Abstract
We enrolled 7 individuals with recurrent symptoms or antigen test conversion following nirmatrelvir-ritonavir treatment. High viral loads (median 6.1 log10 copies/mL) were detected after rebound for a median of 17 days after initial diagnosis. Three had culturable virus for up to 16 days after initial diagnosis. No known resistance-associated mutations were identified.
Keywords: COVID-19, therapeutics, anti-virals, nirmatrelvir-ritonavir, virology
BACKGROUND
Nirmatrelvir-ritonavir, which inhibits the main viral protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been shown to reduce hospitalization in high-risk patients with early-stage, symptomatic coronavirus disease 2019 (COVID-19) infection [1]. The US Food and Drug Administration granted emergency use authorization (EUA) status for its use in December 2021, and it is currently a preferred therapy for ambulatory individuals with COVID-19 at high risk of severe disease [2]. As nirmatrelvir-ritonavir has entered into broad clinical use, reports have emerged of recurrent symptoms in a subset of treated individuals who had initial symptomatic improvement [3, 4]. However, the mechanism and viral characteristics of symptomatic relapse after nirmatrelvir-ritonavir therapy remain unclear. We sought to characterize the virology of rebound after nirmatrelvir-ritonavir with longitudinal sampling of individuals and testing of nasal swabs by viral load quantification, viral culture, and whole genome viral sequencing.
METHODS
Study Participants
The Post-vaccination Viral Characteristics Study (POSITIVES) cohort is a longitudinal study of individuals with COVID-19 infection that aims to characterize virologic and immunologic aspects of infection [5–7]. To characterize the virologic features of relapse after nirmatrelvir-ritonavir treatment, we selectively enrolled ambulatory individuals recently treated with five days of nirmatrelvir-ritonavir with recurrent symptoms after initial resolution or recurrent antigen test positivity after testing negative during or after their treatment course. Individuals in the Mass General Brigham health system were referred to our study team by healthcare providers and contacted by phone for informed consent at the time of symptom recurrence. We recorded information from participants and medical chart review about initial COVID-19 diagnostics and nirmatrelvir-ritonavir treatment course, home-based rapid antigen test results, and past medical history, including the presence of immunosuppressing conditions. Following enrollment, anterior nasal (AN) swabs were collected and placed into universal viral transport media (Becton Dickinson, Franklin Lakes, New Jersey, USA) 3 days per week until polymerase chain reaction (PCR) negativity. Swabs were simultaneously analyzed for viral RNA level by quantitative real-time polymerase chain reaction (qRT-PCR), viral whole genome sequencing, semiquantitative viral culture, and laboratory-based rapid antigen testing.
Viral Load Quantification
Viral transport media was centrifuged for 2 hours at 21 000 × g and 4°C to pellet virions. TRIzol-LS™ Reagent (ThermoFisher) was then added to the pellets, and samples were subsequently incubated on ice for 10 minutes. Chloroform (MilliporeSigma) was added to each sample, and the resulting mixtures were then vortexed and centrifuged for 15 minutes at 21 000 × g and 4°C. The clear aqueous layer was collected and concentrated using isopropanol precipitation. RNA was washed with cold 70% ethanol before being resuspended in DEPC-treated water (ThermoFisher). SARS-CoV-2 viral RNA was tested with a qPCR assay using the US Centers for Disease Control and Prevention (CDC) 2019 nCoV_N1 primer and probe set and quantified using a standard curve. Full details of assay development and validation have been described previously [8].
SARS-CoV-2 Culture
Viral culture was performed in the BSL3 laboratory of the Ragon Institute of Massachusetts General Hospital (MGH), Massachusetts Institute of Technology (MIT), and Harvard. Viral culture was assessed semi-quantitatively by median tissue culture infectious dose assay (TCID50) as previously reported [5, 7]. Briefly, aliquoted viral transport media were filtered with 0.45–0.65 µm centrifugal filters and added to Vero-E6 cells plated in DMEM culture media supplemented with 2% fetal bovine serum, HEPES, antibiotic-antimycotic solution and 5 µg/mL of polybrene. Each sample was added to the cells in 4 replicates and serially diluted 6 times in 5-fold increments in 96-well format. The infection was performed by spinfection for 1 hour at 2000 × g at 37°C. The cytopathic effect (CPE) was scored 7 days postinfection with light microscopy and TCID50/mL titers were calculated using the Reed-Muench method. The supernatant of wells showing CPE was harvested for RNA extraction and viral sequence confirmation.
SARS-CoV-2 Whole Genome Sequencing
We sequenced SARS-CoV-2 genomes using the Illumina COVIDSeq Test protocol as previously described [5]. We constructed sequencing libraries using the Illumina Nextera XT Library Prep Kit and sequenced them using an Illumina NextSeq 2000 instrument. Complete genomes (>24 000 assembled base pair length) were assigned a Pango lineage using pangolin v4.0.6. All samples were deposited to GenBank (Bioproject Accession PRJNA759255) and GISAID. Notable amino acid mutations and sequence quality were analyzed using Nextclade v1.14.1.
Antigen Testing Using Abbott BinaxNow SARS-CoV-2 Rapid Antigen Assay
In addition to rapid antigen tests conducted by participants prior to enrollment, we performed rapid antigen tests on study specimens. Frozen viral transport media aliquots were thawed on ice and 50 μL was transferred to a test tube. Swabs from BinaxNow antigen kits (Abbott, Chicago, Illinois, USA) were immersed into the liquid until fully absorbed and then tested according to manufacturer’s instructions as previously described [9]. After 15 minutes, results were interpreted as positive, negative, or discordant by a reader blinded to the viral load result.
Ethical Considerations
Study procedures were approved by the human subjects review committee at Mass General Brigham and all participants gave informed consent to participate.
RESULTS
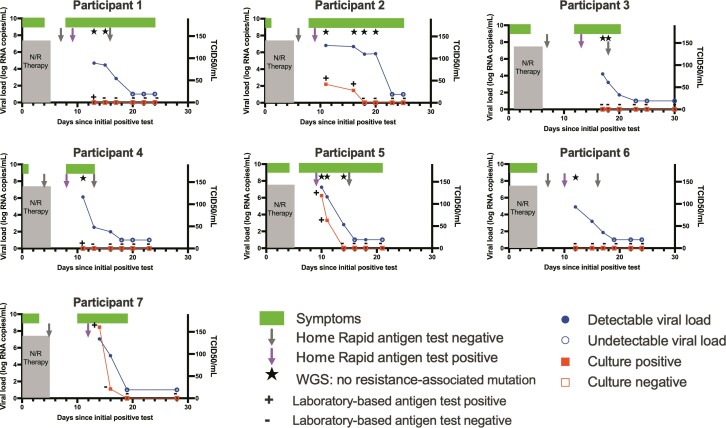
Participant demographic details are provided in Supplementary Table 1. All 7 participants were fully vaccinated and had received at least 1 booster dose. One of the 7 had an immunosuppressing condition and was on monthly intravenous immunoglobulin therapy. All 7 participants reported symptom improvement and conversion to negative home-based antigen testing following treatment with nirmatrelvir-ritonavir. Six of 7 had symptom recurrence, and 1 had repeat antigen test positivity during an asymptomatic screen. Symptoms recurred a median of 9 days after initial positive test or 4 days after completion of the nirmatrelvir-ritonavir course. At study enrollment, all 7 participants had a detectable viral load (median 6.1 log10 copies per mL [range 4.2–7.3]). A detectable viral load was identified for a median of 12 days (range 9–15) after completion of nirmatrelvir-ritonavir (Figure 1). Enrollment sample viral cultures were positive in 3 of 7 individuals. In these 3 individuals, cultures were positive until 5, 11, and 11 days after completion of the course of nirmatrelvir-ritonavir, respectively.
Figure 1.
Virologic and clinical course of individuals with rebound of COVID-19 following nirmatrelvir-ritonavir treatment. Abbreviations: COVID-19, coronavirus disease 2019; N/R, nirmatrelvir-ritonavir; WGS, whole genome sequencing.
We sequenced virus in 6 of the 7 participants after completion of therapy. We found no known resistance-associated mutations in nsp5 encoding the main SARS-CoV-2 protease (Supplementary Figure 1) or in any of the protease cleavage sites.
Finally, when comparing results of viral cultures and laboratory-based antigen testing of study specimens, we found high concordance between laboratory-based antigen and viral culture testing (92%, 24/26). Although there were two specimens that were antigen positive and culture negative, no specimens were antigen negative and culture positive.
DISCUSSION
We found that virologic rebound after nirmatrelvir-ritonavir therapy for early stage COVID-19 infection is associated with high viral load and, in a subset of individuals, culturable virus. We identified live virus up to 11 days after completion of nirmatrelvir-ritonavir therapy (16 days from the pretreatment PCR test). By contrast, we recently reported that untreated outpatients infected with the Omicron variant SARS-CoV-2 shed viable virus for a median of 5 days after an initial positive test [7]. These data reinforce the importance of testing and isolation guidelines for individuals with recurrent symptoms after nirmatrelvir-ritonavir treatment, irrespective of intermediate negative antigen testing or initial symptom resolution. Because live viral shedding can occur at the time of relapse, restarting monitoring and isolation from the time of relapse may be warranted.
Although field-based testing is needed to confirm our findings, we found high concordance between laboratory-based rapid antigen testing and culture positivity, with no specimens that were antigen negative and culture positive. Consequently, antigen test-based monitoring of individuals with relapse after therapy holds promise as a means of signaling a safe release from isolation.
Finally, we did not identify emergence of resistance-associated polymorphisms in any of the 6 specimens that were sequenced. Our findings add support to previous studies that drug resistance does not appear to be a significant contributor to relapse [3, 4, 10] and suggest that nirmatrelvir-ritonavir may retain activity in most cases of symptom recrudescence. Viral relapse in the absence of drug resistance suggests that a longer duration of therapy could be explored to help prevent this phenomenon.
Our study should be interpreted in the context of limitations. Most notably, these data were derived from a small case series so precise estimates of culture positivity, duration of viral shedding, or incidence of drug resistance cannot be made. We also enrolled individuals after clinical rebound and therefore cannot determine the incidence of this syndrome among individuals taking nirmatrelvir-ritonavir treatment. Finally, we use viral culture as a proxy measure of contagiousness but cannot quantify the risk of transmission for this patient population.
In summary, we found evidence of high viral load and, in some cases, culturable virus among individuals with recurrent clinical disease after nirmatrelvir-ritonavir therapy for COVID-19. Culturable virus was present for up to 2 weeks after completion of therapy. Consideration should be given to revising public health guidelines to specifically recommend repeat testing and isolation in these cases. Future work is needed to better understand the incidence, mechanisms, clinical significance, and public health consequences of symptomatic relapse after nirmatrelvir-ritonavir.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Julie Boucau, Ragon Institute of Massachusetts General Hospital (MGH), Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, Massachusetts, USA.
Rockib Uddin, Massachusetts General Hospital, Boston, Massachusetts, USA.
Caitlin Marino, Ragon Institute of Massachusetts General Hospital (MGH), Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, Massachusetts, USA.
James Regan, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
James P Flynn, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Manish C Choudhary, Brigham and Women’s Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Geoffrey Chen, Massachusetts General Hospital, Boston, Massachusetts, USA.
Ashley M Stuckwisch, Massachusetts General Hospital, Boston, Massachusetts, USA.
Josh Mathews, Massachusetts General Hospital, Boston, Massachusetts, USA.
May Y Liew, Massachusetts General Hospital, Boston, Massachusetts, USA.
Arshdeep Singh, Massachusetts General Hospital, Boston, Massachusetts, USA.
Zahra Reynolds, Massachusetts General Hospital, Boston, Massachusetts, USA.
Surabhi L Iyer, Massachusetts General Hospital, Boston, Massachusetts, USA.
Grace C Chamberlin, Massachusetts General Hospital, Boston, Massachusetts, USA.
Tammy D Vyas, Massachusetts General Hospital, Boston, Massachusetts, USA.
Jatin M Vyas, Massachusetts General Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Sarah E Turbett, Massachusetts General Hospital, Boston, Massachusetts, USA.
Jonathan Z Li, Brigham and Women’s Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Jacob E Lemieux, Massachusetts General Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA; Broad Institute, Cambridge, Massachusetts, USA.
Amy K Barczak, Ragon Institute of Massachusetts General Hospital (MGH), Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, Massachusetts, USA; Massachusetts General Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Mark J Siedner, Massachusetts General Hospital, Boston, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Notes
Acknowledgments. The authors would like to thank Vamsi Thiriveedhi, Ha Eun Cho, and Seamus Carroll for assistance with sequencing. They would also like to thank the participants for their time and participation, as well as the many physicians for referral of patients to our study.
Financial support. This study was supported by the Massachusetts Consortium for Pathogen Readiness (grants to J. Z. L., J. E. L., M. J. S., and A. K. B.) and the Massachusetts General Hospital Department of Medicine (grant to T. D. V.). The BSL3 laboratory where viral culture work was performed is supported by the Harvard Center for AIDS Research (CFAR) (US National Institutes of Health grant number P30 AI060354).
Potential conflicts of interest. J. E. L. reports grant to institution from the Centers for Disease Control and Prevention (CDC) outside of the submitted work; consulting fees from Sherlock Biosciences paid to individual; and honoraria paid to individual from Emerson Hospital and Virology Education. S. E. T. reports grants or contracts from the CDC for COVID-related work paid to the institution outside of the submitted work and travel support from the Duke Clinical Research Institute paid to self. All other authors report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hammond J, Leister-Tebbe H, Gardner A, et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med 2022; 386:1397–408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Therapeutic Management of Nonhospitalized Adults With COVID-19. Available at: https://www.covid19treatmentguidelines.nih.gov/management/clinical-management/nonhospitalized-adults--therapeutic-management/. Accessed 20 May 2022.
- 3. Carlin A, Clark A, Chaillon A, et al. Virologic and immunologic characterization of COVID-19 recrudescence after nirmatrelvir/ritonavir treatment. 2022; rs.3.rs:1662783. doi: 10.21203/rs.3.rs-1662783/v1. [DOI] [Google Scholar]
- 4. Charness M, Gupta K, Stack G, et al. Rapid relapse of symptomatic Omicron SARS-CoV-2 infection following early suppression with nirmatrelvir/ritonavir. 2022. Available at: 10.21203/rs.3.rs-1588371/v3. Accessed 3 June 2022. [DOI]
- 5. Siedner MJ, Boucau J, Gilbert RF, et al. Duration of viral shedding and culture positivity with post-vaccination SARS-CoV-2 Delta variant infections. JCI Insight 2021; 7:e155483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seaman MS, Siedner MJ, Boucau J, et al. Vaccine breakthrough infection with the SARS-CoV-2 Delta or Omicron (BA.1) Variant leads to distinct profiles of neutralizing antibody responses. medRxiv 2022. doi: 10.1101/2022.03.02.22271731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boucau J, Marino C, Regan J, et al. Duration of viable virus shedding in SARS-CoV-2 Omicron variant infection. medRxiv 2022. 2022.03.01.22271582. doi: 10.1101/2022.03.01.22271582. [DOI] [Google Scholar]
- 8. Fajnzylber J, Regan J, Coxen K et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Regan J, Flynn JP, Choudhary MC, et al. Detection of the omicron variant virus with the Abbott BinaxNow SARS-CoV-2 rapid antigen assay. Open Forum Infect Dis 2022; 9:ofac022. doi: 10.1093/ofid/ofac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fact Sheet For Healthcare Providers: Emergency Use Authorization for Paxlovid. Available at: https://www.fda.gov/media/155050/download. Accessed 20 May 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.