Abstract
Epidemiological studies show that COVID-19 variants-of-concern, like Delta and Omicron, pose different risks for severe disease, but they typically lack sequence-level information for the virus. Studies which do obtain viral genome sequences are generally limited in time, location, and population scope. Retrospective meta-analyses require time-consuming data extraction from heterogeneous formats and are limited to publicly available reports. Fortuitously, a subset of GISAID, the global SARS-CoV-2 sequence repository, includes “patient status” metadata that can indicate whether a sequence record is associated with mild or severe disease. While GISAID lacks data on comorbidities relevant to severity, such as obesity and chronic disease, it does include metadata for age and sex to use as additional attributes in modeling. With these caveats, previous efforts have demonstrated that genotype-patient status models can be fit to GISAID data, particularly when country-of-origin is used as an additional feature. But are these models robust and biologically meaningful? This paper shows that, in fact, temporal and geographic biases in sequences submitted to GISAID, as well as the evolving pandemic response, particularly reduction in severe disease due to vaccination, create complex issues for model development and interpretation. This paper poses a potential solution: efficient mixed effects machine learning using GPBoost, treating country as a random effect group. Training and validation using temporally split GISAID data and emerging Omicron variants demonstrates that GPBoost models are more predictive of the impact of spike protein mutations on patient outcomes than fixed effect XGBoost, LightGBM, random forests, and elastic net logistic regression models.
Keywords: Viral genomics, COVID-19, SARS-CoV-2, Bioinformatics, Machine learning
1. Introduction
Throughout the COVID-19 pandemic, SARS-CoV-2 has mutated in ways that have significantly impacted pathogenesis. Epidemiological studies can show different risks of severe disease due to different COVID-19 variants, such as Delta and Omicron, but typically lack resolution at the level of specific combinations of changes in viral genome sequences. The emergence of the COVID-19 pandemic, however, has coincided with the widespread availability of lower cost, rapid whole genome sequencing. As of writing, over 10 million SARS-CoV-2 sequences were available to researchers from the GISAID website (http://www.gisaid.org) [1], [2]. GISAID includes a metadata field for “patient status” for a subset of sequences, which represents a potentially unparalleled resource for genetic analysis. If its potential can be unlocked, GISAID could provide the data necessary to develop a model of disease severity based on viral genotype and the limited patient characteristics available on GISAID, age and gender.
Clinical data which differentiate SARS-CoV-2 genotype generally do so at the level of lineages using the Pango nomenclature [3], [4], or most commonly variants of concern (VOC) [5] based on those lineages, such as Alpha (Pango lineage designation B.1.1.7), Beta (B.1.351), Delta (B.1.167.2), and Omicron (BA.1 and BA.2). VOC designations generally refer not only to the original lineage but other “sublineages”, such as AY.x sublineages of Delta and Omicron sublineages, such as BA2.12.1, B.4, and B.5. Studies have shown differences in case outcomes between lineages, supported in at least some cases with in vitro or animal model evidence for changes in virulence. The Delta variant had clear-cut increases in transmissibility and virulence, as indicated by both epidemiological estimates [6] and laboratory studies that show increased fitness over previous variants, including enhanced viral replication due to modification in the furin cleavage site of the spike protein [7], [8]. Alpha, also appears to have resulted in more severe disease than the ancestral genome [9], [10], [11], [12]. Overall, while Alpha resulted in elevated hospitalizations, ICU admissions, and other markers of severe outcomes as compared to ancestral lineages, Delta appears to have been yet more severe than Alpha [13], [14], [15], [16]. By contrast, following Delta, Omicron has appeared to result in less severe disease, even when controlling for vaccination [17]. Lower risks of hospitalization, and in particular shorter hospital stays, reduced ICU admissions, and less use of ventilation rates, particularly as compared to Delta, have been shown in studies from Denmark [18], the United States, [19], [20], and the United Kingdom [21], [22]. Epidemiological data for Omicron are consistent with in vitro and animal studies, which have shown a reduction in lower lung infectivity, deficient cell entry, and a reduction in syncytium formation due to reduced ability of the spike protein to mediate plasma membrane fusion [23], [24], [25], [26].
The aforementioned differences in case outcome are between apparently sequential emergent SARS-CoV-2 genetic lineages. In fact, however, changes to SARS-CoV-2 properties often implicate combinations of multiple mutations that emerge simultaneously—and then sometimes revert in whole or in part as the virus continues to evolve [27], [28]. For example, Delta, originally Pango-designated B.1.617.2, has spawned complex sublineages (generally identified by an AY prefix) with distinct immune evasion and virulence properties—and can genetically share more in common with other lineages than the one from which they apparently branched [29], [30]. During a long-term infection, a spike protein may emerge with multiple variations, i.e., a “long branch” divergence from the phylogenetic tree, a process hypothesized as the origin of the Omicron variant of concern (initially identified as Pango lineage B.1.592, and soon redesignated as BA.1 and BA.2) [31]. As Omicron has become dominant, subsequent Omicron subvariants have emerged, including subvariants of BA.1 and BA.2, as well as BA.3, BA.4, and BA.5 [32]. These Omicron subvariants are characterized by apparent recombination of mutations, as well as the appearance of mutations that look similar to those in previous variants, such as Alpha [33]. Recombinant variants of Delta and Omicron have also been identified in larger numbers; these are designated in the Pango scheme with an initial “X”, e.g., XD and XE [34].
Conventional techniques for studying the effect of SARS-CoV-2 genotype on COVID-19 mortality and symptom severity have included analysis of single nucleotide polymorphisms (SNP) and genome wide association studies [35], [36], [37], literature meta-analysis [38]. Individual studies have been limited in the number of patients, and meta-analyses are time-consuming, complicated by having to access underlying data, and generally exclude unpublished data. The viral sequences from many such published and unpublished studies have been deposited in the GISAID global SARS-CoV-2 sequence repository, along with data for patient status. Various groups have investigated the incidence and prevalence of mutations in GISAID entries with clinical metadata [39], [40]. While these studies have yielded some potential candidate mutations, the data are often conflicting or not necessarily consistent with epidemiological or laboratory observations. For example, some of the latter studies showed a link between the D614G mutation, which emerged early on as cases spread from Asia to Europe and North America, with increased disease burden. But another study of case fatality rates by region did not find a correlation with the dominant clade in that region [41].
In general, efforts to build logistic regression and other statistical models to predict mild versus severe disease on GISAID data have shown that much of the explanatory power is provided by patient age, gender, and region of origin, rather than clade or lineage [42]. However, it has been shown that adding sequence data to a logistic regression method can produce a more accurate prediction of severe versus mild disease than one with only age, gender, and region, although the difference was not particularly large [43]. Moreover, an updated of the latter model trained and tested on more recent sequence data resulted in deteriorated performance, even when employing a more accurate random forests classifier method [44]. Another group employed a powerful gradient-boosted decision tree ensemble classifier method, XGBoost, and found that models evaluated using temporally split data, i.e. trained on earlier sequences and tested on later-emerging sequences, substantially outperformed models evaluated using cross-validation, in which the training and test samples are randomly selected [45]. The authors analyzed the trained models to identify mutations associated with increased severe disease risk. However, key findings such as the V1176F mutation, while present in VOC, have not been specifically linked to disease severity in laboratory or epidemiological studies. Other methods have been employed, including deep neural networks [46], [47] and Bayesian multinomial logistic regression to infer growth rate, and thus viral fitness, from individual sequence mutations [48]. However, there is no consensus modeling method for analyzing GISAID data, and the complexity of the data has not been fully analyzed.
The modeling approach in this paper begins by first evaluating the trends and structure of GISAID data—a critical step for developing robust genotype-phenotype models. There are two issues that impact the use of GISAID as a data source: (1) The nature of the pandemic has changed over time, with more screening of asymptomatic or mild cases, as well as improvements in therapeutics and widespread vaccination. (2) While unprecedented numbers of SARS-CoV-2 sequences have been deposited, that still represents only a sampling of viral infections worldwide. For example, as of April 2022, over half of all sequences in GISAID are from the United States and United Kingdom [49]. The set of sequences with GISAID patient metadata which we were able to curate (excluding illegible or unknown metadata fields) are an even smaller subsample. In practice, other work has shown that models trained on earlier records do not perform well on later sequence records [44], [45]. In part due to the evolution of novel mutations, but it may also have to do with changes in the nature of what kind of sequences are submitted to GISAID over time. For example, in our previous work, we showed that, through September 2021, there had been a consistent increase in “mild” cases observed in the database [47]. In the analysis shown here, we identify that the latter temporal trends may have now stabilized, but that heterogeneity in the geographic origin of samples may be an important confounder.
Notably, other potential risk factors, such as obesity or chronic disease, are not provided in GISAID metadata. Moreover, while vaccination status is a metadata field, only very few samples include an entry for it. As a result, modeling efforts based on GISAID data will always lack information on known co-founders. Even so, GISAID does have many more samples than targeted studies that do include information about comorbidities, which at least mitigates potential data bias. Also, training om data after vaccination is more widespread, as shown here, can mitigate biases due to vaccination status. That said, the foregoing caveats are important for the work presented here as well as all efforts to model the effect of viral genotype on disease severity.
Taking the aforementioned observations and caveats into account, in this paper we examine the overall data set. Then, we identify a timeframe for model training that can result in more robust models for evaluation, and hypothesize that including geographic origin through the “country” metadata field in a mixed effects model will result in more robust models as well. We propose to use a recently-developed mixed effects machine learning method, GPBoost, which incorporates decision tree-boosting to efficiently train on large data sets with many features [50]. GPBoost is compared to conventional methods, including logistic regression, as well as ensemble decision-tree based methods, Random Forests [51], XGBoost [52], and LightGBM [53]. The best-performing methods, GPBoost, XGBoost, and LightGBM, are interpreted using SHAP (SHapley Additive exPlanations) [54], which has previously been used to interpret XGBoost models [45]. The modeling methodologies are evaluated on the spike protein sequence, as it binds to host cell receptors, mediates cell entry, is a key target for the immune response, and has a high rate of mutation [55], [56], [57], [58]. Analyzing only the spike protein sequence further reduces the risk of overfitting and make models more computationally tractable.
2. Methods
2.1. Spike protein sequence collection and pre-processing
Spike protein sequences are obtained from a FASTA file available from the GISAID database (http://www.gisaid.org). The data for this study were downloaded on sequences that were submitted to and processed by GISAID as of April 15, 2022. Based on the metadata for collection date, the latest-collected sample in this data set was from April 10, 2022. GISAID performs various data curation tasks; of relevance here, Spike protein sequences are preprocessed by GISAID by multiple sequencing alignment, identifying ORFs, and translating nucleotide sequences to obtain protein FASTA files [2]. The FASTA file is parsed to obtain only those sequences for which patient metadata are available. (The section below details how patient metadata are obtained). The acknowledgment table for the sequences used in this study may be found at https://doi.org/10.55876/gis8.220606hk.
Many of the spike sequences are truncated due to sequencing gaps and errors. Therefore, the Spike protein sequences from the FASTA file are aligned with respect to the consensus Spike reference sequence (Wuhan-Hu-1 isolate) obtained by multiple sequence alignment of early genome sequences [59]. The alignments are generated using the local pairwise Striped Smith–Waterman (SSW) method [60], [61], with BLOSUM62, implemented with the scikit-bio package in Python 3.8 [62]. Aligned sequences shorter than the reference (1273 residues) are front and/or end padded with a “*”, and otherwise all indels are at positions corresponding to the reference. To preserve as many samples as possible, there is no filtering sequences with “*” (mask) or “X” (ambiguous amino acid).
2.2. Patient status metadata collection and pre-processing
The GISAID database provides an option to identify sequence records that include “patient metadata” and to download the metadata file with that information. This study includes the data from the records available for sequences collected by April 15, 2020: 414,297 records in total. (By comparison, at that time, the aforementioned Spike protein sequence FASTA file, that are from studies with and without metadata, included over 5 million sequences.) After metadata exclusions are applied (described in the following paragraphs), 163,496 samples remain available for machine learning. These records include an entry for “patient status” as well as metadata fields generally available for all SARS-CoV-2 sequences on GISAID, which include inter alia host, the continent/country/region of collection, Pango nomenclature lineage, NextStrain clade, sample collection and submission date, patient age, and patient gender. As an initial matter, all samples for which the host is not identified as “Human” are removed from the dataset.
The patient metadata consists of a single field with text provided by the submitter of the sequence. There are many different kinds of entries, including misspellings. As a preliminary step, these metadata entries are translated to a “Status”. The “Status” translates different entries which may consist of different spellings or synonyms for the same activity, such as cases obtained by screening asymptomatic carriers. The table includes examples of entries assigned to these categories. Table 1 shows all of the unique metadata entries in the full patient metadata set (414,297 records) along with the corresponding “Status” designation. The resulting status is then categorized, generally following the commonly used case classification such as those defined by the United States National Institutes for Health (NIH) COVID-19 guidelines [63]. 1 shows the categories and the status designation. For example, sequences with metadata indicating ICU admission or mechanical ventilation are categorized as “Severe”. Some metadata entries are categorized as “Unknown” even if they are not explicitly entered as such, as they do not contain information about the patient’s status, for example some appear to refer to the age of the patient or to the location where the sample was taken. Metadata entries of “recovered” were also placed in the unknown category, as there was no indication of the severity of prior illness. Notably, the “Asymptomatic” category is defined to also include paucisymptomatic cases which are not expressly defined as “Mild”. As such, there will be some overlap between those two categories.
The categories are then assigned to “Mild”, “Severe”, and “Unknown” classes, according to the NIH categories where there is sufficient information. 1 shows the class assignments for each category. For example, it is not clear whether “Alive” indices alive, but in an ICU, or alive and with mild symptoms. Accordingly, the “Alive” category is assigned to “Unknown”. Similarly, a “Symptomatic” or “Alive” patient may have severe symptoms or have been hospitalized; therefore, the “Symptomatic” category is thus assigned to “Unknown”. The “Released” category indicates release from prior hospitalization, and, therefore, “Released” is classified the same as “Hospitalized”. Cases in the “Screening” category are classified as “Mild”. These are cases with metadata entries such as “random screening”, “community screening”, and “airport screening;” as such, they are assumed to be from asymptomatic, or, at minimum, ambulatory individuals who were not hospitalized for COVID-19 symptoms at the time of sequencing. Cases in the Unknown class are dropped from the analysis.
The models described in this study also utilize metadata fields for “age” and “gender”. Notably, the “gender” field includes entries that suggest it is being used interchangeably for gender and sex. For the purpose of simplicity and to align with the GISAID field names, the term “gender” is used in this paper. With respect to the gender field, any entry that is cognizable as Male or Female (e.g., misspellings, foreign language words such as “Homme”, which is French for “man”, etc.) are classified accordingly. Any other entry is classified as “Unknown” and excluded from analysis. The “age” metadata entries are assigned to an integer age where possible. Where the “age” entry is provided as a range, e.g., “‘21-30”, it is assigned to the mid-point, e.g., 25. Where the “age” field entry is “unknown” or a value that cannot be translated to an integer, the sample is excluded.
2.3. Machine learning
Five machine learning methods are used: (1) logistic regression with elastic net regularization [64] (referred to as “elastic net” or “logistic regression” in this paper), which has previously been utilized for genetic association studies [65], [66]; (2) the random forests (RF) ensemble tree-based method [51], which is used to classify SARS-CoV-2 sequences to Pango lineages [3]; (3) eXtreme Gradient Boosting (XGBoost) [52], a decision tree-based ensemble learning method which has been used for SARS-CoV-2 nucleotide sequence classification [45] and which our group and others have previously used to classify protein sequences [67], [68]; (4) LightGBM, which is a gradient-boosting method developed by Microsoft that grows trees leaf-wise, unlike XGBoost, which grows trees depth-wise, thus running much more efficiently while achieving comparable results [53], [69]; and (5) GPBoost, which trains a mixed effects model including both features, implemented using decision trees (trained using LightGBM), and random effects at the group level [50].
2.3.1. Mixed effects models
Linear mixed effects models have been used for analyzing genetic studies where there are group-level random effects, such as in longitudinal studies and other sampling studies where there may be batch effects due to different laboratory methods being used for samples taken at different locations or times [70], [71], [72], [73]. Eq. (1) shows the general matrix formulation for a mixed effects model.
| (1) |
is the row-size evaluation of function F, and . and are fixed effects and random effects predictor variable matrices respectively, i.e., the rows of are predictor variables for observations (columns of ). In this study, the random effects vector is assumed to contain grouped (i.e. clustered) random effects. In this case, the columns of will be one-hot encoded (i.e., will be an incidence matrix with 1s and 0s) with the categorical variables that define the structure of groups. Assuming that the fixed effects model is linear, then Eq. (1) may be written in terms of groups as shown in Eq. (2).
| (2) |
In Eq. (2), the linear model for is given as is the model matrix for fixed effects for observations in the th group, where there are features, is a vector of model coefficients, and is the vector of errors for the th group. is now a model matrix of random effects for the th group, where is the vector of random effect coefficients for the th group.
In this paper, groups of random effects are identified by country metadata. The means of and , an unknown vector of random errors, are 0; accordingly, we take the mean of the model response in order to evaluate its predictions. We implement mixed effects machine learning with GPBoost, which has been made publicly available at https://github.com/fabsig/GPBoost. GPBoost is a highly efficient package for fitting mixed effects models to data, as it utilizes LightGBM tree-boosting to model fixed effects [50]. Further elaboration of the mathematical foundation of mixed effects models relevant to this paper can be found in [50]. GPBoost is thus able to handle the large feature set required to include the full spike protein sequence.
2.3.2. Feature representation
The input for machine learning are features vectors of integers for each sample, and training labels set at 0 (for Mild) and 1 (for Severe). Features are obtained as follows. After the alignment procedure described above, all of the resulting sequences have 1273 characters (amino acids, deletions, or masks). The sequences are tokenized, converting each character, including the deletion symbol “-”, to a distinct nonzero integer. A position with padding mask “*” or ambiguous amino acids represented as X, B, J, or Z are considered to be missing data. Accordingly, they are a value of NAN for XGBoost, LightGBM, and GPBoost, which can then treat them as missing values; or, they are assigned a value of 0 for logistic regression and Random Forests, which cannot handle missing data. The age is represented as an integer, as describe above, and gender is treated as 0 and 1. In total, then, there are 1275 features: 1273 amino acid positions, age, and gender. As described in the paper, we also tested using the metadata for “Country” of origin of a case as a feature (increasing the number of features to 1276), or in the case of GPBoost, as a grouping of random effects in a mixed effects model. In that case, the “Country” was tokenized and represented as an integer using scikit-learn.
2.3.3. Model interpretation to obtain feature importance
The feature significance shown in the Results section for XGBoost, LightGBM, and GPBoost were obtained from SHAP (Shapley Additive eXplanations) values of terms for the test data set using the TreeExplainer method within the SHAP module (https://shap.readthedocs.io/) in Python 3.7 [54]. Among the principal reasons for selecting GPBoost to implement mixed effects machine learning was its compatibility with SHAP for interpretation [50], [74]. Feature importance can also be derived for the aforementioned ensemble decision tree methods by computing, for example, the number of times a feature is used to split trees, or the gain in score towards the objective function obtained by splitting trees based on a feature [75], [76]. However, we found no substantive differences between the features identified as significant using SHAP and those computed based on decision tree characteristics; moreover, SHAP not only estimates feature significance, but can also estimate whether a feature value tends to result in one classification or another.
2.3.4. Hyperparameter tuning and model implementation
For the results of this paper, training and testing data splits were determined by sample collection date as described in the Results section. Hyperparameter tuning was performed using a data set consisting of 60,196 samples collected between May 6, 2021 through November 2, 2021. Five-fold cross-validation was used to define training and testing splits, and the mean class prediction accuracy on the testing sets across three runs of the algorithm was computed for each hyperparameter combination. The hyperparameter combination with the highest accuracy was selected for the data presented in the Results. Other hyperparameter combinations were tested on that data and were not found to perform better than those that were used. The hyperparameters for the respective methods are as follows. Where not provided here, hyperparameters were set at their default values.
-
•
GPBoost. The number of boosters was set at 2000 (values from 500 through 3000 were tested), maximum tree depth set to 30 (values from 10 to 50, as well as unlimited, were tested), maximum number of leaves set to 20 (tested 10 to 50), and learning rate set to 0.01 (tested 0.001 to 0.1).
-
•
LightGBM. The number of boosters was set at 2000 (tested 500 to 3000), maximum tree depth set to 30 (tested 10 to 50 and unlimited), maximum number of leaves set to 20 (tested 10 to 50), and learning rate set to 0.01 (tested 0.001 to 0.1).
-
•
XGBoost. The number of estimators was set at 2000 (tested 500 to 3000), maximum tree depth set to 20 (tested 10 to 50 and unlimited), lambda regularization set at 2.0 (tested 0.0 to 3.0), gamma set to 1.0 (tested 0.0 to 2.0), and learning rate set to 0.01 (tested 0.001 to 0.1).
-
•
Elastic Net. The l1 ratio is set at 0.65 (tested 0.4 to 1.0), C is set to 0.1 (tested 0.01 to 0.8), and the maximum number of iterations was set to 1000 (tested 200 to 2000).
-
•
Random Forests. The number of estimators is set at 500 (tested 200 to 2000), the maximum depth is set at unlimited (tested 10–50 and unlimited), the minimum number of samples required in a leaf node is set at 1 (tested 1–3), and the minimum number of samples required to split an internal node is set to 2 (tested 1–5).
The results in this paper were obtained using Python 3.7.13 or 3.94, scikit-learn package version 1.0.2 [62] (for elastic net and random forests methods), and the Python implementations for xgboost version 1.6.1, gpboost version 0.7.6.2, lightgbm version 2.2.3, and shap version 0.40.0. Training and hyperparameter tuning were performed on the Drexel University Research Computing Facility’s Picotte high performance cluster using multithreaded implementations of the methods on Dell PowerEdge R640 servers with Intel® Xeon® Platinum 8268 CPUs. Model evaluation and visualization were performed in the Google Colab environment.
2.4. Resource availability
2.4.1. Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dr. Bahrad A. Sokhansanj (bahrad@molhealtheng.com).
2.4.2. Data and code availability
-
•
The datasets analyzed for this study were downloaded from GISAID EpiCoV database pursuant to the GISAID terms of use. They are availabile for download to users who register with GISAID at the website http://wwww.gisaid.org. The list of GISAID accession numbers used for this paper and data acknowledgments are available at https://epicov.org/epi3/epi_set/EPI_SET_20220606hk or https://doi.org/10.55876/gis8.220606hk.
-
•
The code used for pre-processing and analysis in this paper has been deposited to and made publicly available from the authors’ GitHub repository, https://github.com/EESI/covid_severity.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
3. Results
3.1. Descriptive analysis of GISAID patient data
To develop an understanding of the GISAID patient data set (i.e. data with metadata subject to the exclusions described in the Methods section), we analyze the trends for severity for the metadata fields available in the GISAID data set: age, gender, sample collection date, and geographic origin of the case. To quantitatively measure severity trends, the classifications for Mild disease is assigned a severity level of 0, and Severe, a severity of 1. (The classifications are derived from patient status metadata based on Supplementary Tables S1 and S2 as described in the Methods.) Thereby, the mean of the severity values can be computed, which equates to the proportion of samples which are classified as Severe cases.
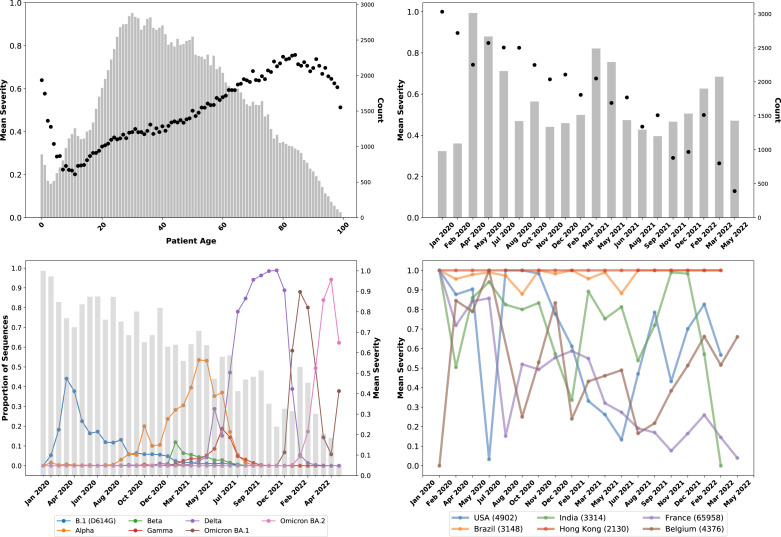
Fig. 1A shows the mean severity for each age from 0 to 100, as well as the number of samples in the GISAID patient data set for each age. In general, the fraction of severe cases increases with age, which has been a consistent feature of the pandemic [77], [78]. Trends are different at the extremes, very young and old ages. Notably, there are far fewer cases at these ages, thus small biases in sample collection can have significant effects. For example, very young patients may be likelier to be observed in a hospital setting than observed in a screening study. Male sex has also been identified as a potential risk factor for more severe outcomes, such as ICU admission and death [79], [80]. GISAID provides sex information in a “gender” metadata field. As shown in Fig. 2A, the relationship between increased age and increased proportion of severe cases are consistent throughout the pandemic.
Fig. 1.
Overview of age, sample collection date, and country metadata trends in GISAID data.(A – Upper Left) Mean case severity, where 0 is Mild and 1 is Severe, which equates to the probability of a severe case) by patient age in the GISAID database. The bars show the count of samples for each age. Increasing age trends with increasing severity, as expected, with differences at extremely low and old ages characterized by low sample counts. (B – Upper Right) Mean clinical severity (probability of severe case) by sample collection date recorded in the GISAID data. For clarity, data have been binned over time periods; the bars indicate the number of samples. Over time, the proportion of severe cases has declined, although that trend has been less consistent since Fall 2021. (C – Lower Left) Proportion of sequences in the GISAID patient data set (sequences with patient metadata) for principal variants, including B.1 (the ancestral lineage with the D614G which emerged in Northern Italy and New York in February–March 2020) and its sublineages, Alpha (B.1.1.7 and “Q” sublineages), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2 and AY sublineages), and the two major Omicron lineages, BA.1 and BA.2 (and their sublineages). The bars indicate the mean case severity for each date bin. The trends of sequential lineage waves in GISAID patient data appear to be consistent with the larger GISAID data set, i.e., showing successive Alpha, Delta, and Omicron waves. (D – Lower Right) Mean case severity of samples separated by GISAID metadata for the country where the sequence was collected for selected countries. The total number of sequences in the GISAID patient data set per country is shown within parentheses in the legend. Fluctuations in severity observed in countries appear due to systemic issues or differences in where samples are collected (e.g., in hospitals or outside settings) at different times.
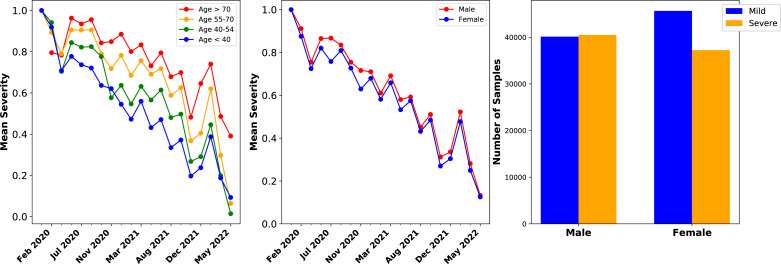
Fig. 2.
Patient age and gender metadata trends in GISAID data.(A – Left) Mean clinical severity over time for patients in different age groups, showing that the overall trends are generally consistent across age groups, with older patients having mean severity as shown in Panel A. (B – Middle) Mean clinical severity, separating male and female samples, showing consistent trends across gender with male patients generally having a somewhat higher ratio of severe cases. (C - Right) Number of mild and severe cases across all samples split by gender, showing that there are more mild cases than severe among samples from female patients.
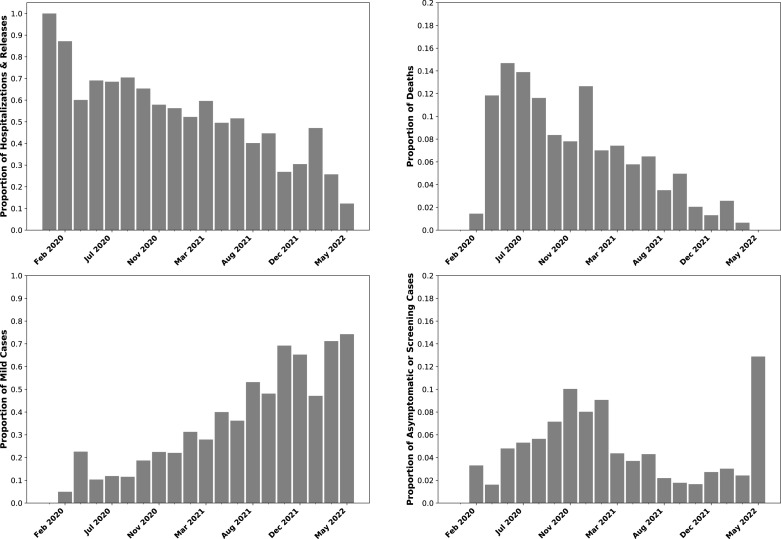
Fig. 1B shows that, in addition to known risk factors, over time the mean case severity significantly decreases. The declining severity trend through 2020 in GISAID data is consistent with a period of improved COVID-19 therapeutics. For example, a Canadian study measured a decrease in case fatality rate (CFR) between the first and second waves prior to any vaccination, even when controlling for age and increased testing [81]. Later reduction is consistent with increased levels of COVID-19 vaccination reducing severe outcomes [82], [83], [84], [85], as well as continued improvements in therapeutics such as monoclonal antibodies [86]. Notably, while the trend shows an overall decrease, which we had previously observed through October 2021, it is not monotonic, which an increase shown in late 2021 and early 2022. Moreover, the absolute level of severe cases suggests that while the trend of decreasing severity is consistent with the global trend of decreasing overall severity, the nature of reported cases also affects the trend. For example, in the initial first binned time periods, there are over 70% severe cases. As illustrated in Fig. 3, the large majority of these are hospitalizations, although approximately 10% of samples are from dead patients according to the metadata entries. Studies of the initial pandemic waves in March–April 2020 did show that a CFR that approached or exceeded 10% [87], [88], [89]. Subsequent analysis estimated that the infection fatality rate (IFR) was likely much closer to 1%–2%, with the elevated CFR being due to underreporting of cases [88], [89]. Among the passengers of the Diamond Princess cruise ship in February 2020, who were all tested, the IFR was 1% (in a population which skewed older). Accordingly, while the proportion of deaths in the GISAID patient data set did reflect community observations, at least in this timeframe, they were elevated due to so many cases being missed. However, the fraction of sequences submitted from dead patients continued to run at 4%–5% even through October 2021. We do observe that many cases have a metadata entry of “alive” or “live”, as a contrast with “dead”. But “alive” or “live” metadata cannot be identified as either Mild and Severe, and are thus excluded from the data set analyzed in this paper and shown in Fig. 1. Sequences from dead patients are thus overrepresented.
Fig. 3.
GISAID patient status metadata trends over time.(A – Upper Left) Fraction of cases categorized as Hospitalized or Released (from hospital) over time, binning dates as indicated by the bars. The definitions of hospitalizations and releases based on patient metadata are provided in 1 and 1. (B – Upper Right) Fraction of samples annotated as being from dead individuals in the GISAID patient status metadata field, binned by date as in Panel A. The cases in Panels A and B are collectively classified as Severe. C - Lower Left Fraction of cases categorized as Mild according to 1. (D – Lower Right) Fraction of cases categorized as Asymptomatic or Screening according to 1. Panels C and D are collectively classified as Mild. The subgroups of Mild and Severe classifications show similar trends, showing that the overall trends in Fig. 1 are not due to changes in how metadata are described and characterized.
Fig. 2B and C show that GISAID gender metadata similarly indicate elevated severe disease among male patients. Samples with Male gender metadata are classified as 49.8% Mild and 50.2% Severe; by comparison, samples with Female gender metadata are 55.1% Mild and 44.9% Severe. An increased proportion of severe disease for older and male patients is consistently observed in GISAID samples collected at different dates over time. Notably, the difference in severity between male and female patients (defined according to gender metadata) was much greater in samples collected up to mid-2021, and has decreased since then. It is unclear whether this reflects a broader trend or is an artifact of where GISAID samples are collected.
Moreover, the number of hospitalizations is much more elevated even than the inflated hospitalization rates observed during the period of significant underreporting in early 2020. As Fig. 3 shows, even by March 2021, over 50% of GISAID samples being collected were from individuals who were either hospitalized or released from hospital, per their patient status metadata. This makes intuitive sense, since sequence samples with clinical information may well come from clinical settings, particularly hospitals. As a result, even though there is a steady increase in cases classified as Mild (see Fig. 3), this is likely at least in part because of a change in settings where sequences with metadata are collected, with more of a mix of outpatient settings. We observed a pronounced spike in the proportion of sequences annotated as Asymptomatic or collected from population screening studies in April–May 2021, from around 3% to 12%, which is consistent with changes in sampling sources.
Fig. 1C shows how the aforementioned distortions in the data can practically impact the development of genotype-severity models. There is a lower observed mean severity during the Delta wave as compared to the timeframe of Delta’s emergence (when other lineages have a significant fraction of samples being collected) and before then, when Alpha was a plurality lineage. While this potentially could be due to continued vaccination resulting in less severe cases overall, and in turn fewer sequences from severe cases in the GISAID patient data set, Fig. 1C shows the trend of reduced severity reverses in the initial Omicron (BA.1) wave. However, vaccination and improved therapeutics, while certainly having resulted in a reduction in severe disease outcomes in general, do not explain the observed reduction of mean severity within the GISAID data set. We can show that as follows: Time-dependent changes in external conditions, such as increased vaccination rates, can be controlled for by looking only at the short timeframe where Alpha and Delta were collected in similar numbers, circa May–June 2021. As Fig. 1C shows, the reduction in severity over time at the Alpha–Delta transition point is abrupt. Significantly, during May–June 2021, Delta samples were 60.7% Mild (5199 total samples) and Alpha samples were 52.2% Mild (8658 total samples). As a result, a model based on the GISAID data set will show that Delta is milder than Alpha, contradicting the epidemiological and laboratory evidence discussed above [13], [14], [15], [16]. It is also not the case that Delta samples during the May-June 2021 timeframe were collected in countries with higher vaccination rates than Alpha samples. The main source of samples during this timeframe for both lineages was France (44% and 42% of Delta and Alpha respectively), and the second largest sources was Mexico (19% and 11%). Notably, in samples from France, Delta was 92.6% Mild and Alpha was 62.6% Mild, while in samples from Mexico, Delta was 27.1% Mild and Alpha was 38.4% Mild. Therefore, the observed decrease in severity from Alpha to Delta, due to this apparent artifact found in data from France, will confound a genotype-patient status model. Mutations associated with Delta will appear to result in reduced severity, in contradiction to epidemiological and other evidence.
Previous efforts in modeling GISAID patient data have found that including the location metadata for the country of origin of the sequence results in a more accurately predictive model [43]. As an initial matter, country and sequence will likely have some correlation, since mutations, including both the lineage an sublineage level, cluster between countries [90], [91]. However, there is likely some variance in patient outcomes between countries due to differences in the enforcement of non-pharmaceutical interventions (NPI) which may be more protective of vulnerable populations, differences in circulating virus and thus hospital burdens, and differences in hospital capacity and standards of care [92], [93], [94]. Fig. 1D shows, however, that inter-country variation is more complex and does not appear to be directly related to the aforementioned factors. Some of the data show consistent trends or levels across all time points. For example, essential all records submitted from Hong Kong, and the overwhelming majority of records from Brazil, are classified as Severe (hospitalizations or deaths). Cases from France, which as discussed above is the largest source of sequences, show a decline over time, although with a lot of fluctuation. Cases from neighboring Belgium, by contrast, have been consistently increasing in severity since Summer 2021. Samples from the United States have increased and decreased in severity with no discernible pattern. In sum, samples originating from different countries follow distinct patterns, which means that including country as a feature will likely improve classification. But the country feature will reflect sampling differences over time between countries—whether the sequences are being submitted mostly from hospital settings, or if that is the case at different points in time. While there is variance between samples from a country that is independent from that of other countries, it is at least in large part due to factors that are not relevant to disease outcome.
Accordingly, we hypothesize that sampling variations can be modeled as a random effect in a linear mixed effects model [72], [73], [95], where country of origin is a random effect group rather than a feature. To test this hypothesis, in the following section we compare mixed effect machine learning to other classification methods for predicting disease severity. The comparison is based on GISAID data from a timeframe when the overall decrease in severity will not confound a model. Otherwise, as discussed above, any model will inevitably make predictions that are not clinically relevant going forward. For example, mutations that emerged in Delta will be found as leading to less severe disease, and thus when they are found in Omicron sublineages, they will be predicted as reducing severity—due to the potential artifact in samples collected during the period when Delta and Alpha coincided discussed above. We therefore limit our analysis of modeling methods to samples collected beginning in July 2021, when the declining trend in cases has stabilized (see Fig. 1B and C). The result is a training data set made up of mostly Delta subvariants, along with a substantial number of Omicron sequences, and smaller numbers of other lineages such as the Gamma (P.1) variant of concern, which has shown some ability to evade neutralizing antibodies [96], [97] and may result in increased disease severity [98]. Doing so helps avoid confounders in analyzing the impact of different mutations and combinations of mutations in Delta and Omicron subvariants, allowing for predictions about whether mutations observed in Delta will result in more severe disease if they occur in future Omicron variants.
3.2. Comparison of machine learning methods
To evaluate our hypothesis that a mixed effect modeling approach can be useful for GISAID patient data, where country should be treated as a random effect group, we evaluate the GPBoost mixed effect machine learning method [50] alongside two popular highly efficient ensemble decision tree methods which employ gradient-boosting, XGBoost and LightGBM [53], random forests [51], a well-established ensemble decision tree method, and conventional logistic regression with elastic net regularization [65]. To compare with GPBoost, two feature sets of models are evaluated: (i) using sequence, age, and gender as features, and (ii) using country metadata as an additional feature.
As discussed above, the following analysis focuses on GISAID data starting from July 2021. Models are trained on the samples collected between July 17, 2021 through December 25, 2021 (68,815 in total). Testing is performed on samples collected entirely after the training period, from December 26, 2021 through the latest-collected sequences from April 10, 2022 (42,420 samples). Although cross-validation is a typical way of evaluating classifiers to avoid overfitting [99], [100], in this data set, as shown in Fig. 1 and discussed above, there will be potential clusters of confounding variables at different times. For example, a narrow range of sequence collection dates may correspond to a study of patients with common characteristics, e.g., patients who are all hospitalized, or mildly symptomatic patients from a screening study. Cross-validation will sample time points evenly, and, as such, a classifier may overfit to patterns within the confounders and then appear to perform better than it otherwise would be on a realistic prediction task. As an alternative, we evaluate the classifiers on temporally split data: i.e., we seek to predict disease severity for sequences from a model trained on samples collected earlier. Previous work has confirmed that temporally split test and training sets provide a more realistic evaluation of classification methods, in that methods perform less well when evaluated on a temporally split validation data set than they do using cross-validation [45]. Notably, there is some class imbalance, which is similar between the training and test data sets: 39.2% of the test samples were severe and 37.4% of training were Severe. Class or sample balancing did not substantially affect the results for the methods which allow it, i.e., not GPBoost (data not shown).
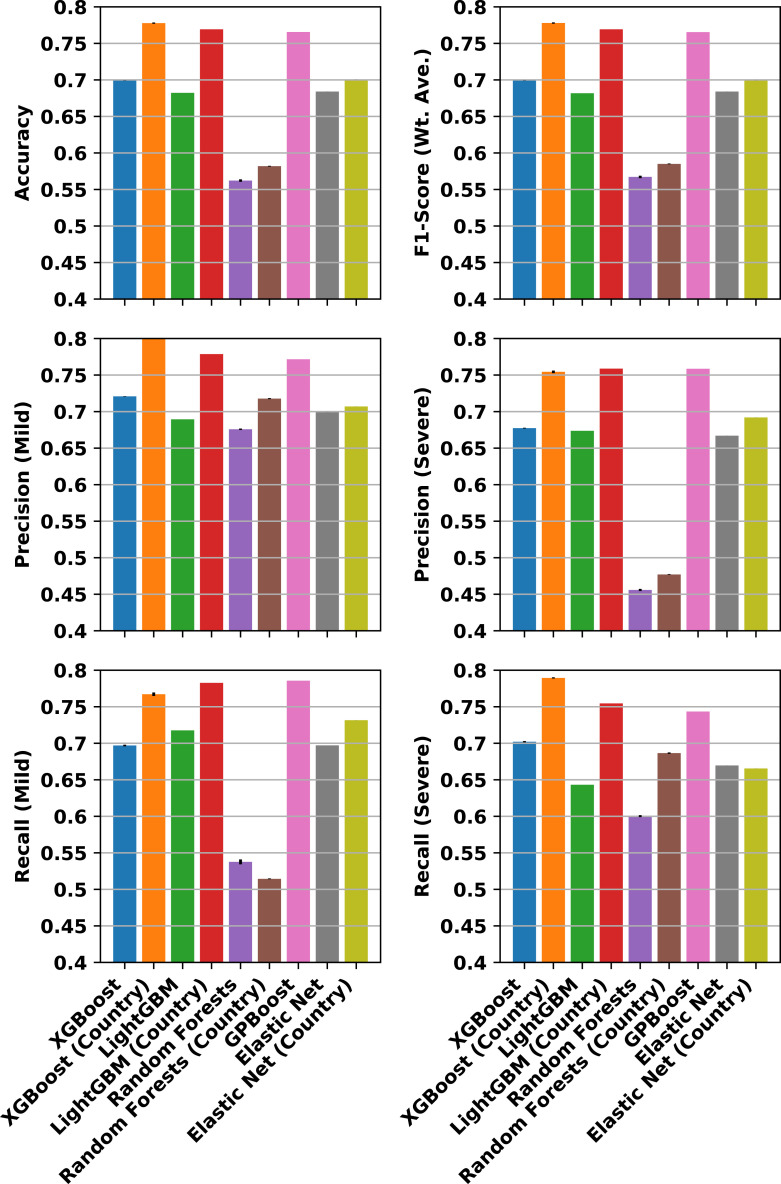
Fig. 4 compares machine learning methods by showing aggregate and class-specific test data classification metrics for models trained using the different methods under evaluation. The aggregate metrics shown in Fig. 4 are the accuracy, which is measured as the number of correct class predictions divided by the number of total predictions, and the balanced (weighted average) F1-score, which reflects the sensitivity and specificity of the predictions, accounting for the aforementioned class imbalance. The balanced F1-score is the harmonic mean of precision (true positives divided by all positive predictions) and recall (true positive rate, i.e., sensitivity). Fig. 4 also shows the class-specific precision and recall, which is a useful measure as to whether methods might underperform on predicting a particular class, such as the minority (Severe) or majority (Mild) class.
Fig. 4.
Comparison of classification metrics for different machine learning methods. Metrics are computed on test samples collected from December 26, 2021 through April 10, 2022, for models trained on samples from July 17 through December 25, 2021. The top row shows, at left, the accuracy of the Mild/Severe classification and, at right, the balanced F1-score, which is the harmonic mean of precision (true positives divided by all positive predictions) and recall (true positive rate, i.e., sensitivity). The middle row shows the precision for the Mild and Severe class predictions separately, and the bottom row shows the recall. Metrics are shown for models trained with country metadata used as a feature and without, as indicated in the labeled axes below, except for GPBoost, which takes into account the country metadata by using it as the groups of random effects. All models otherwise use age, gender, and each sequence position as a feature. Error bars show the standard deviations across three runs with different random number seeds, and in some cases are not visible. Statistics for GPBoost are computed based on the mean of the response. GPBoost and LightGBM/XGBoost including country as a feature consistently outperform other methods.
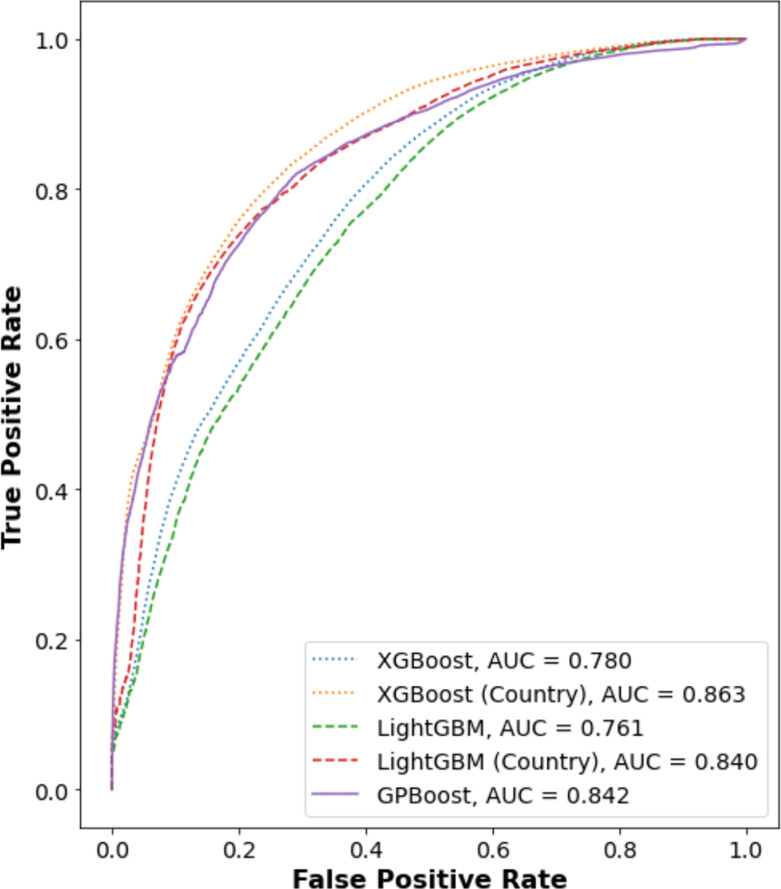
Although the performance of the methods varies depending on metric, two trends are clear. First, the best-performing methods are consistently (1) the high-performance gradient boosting decision tree methods, XGBoost, LightGBM, and GPBoost, with class prediction accuracies above 75%. Second, the best-performing methods account for country—either as an independent feature, in the case of XGBoost and LightGBM, or as group level random effects for the mixed effects model trained by GPBoost. Notably, these three methods, unlike classical regression methods, can handle missing information for sequence positions, which are allowed to increase the number of data for training. Fig. 5 further shows the receiver operator characteristic (ROC) curves for the best-performing methods, and reports the area under the curve (AUC), which provides a metric for comparing model performance. Using the AUC metric, XGBoost with country as an independent feature has the highest AUC. It is important to keep in mind, however, that as shown in Fig. 1D, a model that includes country as a feature may be overfitting to consistent sample collection biases. Notably, GPBoost outperforms LightGBM and XGBoost when country is not an independent feature of the latter two models. GPBoost also has a higher AUC than LightGBM and only marginally lower than that of XGBoost.
Fig. 5.
Receiver operator characteristic curves for best-performing modeling methods. ROC curves were obtained using the scikit-learn package version 1.0.2 [62] for test samples and trained models as described for Fig. 4 for XGBoost and LightGBM (with and without country metadata) and GPBoost (using country as a random effects group). The data are shown for one run; run-to-run variation was found to be insignificant. GPBoost performs better than either LightGBM or XGBoost, unless country metadata are used for the latter methods.
To further compare the performance of the best-performing models, they can be tested on their ability to predict whether specific sequence mutations affect the relative risk of severe disease. This analysis focuses on two specific spike protein site mutations for which there is substantial evidence from both epidemiological and laboratory studies for increased disease severity: a leucine-to-arginine mutation at position 452 (L452R) and a proline to arginine mutation at position 681 (P681R). These mutations are characteristic of the Delta variant [101]. SARS-CoV-2 with P681R has been found to have higher spike protein cleavage and viral fusogenicity in vitro, and result in higher pathogenicity in a Syrian hamster animal model [102]. Another study introducing P681R on an Omicron background showed an increase in fusogenicity and synctitia formation, which have been correlated to pathogenicity [103]. The L452R mutation has been found to also increase viral fusogenicity in vitro, and to result in increased infectivity in a mouse lung cell model [104]. Another in vitro study has also shown that L452R resulted in increased spike protein stability, viral fusogenicity and infectivity, and, in turn, increased viral replication [105]. And, the Delta variant, which is characterized by the P681R and L452R mutations, was to result in an increased risk of hospitalizations in epidemiological studies in Denmark [13], England [16], and Canada [14]. Accordingly, a model would be expected to show that a L452R or P681R mutation will result in greater severity.
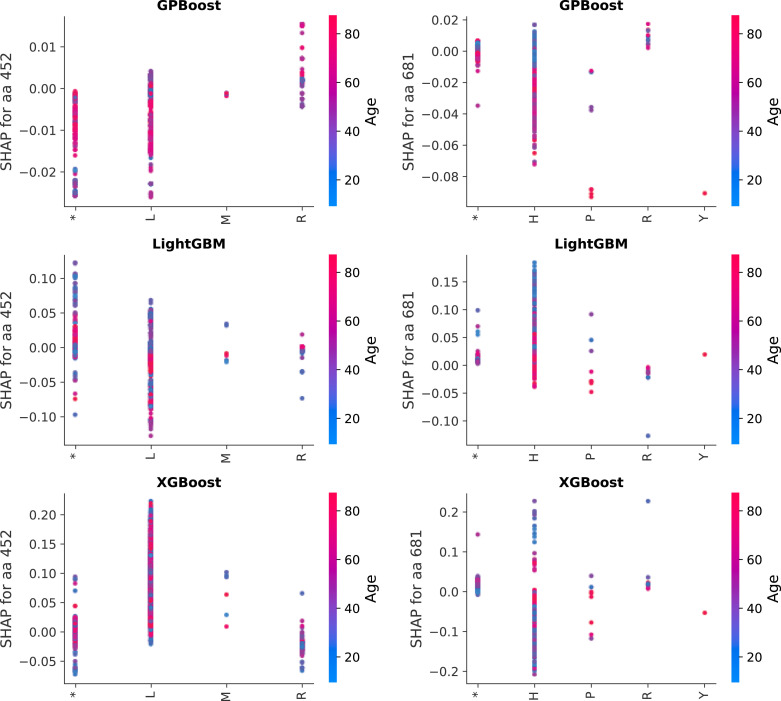
As an additional validation study, therefore, machine learning models are evaluated on whether they are more likely to a predict a Severe classification in the presence of L452R or P681R sequence changes. However, the methods being compared here are decision tree-based methods, which unlike classical logistic regression do not generate coefficients that can be used to analyze individual features. The impact of specific feature changes may be estimated instead. In particular, SHAP values can be utilized in conjunction to provide an estimate of the log-odds for a Severe case given a particular feature value [54]. SHAP values are typically generated for a subset of samples, as it is a computationally intensive process. Fig. 6 shows SHAP dependency plots for samples collected from March 8 through April 10, 2022 (5918 samples). The points in the plots represents the estimated SHAP value (log-odds for a Severe case) for each sample; the color indicates the age of the patient for the sample. This means that SHAP dependency plots show how a specific feature interacts with another feature: the age of the patient in Fig. 6. (As indicated in Fig. 1, age has a significant correlation with disease severity in GISAID patient data, as well as in real-world epidemiological studies.) The SHAP dependency plots represent the potential sequence features at spike protein positions 452 and 681: L (ancestral), leucine, M, methionine, and R, arginine for position 452, and P (ancestral), proline, H, histidine, R, and Y, tyrosine for position 681. (P681H is a common mutation founds in Omicron sequences [106]). The ‘*’ character indicates that there was a missing amino acid at that position in the sampled sequence, likely due to sequencer error, which is treated as missing data by the respective methods. As Fig. 6 shows, GPBoost is the only method which shows an increased SHAP value, or estimated log-odds of a Severe outcome, for the L452R and P681R mutations.
Fig. 6.
Comparison of SHAP dependence plots to severity for sequence positions 452 and 681 for the best-performing models. LightGBM and XGBoost with country as a feature are compared to the GPBoost mixed effect model, trained aon data as described in 4. The predicted SHAP values for each of the samples used to generate the SHAP estimate (sequences collected from March 8 through April 10, 2022) are plotted for the 452 and 281 sequence positions in the left and right columns respectively, showing the SHAP values for predictions with sequences of the indicated amino acid at that position, i.e. L (ancestral), leucine, M, methionine, and R, arginine for residue 452; P (ancestral), H, R, and Y for residue 681; and ‘*’ for missing amino acid in the sample. A positive SHAP value indicates that an amino acid change is positively related to increased severity. The interaction of the patient age feature is shown by the coloring of the points, where more red points are from older patients and blue points from younger. GPBoost indicates increased severity as expected from validated experiments of L R for this time period.
3.3. Predicting the potential severity of emerging omicron variants
A key objective for training a sequence-phenotype model is to be able to predict how novel combinations of mutations – such as the reemergence of a mutation found in a separate lineage – could affect pathogenicity and clinical outcomes. Here, the potential utility of a spike protein sequence-clinical severity prediction model trained on GISAID data is demonstrated for Omicron lineages emerging as significant threats as of May 2022: BA.4 and BA.5, which had become the predominant variants in South Africa and found to be rapidly growing in Portugal [107], and BA.2.12.1, which had accounted for substantial case growth in the United States [108].
The predicted relative severity resulting from different spike sequences may be compared by looking at the relative raw (unrounded) prediction of the model. In the context of this paper, the probability on the logistic curve fit by the model that the binary classification will be 0 or 1. In practice, the class prediction is provided by rounding the model output to 0 (Mild) or 1 (Severe), i.e., to generate the classification metrics shown in Fig. 4. However, as explained above, GISAID data do not provide a realistic measurement of the actual observed probability of severe outcomes, as there are far more hospitalized and deceased patients than real-world hospitalization and CFR data indicate. The quantitative model predictions should be interpreted, therefore, in a relative manner. Accordingly, the raw model output can help in providing relative predictions, but should not be interpreted as an absolute probability of severe disease. In sum, predictions for the aforementioned emerging sublineages may be compared against the predictions for the original Omicron sublineages, BA.1 and BA.2.
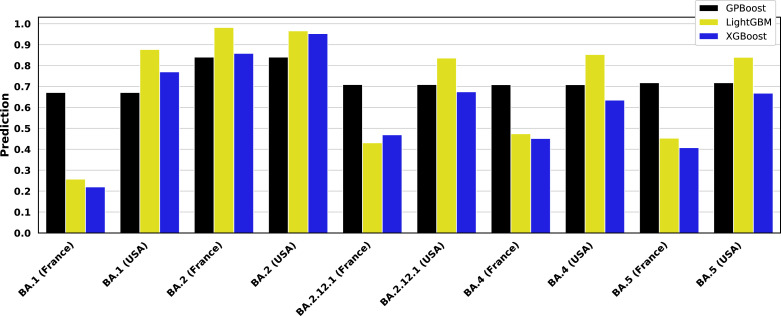
Fig. 7 shows the output of the trained GPBoost, LightGBM, and XGBoost models, where the latter two include country as a feature, as shown above in Fig. 6. The sequences used to generate the predictions in Fig. 7 are the most common of those variants found in the GISAID patient data set used in this paper (collected before April 15, 2022), with GISAID accession numbers as provided in the figure caption. An additional BA.2.12.1 sequence collected after the data set used in this paper was separately retrieved from GISAID (accession number EPI_ISL_12048110). As Fig. 7 illustrates, Country has a substantial impact on the predictions made using LightGBM and XGBoost. This sharply limits the utility of LightGBM and XGBoost models as predictive tools.
Fig. 7.
Predictions of Omicron subvariant severity. Trained GPBoost, LightGBM, and XGBoost models are simulated for representative BA.1, BA.2, BA.2.12.1, BA.4, and BA.5 sequences from a 60 year-old male patient obtained in the United States, France, and Mexico. The GISAID accession numbers of the sequences for the sequences are: EPI_ISL_6590782 (BA.1), EPI_ISL_7852877 (BA.2), EPI_ISL_12048110 (BA.2.12.1), EPI_ISL_11674447 (BA.4), and EPI_ISL_12029894 (BA.5). The predictions shown here are for models trained on training data as shown in Fig. 4, Fig. 6 where country is a feature for LightGBM and XGBoost. The GPBoost predictions shown here are for the mean of the model response, and it does not vary by country, since country is not a fixed effect in the mixed effects model trained using GPBoost. By contrast, LightGBM and XGBoost predictions fluctuate significantly by simulated country. Emerging Omicron subvariants are uniformly predicted to be more severe than BA.1.
While it is possible to standardize the country and view the prediction relatively, as shown in 7, the relative difference between variants differs greatly between countries. For a simulated patient sample from the United States, the variants have nearly identical (and very high) predictions, while the predictions for simulated samples from France vary differently, with much more dynamic range. Samples from Mexico are in between. GPBoost models, by contrast, do not vary between countries. The mixed effects model trained by GPBoost does not account for country in grouping only random effects. By considering only the mean model response, random effects cancel each other out, and there is only one prediction for any country. Given that, as shown in Fig. 1D, the differences between countries are apparently unrelated to actual local conditions, such as access to treatment, a country-neutral prediction provides a more realistic, and likely more relevant, of the relative increase in severe disease risk associated with a new SARS-CoV-2 variant.
Accounting for the variation between countries, Fig. 7 generally shows that BA.2, BA.2.12.1, BA.4, and BA.5 all have higher predicted severity than BA.1. Notably, a study of infectivity in mouse and hamster models suggested that there is no difference in infectivity, replication, and pathogenicity between BA.1 and BA.2 virus [109]. Another study, however, found greater fusogenicity and replication in nasal epithelial cells studied in vitro, as well as more pathogenicity in a hamster model for BA.2 as compared to BA.1 [110]. Moreover, a recently published population study in England reports that individuals infected with BA.2 reported more symptoms than those with BA.1 [111]. Another study of patients in Italy also reported more symptomatic disease when infected with BA.2 rather than BA.1 [112].
The SHAP method used for feature analysis above can be used to examine in detail how specific features influence the prediction for emerging variants as well [54]. Fig. 8 shows exemplary SHAP visualization for the GPBoost prediction of the representative BA.2.12.1 sequences shown in Fig. 7 (GISAID accession EPI_ISL_12048110), simulated for a 30 year old male patient. The plot shows how key features tend to make a prediction of greater severity (indicated by an increasing value) or lower severity (decreasing value). In the case of this younger patient, for example, the Age feature tends to reduce the predicted severity. Notably, Fig. 8 suggests that three mutations characteristic of BA.2 influence an increase in predicted severity for BA.2.12.1: a deletion at positions 24 through 26, S371F (serine to phenylalanine), and R408S (arginine to serine) [113]. S371F is a mutation in the receptor binding domain (RBD) of the spike protein which has been shown to be evasive to antibodies [114], [115]. While an antibody evasive mutant might not necessarily confer greater severity on an immunonaive patient, given the high rates of vaccination and/or prior infection now, a model based on contemporary GISAID data can be expected to shown greater severity for immune escape variants. While the impact is smaller than for those features shown in Fig. 8, analysis of SHAP values shows that another immune escape change found in BA2.12.1, E484 A, also tends to elevate the severity prediction [116]. Similarly, BA.4 and BA.5 have been found to be more immunoevasive in BA.1, which may also result in increased severe disease among populations with acquired immunity [117], [118].
Fig. 8.
SHAP force plot showing impact of features on BA.2.12.1 severity prediction by GPBoost. The “force plot” is a visualization which shows, based on SHAP values estimating the log-odds contribution of features to the model prediction, how much a specific feature tends to weigh the decision between binary classes. This plot is based on a simulated 30 year old male patient, and thus the Age feature tends to weigh the model towards a Mild prediction for this sample. Other features tend to weigh towards a more Severe prediction, such as mutations at sites characteristic of BA.2, including positions 371 and 408.
4. Discussion
Global genome repositories like GISAID have the potential to be an unparalleled resource for understanding and quantitatively modeling genotype-phenotype relationships. As the foremost repository for SARS-CoV-2 genome sequences, GISAID offers the largest possible potential data set with the greatest global reach. As a result, GISAID can solve one of the key challenges with biomedical modeling problems: small data set sizes which make them particularly vulnerable to overfitting, because it is often difficult and costly to obtain experimental data [119], [120], [121]. The best (and perhaps only real) solution to overfitting is to have more data to develop models. Conventional meta-analyses require searching for relevant studies and parsing through papers with often inconsistent formats and data reporting methods, and they are also limited to published or otherwise documented studies. However, because repositories are generally incorporating multiple studies collected from different sites and under different conditions, heterogeneity is still the key challenge [122]. As Fig. 1 and the accompanying text explain, heterogeneity is a critical problem with GISAID data. The challenges of GISAID source data heterogeneity are particularly exacerbated by the very limited metadata associated with patient samples, even for the small subset for which patient status metadata are available at all. Sequence repository data will be more useful as efforts continue to grow to collect and curate important information about the sample and establish minimum information standards [123]. The results in this paper demonstrate that, accounting for the aforementioned caveats, useful information can be obtained by analyzing the GISAID patient data set. There are three key problems with the data set that analytical and modeling methods need to address.
First, it is hard to robustly define mild and severe cases based on patient status metadata. As an initial matter, metadata entries are often inconsistent between different entries or noisy and hard to interpret reliable (see, e.g., Supplementary Table S1). This paper takes a hierarchical approach to defining mild and severe cases, based on established clinical definitions [63], as described in Supplementary Table S2. However, because of confounding variables like vaccination, therapeutic availability, and prior infection, it has become difficult to estimate the “intrinsic” severity of variants [124]. A particular challenging issue concerns whether hospitalizations should be considered as mild or severe cases, especially given how prevalent they are in the GISAID data set (see, e.g., Fig. 3 and accompanying text). While this paper treats them as severe cases, that definition has become increasingly unreliable as the vaccination has become more prevalent. Studies from multiple sites suggests that as vaccination has increased, more hospitalization patients classified have only tested positive on admission but have mild or no symptoms [125], [126], [127], [128]. Moreover, the kinds of sequence variation that lead to more severe clinical outcomes may change due to vaccination. As suggested in Fig. 8 and accompanying text, as the overwhelming majority of individuals in many regions have at least some immunity due to vaccination and/or prior infection, immune escape variants may result inn severe disease because they can evade immune responses that would otherwise rapidly clear the virus and prevent infection. However, such variants may not result in more pathogenicity in immunonaive hosts, and thus would not show more severe outcomes earlier in the pandemic.
Second, conditions have changed over time, as shown in the reduction of case severity over time shown in Fig. 1, which consists of reductions in the proportions of both hospitalizations and deaths (see Fig. 3). While trends of decreasing severity are consistent with improved patient outcomes due to vaccination and improved therapeutics [81], [82], [83], [84], [85], [86], they may also reflect changes in sequence collection practices, such as obtaining more sequences and performing more studies based on screening the general public outside of hospital settings. These artifacts can have significant impacts on modeling studies. For example, the models derived in this study similarly indicate that the E1258D spike protein mutation has a significant impact on increasing severity. In addition to our group’s previous work, another independent investigation of GISAID terminating in Fall 2021 showed E1258D as the strongest sequence feature in determining the severity prediction [44], [47]. While E1258D was observed in one publication as an observed result of a missense mutation, that study did not show any effects for that mutation on increased pathogenicity [129]. In fact, E1258D is only found in 1898 of the over 160,000 samples analyzed in this paper. Of those samples, 1849, or 97.4% originated from Mexico, of which 1772 were hospitalized or released from hospital (i.e. considered severe in most studies), and 76 were deceased. Significantly, the metadata were all consistent, including at the level of capitalization, whereas metadata entries generally showed a high degree of heterogeneity. (Supplementary Table S1 shows all unique entries.) Therefore, it is highly likely that E1258D is either sequence artifact, particularly as it is in the cytoplasmic tail of the spike protein and the result of a missense mutation, and thus potentially an unreliable site for interpreting short-read next generation sequencing technologies [130], [131]. In sum, caution must be employed in interpreting any features identified as important.
Third, the region from which sequences are collected can have a significant impact on data due to systematic bias. As the E1258D feature demonstrates, large-scale studies in particular regions may interpret sequence data in such a way that can identify a spurious variant if it is inconsistent with other studies. As Fig. 1D shows, even though it seems logical to ascribe regional differences in clinical outcomes to factors like vaccination, fluctuations at the country level are either virtually constant or otherwise have no consistent pattern. As such, country-level seem more reflective of how samples are collected and metadata are annotated within countries, which motivates the use of mixed effect models as has been previously used for genotype-phenotype modeling where sample batches affect the data [70], [71], [72], [73]. The results in this paper demonstrate that a mixed effect machine learning approach in which countries are groups for random effects can be successful in developing a predictive model. The GPBoost method [50] proves to be fast, effective, and robust to missing data, which suggests that it should be more widely utilized in modeling genetic variation. Notably, as Fig. 4 illustrates, using country as a feature does result in much more accurate models. However, these models are overfitting to country-level trends, as evinced by the predictions graphed in Fig. 7, which show dramatic differences in predictions between different countries. As such, while previous studies of GISAID data have shown that including country metadata as a feature in models provides greater explanatory power [42], [43], [46], any resulting models are likely overfitting as they are here and will have difficulty being generalized to real-world predictions. Accordingly, while region-level features may appear to result in superior models, they risk creating artifacts. For example, as shown in this paper, including country as a feature results in predictions for the impact of L452R and P681R mutations at odds with epidemiological and in vitro evidence (see Fig. 6). The challenges of country-level variation are heightened by substantial regional imbalances in the GISAID patient data set. The entire GISAID database is fundamentally biased towards Europe, North America, and select countries in Asia and elsewhere, with over half the sample originating in either the United Kingdom or United States as of January 2022 [49]. Within the subset of data with patient status metadata, the biases are similarly idiosyncratic; for example, over 40% of the training and test samples shown in this paper were obtained from France.
In addition to the foregoing issues, the work in this paper has further limitations in scope. GISAID patient metadata omit information about comorbidities known to increase the risk of severe clinical outcomes and mortality, such as chronic disease and obesity [132], [133]. Studies have shown that host (patient) genetics may also be significant determinants of infection outcomes. a [134], [135], [136] Indeed, a recent study showed many genetic correlates of severe COVID-19 that were also correlates for other chronic conditions associated with heightened severity, with a particular focus on immune-mediated conditions [137]. Epigenetic factors may also be significant [138], as well as the host transcriptome [139]. In addition, to make the work shown here more tractable, we focus on the spike protein. However, there is some evidence that a mutation in the nucleocapsid gene may account for some of Delta’s increased severity [140]. Finally, the methods described herein rely on training or fitting to existing databases. Entirely novel mutations will not be accounted for and may result in unpredictable outcomes. However, it may be possible to train models on the predicted or in vitro studies of novel mutations that could emerge in the future, such as those identified in deep mutational scanning and other exploration of the mutational landscape [141], [142], [143], [144]. In sum, while there are important caveats to utilizing the GISAID data set as a resource for modeling clinical outcomes based on viral genotypes, it provides the most diverse and largest data set possibility. Any other meta-analyses will inherently suffer from the same kinds of data heterogeneity, and will necessarily be more limited as there is data in GISAID beyond that contained in published reports. The relative success of a mixed effect modeling approach suggests that refining the modeling of group level random effects or otherwise incorporate hidden variables are necessary to account for structural issues in the data. Moreover, having established a proof of concept in this study using logistic regression and boosted decision trees, future work can explore the potential application of deep learning methods, which have proven to be highly useful to genetic sequence to function modeling in other contexts [49], [145], [146], [147].
5. Conclusion
Despite increasingly widespread vaccination and development of new antiviral therapies, COVID-19 continues to represent a significant threat to human health. The virus also continues to be highly unpredictable. Significant genetic variants of SARS-CoV-2 continue to proliferate, and the risk of severe disease in an emerging variant is a particular concern. A critical tool in staying ahead of the virus can be a predictive model for the risks of severe disease based on viral genotype. Potentially predictive genotype-disease severity models depend on a substantial amount of patient data, which exceeds the capability of conventional epidemiological studies and meta-analyses. Patient data within GISAID, the primary global SARS-CoV-2 sequence repository, therefore, represents a key resource for building predictive models. Unfortunately, GISAID patient metadata are limited, both in number and quality; for example, there is no data on comorbidities or vaccination status. Despite these caveats, it has been previously shown that GISAID patient metadata can be used to develop predictive models. However, until this paper, there has not been a rigorous analysis of potential confounders within the data which may prevent such models from being clinically useful.
As shown in this paper, there are temporal trends in sample collection biases which must be accounted for in model training. Moreover, there are significant differences in sample collection biases between countries. Models are more predictive if they take country-of-origin of sequences into account, but such models are likely overfitting to artifacts in how samples are collected in different countries. This study demonstrates that a superior approach to accounting for variation between the country-of-origin of viral sequence and patient data is to employ mixed effects modeling, where country is treated as a random effect group. Mixed effects modeling can be efficiently implemented for the large number of sequence features analyzed in this paper by using the recently developed GPBoost package, which uses gradient boosted decision trees for fixed effects with performance comparable to XGBoost and conventional LightGBM. This study also presents a novel way to validate genotype-disease severity models for COVID-19: interpreting models to determine whether they are able to show that they can predict the effect of known mutations which affect disease severity. This kind of validation further reinforces the potential superiority of mixed effects methods over conventional logistic regression and boosted decision tree methods. Finally, trained GPBoost genotype-severity models are shown to be able to predict severity of emerging SARS-CoV-2 Omicron variants. For example, the GPBoost model presented in this paper predicts that BA.2 and subsequent Omicron variants may pose a greater risk of severe disease than Omicron BA.1, in line with preliminary epidemiological evidence.
CRediT authorship contribution statement
Bahrad A. Sokhansanj: Conceptualization of this study, Data curation, Methodology, Data analysis, Software, Writing – original draft. Gail L. Rosen: Conceptualization of this study, Data analysis, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge the following Authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based. A table of acknowledgments is located at https://epicov.org/epi3/epi_set/EPI_SET_20220606hk (DOI link: https://doi.org/10.55876/gis8.220606hk). GLR received U.S. National Science Foundation (NSF) grants #1919691, #1936791, and #2107108. The funders had no role in study design, deciding to publish, collecting or analyzing data, or preparing the manuscript. Work reported here was run on hardware supported by Drexel’s University Research Computing Facility.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105969.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Supplementary Table S1 showing a mapping scheme used to encode raw patient status metadata.
Supplementary Table S2 showing metadata entries mapped to an encoded patient status and resulting disease severity.
Supplementary Table S3 showing lineage counts for samples collected from July 17 through December 25, 2021.
References
- 1.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W., Maurer-Stroh S., GISAID Core Curation Team GISAID’s role in pandemic response. China CDC Wkly. 2021;3(49):1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., Yeats C., du Plessis L., Maloney D., Medd N., Attwood S.W., Aanensen D.M., Holmes E.C., Pybus O.G., Rambaut A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parums D.V. Editorial: revised world health organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med. Sci. Monit. : Int. Med. J. Exp. Clin. Res. 2021;27:e933622–1–e933622–2. doi: 10.12659/MSM.933622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Rocklöv J. The reproductive number of the delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021;28(7):taab124. doi: 10.1093/jtm/taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Liu J., Johnson B.A., Xia H., Ku Z., Schindewolf C., Widen S.G., An Z., Weaver S.C., Menachery V.D., Xie X., Shi P.-Y. 2021. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., Pandey R., Brown J., Zhou J., Goonawardane N., Mishra S., Whittaker C., Mellan T., Marwal R., Datta M., Sengupta S., Ponnusamy K., Radhakrishnan V.S., Abdullahi A., Charles O., Chattopadhyay P., Devi P., Caputo D., Peacock T., Wattal C., Goel N., Satwik A., Vaishya R., Agarwal M., Mavousian A., Lee J.H., Bassi J., Silacci-Fegni C., Saliba C., Pinto D., Irie T., Yoshida I., Hamilton W.L., Sato K., Bhatt S., Flaxman S., James L.C., Corti D., Piccoli L., Barclay W.S., Rakshit P., Agrawal A., Gupta R.K. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ (Clin. Res. Ed.) 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J., CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538):eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., Bergstrom M., Spyer M.J., Miralhes P.B., Grant P., Kirk S., Valerio C., Mangera Z., Prabhahar T., Moreno-Cuesta J., Arulkumaran N., Singer M., Shin G.Y., Sanchez E., Paraskevopoulou S.M., Pillay D., McKendry R.A., Mirfenderesky M., Houlihan C.F., Nastouli E. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021;21(9):1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giles B., Meredith P., Robson S., Smith G., Chauhan A., PACIFIC-19 and COG-UK research groups The SARS-CoV-2 B.1.1.7 variant and increased clinical severity-the jury is out. Lancet Infect. Dis. 2021;21(9):1213–1214. doi: 10.1016/S1473-3099(21)00356-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bager P., Wohlfahrt J., Rasmussen M., Albertsen M., Krause T.G. Hospitalisation associated with SARS-CoV-2 delta variant in Denmark. Lancet Infect. Dis. 2021;21(10):1351. doi: 10.1016/S1473-3099(21)00580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisman D.N., Tuite A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: A retrospective cohort study in Ontario, Canada. CMAJ. 2021;193(42):E1619–E1625. doi: 10.1503/cmaj.211248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paredes M.I., Lunn S.M., Famulare M., Frisbie L.A., Painter I., Burstein R., Roychoudhury P., Xie H., Mohamed Bakhash S.A., Perez R., Lukes M., Ellis S., Sathees S., Mathias P.C., Greninger A., Starita L.M., Frazar C.D., Ryke E., Zhong W., Gamboa L., Threlkeld M., Lee J., Nickerson D.A., Bates D.L., Hartman M.E., Haugen E., Nguyen T.N., Richards J.D., Rodriguez J.L., Stamatoyannopoulos J.A., Thorland E., Melly G., Dykema P.E., MacKellar D.C., Gray H.K., Singh A., Peterson J.M., Russell D., Torres L.M., Lindquist S., Bedford T., Allen K.J., Oltean H.N. 2021. Associations between SARS-CoV-2 variants and risk of COVID-19 hospitalization among confirmed cases in Washington State: A retrospective cohort study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., Seaman S.R., Harris R.J., Hope R., Lopez-Bernal J., Gallagher E., Charlett A., Angelis D.D., Presanis A.M., Dabrera G., Koshy C., Ash A., Wise E., Moore N., Mori M., Cortes N., Lynch J., Kidd S., Fairley D., Curran T., McKenna J., Adams H., Fraser C., Golubchik T., Bonsall D., Hassan-Ibrahim M., Malone C., Cogger B., Wantoch M., Reynolds N., Warne B., Maksimovic J., Spellman K., McCluggage K., John M., Beer R., Afifi S., Morgan S., Marchbank A., Price A., Kitchen C., Gulliver H., Merrick I., Southgate J., Guest M., Munn R., Workman T., Connor T., Fuller W., Bresner C., Snell L., Patel A., Charalampous T., Nebbia G., Batra R., Edgeworth J., Robson S., Beckett A., Aanensen D., Underwood A., Yeats C., Abudahab K., Taylor B., Menegazzo M., Clark G., Smith W., Khakh M., Fleming V., Lister M., Howson-Wells H., Berry L., Boswell T., Joseph A., Willingham I., Jones C., Holmes C., Bird P., Helmer T., Fallon K., Tang J., Raviprakash V., Campbell S., Sheriff N., Blakey V., Williams L.-A., Loose M., Holmes N., Moore C., Carlile M., Wright V., Sang F., Debebe J., Coll F., Signell A., Betancor G., Wilson H., Eldirdiri S., Kenyon A., Davis T., Pybus O., du Plessis L., Zarebski A., Raghwani J., Kraemer M., Francois S., Attwood S., Vasylyeva T., Zamudio M.E., Gutierrez B., Torok M.E., Hamilton W., Goodfellow I., Hall G., Jahun A., Chaudhry Y., Hosmillo M., Pinckert M., Georgana I., Moses S., Lowe H., Bedford L., Moore J., Stonehouse S., Fisher C., Awan A., BoYes J., Breuer J., Harris K., Brown J., Shah D., Atkinson L., Lee J., Storey N., Flaviani F., Alcolea-Medina A., Williams R., Vernet G., Chapman M., Levett L., Heaney J., Chatterton W., Pusok M., Xu-McCrae L., Smith D., Bashton M., Young G., Holmes A., Randell P., Cox A., Madona P., Bolt F., Price J., Mookerjee S., Ragonnet-Cronin M., Nascimento F.F., Jorgensen D., Siveroni I., Johnson R., Boyd O., Geidelberg L., Volz E., Rowan A., Taylor G., Smollett K., Loman N., Quick J., McMurray C., Stockton J., Nicholls S., Rowe W., Poplawski R., McNally A., Nunez R.M., Mason J., Robinson T., O’Toole E., Watts J., Breen C., Cowell A., Sluga G., Machin N., Ahmad S., George R., Halstead F., Sivaprakasam V., Hogsden W., Illingworth C., Jackson C., Thomson E., Shepherd J., Asamaphan P., Niebel M., Li K., Shah R., Jesudason N., Tong L., Broos A., Mair D., Nichols J., Carmichael S., Nomikou K., Aranday-Cortes E., Johnson N., Starinskij I., Filipe A.d.S., Robertson D., Orton R., Hughes J., Vattipally S., Singer J., Nickbakhsh S., Hale A., Macfarlane-Smith L., Harper K., Carden H., Taha Y., Payne B., Burton-Fanning S., Waugh S., Collins J., Eltringham G., Rushton S., O’Brien S., Bradley A., Maclean A., Mollett G., Blacow R., Templeton K., McHugh M., Dewar R., Wastenge E., Dervisevic S., Stanley R., Meader E., Coupland L., Smith L., Graham C., Barton E., Padgett D., Scott G., Swindells E., Greenaway J., Nelson A., McCann C., Yew W., Andersson M., Peto T., Justice A., Eyre D., Crook D., Sloan T., Duckworth N., Walsh S., Chauhan A., Glaysher S., Bicknell K., Wyllie S., Elliott S., Lloyd A., Impey R., Levene N., Monaghan L., Bradley D., Wyatt T., Allara E., Pearson C., Osman H., Bosworth A., Robinson E., Muir P., Vipond I., Hopes R., Pymont H., Hutchings S., Curran M., Parmar S., Lackenby A., Mbisa T., Platt S., Miah S., Bibby D., Manso C., Hubb J., Chand M., Dabrera G., Ramsay M., Bradshaw D., Thornton A., Myers R., Schaefer U., Groves N., Gallagher E., Lee D., Williams D., Ellaby N., Harrison I., Hartman H., Manesis N., Patel V., Bishop C., Chalker V., Ledesma J., Twohig K., Holden M., Shaaban S., Birchley A., Adams A., Davies A., Gaskin A., Plimmer A., Gatica-Wilcox B., McKerr C., Moore C., Williams C., Heyburn D., Lacy E.D., Hilvers E., Downing F., Shankar G., Jones H., Asad H., Coombes J., Watkins J., Evans J., Fina L., Gifford L., Gilbert L., Graham L., Perry M., Morgan M., Bull M., Cronin M., Pacchiarini N., Craine N., Jones R., Howe R., Corden S., Rey S., Kumziene-SummerhaYes S., Taylor S., Cottrell S., Jones S., Edwards S., O’Grady J., Page A., Mather A., Baker D., Rudder S., Aydin A., Kay G., Trotter A., Alikhan N.-F., Martins L.d.O., Le-Viet T., Meadows L., Casey A., Ratcliffe L., Simpson D., Molnar Z., Thompson T., Acheson E., Masoli J., Knight B., Ellard S., Auckland C., Jones C., Mahungu T., Irish-Tavares D., Haque T., Hart J., Witele E., Fenton M., Dadrah A., Symmonds A., Saluja T., Bourgeois Y., Scarlett G., Loveson K., Goudarzi S., Fearn C., Cook K., Dent H., Paul H., Partridge D., Raza M., Evans C., Johnson K., Liggett S., Baker P., Bonner S., Essex S., Lyons R., Saeed K., Mahanama A., Samaraweera B., Silveira S., Pelosi E., Wilson-Davies E., Williams R., Kristiansen M., Roy S., Williams C., Cotic M., Bayzid N., Westhorpe A., Hartley J., Jannoo R., Lowe H., Karamani A., Ensell L., Prieto J., Jeremiah S., Grammatopoulos D., Pandey S., Berry L., Jones K., Richter A., Beggs A., Best A., Percival B., Mirza J., Megram O., Mayhew M., Crawford L., Ashcroft F., Moles-Garcia E., Cumley N., Smith C., Bucca G., Hesketh A., Blane B., Girgis S., Leek D., Sridhar S., Forrest S., Cormie C., Gill H., Dias J., Higginson E., Maes M., Young J., Kermack L., Gupta R., Ludden C., Peacock S., Palmer S., Churcher C., Hadjirin N., Carabelli A., Brooks E., Smith K., Galai K., McManus G., Ruis C., Davidson R., Rambaut A., Williams T., Balcazar C., Gallagher M., O’Toole A., Rooke S., Hill V., Williamson K., Stanton T., Michell S., Bewshea C., Temperton B., Michelsen M., Warwick-Dugdale J., Manley R., Farbos A., Harrison J., Sambles C., Studholme D., Jeffries A., Jackson L., Darby A., Hiscox J., Paterson S., Iturriza-Gomara M., Jackson K., Lucaci A., Vamos E., Hughes M., Rainbow L., Eccles R., Nelson C., Whitehead M., Turtle L., Haldenby S., Gregory R., Gemmell M., Wierzbicki C., Webster H., de Silva T., Smith N., Angyal A., Lindsey B., Groves D., Green L., Wang D., Freeman T., Parker M., Keeley A., Parsons P., Tucker R., Brown R., Wyles M., Whiteley M., Zhang P., Gallis M., Louka S., Constantinidou C., Unnikrishnan M., Ott S., Cheng J., Bridgewater H., Frost L., Taylor-Joyce G., Stark R., Baxter L., Alam M., Brown P., Aggarwal D., Cerda A., Merrill T., Wilson R., McClure P., Chappell J., Tsoleridis T., Ball J., Buck D., Todd J., Green A., Trebes A., MacIntyre-Cockett G., de Cesare M., Alderton A., Amato R., Ariani C., Beale M., Beaver C., Bellis K., Betteridge E., Bonfield J., Danesh J., Dorman M., Drury E., Farr B., Foulser L., Goncalves S., Goodwin S., Gourtovaia M., Harrison E., Jackson D., Jamrozy D., Johnston I., Kane L., Kay S., Keatley J.-P., Kwiatkowski D., Langford C., Lawniczak M., Letchford L., Livett R., Lo S., Martincorena I., McGuigan S., Nelson R., Palmer S., Park N., Patel M., Prestwood L., Puethe C., Quail M., Rajatileka S., Scott C., Shirley L., Sillitoe J., Chapman M.S., Thurston S., Tonkin-Hill G., Weldon D., Rajan D., Bronner I., Aigrain L., Redshaw N., Lensing S., Davies R., Whitwham A., Liddle J., Lewis K., Tovar-Corona J., Leonard S., Durham J., Bassett A., McCarthy S., Moll R., James K., Oliver K., Makunin A., Barrett J., Gunson R. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022;22(1):35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies M.-A., Kassanjee R., Rosseau P., Morden E., Johnson L., Solomon W., Hsiao N.-Y., Hussey H., Meintjes G., Paleker M., Jacobs T., Raubenheimer P., Heekes A., Dane P., Bam J.-L., Smith M., Preiser W., Pienaar D., Mendelson M., Naude J., Schrueder N., Mnguni A., Roux S.L., Murie K., Prozesky H., Mahomed H., Rossouw L., Wasserman S., Maughan D., Boloko L., Smith B., Taljaard J., Symons G., Ntusi N., Parker A., Wolter N., Jassat W., Cohen C., Lessells R., Wilkinson R.J., Arendse J., Kariem S., Moodley M., Vallabhjee K., Wolmarans M., Cloete K., Boulle A., Africa, On behalf of the Western Cape and South African National Departments of Health in collaboration with the National Institute for Communicable Diseases in South . 2022. Outcomes of laboratory-confirmed SARS-CoV-2 infection in the Omicron-driven fourth wave compared with previous waves in the Western Cape Province, South Africa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bager P., Wohlfahrt J., Bhatt S., Stegger M., Legarth R., Møller C.H., Skov R.L., Valentiner-Branth P., Voldstedlund M., Fischer T.K., Simonsen L., Kirkby N.S., Thomsen M.K., Spiess K., Marving E., Larsen N.B., Lillebaek T., Ullum H., Mølbak K., Krause T.G., Edslev S.M., Sieber R.N., Ingham A.C., Overvad M., Gram M.A., Lomholt F.K., Hallundbæk L., Espensen C.H., Gubbels S., Karakis M., Møller K.L., Olsen S.S., Harboe Z.B., Johannesen C.K., van Wijhe M., Holler J.G., Dessau R.B.C., Friis M.B., Fuglsang-Damgaard D., Pinholt M., Sydenham T.V., Coia J.E., Marmolin E.S., Fomsgaard A., Fonager J., Rasmussen M., Cohen A. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: An observational cohort study. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Berger N.A., Davis P.B., Kaelber D.C., Volkow N.D., Xu R. 2022. Comparison of outcomes from COVID infection in pediatric and adult patients before and after the emergence of Omicron. [DOI] [Google Scholar]
- 20.Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. 2022. Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS-CoV-2 variant in Southern California. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson N., Ghani A., Hinsley W., Volz E. 2021. Report 50 - Hospitalisation risk for Omicron cases in England. http://www.imperial.ac.uk/medicine/departments/school-public-health/infectious-disease-epidemiology/mrc-global-infectious-disease-analysis/covid-19/report-50-severity-omicron/ [Google Scholar]
- 22.Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., Hinsley W., Bernal J.L., Kall M., Bhatt S., Blomquist P., Zaidi A., Volz E., Aziz N.A., Harman K., Funk S., Abbott S., Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., Hinsley W., Bernal J.L., Kall M., Bhatt S., Blomquist P., Zaidi A., Volz E., Aziz N.A., Harman K., Funk S., Abbott S., Hope R., Charlett A., Chand M., Ghani A.C., Seaman S.R., Dabrera G., Angelis D.D., Presanis A.M., Thelwall S., Hope R., Charlett A., Chand M., Ghani A.C., Seaman S.R., Dabrera G., Angelis D.D., Presanis A.M., Thelwall S. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants in England: A cohort study. Lancet. 2022;399(10332):1303–1312. doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng B., Ferreira I., Abdullahi A., Kemp S.A., Goonawardane N., Papa G., Fatihi S., Charles O., Collier D., Choi J., Lee J.H., Mlcochova P., James L., Doffinger R., Thukral L., Sato K., Gupta R.K., CITIID-NIHR BioResource COVID-19 Collaboration, The Genotype to Phenotype Japan (G2P-Japan) Consortium . 2021. SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. [DOI] [Google Scholar]
- 24.Zhao H., Lu L., Peng Z., Chen L.-L., Meng X., Zhang C., Ip J.D., Chan W.-M., Chu A.W.-H., Chan K.-H., Jin D.-Y., Chen H., Yuen K.-Y., To K.K.-W. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. 2022;11(1):277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelnabi R., Foo C.S.-Y., Zhang X., Lemmens V., Maes P., Slechten B., Raymenants J., Andre E., Weynand B., Dallmeier K., Neyts J. 2021. The Omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan K.A., Watson R.J., Bewley K.R., Burton C.A., Carnell O., Cavell B.E., Challis A.R., Coombes N.S., Emery K., Fell R., Fotheringham S.A., Gooch K.E., Gowan K., Handley A., Harris D.J., Humphreys R., Johnson R., Knott D., Lister S., Morley D., Ngabo D., Osman K.L., Paterson J., Penn E.J., Pullen S.T., Richards K.S., Shaik I., Summers S., Thomas S.R., Weldon T., Wiblin N.R., Vipond R., Hallis B., Funnell S.G.P., Hall Y. 2021. Convalescence from prototype SARS-CoV-2 protects Syrian hamsters from disease caused by the Omicron variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 28.Tasakis R.N., Samaras G., Jamison A., Lee M., Paulus A., Whitehouse G., Verkoczy L., Papavasiliou F.N., Diaz M. SARS-CoV-2 variant evolution in the United States: High accumulation of viral mutations over time likely through serial Founder Events and mutational bursts. PLOS ONE. 2021;16(7) doi: 10.1371/journal.pone.0255169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baj A., Novazzi F., Drago Ferrante F., Genoni A., Tettamanzi E., Catanoso G., Dalla Gasperina D., Dentali F., Focosi D., Maggi F. Spike protein evolution in the SARS-CoV-2 Delta variant of concern: A case series from Northern Lombardy. Emerg. Microb. Infect. 2021;10(1):2010–2015. doi: 10.1080/22221751.2021.1994356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baj A., Novazzi F., Pasciuta R., Genoni A., Ferrante F.D., Valli M., Partenope M., Tripiciano R., Ciserchia A., Catanoso G., Focosi D., Maggi F. Breakthrough infections of E484K-Harboring SARS-CoV-2 Delta Variant, Lombardy, Italy. Emerg. Infect. Diseases. 2021;27(12):3180–3182. doi: 10.3201/eid2712.211792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L., Zody M.C., Di Germanio C., Martinelli R., Mediavilla J.R., Cunningham M.H., Composto K., Chow K.F., Kordalewska M., Corvelo A., Oschwald D.M., Fennessey S., Zetkulic M., Dar S., Kramer Y., Mathema B., Germer S., Stone M., Simmons G., Busch M.P., Maniatis T., Perlin D.S., Kreiswirth B.N. Emergence of multiple SARS-CoV-2 antibody escape variants in an immunocompromised host undergoing convalescent plasma treatment. mSphere. 2021;6(4) doi: 10.1128/mSphere.00480-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora P., Zhang L., Rocha C., Sidarovich A., Kempf A., Schulz S., Cossmann A., Manger B., Baier E., Tampe B., Moerer O., Dickel S., Dopfer-Jablonka A., Jäck H.-M., Behrens G.M.N., Winkler M.S., Pöhlmann S., Hoffmann M. Comparable neutralisation evasion of SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022:S1473–3099(22)00224–9. doi: 10.1016/S1473-3099(22)00224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., Ren Y., Quan L., Zhao W., Seto D., Chodosh J., Luo Z., Wu J., Zhang Q. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Target. Therapy. 2022;7(1):138. doi: 10.1038/s41392-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty C., Bhattacharya M., Sharma A.R., Dhama K. Recombinant SARS-CoV-2 variants XD, XE, and XF: The emergence of recombinant variants requires an urgent call for research - Correspondence. Int. J. Surg. (London, England) 2022;102 doi: 10.1016/j.ijsu.2022.106670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dite G.S., Murphy N.M., Allman R. Development and validation of a clinical and genetic model for predicting risk of severe COVID-19. Epidemiol. Infect. 2021;149 doi: 10.1017/S095026882100145X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dite G.S., Murphy N.M., Allman R. An integrated clinical and genetic model for predicting risk of severe COVID-19: A population-based case-control study. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiewsakun P., Wongtrakoongate P., Thawornwattana Y., Hongeng S., Thitithanyanont A. SARS-CoV-2 genetic variations associated with COVID-19 severity. MedRxiv. 2020 doi: 10.1099/mgen.0.000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SeyedAlinaghi S., Mirzapour P., Dadras O., Pashaei Z., Karimi A., MohsseniPour M., Soleymanzadeh M., Barzegary A., Afsahi A.M., Vahedi F., Shamsabadi A., Behnezhad F., Saeidi S., Mehraeen E., Jahanfar S. Characterization of SARS-CoV-2 different variants and related morbidity and mortality: A systematic review. Eur. J. Med. Res. 2021;26(1):51. doi: 10.1186/s40001-021-00524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas S.K., Mudi S.R. Spike protein D614G and RdRp P323L: The SARS-CoV-2 mutations associated with severity of COVID-19. Genom. Inform. 2020;18(4) doi: 10.5808/GI.2020.18.4.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskar R., Ali S. Differential mutation profile of SARS-CoV-2 proteins across deceased and asymptomatic patients. Chem. Biol. Interact. 2021;347 doi: 10.1016/j.cbi.2021.109598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clauwaert J., Menschaert G., Waegeman W., Dumonteil E., Fusco D., Drouin A., Herrera C., Esper F.P., Cheng Y.-W., Adhikari T.M., Tu Z.J., Li D., Li E.A., Farkas D.H., Procop G.W., Ko J.S., Chan T.A., Jehi L., Rubin B.P., Li J., Fisman D.N., Tuite A.R., Hamed S.M., Elkhatib W.F., Khairalla A.S., Noreddin A.M., Sarkar R., Chawla-Sarkar M., Majumdar S., Lo M., Chattopadhyay S., Schmidt F., Weisblum Y., Rutkowska M., Poston D., DaSilva J., Zhang F., Bednarski E., Cho A., Schaefer-Babajew D.J., Gaebler C., Caskey M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Geographical and temporal distribution of SARS-CoV-2 globally: An attempt to correlate case fatality rate with the circulating dominant SARS-CoV-2 clades. MedRxiv. 2021;193(42) 2021.05.25.21257434. [Google Scholar]
- 42.Hamed S.M., Elkhatib W.F., Khairalla A.S., Noreddin A.M. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci. Rep. 2021;11(1):8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss J.D., Skarzynski M., McAuley E.M., Maier E.J., Gibbons T., Fries A.C., Chapleau R.R. Variants in SARS-CoV-2 associated with mild or severe outcome. Evol. Med. Public Health. 2021;9(1):267–275. doi: 10.1093/emph/eoab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal R., Leblond T., McAuley E.M., Maier E.J., Skarzynski M., Voss J.D., Sozhamannan S. 2022. Linking genotype to phenotype: Further exploration of mutations in SARS-CoV-2 associated with mild or severe outcomes - SARS-CoV-2 coronavirus. https://virological.org/t/linking-genotype-to-phenotype-further-exploration-of-mutations-in-sars-cov-2-associated-with-mild-or-severe-outcomes/794. [Google Scholar]
- 45.Nagpal S., Pinna N.K., Srivastava D., Singh R., Mande S.S. 2021. (Machine) learning the mutation signatures of SARS-CoV-2: A primer for predictive prognosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawmya S., Saha A., Tasnim S., Toufikuzzaman M., Anjum N., Rafid A.H.M., Rahman M.S., Rahman M.S. 2021. Analyzing hCov genome sequences: Predicting virulence and mutation. [DOI] [Google Scholar]
- 47.Sokhansanj B.A., Zhao Z., Rosen G.L. 2021. Interpretable and predictive deep modeling of the SARS-CoV-2 spike protein sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obermeyer F., Schaffner S.F., Jankowiak M., Barkas N., Pyle J.D., Park D.J., MacInnis B.L., Luban J., Sabeti P.C., Lemieux J.E. Analysis of 2.1 million SARS-CoV-2 genomes identifies mutations associated with transmissibility. medRxiv. 2021 doi: 10.1126/science.abm1208. 2021.09.07.21263228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokhansanj B.A., Rosen G.L. Mapping data to deep understanding: Making the most of the deluge of SARS-CoV-2 genome sequences. mSystems. 2022;7(2):e00035–22. doi: 10.1128/msystems.00035-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigrist F. 2021. Gaussian process boosting. arXiv:2004.02653 [cs, stat] [Google Scholar]
- 51.Goldstein B.A., Polley E.C., Briggs F.B.S. Random forests for genetic association studies. Stat. Appl. Genet. Mol. Biol. 2011;10(1):32. doi: 10.2202/1544-6115.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery; New York, NY, USA: 2016. XGBoost: A scalable tree boosting system; pp. 785–794. (KDD ’16). [DOI] [Google Scholar]
- 53.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., Ye Q., Liu T.-Y. Advances in Neural Information Processing Systems, Vol. 30. Curran Associates, Inc.; 2017. LightGBM: A highly efficient gradient boosting decision tree; pp. 3146–3154. [Google Scholar]
- 54.Lundberg S.M., Lee S.-I. Advances in Neural Information Processing Systems, Vol. 30. Curran Associates, Inc.; 2017. A unified approach to interpreting model predictions; pp. 4768–4777. [Google Scholar]
- 55.Pillay T.S. Gene of the month: The 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73(7):366. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 56.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren L., Zhang Y., Li J., Xiao Y., Zhang J., Wang Y., Chen L., Paranhos-Baccalà G., Wang J. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015;5(1):11451. doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The scikit-bio development team . 2020. Scikit-bio: A bioinformatics library for data scientists, students, and developers. [Google Scholar]
- 61.Zhao M., Lee W.-P., Garrison E.P., Marth G.T. SSW library: An SIMD Smith-Waterman C/C++ Library for use in genomic applications. PLOS ONE. 2013;8(12) doi: 10.1371/journal.pone.0082138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay E. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 63.National Institutes of Health . 2021. Clinical spectrum of SARS-CoV-2 infection. [Google Scholar]
- 64.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005;67(2):301–320. [Google Scholar]
- 65.Waldmann P., Mészáros G., Gredler B., Fürst C., Sölkner J. Evaluation of the lasso and the elastic net in genome-wide association studies. Front. Genet. 2013;4 doi: 10.3389/fgene.2013.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Goethem N., Robert A., Bossuyt N., Van Poelvoorde L.A.E., Quoilin S., De Keersmaecker S.C.J., Devleesschauwer B., Thomas I., Vanneste K., Roosens N.H.C., Van Oyen H. Evaluation of the added value of viral genomic information for predicting severity of influenza infection. BMC Infect. Dis. 2021;21(1):785. doi: 10.1186/s12879-021-06510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J., Gribskov M. IRESpy: An XGBoost model for prediction of internal ribosome entry sites. BMC Bioinformatics. 2019;20(1):409. doi: 10.1186/s12859-019-2999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.ValizadehAslani T., Zhao Z., Sokhansanj B.A., Rosen G.L. Amino acid K-mer feature extraction for quantitative antimicrobial resistance (AMR) prediction by machine learning and model interpretation for biological insights. Biology. 2020;9(11):E365. doi: 10.3390/biology9110365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang X., Li F., Chen J., Li J., Wu H., Li S., Song J., Liu Q. Large-scale comparative review and assessment of computational methods for anti-cancer peptide identification. Brief. Bioinform. 2021;22(4):bbaa312. doi: 10.1093/bib/bbaa312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benson A.K., Kelly S.A., Legge R., Ma F., Low S.J., Kim J., Zhang M., Oh P.L., Nehrenberg D., Hua K., Kachman S.D., Moriyama E.N., Walter J., Peterson D.A., Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA. 2010;107(44):18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X., Guo B., Yi N. Zero-inflated Gaussian mixed models for analyzing longitudinal microbiome data. PLoS ONE. 2020;15(11) doi: 10.1371/journal.pone.0242073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang Y., Chen J., Chen W. Controlling batch effect in epigenome-wide association study. Methods Mol. Biol. (Clifton, N.J.) 2022;2432:73–84. doi: 10.1007/978-1-0716-1994-0_6. [DOI] [PubMed] [Google Scholar]
- 73.Ngufor C., Van Houten H., Caffo B.S., Shah N.D., McCoy R.G. Mixed effect machine learning: A framework for predicting longitudinal change in hemoglobin A1c. J. Biomed. Inform. 2019;89:56–67. doi: 10.1016/j.jbi.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou F., Alsaid A., Blommer M., Curry R., Swaminathan R., Kochhar D., Talamonti W., Tijerina L. Predicting driver fatigue in monotonous automated driving with explanation using gpboost and SHAP. Int. J. Human Comput. Interact. 2022;38(8):719–729. [Google Scholar]
- 75.Ramraj S., Uzir N., Sunil R., Banerjee S. Experimenting XGBoost algorithm for prediction and classification of different datasets. Int. J. Control Theory Appl. 2016;9:651–662. [Google Scholar]
- 76.Elith J., Leathwick J.R., Hastie T. A working guide to boosted regression trees. J. Anim. Ecol. 2008;77(4):802–813. doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- 77.Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., Bonanomi E., Cabrini L., Carlesso E., Castelli G., Cattaneo S., Cereda D., Colombo S., Coluccello A., Crescini G., Forastieri Molinari A., Foti G., Fumagalli R., Iotti G.A., Langer T., Latronico N., Lorini F.L., Mojoli F., Natalini G., Pessina C.M., Ranieri V.M., Rech R., Scudeller L., Rosano A., Storti E., Thompson B.T., Tirani M., Villani P.G., Pesenti A., Cecconi M., COVID-19 Lombardy ICU Network Risk factors associated with mortality among patients with COVID-19 in intensive care units in Lombardy, Italy. JAMA Internal Med. 2020;180(10):1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holt H., Talaei M., Greenig M., Zenner D., Symons J., Relton C., Young K.S., Davies M.R., Thompson K.N., Ashman J., Rajpoot S.S., Kayyale A.A., El Rifai S., Lloyd P.J., Jolliffe D., Timmis O., Finer S., Iliodromiti S., Miners A., Hopkinson N.S., Alam B., Lloyd-Jones G., Dietrich T., Chapple I., Pfeffer P.E., McCoy D., Davies G., Lyons R.A., Griffiths C., Kee F., Sheikh A., Breen G., Shaheen S.O., Martineau A.R. Risk factors for developing COVID-19: A population-based longitudinal study (COVIDENCE UK) Thorax. 2021:thoraxjnl–2021–217487. doi: 10.1136/thoraxjnl-2021-217487. [DOI] [PubMed] [Google Scholar]
- 79.Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nature Commun. 2020;11(1):6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukherjee S., Pahan K. Is COVID-19 gender-sensitive? J. Neuroimmune Pharmacol.: Off. J. Soc. NeuroImmune Pharmacol. 2021;16(1):38–47. doi: 10.1007/s11481-020-09974-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hsu S.H., Chang S.-H., Gross C.P., Wang S.-Y. Relative risks of COVID-19 fatality between the first and second waves of the pandemic in Ontario, Canada. Int. J. Infect. Dis.: IJID : Off. Publ. Int. Soc. Infect. Dis. 2021;109:189–191. doi: 10.1016/j.ijid.2021.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopez Bernal J., Andrews N., Gower C., Robertson C., Stowe J., Tessier E., Simmons R., Cottrell S., Roberts R., O’Doherty M., Brown K., Cameron C., Stockton D., McMenamin J., Ramsay M. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on Covid-19 related symptoms, hospital admissions, and mortality in older adults in England: Test negative case-control study. BMJ (Clin. Res. Ed.) 2021;373:n1088. doi: 10.1136/bmj.n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akpolat T., Uzun O. Reduced mortality rate after coronavac vaccine among healthcare workers. J. Infect. 2021;83(2):e20–e21. doi: 10.1016/j.jinf.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K., Southern J., Swerdlow D.L., Jodar L., Levy Y., Alroy-Preis S. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet (London, England) 2021;397(10287):1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grima A.A., Murison K.R., Simmons A.E., Tuite A.R., Fisman D.N. Relative virulence of SARS-CoV-2 among vaccinated and unvaccinated individuals hospitalized with SARS-CoV-2. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2022:ciac412. doi: 10.1093/cid/ciac412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aggarwal N.R., Beaty L.E., Bennett T.D., Carlson N.E., Davis C.B., Kwan B.M., Mayer D.A., Ong T.C., Russell S., Steele J., Wogu A.F., Wynia M.K., Zane R.D., Ginde A.A. Real world evidence of the neutralizing monoclonal antibody sotrovimab for preventing hospitalization and mortality in COVID-19 outpatients. MedRxiv: Prepr. Serv. Health Sci. 2022 doi: 10.1093/infdis/jiac206. 2022.04.03.22273360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323(18):1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 88.Mahajan S., Caraballo C., Li S.-X., Dong Y., Chen L., Huston S.K., Srinivasan R., Redlich C.A., Ko A.I., Faust J.S., Forman H.P., Krumholz H.M. SARS-CoV-2 infection hospitalization rate and infection fatality rate among the non-congregate population in connecticut. Am. J. Med. 2021;134(6):812–816.e2. doi: 10.1016/j.amjmed.2021.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang W., Kandula S., Huynh M., Greene S.K., Van Wye G., Li W., Chan H.T., McGibbon E., Yeung A., Olson D., Fine A., Shaman J. Estimating the infection-fatality risk of SARS-CoV-2 in New York City during the spring 2020 pandemic wave: A model-based analysis. Lancet Infect. Dis. 2021;21(2):203–212. doi: 10.1016/S1473-3099(20)30769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Z., Sokhansanj B.A., Malhotra C., Zheng K., Rosen G.L. Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput. Biol. 2020;16(9) doi: 10.1371/journal.pcbi.1008269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Negi S.S., Schein C.H., Braun W. Regional and temporal coordinated mutation patterns in SARS-CoV-2 spike protein revealed by a clustering and network analysis. Sci. Rep. 2022;12(1):1128. doi: 10.1038/s41598-022-04950-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monod M., Blenkinsop A., Xi X., Hebert D., Bershan S., Tietze S., Baguelin M., Bradley V.C., Chen Y., Coupland H., Filippi S., Ish-Horowicz J., McManus M., Mellan T., Gandy A., Hutchinson M., Unwin H.J.T., van Elsland S.L., Vollmer M.A.C., Weber S., Zhu H., Bezancon A., Ferguson N.M., Mishra S., Flaxman S., Bhatt S., Ratmann O. Age groups that sustain resurging COVID-19 epidemics in the United States. Science. 2021;371(6536) doi: 10.1126/science.abe8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10(1):14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Z., Chong K.C., Wong M.C.S., Boon S.S., Huang J., Wang M.H., Ng R.W.Y., Lai C.K.C., Chan P.K.S. A global analysis of replacement of genetic variants of SARS-CoV-2 in association with containment capacity and changes in disease severity. Clin. Microbiol. Infect.: Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021;27(5):750–757. doi: 10.1016/j.cmi.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oberg A.L., Mahoney D.W. Linear mixed effects models. Methods Mol. Biol. (Clifton, N.J.) 2007;404:213–234. doi: 10.1007/978-1-59745-530-5_11. [DOI] [PubMed] [Google Scholar]
- 96.Lazarevic I., Pravica V., Miljanovic D., Cupic M. Immune evasion of SARS-CoV-2 emerging variants: What have we learnt so far? Viruses. 2021;13(7):1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Noori M., Nejadghaderi S.A., Arshi S., Carson-Chahhoud K., Ansarin K., Kolahi A.-A., Safiri S. Potency of BNT162b2 and mRNA-1273 vaccine-induced neutralizing antibodies against severe acute respiratory syndrome-CoV-2 variants of concern: A systematic review of in vitro studies. Rev. Med. Virol. 2022;32(2) doi: 10.1002/rmv.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nonaka C.K.V., Gräf T., Barcia C.A.d.L., Costa V.F., de Oliveira J.L., Passos R.d.H., Bastos I.N., de Santana M.C.B., Santos I.M., de Sousa K.A.F., Weber T.G.L., de Siqueira I.C., Rocha C.A.G., Mendes A.V.A., Souza B.S.d.F. SARS-CoV-2 variant of concern P.1 (Gamma) infection in Young and middle-aged patients admitted to the intensive care units of a single hospital in Salvador, Northeast Brazil, February 2021. Int. J. Infect. Dis. 2021;111:47–54. doi: 10.1016/j.ijid.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Albaradei S., Thafar M., Alsaedi A., Van Neste C., Gojobori T., Essack M., Gao X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput. Struct. Biotechnol. J. 2021;19:5008–5018. doi: 10.1016/j.csbj.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Domingos P. A few useful things to know about machine learning. Commun. ACM. 2012;55(10):78–87. [Google Scholar]
- 101.Dhawan M., Sharma A., Priyanka n., Thakur N., Rajkhowa T.K., Choudhary O.P. Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions. Human Vaccines Immunother. 2022 doi: 10.1080/21645515.2022.2068883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., Iwatsuki-Horimoto K., Ito M., Yamayoshi S., Ozono S., Butlertanaka E.P., Tanaka Y.L., Shimizu R., Shimizu K., Yoshimatsu K., Kawabata R., Sakaguchi T., Tokunaga K., Yoshida I., Asakura H., Nagashima M., Kazuma Y., Nomura R., Horisawa Y., Yoshimura K., Takaori-Kondo A., Imai M., Nakagawa S., Ikeda T., Fukuhara T., Kawaoka Y., Sato K., The Genotype to Phenotype Japan (G2P-Japan) Consortium . 2021. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. [DOI] [Google Scholar]
- 103.Kuzmina A., Atari N., Ottolenghi A., Korovin D., Lass I.C., Rosental B., Rosenberg E., Mandelboim M., Taube R. 2022. P681 mutations within the polybasic motif of spike dictate fusogenicity and syncytia formation of SARS CoV-2 variants. [DOI] [Google Scholar]
- 104.Zhang Y., Zhang T., Fang Y., Liu J., Ye Q., Ding L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct. Target. Ther. 2022;7(1):1–3. doi: 10.1038/s41392-022-00941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., Yue Y., Shimizu R., Ito J., Torii S., Yonekawa A., Shimono N., Nagasaki Y., Minami R., Toya T., Sekiya N., Fukuhara T., Matsuura Y., Schreiber G., Ikeda T., Nakagawa S., Ueno T., Sato K., Genotype to Phenotype Japan (G2P-Japan) Consortium SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bansal K., Kumar S. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. Virus Res. 2022;315 doi: 10.1016/j.virusres.2022.198765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schnirring L. ECDC ups BA.4, BA.5 to variants of concern, warns of case rises. CIDRAP. 2022 [Google Scholar]
- 108.Maxmen A. Why call it BA.2.12.1? A guide to the tangled Omicron family. Nature. 2022 doi: 10.1038/d41586-022-01466-9. [DOI] [PubMed] [Google Scholar]
- 109.Uraki R., Kiso M., Iida S., Imai M., Takashita E., Kuroda M., Halfmann P.J., Loeber S., Maemura T., Yamayoshi S., Fujisaki S., Wang Z., Ito M., Ujie M., Iwatsuki-Horimoto K., Furusawa Y., Wright R., Chong Z., Ozono S., Yasuhara A., Ueki H., Sakai-Tagawa Y., Li R., Liu Y., Larson D., Koga M., Tsutsumi T., Adachi E., Saito M., Yamamoto S., Hagihara M., Mitamura K., Sato T., Hojo M., Hattori S.-I., Maeda K., Valdez R., Okuda M., Murakami J., Duong C., Godbole S., Douek D.C., Maeda K., Watanabe S., Gordon A., Ohmagari N., Yotsuyanagi H., Diamond M.S., Hasegawa H., Mitsuya H., Suzuki T., Kawaoka Y., IASO study team Characterization and antiviral susceptibility of SARS-CoV-2 omicron/BA.2. Nature. 2022 doi: 10.1038/s41586-022-04856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., Suzuki R., Kishimoto M., Kosugi Y., Kobiyama K., Hara T., Toyoda M., Tanaka Y.L., Butlertanaka E.P., Shimizu R., Ito H., Wang L., Oda Y., Orba Y., Sasaki M., Nagata K., Yoshimatsu K., Asakura H., Nagashima M., Sadamasu K., Yoshimura K., Kuramochi J., Seki M., Fujiki R., Kaneda A., Shimada T., Nakada T.-a., Sakao S., Suzuki T., Ueno T., Takaori-Kondo A., Ishii K.J., Schreiber G., Sawa H., Saito A., Irie T., Tanaka S., Matsuno K., Fukuhara T., Ikeda T., Sato K., The Genotype to Phenotype Japan (G2P-Japan) Consortium . 2022. Virological characteristics of SARS-CoV-2 BA.2 variant. [DOI] [Google Scholar]
- 111.Whitaker M., Elliott J., Bodinier B., Barclay W., Ward H., Cooke G., Donnelly C.A., Chadeau-Hyam M., Elliott P. 2022. Variant-specific symptoms of COVID-19 among 1,542,510 people in England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Loconsole D., Centrone F., Sallustio A., Accogli M., Casulli D., Sacco D., Zagaria R., Morcavallo C., Chironna M. Characteristics of the first 284 patients infected with the SARS-CoV-2 omicron BA.2 subvariant at a single center in the apulia region of Italy, January–March 2022. Vaccines. 2022;10(5):674. doi: 10.3390/vaccines10050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu J., Collier A.-r.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., Hachmann N.P., Surve N., Nampanya F., Chandrashekar A., Barouch D.H. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386(16):1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K.-H., Yuen T.T.-T., Yoon C., To K.K.-W., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K.-Y., Ho D.D. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602(7898):676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 115.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.-W., Huang Y., Wang M., Luo Y., Yu J., Chu H., Chik K.K.-H., Yuen T.T.-T., Yin M.T., Sobieszczyk M.E., Huang Y., Yuen K.-Y., Wang H.H., Sheng Z., Ho D.D. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604(7906):553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vogt A.-C.S., Augusto G., Martina B., Chang X., Nasrallah G., Speiser D.E., Vogel M., Bachmann M.F., Mohsen M.O. Increased receptor affinity and reduced recognition by specific antibodies contribute to immune escape of SARS-CoV-2 variant omicron. Vaccines. 2022;10(5):743. doi: 10.3390/vaccines10050743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.-K., Adams-Quack P., Bacher M., Finlayson A., Ozhelvaci O., Vogler I., Grikscheit K., Hoehl S., Goetsch U., Ciesek S., Türeci O., Sahin U. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Q., Guo Y., Iketani S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., Shah J.G., Nguyen N., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D.D. 2022. SARS-CoV-2 omicron BA.2.12.1, BA.4, and BA.5 subvariants evolved to extend antibody evasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andreassen C.N. A simulated SNP experiment indicates a high risk of over-fitting and false positive results when a predictive multiple SNP model is established and tested within the same dataset. Radiother. Oncol.: J. Eur. Soc. Ther. Radiol. Oncol. 2015;114(3):310–313. doi: 10.1016/j.radonc.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 120.Jones D.T. Setting the standards for machine learning in biology. Nat. Rev. Mol. Cell Biol. 2019;20(11):659–660. doi: 10.1038/s41580-019-0176-5. [DOI] [PubMed] [Google Scholar]
- 121.Takahashi Y., Ueki M., Tamiya G., Ogishima S., Kinoshita K., Hozawa A., Minegishi N., Nagami F., Fukumoto K., Otsuka K., Tanno K., Sakata K., Shimizu A., Sasaki M., Sobue K., Kure S., Yamamoto M., Tomita H. Machine learning for effectively avoiding overfitting is a crucial strategy for the genetic prediction of polygenic psychiatric phenotypes. Transl. Psychiatry. 2020;10(1):1–11. doi: 10.1038/s41398-020-00957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mikolajewicz N., Komarova S.V. Meta-analytic methodology for basic research: A practical guide. Front. Physiol. 2019;10 doi: 10.3389/fphys.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schriml L.M., Chuvochina M., Davies N., Eloe-Fadrosh E.A., Finn R.D., Hugenholtz P., Hunter C.I., Hurwitz B.L., Kyrpides N.C., Meyer F., Mizrachi I.K., Sansone S.-A., Sutton G., Tighe S., Walls R. COVID-19 pandemic reveals the peril of ignoring metadata standards. Sci. Data. 2020;7(1):188. doi: 10.1038/s41597-020-0524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhattacharyya R.P., Hanage W.P. Challenges in inferring intrinsic severity of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386(7) doi: 10.1056/NEJMp2119682. [DOI] [PubMed] [Google Scholar]
- 125.Calderwood M.S., Deloney V.M., Anderson D.J., Cheng V.C.-C., Gohil S., Kwon J.H., Mody L., Monsees E., Vaughn V.M., Wiemken T.L., Ziegler M.J., Lofgren E. Policies and practices of SHEA research network hospitals during the COVID-19 pandemic. Infect. Control Hosp. Epidemiol. 2020;41(10):1127–1135. doi: 10.1017/ice.2020.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.N. Fillmore, J. La, C. Zheng, S. Doron, N. Do, P. Monach, W. Branch-Elliman, The COVID-19 Hospitalization Metric in the Pre- and Post-Vaccination Eras as a Measure of Pandemic Severity: A Retrospective, Nationwide Cohort Study, Preprint, 2021, 10.21203/rs.3.rs-898254/v1, In Review. [DOI] [PMC free article] [PubMed]
- 127.Kushner L.E., Schroeder A.R., Kim J., Mathew R. “For COVID” or “with COVID”: Classification of SARS-CoV-2 hospitalizations in children. Hosp. Pediatr. 2021;11(8):e151–e156. doi: 10.1542/hpeds.2021-006001. [DOI] [PubMed] [Google Scholar]
- 128.Webb N.E., Osburn T.S. Characteristics of hospitalized children positive for SARS-CoV-2: Experience of a large center. Hosp. Pediatr. 2021;11(8):e133–e141. doi: 10.1542/hpeds.2021-005919. [DOI] [PubMed] [Google Scholar]
- 129.Rocheleau L., Laroche G., Fu K., Stewart C.M., Mohamud A.O., Côté M., Giguère P.M., Langlois M.-A., Pelchat M. Identification of a high-frequency intrahost SARS-CoV-2 spike variant with enhanced cytopathic and fusogenic effects. MBio. 2021 doi: 10.1128/mBio.00788-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jacot D., Pillonel T., Greub G., Bertelli C. Assessment of SARS-CoV-2 genome sequencing: Quality criteria and low-frequency variants. J. Clin. Microbiol. 2021;59(10) doi: 10.1128/JCM.00944-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lagerborg K.A., Normandin E., Bauer M.R., Adams G., Figueroa K., Loreth C., Gladden-Young A., Shaw B.M., Pearlman L.R., Berenzy D., Dewey H.B., Kales S., Dobbins S.T., Shenoy E.S., Hooper D., Pierce V.M., Zachary K.C., Park D.J., MacInnis B.L., Tewhey R., Lemieux J.E., Sabeti P.C., Reilly S.K., Siddle K.J. Synthetic DNA spike-ins (SDSIs) enable sample tracking and detection of inter-sample contamination in SARS-CoV-2 sequencing workflows. Nat. Microbiol. 2022;7(1):108–119. doi: 10.1038/s41564-021-01019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ejaz H., Alsrhani A., Zafar A., Javed H., Junaid K., Abdalla A.E., Abosalif K.O.A., Ahmed Z., Younas S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health. 2020;13(12):1833–1839. doi: 10.1016/j.jiph.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dessie Z.G., Zewotir T. Mortality-related risk factors of COVID-19: A systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect. Dis. 2021;21(1):855. doi: 10.1186/s12879-021-06536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang S.-W., Wang S.-F. SARS-CoV-2 entry related viral and host genetic variations: Implications on COVID-19 severity, immune escape, and infectivity. Int. J. Mol. Sci. 2021;22(6):3060. doi: 10.3390/ijms22063060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mohammadpour S., Torshizi Esfahani A., Halaji M., Lak M., Ranjbar R. An updated review of the association of host genetic factors with susceptibility and resistance to COVID-19. J. Cell. Physiol. 2021;236(1):49–54. doi: 10.1002/jcp.29868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fricke-Galindo I., Falfán-Valencia R. Genetics insight for COVID-19 susceptibility and severity: A review. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.622176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Verma A., Tsao N.L., Thomann L.O., Ho Y.-L., Iyengar S.K., Luoh S.-W., Carr R., Crawford D.C., Efird J.T., Huffman J.E., Hung A., Ivey K.L., Levin M.G., Lynch J., Natarajan P., Pyarajan S., Bick A.G., Costa L., Genovese G., Hauger R., Madduri R., Pathak G.A., Polimanti R., Voight B., Vujkovic M., Zekavat S.M., Zhao H., Ritchie M.D., Chang K.-M., Cho K., Casas J.P., Tsao P.S., Gaziano J.M., O’Donnell C., Damrauer S.M., Liao K.P., VA Million Veteran Program COVID-19 Science Initiative A phenome-wide association study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the million veteran program. PLOS Genet. 2022;18(4) doi: 10.1371/journal.pgen.1010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chlamydas S., Papavassiliou A.G., Piperi C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics. 2021;16(3):263–270. doi: 10.1080/15592294.2020.1796896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Islam A.B.M.M.K., Khan M.A.-A.-K., Ahmed R., Hossain M.S., Kabir S.M.T., Islam M.S., Siddiki A.M.A.M.Z. Transcriptome of nasopharyngeal samples from COVID-19 patients and a comparative analysis with other SARS-CoV-2 infection models reveal disparate host responses against SARS-CoV-2. J. Transl. Med. 2021;19:32. doi: 10.1186/s12967-020-02695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao H., Nguyen A., Wu D., Li Y., Hassan S.A., Chen J., Shroff H., Piszczek G., Schuck P. Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein. PNAS Nexus. 2022:pgac049. doi: 10.1093/pnasnexus/pgac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Puray-Chavez M., LaPak K.M., Schrank T.P., Elliott J.L., Bhatt D.P., Agajanian M.J., Jasuja R., Lawson D.Q., Davis K., Rothlauf P.W., Liu Z., Jo H., Lee N., Tenneti K., Eschbach J.E., Mugisha C.S., Cousins E.M., Cloer E.W., Vuong H.R., VanBlargan L.A., Bailey A.L., Gilchuk P., Crowe J.E., Diamond M.S., Hayes D.N., Whelan S.P.J., Horani A., Brody S.L., Goldfarb D., Major M.B., Kutluay S.B. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021;36(2) doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P., Carnahan R.H., Crowe J.E., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Torrens-Fontanals M., Peralta-García A., Talarico C., Guixà-González R., Giorgino T., Selent J. SCoV2-MD: A database for the dynamics of the SARS-CoV-2 proteome and variant impact predictions. Nucleic Acids Res. 2022;50(D1):D858–D866. doi: 10.1093/nar/gkab977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaur A., Chauhan A.S., kumar Aggarwal A. Prediction of enhancers in DNA sequence data using a hybrid CNN-DLSTM model. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022:1. doi: 10.1109/TCBB.2022.3167090. [DOI] [PubMed] [Google Scholar]
- 146.Kaur A., Chauhan A.S., kumar Aggarwal A. Dynamic deep genomics sequence encoder for managed file transfer. IETE J. Res. 2022 [Google Scholar]
- 147.Bileschi M.L., Belanger D., Bryant D.H., Sanderson T., Carter B., Sculley D., Bateman A., DePristo M.A., Colwell L.J. Using deep learning to annotate the protein universe. Nature Biotechnol. 2022 doi: 10.1038/s41587-021-01179-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 showing a mapping scheme used to encode raw patient status metadata.
Supplementary Table S2 showing metadata entries mapped to an encoded patient status and resulting disease severity.
Supplementary Table S3 showing lineage counts for samples collected from July 17 through December 25, 2021.
Data Availability Statement
-
•
The datasets analyzed for this study were downloaded from GISAID EpiCoV database pursuant to the GISAID terms of use. They are availabile for download to users who register with GISAID at the website http://wwww.gisaid.org. The list of GISAID accession numbers used for this paper and data acknowledgments are available at https://epicov.org/epi3/epi_set/EPI_SET_20220606hk or https://doi.org/10.55876/gis8.220606hk.
-
•
The code used for pre-processing and analysis in this paper has been deposited to and made publicly available from the authors’ GitHub repository, https://github.com/EESI/covid_severity.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.