unlikely that the revertants gained novel Lon-like proteolytic activities that can degrade RcsA. All the class I revertants and several class II revertants were further examined to analyze the mechanisms underlying the defective growth of the parental multiple-protease mutant (KY2350).
Identification of suppressors in class I revertants.
Two distinct possibilities as to the nature of the suppressors were considered. First, a suppressor may represent a mutation that reduces the level or activity of a common substrate of the proteases whose excessive accumulation inhibits cell growth. Such a suppressor would be recessive to the wild-type allele. Second, a suppressor may represent a dominant mutation that renders a multiple-protease mutant resistant to inhibition by excess protease substrates. To discriminate between these possibilities, DNA extracted from the parental mutant (KY2350) was partially digested with restriction enzyme Sau3AI (average fragment size, 7 kb) and ligated with BamHI-treated plasmid pKV1238, and the resulting DNA library was introduced into a class I revertant (KY2691). Although approximately 5,000 ampicillin-resistant transformants obtained at 42°C were tested for cold sensitivity by replica plating, none of them was cold sensitive. On the other hand, when a DNA library from strain KY2691 was introduced into strain KY2350, many transformants were obtained at 30°C on L agar containing ampicillin. These results suggested that the suppressor in revertant KY2691 represents a dominant mutation.
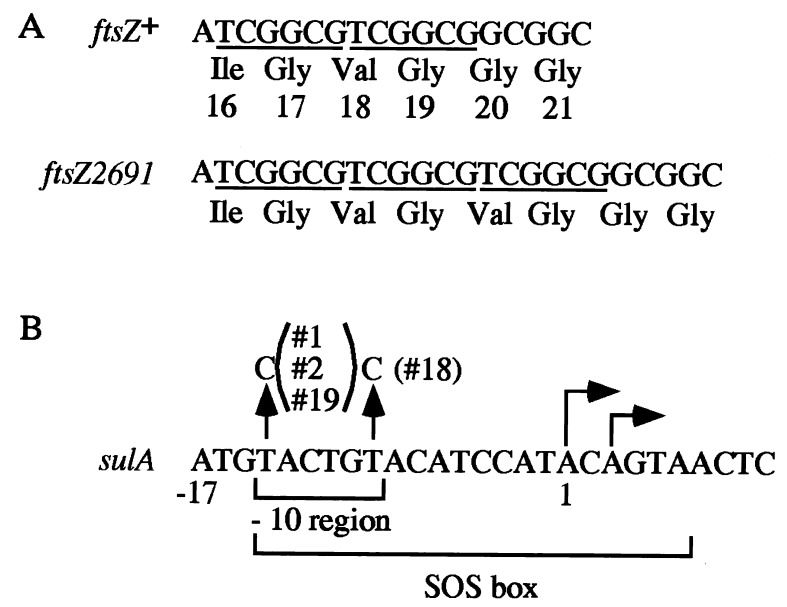
Restriction analysis of several plasmids which conferred upon KY2350 cells the ability to grow at 30°C revealed that all contained the same DNA fragment derived from KY2691. Nucleotide sequencing revealed the presence of a 3′ portion of ftsA and the entire ftsZ gene, suggesting that the suppressor affected FtsZ, which plays a critical role in cell division. Indeed, an insertion of 6 bp (TCGGCG) found near the 5′ end of the ftsZ coding region (ftsZ2691; Fig. 1A) resulted in the addition of two amino acid residues. This result was reminiscent of those of previous work on suppressors of lon mutants that were hypersensitive to DNA damage due to the accumulation of cell division inhibitor SulA; such suppressors affected either sulA/sfiA and produced inactive division inhibitor SulA or ftsZ/sulB/sfiB and produced FtsZ which was refractory to SulA inhibition (1, 8, 10, 11, 13, 21, 22, 30). The above results therefore suggested specifically that excessive SulA accumulated in KY2350 cells in the absence of DNA-damaging treatments and that ftsZ2691 rendered the FtsZ protein resistant to the SulA inhibitor at a low temperature. In fact, ftsZ2691 was found to be identical to ftsZ9, known to produce FtsZ that cannot interact with SulA (20). When the ftsZ2691 mutation was transduced into the wild type (MG1655) by selection for the nearby leu::Tn5 marker (44), the expected fraction (30%) of transductants showed little growth at 45 or 46°C (efficiency of plating, 3.4 × 10−4); marked filamentation (>70%) occurred after 60 min of incubation in liquid media. Thus, the temperature-sensitive growth of the class I revertant KY2691 can be ascribed to the ftsZ2691 mutation itself. Two other class I revertants also contained a mutation identical to ftsZ2691; such recurrent mutations might be related to the fact that we initially picked only fast-growing revertants.
FIG. 1.
Nucleotide (or amino acid) alterations caused by suppressor mutations isolated in this study. (A) The ftsZ2691 allele of a class I revertant contained a 6-bp insertion near the 5′ end of the ftsZ coding region: TCGGCG (underlined) was repeated three times, instead of twice in the wild type. Numbers represent amino acid residues, starting from the N-terminal methionine (not shown). (B) All four class II revertants tested contained a mutation within the promoter-operator region of sulA. Arrows pointing up indicate mutational changes observed, and numbers in parentheses indicate independent revertants. The transcription start sites (6) are indicated by arrows pointing to the right; the first base of the longer transcript is base 1.
Identification of suppressors in class II revertants.
Since it seemed likely that the class II revertants carry a suppressor mutation at or around sulA, we determined the nucleotide sequence of the sulA-containing DNA fragment derived from several independent revertants. As expected, all the revertants tested contained a T-to-C transition at the −10 promoter region of sulA (Fig. 1B). sulA is a typical SOS gene whose expression is normally repressed by the LexA repressor by binding to the operator commonly referred to as an SOS box (42). Since the SOS box of sulA overlaps with the −10 promoter region (6) and since the suppressor altered the consensus sequence of the operator, the suppressor could affect either the promoter or the operator or both. However, as shown below, the mutation primarily reduced promoter activity, resulting in reduced synthesis of SulA and increased survival at a low temperature (30°C) and at a very high temperature (45°C) as well. To confirm such a possibility, a sulA::Tn5 null mutation (9) was introduced into the parental ΔhslVU Δ(clpPX-lon) mutant (KY2350) at the permissive temperature (42°C). The resulting triple mutant (KY3052) could grow at both 30 and 45°C, although the growth at 45°C was slightly slower than that of the above revertants carrying the sulA-repressing promoter mutation. This result indicated that excessive SulA function is mainly responsible for the growth defects of the parental mutant. All the results presented here are in good agreement with the known properties of lon suppressors revealed under conditions of induced DNA damage. The revertant carrying sulA2981 (#1 in Fig. 1B) was designated KY2981 and was used for most subsequent experiments.
SulA accumulates in the multiple-protease mutant and in class I revertants.
In view of the above findings, the cellular levels of SulA in several mutants and revertants were determined by immunoblotting. SulA was hardly detected in extracts from the wild type (MG1655) or the ΔhslVU (KY2966) or Δ(clpPX-lon) (KY2347) mutant grown at 42°C, whereas an appreciable amount of SulA was found in the parental double-deletion mutant (KY2350) (Fig. 2A). This result strongly suggested that the HslVU protease can degrade SulA and that the increased level of SulA in the parental mutant is primarily responsible for its inability to grow at a low temperature. The SulA level found in the class I revertant (KY2691) was comparable to or higher than that found in the parental mutant (KY2350) at 42°C, consistent with the nature of the suppressor involved. When grown at 30°C, none of the strains tested, including the class I revertant (KY2691), produced detectable amounts of SulA. However, when cells of KY2350 grown at 42°C were shifted to 30°C, an appreciable fraction of cells (ca. 30%) elongated within 60 min; this fraction increased to 50% after 120 min, suggesting that the increased SulA level due to multiple protease deficiencies most probably explained the lack of growth at a low temperature (30°C). It seemed possible that cells exhibit greater sensitivity to SulA or hyperactive SulA at a low temperature. The SulA-FtsZ interaction may actually be stronger at a low temperature (4). In contrast, such filamentation was not observed when KY2350 cells were shifted from 42°C to 45°C. Since the sulA-repressing promoter mutation (class II revertants; see below) as well as the sulA null mutation restored the growth of KY2350 cells at both 30 and 45°C, it appeared evident that the growth defect of KY2350 was due to excess SulA and the consequent division inhibition.
FIG. 2.
Immunoblotting of SulA in the representative protease mutants studied, with or without suppressors. (A) Cells were grown in L broth at 42 or 30°C to the mid-log phase, and whole-cell proteins were prepared and analyzed by SDS-PAGE (13% polyacrylamide gel) followed by immunoblotting. (B) Cells were grown in L broth at 42°C and treated with mitomycin C. Samples were taken before (−) and 30 min after (+) the addition of mitomycin C. Whole-cell proteins were prepared and analyzed as described for panel A. Asterisks indicate a nonspecific band immediately below SulA. MG1655, wild type; KY2966, ΔhslVU; KY2347, Δ(clpPX-lon); KY2350, ΔhslVU Δ(clpPX-lon); KY2691 and KY2350, ftsZ2691; KY2981 and KY2350, sulA2981.
To further substantiate the above findings, cells were treated with a DNA-damaging agent (mitomycin C) to facilitate the detection of SulA. After 30 min of incubation with mitomycin C (2.5 μg/ml), SulA was detected even in the wild type at 42°C. The mitomycin C-induced SulA levels in the multiple-protease mutant (KY2350) and in the class I revertant (KY2691) were much higher than that in the wild type (Fig. 2B), in agreement with the results shown in Fig. 2A. In contrast, the SulA level in the class II revertant (KY2981) was detectable upon mitomycin C treatment but was much lower than that in the wild type, suggesting that the sulA2981 mutation reduced promoter activity while maintaining at least some operator activity.
HslVU degrades SulA in strains lacking Lon and Clp proteases.
The above results suggested that HslVU as well as the Lon protease is involved in the turnover of SulA in E. coli. To further examine this possibility, the SulA levels in hslVU+ (KY2347) and ΔhslVU (KY2350) strains, both lacking Lon and Clp protease activities, were compared. Upon induction with mitomycin C, the SulA levels increased rapidly and markedly in both strains, but the level of accumulation was appreciably higher in the ΔhslVU mutant (KY2350) (Fig. 3A). To compare the stability of SulA under these conditions, spectinomycin (1 mg/ml) was added to stop protein synthesis 30 min after the addition of mitomycin C, and samples taken at intervals were analyzed for remaining SulA levels by immunoblotting (Fig. 3B). The half-life of SulA was about 20 min in the hslVU+ strain (KY2347), comparable to that reported previously for a lon mutant (32), but was much longer (>60 min) in the ΔhslVU mutant (KY2350). Similar results were obtained when spectinomycin was added 15 min after mitomycin C induction (data not shown). It thus seemed apparent that HslVU participates in the in vivo turnover of SulA, at least in cells lacking Lon and Clp proteases. The data also suggested that the almost normal growth and survival of the hslVU+ Δ(clpPX-lon) mutant at a low temperature depend on the (proteolytic) effect of HslVU on SulA.
FIG. 3.
Stability of mitomycin C-induced SulA in Δ(clpPX-lon) and ΔhslVU Δ(clpPX-lon) mutants. (A) Time course of accumulation of SulA upon addition of mitomycin C. Cells were grown in L broth at 42°C, and mitomycin C was added at time zero. Samples were taken at intervals, and whole-cell proteins were analyzed as described in the legend to Fig. 2A. (B) Stability of mitomycin C-induced SulA. Cells were grown in L broth at 42°C and treated with mitomycin C for 30 min, and spectinomycin (1 mg/ml) was added at time zero. Samples were taken at the times indicated, and the remaining SulA level was determined by immunoblotting. KY2347, Δ(clpPX-lon); KY2350, ΔhslVU Δ(clpPX-lon).
We next investigated the effect of the ΔhslVU mutation on SulA levels in lon+ strains. When mitomycin C was added to cells of the wild type (MG1655) or the isogenic ΔhslVU mutant (KY2966), the SulA levels increased in both strains. The SulA level in the wild type increased very rapidly and reached the maximum within 15 min, followed by a gradual decrease, whereas the SulA level increased more slowly in KY2966 and reached the maximum at about 30 min (Fig. 4). Mitomycin C-induced SulA was almost equally unstable (half-life, 1 to 2 min) in both strains (data not shown). This finding first appeared paradoxical but actually was not unexpected, because the ΔhslVU mutation was previously shown to increase ς32 and heat shock protein levels as well (25). Thus, the slower appearance of SulA in the ΔhslVU mutant seemed to be explained by the increased level of Lon protease. As shown in Fig. 4, the levels of Lon and ς32 in the wild type were low during steady-state growth at 42°C and rapidly increased upon the addition of mitomycin C. In contrast, the levels of Lon and ς32 in the ΔhslVU mutant were constitutively high and were comparable to those in mitomycin C-treated wild-type cells. The above interpretation was also consistent with the finding that when excess Lon was supplied to wild-type cells by means of pKV1238-lon, mitomycin C induction of SulA was hardly observed (data not shown).
FIG. 4.
Cellular levels of SulA, Lon, and ς32 upon mitomycin C treatment of the wild type (MG1655) and the ΔhslVU mutant (KY2966). Cells were grown in L broth at 42°C, mitomycin C was added, and samples taken at intervals were analyzed by SDS-PAGE and immunoblotting as described in the legend to Fig. 2A. Asterisks indicate a nonspecific band.
HslVU protease degrades SulA in vitro.
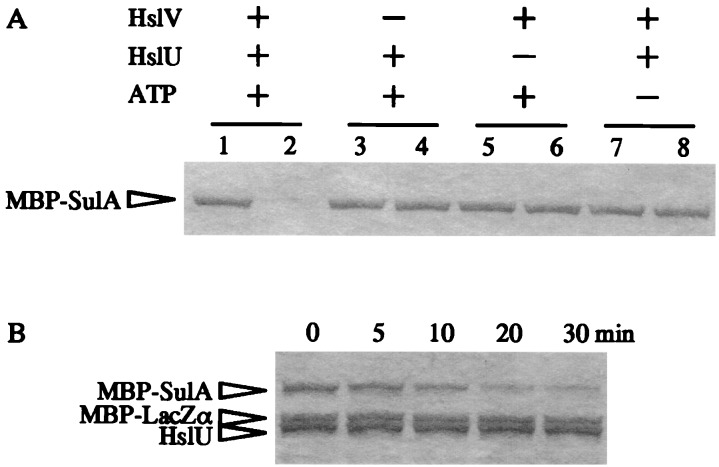
Finally, we tested whether purified HslVU protease directly degrades SulA in vitro by using an MBP-SulA fusion protein as a substrate. This fusion protein was known to function as a cell division inhibitor in vivo, like authentic SulA, and the purified fusion protein could bind to FtsZ and was degraded by Lon in vitro (18, 19, 36). When separately purified HslV, HslU, and MBP-SulA proteins were mixed and incubated at 37°C in the presence or absence of ATP, the MBP-SulA fusion protein was degraded only in the presence of both HslV and HslU in an ATP-dependent manner (Fig. 5A). The degradation of the MBP-SulA fusion protein seemed to be specific, since the MBP-LacZα fusion protein was hardly affected under the conditions in which MBP-SulA was rapidly degraded (Fig. 5B).
FIG. 5.
In vitro degradation of SulA by purified HslVU protease. (A) Purified HslV (0.96 μg), HslU (2.4 μg), and MBP-SulA (2.4 μg) were mixed in a reaction mixture (60 μl) with or without 4 mM ATP as described in Materials and Methods. Samples were analyzed by SDS-PAGE before (lanes 1, 3, 5, and 7) or after (lanes 2, 4, 6, and 8) incubation at 37°C for 2 h. (B) Time course of degradation. HslV, HslU, MBP-SulA, and MBP-LacZα (2.4 μg) were mixed essentially as described for panel A and incubated at 37°C in the presence of 4 mM ATP. Samples were withdrawn at the indicated times before (0 min) and after incubation at 37°C and analyzed by SDS-PAGE.
DISCUSSION
The present study of pseudorevertants isolated from a multiple-protease mutant revealed that the inability of the latter mutant (KY2350) to grow at a low temperature is probably due to the accumulation of cell division inhibitor SulA (Fig. 2). Evidence suggested that at least in the absence of Lon and Clp proteases, HslVU can degrade SulA, thus functionally substituting for Lon (and possibly Clp). It became evident that the amount of SulA, which is very low in the wild type, is enhanced to a level sufficient to inhibit cell growth in the parental double-deletion mutant grown at restrictive temperatures. In other words, unlike the SulA level in the lon mutant, which exhibits defective growth only upon DNA-damaging treatment, the SulA level in the multiple-protease mutant is elevated during normal growth, apparently independent of DNA damage. The possibility that the multiple-protease mutations caused constitutively high expression of the SOS regulon and indirectly enhanced the SulA level seemed unlikely, since the level of RecA, one of the SOS gene products, appeared to remain unaffected, as judged by staining of the protein after SDS-PAGE (data not shown).
The in vivo comparison of SulA levels and stability in the hslVU+ and ΔhslVU strains lacking Lon (and Clp) (Fig. 3) as well as in vitro proteolysis experiments with purified proteins (Fig. 5) established that HslVU protease can degrade SulA, at least under the conditions used here. The fact that HslVU is essential for growth at a low temperature (and at a very high temperature) in the absence of Lon and Clp proteases suggests that even in the lon+ strain, HslVU plays a significant role in modulating the SulA level, at least in the presence of limited levels of active Lon (and Clp) proteases possibly resulting from titration by excess potential substrates. It was recently reported that the overexpression of HslVU endows the lon mutant with marked resistance to DNA-damaging agents, suggesting that HslVU can functionally replace Lon, at least partially (27). On the other hand, our results with the lon+ strains showed a lower (rather than a higher) accumulation of SulA in the ΔhslVU mutant than in the hslVU+ control during the early phase of induction with mitomycin C (Fig. 4), suggesting that the role of HslVU in SulA turnover during the steady-state growth of wild-type E. coli is limited.
Similar results on the role of HslVU in SulA degradation were obtained by Wu et al. (45), who compared the SulA levels among lon, hslV, and hslU single mutants and lon hslV and lon hslU double mutants. However, the fact that the level of Lon increases appreciably in the ΔhslVU mutant (Fig. 4) makes it difficult to evaluate quantitatively the potential contribution of the HslVU protease to SulA degradation by such comparisons alone. The possible contribution of Clp proteases to SulA degradation was suggested by the observation that the low-copy-number plasmid expressing clpPX (pKV1238-clpPX) could suppress the growth defect of the multiple-deletion mutant at 30°C, although the extent of suppression was lower than that with pKV1238-lon or pKV1238-hslVU (data not shown). However, the level of SulA that accumulated in the double-deletion mutant carrying pKV1238-clpPX upon mitomycin C induction was not significantly lower than that in the same mutant carrying the vector alone (data not shown). Thus, the contribution of Clp proteases to SulA degradation appears to be small, if significant at all.
Since all the known ATP-dependent proteases (except ClpA) are heat shock proteins under ς32 control (15) and many proteases (except ClpXP) appear to participate significantly in the turnover of ς32 (25), a decrease in the level of any of these proteases that could result from titration by excess substrates can be compensated for by enhanced synthesis of these proteases by an autoregulatory mechanism through the stabilization of ς32. When the substrate levels are sufficiently reduced, the proteases become present in a relative excess, and the synthesis of the proteases is repressed through the destabilization of ς32. Such a negative feedback circuit may be illustrated in part by the results shown in Fig. 4. The treatment of wild-type cells with mitomycin C initially enhanced the level of SulA; this effect was followed shortly by an increase in the levels of ς32 and Lon and by a subsequent decrease in the levels of SulA and ς32. However, other unstable proteins, besides SulA, that are induced by mitomycin C (e.g., UmuC and UmuD) may also play roles in modulating protease levels. In addition to the proteases, the DnaK chaperone team (DnaK, DnaJ, and GrpE) is known to be required for the rapid degradation of ς32 and other proteins, including abnormal proteins (23, 37, 38). These protein substrates can therefore titrate the DnaK chaperones as well as proteases away from ς32, resulting in increased stability and level of ς32, which in turn can trigger the induction of heat shock proteins, including proteases and chaperones (3, 7).
The coordinated synthesis of ATP-dependent proteases through the stabilization of ς32 would appear to be further strengthened if the substrate specificity of the proteases overlapped appreciably, because such a situation should effectively accelerate the degradation of the critical common substrates. Our results suggest that the regulation of at least two proteases (Lon and HslVU) involving the stabilization of ς32 is likely to operate in modulating the cellular level of the inhibitor SulA. Besides SulA examined in this study, Xis of phage λ (29), SsrA-tagged proteins (14, 16), and most abnormal proteins (12) are thought to be degraded by more than one protease. In all these cases, coordinated and interdependent regulation among ATP-dependent proteases is expected to operate through stability control of ς32. The participation of multiple proteases in modulating the stability of ς32 is likely to play an important role in the maintenance of appropriate levels and activities of proteases under a variety of physiological and environmental conditions.
ACKNOWLEDGMENTS
We are grateful to W.-F. Wu and S. Gottesman for communicating results prior to publication and to M. Maurizi and S. Gottesman for kind gifts of antisera. We thank M. Nakayama, M. Ueda, and H. Kanazawa for technical assistance.
REFERENCES
- 1.Bi E, Lutkenhaus J. Analysis of ftsZmutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B. Regulation of the Escherichia coliheat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 4.Canceill D, Dervyn E, Huisman O. Proteolysis and modulation of the activity of the cell division inhibitor SulA in Escherichia coli lonmutants. J Bacteriol. 1990;172:7297–7300. doi: 10.1128/jb.172.12.7297-7300.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang S-E, Burland V, Plunkett III G, Daniels D L, Blattner F R. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 6.Cole S T. Characterisation of the promoter for the LexA regulated sulA gene of Escherichia coli. Mol Gen Genet. 1983;189:400–404. doi: 10.1007/BF00325901. [DOI] [PubMed] [Google Scholar]
- 7.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 8.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZthat confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;175:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 10.Gayda R C, Yamamoto L T, Markovitz A. Second-site mutations in capR (lon) strains of Escherichia coliK-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J Bacteriol. 1976;127:1208–1216. doi: 10.1128/jb.127.3.1208-1216.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George J, Castellazzi M, Buttin G. Prophage induction and cell division in E. coli. III. Mutations sfiA and sfiB restore division in tif and lon strains and permit the expression of mutator properties of tif. Mol Gen Genet. 1975;140:309–332. [PubMed] [Google Scholar]
- 12.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by Lon mutants of Escherichia coliK-12. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman S, Roche E, Zhou Y-N, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 16.Herman C, Thevenet D, Bouloc P, Walker G C, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coliprotease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman C, Thevenet D, D’Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashitani A, Higashitani N, Horiuchi K. A cell division inhibitor SulA of Escherichia colidirectly interacts with FtsZ through GTP hydrolysis. Biochem Biophys Res Commun. 1995;209:198–204. doi: 10.1006/bbrc.1995.1489. [DOI] [PubMed] [Google Scholar]
- 19.Higashitani A, Ishii Y, Kato Y, Horiuchi K. Functional dissection of a cell-division inhibitor, SulA, of Escherichia coliand its negative regulation by Lon. Mol Gen Genet. 1997;254:351–357. doi: 10.1007/s004380050426. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huisman O, D’Ari R, George J. Further characterization of sfiA and sfiB mutations in Escherichia coli. J Bacteriol. 1980;144:185–191. doi: 10.1128/jb.144.1.185-191.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson B F. Fine structure mapping and properties of mutations suppressing the lon mutation in Escherichia coliK-12 and B strains. Genet Res. 1977;30:273–286. doi: 10.1017/s0016672300017687. [DOI] [PubMed] [Google Scholar]
- 23.Jubete Y, Maurizi M R, Gottesman S. Role of the heat shock protein DnaJ in the Lon-dependent degradation of naturally unstable proteins. J Biol Chem. 1996;271:30798–30803. doi: 10.1074/jbc.271.48.30798. [DOI] [PubMed] [Google Scholar]
- 24.Kanemori, M. Unpublished data.
- 25.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of ς32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessel M, Wu W, Gottesman S, Kocsis E, Steven A C, Maurizi M R. Six-fold rotational symmetry of ClpQ, the E. colihomolog of the 20S proteasome, and its ATP-dependent activator, ClpY. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 27.Khattar M M. Overexpression of the hslVU operon suppresses SOS-mediated inhibition of cell division in Escherichia coli. FEBS Lett. 1997;414:402–404. doi: 10.1016/s0014-5793(97)01024-7. [DOI] [PubMed] [Google Scholar]
- 28.Kohara Y, Akiyama K, Isono K. The physical map of the whole Escherichia colichromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 29.Leffers G G, Jr, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutkenhaus J F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983;154:1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Missiakas D, Schwager F, Betton J-M, Georgopoulos C, Raina S. Identification and characterization of HslV HslU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 32.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: the longene controls the stability of SulA protein. Proc Natl Acad Sci USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohrwild M, Coux O, Huang H-C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. HslV-HslU: a novel ATP-dependent protease complex in Escherichia colirelated to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H-C, Engel A, Baumeister W, Goldberg A L. The ATP-dependent HslVU protease from Escherichia coliis a four-ring structure resembling the proteasome. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sonezaki S, Ishii Y, Okita K, Sugino T, Kondo A, Kato Y. Overproduction and purification of SulA fusion protein in Escherichia coliand its degradation by Lon protease in vitro. Appl Microbiol Biotechnol. 1995;43:304–309. doi: 10.1007/BF00172829. [DOI] [PubMed] [Google Scholar]
- 37.Straus D B, Walter W A, Gross C A. Escherichia coliheat shock gene mutants are defective in proteolysis. Genes Dev. 1988;2:1851–1858. doi: 10.1101/gad.2.12b.1851. [DOI] [PubMed] [Google Scholar]
- 38.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 39.Tobe T, Ito K, Yura T. Isolation and physical mapping of temperature-sensitive mutants defective in heat-shock induction of proteins in Escherichia coli. Mol Gen Genet. 1984;195:10–16. doi: 10.1007/BF00332716. [DOI] [PubMed] [Google Scholar]
- 40.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Cabassa A S, Gottesman S. Capsule synthesis in Escherichia coliK-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 43.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whalen W A, Berg C M. Gratuitous repression of avtA in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984;158:571–574. doi: 10.1128/jb.158.2.571-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W-F, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coliLon and the ClpYQ (HslUV protease. J Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M-S, Tanaka K, Goldberg A L, Chung C H. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 47.Yura T, Nagai H, Mori H. Regulation of heat shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]