Abstract
Background
Immune dysregulation is a major factor in the development of severe coronavirus disease 2019 (COVID-19). The homeostatic chemokines CCL19 and CCL21 have been implicated as mediators of tissue inflammation, but data on their regulation in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is limited. We thus investigated the levels of these chemokines in COVID-19 patients.
Methods
Serial blood samples were obtained from patients hospitalized with COVID-19 (n = 414). Circulating CCL19 and CCL21 levels during hospitalization and 3-month follow-up were analyzed. In vitro assays and analysis of RNAseq data from public repositories were performed to further explore possible regulatory mechanisms.
Results
A consistent increase in circulating levels of CCL19 and CCL21 was observed, with high levels correlating with disease severity measures, including respiratory failure, need for intensive care, and 60-day all-cause mortality. High levels of CCL21 at admission were associated with persisting impairment of pulmonary function at the 3-month follow-up.
Conclusions
Our findings highlight CCL19 and CCL21 as markers of immune dysregulation in COVID-19. This may reflect aberrant regulation triggered by tissue inflammation, as observed in other chronic inflammatory and autoimmune conditions. Determination of the source and regulation of these chemokines and their effects on lung tissue is warranted to further clarify their role in COVID-19.
Clinical Trials Registration
NCT04321616 and NCT04381819.
Keywords: SARS-CoV-2, chemokine, predictive markers, respiratory distress syndrome, viral infection
Increased circulating levels of chemokines CCL19 and CCL21 were observed in patients hospitalized with COVID-19. High levels were associated with adverse outcomes, including respiratory failure, need for ICU support, and 60-day all-cause mortality. Additionally, high CCL21 on admission correlated with impairment of pulmonary function at 3-month outpatient follow-up.
It is increasingly apparent that the induction of overwhelming systemic inflammatory responses is associated with severe clinical manifestations and unfavorable outcomes in coronavirus disease 2019 (COVID-19). While early triggering of immune defense mechanisms is crucial for an effective elimination of viral particles in the initial stages of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [1, 2], persistent and dysregulated systemic inflammatory responses are detrimental [3, 4]. Although different forms of immune dysregulation involving both the innate and adaptive immune system have been reported [5–9], the drivers of the extensive and persistent immune activation in severe COVID-19 remain unclear.
Interference with chemokine responses forms the basis of effective immune evasion strategies employed by many viruses. Most studies have focused on inflammatory chemokines, which are recognized as integral parts of the inflammatory signaling cascades triggered by tissue injury and invading pathogens. In contrast, the homeostatic chemokines CCL19 and CCL21 are constitutively expressed in secondary lymphoid organs. There they promote the homing of T cells and dendritic cells (DCs) that express the common receptor CCR7 and the more recently discovered CCR10 [10]. In addition to this homeostatic function, elevated circulating levels of CCL19 and CCL21 are found in several acute and chronic inflammatory conditions, including autoimmune diseases, atherosclerosis, and various infections [11–14]. Moreover, these chemokines have effects on vascular smooth muscle cells and extracellular matrix remodeling [15], and are associated with pulmonary hypertension in systemic sclerosis [16]. These findings suggest a role of dysregulated CCL19 and CCL21 signaling in inflammatory pulmonary disorders. Studies on CCL19 and CCL21 in COVID-19 are limited, but the ectopic cellular expression of the ORF7a protein encoded by this virus was found to induce secretion of multiple chemokines, including CCL21, in cultured HeLa cells [17].
The present study aimed to investigate the association between CCL19/CCL21 and disease severity, 60-day total mortality, and pulmonary sequelae in COVID-19. CCL19 and CCL21 levels were analyzed in 2 Norwegian prospective cohort studies of hospitalized COVID-19 patients spanning 3 waves of the COVID-19 pandemic.
METHODS
Study Design and Participants
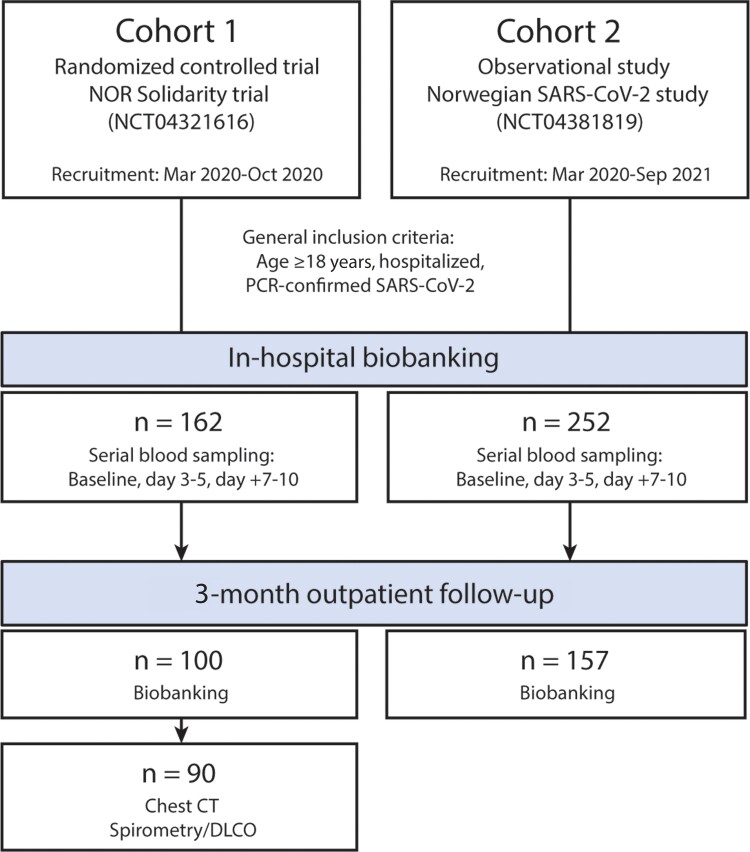
Data from 2 prospective cohort studies were pooled and assessed in our study. A flow chart of the study design is provided in Figure 1. Cohort 1 was the NOR Solidarity trial (NCT04321616), a multicenter, open-label, adaptive randomized controlled trial evaluating the effect of antiviral agents (hydroxychloroquine and remdesivir) in COVID-19 patients admitted to 23 Norwegian hospitals. Study interventions in this trial did not show significant effects on clinical outcomes or viral clearance [18, 19]. The study was approved by the Committee for Medical Research Ethics Region Southeast Norway (approval no. 118684) and the Norwegian Medicines Agency (20/04950-23). Cohort 2 was the Norwegian SARS-CoV-2 study (NCT04381819), an observational study of COVID-19 patients admitted to 5 Norwegian hospitals, conducted as part of an International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) WHO Clinical Characterization Protocol study [20]. The study was approved by the Regional Committees for Medical Research Ethics Southeast Norway (106624 and 2019/306). All patients aged ≥18 years admitted to the hospital with polymerase chain reaction (PCR)-confirmed SARS-2-CoV-2 infection were eligible for inclusion. Blood samples were obtained from each patient within 48 hours of admission and up to 10 days during hospitalization, as well as at 3-month follow-up in a subset of patients. All participants gave informed consent prior to inclusion, either directly or through a legally authorized representative.
Figure 1.
Flow chart showing the study population, with clinical data and blood samples collected from 2 cohort studies. Further details of the 2 trials are provided in the “Methods” section. Blood samples were collected at 3 time points during hospitalization, and at outpatient follow-up after 3 months. A subset of patients from cohort 1 also underwent pulmonary function assessment and chest CT imaging at follow-up. Abbreviations: CT, computed tomography; Dlco, diffusing capacity of the lungs for carbon monoxide; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Patients in cohort 1 were included from March to October 2020, while patients in cohort 2 were included from March 2020 to September 2021. The study period thus spanned the first 3 waves of the COVID-19 pandemic in Norway: 18 Mar 2020 to 23 July 2020 (wave 1), 24 July 2020 to 17 February 2021 (wave 2), and 18 February 2021 to 31 July 2021 (wave 3) [21]. From February 2021, Alpha SARS-CoV-2 was the dominating variant, which was superseded by the Delta variant in July 2021 [22, 23].
Intervention and Outcomes
In cohort 1 (n = 162), participants were randomized to either (1) local standard of care (SoC); (2) SoC plus oral hydroxychloroquine; or (3) SoC plus intravenous remdesivir as described [18]. Previous studies showed no effects of these treatment modalities [18]. Data from the intervention arms in cohort 1 were therefore pooled together with samples from cohort 2 (n = 252) to examine whether levels of CCL19 and CCL21 were associated with disease severity. Severe COVID-19 was defined as 1 or more of the following: (1) development of acute respiratory failure (RF) defined as Po2/FiO2 ratio <26.6 kPa (<200 mmHg) during hospitalization; (2) requirement for intensive care unit (ICU) support during hospitalization; and (3) 60-day postadmission mortality. In addition, for cohort 1, patients were evaluated 3 months after inclusion with pulmonary function testing as detailed below.
Blood Sampling Protocol and Biochemical Analyses
Plasma (cohort 1) or serum (cohort 2) was stored at −80°C, and thawed < 3 times. For reference, circulating CCL19 and CCL21 were also analyzed in plasma from 24 age- and sex-matched healthy controls (mean age 55 years [SD 12 years]; 55% men).
Plasma and serum levels of CCL19 and CCL21 were measured by enzyme immunoassays using commercially available antibodies (R&D Systems). Intra-/interassay coefficients of variation were <10%. Comparing within-patient differences in serum versus plasma in 16 healthy controls, we observed no difference for CCL19 (P = .80) or CCL21 (P = .18).
Three-Month Follow-Up
In total, 257 participants (cohort 1, n = 100; cohort 2, n = 157) attended outpatient follow-up that included blood sampling for routine clinical biochemistry and biobanking. The timing of this 3-month follow-up was different in the 2 cohorts, defined by inclusion date in cohort 1, and date of hospital discharge in cohort 2. In cohort 1, lung function testing (n = 90), consisting of spirometry, and diffusing capacity of the lungs (Dlco) was also performed [24]. The predicted percentage of Dlco and the lower limit of normal were calculated according to the Global Lung Function Initiative Network guidelines [24].
In Vitro SARS-CoV-2 Stimulation of Dendritic Cells
Human peripheral blood mononuclear cells (PBMC) were isolated from buffy coats of healthy donors, differentiated into DCs, and stimulated with inactivated SARS-CoV-2 as detailed in Supplementary material.
Statistics
Clinical characteristics of the participants were compared using Student t test or Mann-Whitney U test depending on variable distribution, or by χ2 for continuous and categorical variables, respectively (Table 1). CCL19 and CCL21 were transformed (log10) for temporal comparisons between groups by linear mixed model analysis. Subject was set as random effect, while time, RF, ICU admission, and 60-day mortality were set as fixed effects (independently and also as interactions). In addition, age, sex, estimated glomerular filtration rate and treatment modalities (study drug for cohort 1, and dexamethasone for cohort 2) were included as independent effects. Data are presented as back-transformed estimated marginal means with 95% confidence intervals (CI). Post hoc analysis (Sequential Sidak test) between groups is reported if the group or group* (ie, interaction term) was significant. Similar models were applied to evaluate the effects of randomized treatment and Dlco in cohort 1 and wave or dexamethasone use in cohort 2.
Table 1.
Baseline Characteristics and Outcomes in 414 Patients Hospitalized for COVID-19 in Norway, Stratified by 2 Large Multicenter Cohorts and Combined
| Parameter | Cohort 1 | Cohort 2 | Combined |
|---|---|---|---|
| n = 162 | n = 252 | n = 414 | |
| Age, y | 59.7 ± 15.4 | 57.0 ± 15.3 | 58.0 ± 15.4 |
| Male sex, No. (%) | 103 (64) | 159 (63) | 262 (63) |
| Body mass index, kg/m2 | 28.2 ± 4.6 | 28.8 ± 5.2 | 28.5 ± 4.9 |
| Treatment group, No. (%) | |||
| SoC | 81 (50) | 254 (100) | 333 (80) |
| SoC + hydroxychloroquine | 43 (27) | 0 (0) | 43 (10) |
| SoC + remdesivir | 38 (24) | 0 (0) | 38 (9) |
| Dexamethasone | 2 (1) | 134 (53)a | 136 (33) |
| Oxygen therapy | 91 (56) | 194 (77)a | 285 (69) |
| Comorbidities, No. (%) | |||
| Chronic cardiac disease | 24 (15) | 47 (19) | 71 (17) |
| Hypertension | 51 (32) | 84 (35) | 135 (34) |
| Chronic pulmonary disease | 31 (20) | 67 (27) | 98 (24) |
| Obesity | 43 (29) | 70 (28) | 113 (28) |
| Diabetes | 27 (17) | 58 (25) | 85 (22) |
| Current smoker | 5 (4) | 16 (7) | 21 (6) |
| Outcomes, No. (%) | |||
| ICU admission | 31 (19) | 79 (31)a | 110 (27) |
| Respiratory failure | 50 (31) | 75 (30) | 125 (31) |
| Deceased at 60 days | 8 (5) | 29 (12) | 37 (9) |
| Po2/FiO2 ratio at admission, kPa | 42.4 (32.4, 49.6) | 40.0 (28.1, 48.3) | 41.3 (30.0, 49.3) |
| Laboratory analysis at admission | |||
| Hemoglobin, g/dL | 13.2 ± 1.5 | 12.9 ± 1.8 | 13.0 ± 1.7 |
| C-reactive protein, mg/L | 70 (35, 136) | 53 (24, 117) | 62 (29, 125) |
| Ferritin, µg/L | 612 (358, 1111) | 617 (297, 1146) | 615 (322, 1127) |
| White blood cell count, × 109/L | 6.5 ± 2.8 | 6.9 ± 3.2 | 6.7 ± 3.1 |
| Neutrophils, × 109/L | 4.8 ± 2.7 | 5.3 ± 3.1 | 5.1 ± 3.0 |
| Lymphocytes, × 109/L | 1.2 ± 0.53 | 1.1 ± 0.5 | 1.1 ± 0.5 |
| eGFR, mL/min/1.73m2 | 87 ± 25 | 90 ± 29 | 89 ± 27 |
Continuous data are given as mean ± SD or median (25th, 75th) percentile.
Abbreviations: eGFR, estimated glomerular filtration rate; ICU, intensive care unit; SoC, standard of care.
P < .05 between cohorts 1 and 2.
Separate linear mixed models for CCL19 and CCL21, with time treated as a factor variable, were employed to model the association between Po2/FiO2 ratio (outcome) and circulating levels of CCL19 and CCL21. A random intercept by subject was used to control for repeated measures, with each subject having between 1 and 3 measured follow-up periods. Associations between admission levels of CCL19 and CCL21 (divided in tertiles) and 60-day mortality were assessed by Kaplan-Meier analysis and Cox regression.
Reanalysis of Public RNAseq Datasets
Publicly available RNASeq datasets were identified via a manual search of the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and details on their analyses are included in the Supplementary material.
RESULTS
Baseline Characteristics and Outcomes of the Study Population
A flow chart of the study design is given in Figure 1. Demographics and clinical characteristics of the cohorts were quite similar (Table 1), except for the use of antiviral agents in cohort 1, and dexamethasone use in cohort 2 after introduction as SoC in the management of severe COVID-19 (autumn 2020). More patients were admitted to ICU in cohort 2. In the combined cohort, 110 patients (27%) were admitted to ICU, and 125 (31%) patients developed RF the first 10 days after admission (Table 1). Sixty-six patients (22%) with RF did not receive treatment in an ICU.
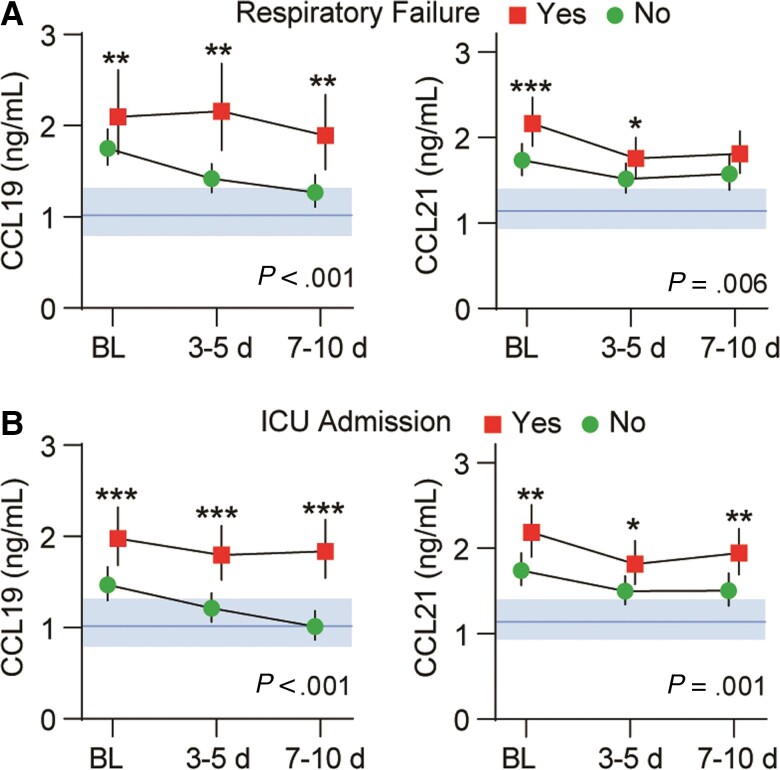
Initial Temporal Profile of Circulating CCL19 and CCL21 and Relation to Respiratory Failure and ICU Admission
As shown in Figure 2A, the levels of both markers were higher in COVID-19 patients than in controls, and significantly higher in RF patients compared to patients without RF. CCL19 remained high in RF patients throughout the observation period, while differences in CCL21 were larger at admission and decreased towards the end of observation. The group effect for higher CCL19 in RF patients within cohorts 1 and 2 separately was P = .051 and P = .002, respectively. Corresponding values for CCL21 were P = .022 and P = .020, respectively. Mixed models regression with Po2/FiO2 ratio as dependent and CCL19 or CCL21 and time as covariates revealed a negative correlation with CCL21 (estimate, −0.16; t = −6.1; P < .001), with a less robust association for CCL19 (estimate, −0.06; t = −2.3; P = .019).
Figure 2.
Intrahospital temporal profile of CCL19 and CCL21 in patients hospitalized with COVID-19 (n = 414) according to (A) respiratory failure (n = 125) or (B) ICU admission (n = 110) during the first 10 days after inclusion. Data is shown as estimated marginal means and 95% CI. The P values reflect the group (outcome) effect from the linear mixed models with subject as random effect, and time and respiratory failure or ICU admission as fixed effects (also as interaction) in addition to age, sex, estimated glomerular filtration rate, and treatment modalities. Shaded areas show reference value range from healthy controls. * P < .05, ** P < .01, *** P < .001 between groups. Abbreviations: BL, baseline; ICU, intensive care unit.
A similar pattern was observed for ICU admission (Figure 2B), with CCL19 and CCL21 remaining elevated throughout the observation period in those admitted to ICU. The overall group effect from the mixed models for higher CCL19 in patients admitted to ICU within cohorts 1 and 2 was P < .001 and P < .001, respectively. Corresponding values for CCL21 were P = .001 and P = .024, respectively.
The dynamics of oropharyngeal SARS-CoV-2 viral load in cohort 1 has previously been reported [18]. Baseline levels of SARS-CoV-2 in oropharynx did not correlate with baseline levels of CCL19 (r = −0.00; P = .94) or CCL21 (r = 0.13; P = .18).
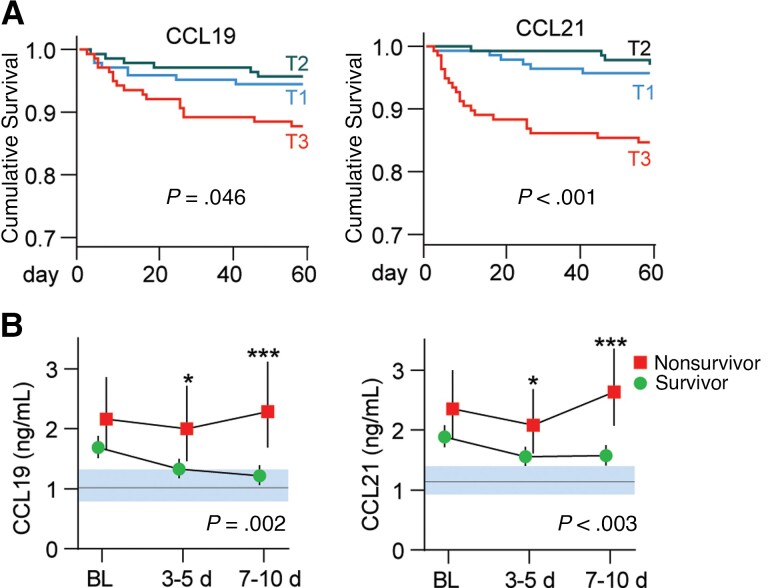
High CCL19 and CCL21 Levels Are Associated With 60-Day Mortality
In the combined cohort, 37 patients died within 60 days of hospital admission (Table 1). Kaplan-Meier analysis of admission levels showed that patients in the upper tertile of CCL19 and CCL21 were at increased risk of death within 60 days (Figure 3A). Evaluated as continuous variables, a 1 SD increase in CCL21 was associated with a 2.46 (95% CI, 1.76–3.42; P < .001) times higher risk of death, while the association with 60-day mortality was not significant for CCL19 (hazard ratio [HR], 1.27; 95% CI, .91–1.76; P = .15). For CCL21, the increased risk of death was present both in patients treated with (HR, 1.93; P = .004) and without dexamethasone (HR, 3.30; P = .001).
Figure 3.
CCL19 and CCL21 and 60-day mortality in patients hospitalized with COVID-19 (n = 414). A, Kaplan-Meier analysis 60-day mortality (n = 37) according to tertiles (T) of CCL19 (T1 ≤ 1.27 ng/mL, T2 1.28–2.09 ng/mL, T3 > 2.10 ng/mL) and CCL21 (T1 ≤ 1.41 ng/mL, T2 1.42–2.44 ng/mL, T3 > 2.45 ng/mL). B, Temporal profile of CCL19 and CCL21 during the first 10 days after inclusion according to 60-day mortality. Data in B is shown as estimated marginal means and 95% CI. The P values reflect the group (outcome) effect from the linear mixed models with subject as random effect, and time and mortality as fixed effects (also as interaction) in addition to age, sex, estimated glomerular filtration rate, and treatment modalities. Shaded areas show reference value range from healthy controls. * P < .05, ** P < .01, *** P < .001 between groups. Abbreviation: BL, baseline.
Evaluation of the temporal profile during the first 10 days after inclusion revealed that patients who died had higher levels of CCL19 and CCL21, with the largest differences at the end of the observation period (Figure 3B).
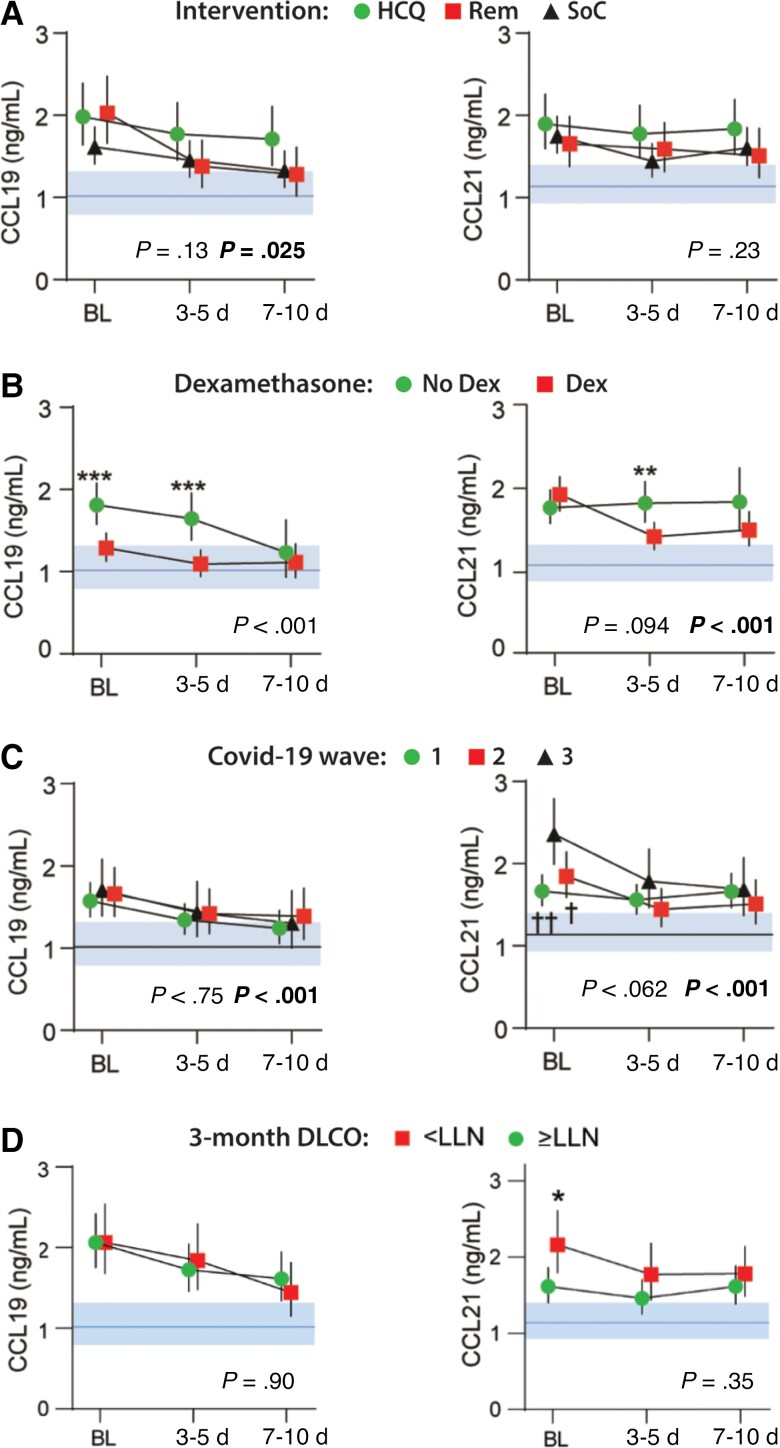
Intrahospital Temporal Profile of CCL19 and CCL21 in Relation to Treatment and COVID-19 Waves
As shown in Figure 4A, no temporal differences in CCL19 or CCL21 were observed according to treatment with hydroxychloroquine, remdesivir, or SoC within cohort 1 (Figure 4A). Dexamethasone use within cohort 2 (Figure 4B) was associated with lower levels of CCL19 and a decrease in CCL21 compared to patients who did not receive glucocorticoid treatment. No overall group effects were observed regarding COVID-19 waves and temporal profile of CCL19 or CCL21, but both chemokines showed an interaction between wave and time (P < .001; Figure 4C). For CCL21 the interaction was driven by higher levels at admission in patients hospitalized during the third wave of the pandemic.
Figure 4.
Intrahospital temporal profile of CCL19 and CCL21 according to (A) treatment with hydroxychloroquine (n = 43) and remdesivir (n = 38) as compared with their respective SoC (n = 81) in cohort 1 (NOR Solidarity trial); (B) dexamethasone treatment; (C) COVID-19 wave; and (D) Dlco below or above LLN at 3-month follow-up. Data is shown as estimated marginal means and 95% CI. The P value indicates group effect, and the bold P value indicates the interaction term between time and group from the linear mixed models with subject as random effect, and time and mortality as fixed effects (also as interaction) in addition to age, sex, and estimated glomerular filtration rate. Shaded areas show reference value range from healthy controls. *P < .01, ***P < .001 between groups; †P < .05, ††P < .01 versus wave 3. Abbreviations: Dlco, diffusing capacity of the lungs for carbon monoxide; LLN, lower limit of normal; HCQ, hydroxychloroquine; REM, remdesivir; SoC, standard of care.
A High Admission Level of CCL21 Is Associated With Impaired Lung Function After 3 Months
At 3-month follow-up, CCL19 and CCL21 levels in the 257 patients assayed were comparable to those of healthy controls (Supplementary Figure 1). However, for the 90 patients who performed pulmonary testing, a high baseline level of CCL21 during hospitalization was associated with impaired Dlco at 3-month follow-up (Figure 4D).
Effect of Inactivated SARS-CoV-2 on the Release of CCL21 and CCL19 in Dendritic Cells
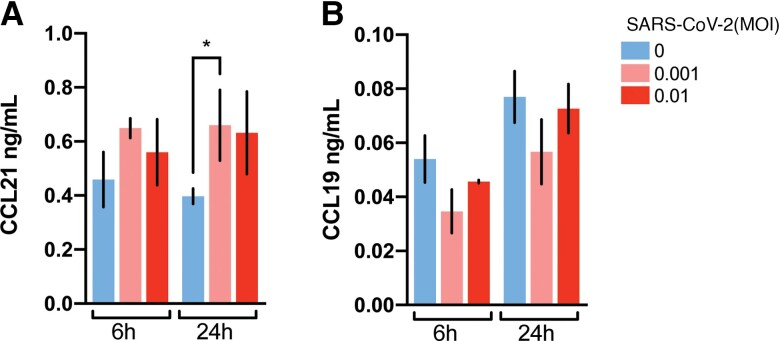
Previous data have suggested that SARS-CoV-2 may trigger immunomodulatory signaling responses in monocyte-derived cells [25]. To determine whether the presence of SARS-CoV-2 particles themselves could induce secretion of homoeostatic chemokines, in vitro differentiated monocyte-derived dendritic cells (moDC) were cultured in the presence of inactivated SARS-CoV-2 virus. Whereas there was a significant increase in the release of CCL21 in the presence of viral particles (Figure 5A), CCL19 secretion from moDC was low and was not significantly affected by the presence of SARS-CoV-2 (Figure 5B).
Figure 5.
In vitro secretion of homeostatic chemokines in SARS-CoV-2–exposed monocyte-derived dendritic cells. Quantitation of secreted CCL21 (A) and CCL19 (B) in cultures of monocyte-derived dendritic cells after exposure to inactivated SARS-CoV-2 viral particles (0.001 or 0.01 multiplicity of infection [MOI]) for 6 and 24 hours. Results are shown as mean ± SD (n = 3 per treatment condition). *P < .05, independent samples t test.
Reanalysis of CCR7/CCL19/CCL21 mRNA Expression From Public RNAseq Data Repositories
To obtain further insight into the regulation of homeostatic chemokines in COVID-19, we reanalyzed the expression of CCL19, CCL21, CCR7, and CCR10 in lung tissue and peripheral blood specimens isolated from COVID-19 patients in RNAseq datasets deposited in public repositories (Supplementary Table 1).
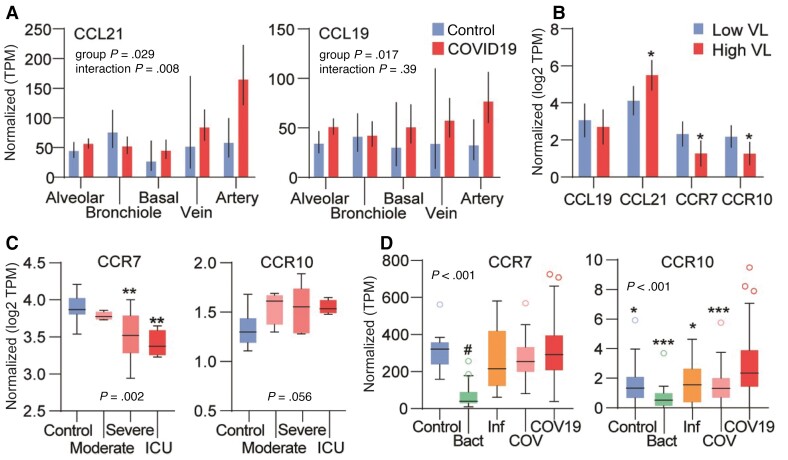
In the first autopsy study (L1) [26], CCL19 and CCL21 mRNA expression levels were modestly increased in lung tissue from COVID-19 patients compared to healthy controls, with particularly high CCL21 expression levels in biopsies containing arterial tissue (Figure 6A). The second autopsy study (L2) [27] showed higher CCL21 mRNA levels in the lungs of patients with high SARS CoV-2 viral load within the lung parenchyma, with lower expression of CCR7 and CCR10 in this subset (Figure 6B). A third autopsy study (L3) detected no differences in CCR7 and CCL21 expression levels compared to healthy lungs [28].
Figure 6.
Regulation of CCR7, CCR10, CCL19, and CCL21 in public RNAseq analysis data of tissues from COVID-19 patients. A, Differences in CCL19 and CCL21 mRNA expression in lung tissue from COVID-19 patients (n = 19) and controls (n = 3). Source data GSE163529. B, mRNA expression in relation to virus load (VL) in COVID-19 patients (GSE150316, n = 15). C, mRNA expression of CCR7 and CCR10 in peripheral blood mononuclear cells isolated from COVID-19 patients (n = 16) grouped by clinical disease severity (moderate, severe disease, and requiring ICU treatment) and age-/sex-matched healthy controls. Source dataset GSE152418. D, Whole blood leukocytes isolated from patients with COVID-19 (COV19), seasonal coronavirus infection (COV; n = 19), influenza (Inf; n = 17), bacterial pneumonia (Bact; n = 20) and matched healthy controls (n = 19). Source dataset GSE161731. Normalized gene expression quantified as transcripts per million (TPM). A and B, P values are from the group and group*tissue location effects from the mixed model analysis (see description of statistics in Supplementary material). C and D, P values are from the Kruskal-Wallis test with asterisks reflecting the results of the post hoc test. *P < .05, **P < .01, ***P < .001; #P < .01 versus other groups.
In studies comparing mRNA expression in PBMCs and whole blood isolated from patients and healthy controls, CCL19 and CCL21 were not detectable or showed very low mRNA counts (Supplementary Table 1, studies PB1-3, WB1–5). CCR7 in PBMCs was lower, while CCR10 tended to be higher in COVID-19 patients compared to controls in study PB1 (Figure 6C) [29]. In another PBMC study (PB2) [30], there was no change in CCR7 expression, while CCR10 was increased in COVID-19 (Supplementary Table 1). In the third PBMC study (PB3) [31] CCR7 expression levels were higher at recovery compared to earlier stages of the disease (Supplementary Table 1). In leukocytes from whole blood, CCR7 was lower in COVID-19 patients compared to controls in one study (WB1) [32] but showed no signs of differential regulation in 4 other studies (WB2–5) (Figure 6D) [33–36]. Two studies (WB3, WB5) [34, 36] reported increased CCR10 levels in COVID-19 patients compared to healthy controls, also compared to patients infected with seasonal coronavirus or with bacterial pneumonia (Figure 6D).
DISCUSSION
Combining data from 2 independent Norwegian multicenter cohorts, we here report high circulating levels of CCL19 and CCL21 on hospital admission and during the in-hospital course of 414 patients with COVID-19, with similar results within each cohort. High levels of both chemokines were associated with adverse outcomes, that is, the degree of RF, need for ICU support, and 60-day all-cause mortality. Finally, high CCL21 on admission correlated with persistent impairment of pulmonary function at 3-month follow-up. Our findings suggest the homeostatic chemokines, particularly CCL21, could be involved in the pathogenesis of COVID-19–related pulmonary pathology and might provide independent prognostic information in hospitalized COVID-19 patients.
High levels of inflammatory cytokines and chemokines and associations with poor outcomes have consistently been reported in hospitalized COVID-19 patients [37], but data on the homeostatic chemokines are scarce. In agreement with our findings, CCL21 was upregulated in patients with thrombotic complications and ranked third as a predictor of mortality amongst 71 cytokines/chemokines assayed [3]. Elevated CCL19 and CCL21 levels in COVID-19 patients could reflect several scenarios: (1) a general increase in homeostatic chemokine secretion in spleen/secondary lymphoid tissue, possibly accentuated by ongoing systemic inflammation or induced by viral antigens; (2) impaired chemokine clearance due to decreased turnover, for example, via CCR10 that has been found upregulated in PBMCs in COVID-19 patients [29, 30]; or (3) increased/ectopic secretion within nonlymphoid tissue (eg, lung). The lack of expression of CCL19 and CCL21 in PBMCs points to affected organs or associated lymphoid tissue as likely sources of increased circulating levels in our COVID-19 cohorts. Supporting a link between pulmonary CCL21 production and impaired lung function, a recent preprint report demonstrated the formation of perivascular foci with strong expression of CCL21 in lung tissue from patients with severe COVID-19 [38]. These areas showed high expression of fibrosis-associated markers, and accumulation of immune cell aggregates with features consistent with tertiary lymphoid structures. CCL19 expression was also elevated in these areas [38].
In agreement with a report showing an early rise in CCL21 [3], peak levels of CCL19 and CCL21 in our study were observed in samples collected within the first 48 hours of hospital admission, suggesting early perturbations in homeostatic chemokines during infection, possibly related to delayed viral clearance or high viral load. Interestingly, in the patients who died within 60 days, the largest difference in CCL21 levels compared with those who survived was seen at the last blood sample taken 7 to 10 days after admission, indicating persistent and increasing CCL21 activation in these patients. Reanalysis of publicly available gene expression datasets revealed indications of differential regulation of CCL21 expression in pulmonary tissue, while CCR7 and CCR10 receptor levels appeared unchanged. In the material from Desai et al [27], higher CCL21 mRNA levels were seen in lungs of patients with high SARS CoV-2 viral load within the lung parenchyma. While we in the present study found no correlation between circulating CCL19 and CCL21 levels and SARS-CoV-2 viral load in oropharyngeal/nasal samples at hospital admission, the upper airway viral load does not necessarily reflect levels of ongoing viral shedding within the lung parenchyma, given considerable topographical differences in viral shedding [39]. Hence, the possibility of a direct role of SARS-CoV-2 in eliciting CCL19 and CCL21 signaling remains unresolved.
Strong, systemic immune responses manifested by increased inflammatory cytokine and chemokine levels are commonly observed in severe cases of acute viral diseases. In severe dengue fever, a secreted form of the viral nonstructural antigen NS1 potentially triggers key aspects of the pathogenesis of severe disease manifestations (dengue hemorrhagic fever) [40]. By triggering leukocyte inflammatory responses and interfering with the integrity of endothelial glycocalyx, NS1 released by infected cells may explain both the endothelial dysfunction and the systemic inflammatory response observed in severe dengue [41]. Understanding of the impact of SARS-CoV-2 antigens on host cells is still evolving, but in vitro studies have identified a multitude of interactions of potential relevance to clinical disease manifestations [42], including disruption of endothelial barrier function [43] and aberrations in immune signaling [44, 45]. Conceivably, persistence or high levels of particular viral antigens in affected tissues could directly impact the pathophysiology of severe disease manifestation in COVID-19. Of particular relevance to the present work, the ORF7a protein of SARS-CoV-2 has been reported to induce the expression of CCL19 and CCL21 in HeLa cells [17]. In our experiments, PBMC-derived DCs exposed to inactivated SARS-CoV-2 viral particles produced significant amounts of CCL21 but not CCL19. Transcriptional changes involving alterations in cytokine release have been observed in monocytes/macrophages exposed to SARS-CoV-2 particles [25]. Although the physiological relevance of these findings remains uncertain, the SARS-CoV-2 virus may trigger distinct inflammatory signaling in monocyte-derived cells that potentially could impact the ensuing immune response. Moreover, our findings may suggest that CCL21 could be directly induced by SARS-CoV-2 and not only be a secondary phenomenon to a general state of systemic inflammation, potentially contributing to the formation of lymphoid and inflammatory tissue within the lungs.
Current knowledge about the direct impact of dexamethasone treatment on the CCR7/CCL19/CCL21 axis is limited. Herein we found that dexamethasone use within cohort 2 was associated with lower levels of CCL19 and CCL21 as compared with nonusers. However, the study was not designed to evaluate the effects of dexamethasone on these markers, and the data should be interpreted with caution. Regarding hydroxychloroquine/remdesivir, we found no effects on CCL19/CCL21 levels. There are no published data to suggest a direct impact of these agents on homeostatic chemokine signaling, although such effects cannot be excluded. It is possible that the inclusion of more targeted immunomodulatory agents in COVID-19 treatment will provide more clarity regarding CCL19/CCL21 regulation. Levels of these chemokines have, to the best of our knowledge, not been reported in published results from clinical trials. The Janus kinase (JAK) pathway inhibitor baricitinib has been approved for use in patients hospitalized with severe COVID-19. Other JAK inhibitors have been shown to inhibit CCR7/CCL19-mediated migration of DCs [46, 47]. It would therefore be of interest to examine if JAK inhibitors such as baricitinib affect the levels and/or function of CCL19/CCL21 in COVID-19.
The present study has some limitations. As mentioned above, the present study was not designed to evaluate the effects of therapeutics used in COVID-19 management, and the impact of such interventions on chemokine levels therefore cannot be excluded. The regulation of secretion of CCL19 and CCL21 under inflammatory conditions is largely unknown. Moreover, the cellular source(s) of CCl19/CCL21 in severe COVID-19 are still uncertain.
In summary, we report a striking increase in the circulating levels of the homeostatic chemokines CCL19 and CCL21 in hospitalized COVID-19 patients, with high levels correlating with progression to severe pulmonary disease, need for ICU support, and 60-day mortality. Furthermore, high admission levels of CCL21 were associated with prolonged impaired pulmonary function 3 months after hospital discharge. Given the key role of these chemokines in lymphoid tissue homeostasis and regulation of adaptive immune responses, potentially promoting lymphoid tissue within the SARS-CoV-2 infected lungs, these findings warrant further investigations to determine the drivers, source, and functional impact of increased levels of CCL19 and CCL21 in COVID-19.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Anders Tveita, Department of Internal Medicine, Bærum Hospital, Vestre Viken Hospital Trust, Gjettum, Norway; Division of Laboratory Medicine, Department of Immunology, Oslo University Hospital, Oslo, Norway.
Sarah Louise Murphy, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway.
Jan Cato Holter, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Anders Benjamin Kildal, Department of Anesthesiology and Intensive Care, University Hospital of North Norway, Tromsø, Norway.
Annika E Michelsen, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway.
Tøri Vigeland Lerum, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Pulmonary Medicine, Oslo University Hospital Ullevål, Oslo, Norway.
Mari Kaarbø, Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Lars Heggelund, Department of Internal Medicine, Drammen Hospital, Vestre Viken Hospital Trust, Drammen, Norway; Department of Clinical Science, Faculty of Medicine, University of Bergen, Bergen, Norway.
Aleksander Rygh Holten, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Acute Medicine, Oslo University Hospital, Oslo, Norway.
Ane-Kristine Finbråten, Department of Medicine, Lovisenberg Diaconal Hospital, Oslo, Norway.
Karl Erik Müller, Department of Internal Medicine, Drammen Hospital, Vestre Viken Hospital Trust, Drammen, Norway.
Alexander Mathiessen, Department of Medicine, Diakonhjemmet Hospital, Oslo, Norway.
Simen Bøe, Department of Anesthesiology and Intensive Care, Hammerfest County Hospital, Hammerfest, Norway.
Børre Fevang, Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway; Section of Clinical Immunology and Infectious Diseases, Oslo University Hospital, Oslo, Norway.
Beathe Kiland Granerud, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Kristian Tonby, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Infectious Diseases, Oslo University Hospital Ullevål, Oslo, Norway.
Andreas Lind, Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Susanne Gjeruldsen Dudman, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Katerina Nezvalova Henriksen, Department of Hematology, Oslo University Hospital, Oslo, Norway; Hospital Pharmacies, South-Eastern Norway Enterprise, Oslo, Norway.
Fredrik Müller, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Microbiology, Oslo University Hospital, Oslo, Norway.
Ole Henning Skjønsberg, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Pulmonary Medicine, Oslo University Hospital Ullevål, Oslo, Norway.
Marius Trøseid, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway; Section of Clinical Immunology and Infectious Diseases, Oslo University Hospital, Oslo, Norway.
Andreas Barratt-Due, Division of Laboratory Medicine, Department of Immunology, Oslo University Hospital, Oslo, Norway; Department of Anesthesia and Intensive Care Medicine, Oslo University Hospital, Oslo, Norway.
Anne Ma Dyrhol-Riise, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Infectious Diseases, Oslo University Hospital Ullevål, Oslo, Norway.
Pål Aukrust, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway; Section of Clinical Immunology and Infectious Diseases, Oslo University Hospital, Oslo, Norway.
Bente Halvorsen, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway.
Tuva Børresdatter Dahl, Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway; Department of Acute Medicine, Oslo University Hospital, Oslo, Norway.
Thor Ueland, Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Research Institute of Internal Medicine, Oslo University Hospital Rikshospitalet, Oslo, Norway; Department of Clinical Medicine, Thrombosis Research and Expertise Center, Arctic University of Norway, Tromsø, Norway.
NOR-SOLIDARITY Consortium and the Norwegian SARS-CoV-2 Study Group Investigators:
Cathrine Austad, Mette Bogen, Anne Hermann, Hanne Opsand, Trude Steinsvik, Bjørn Martin Woll, Erik Egeland Christensen, Kristin Eftestøl, Liv Hesstvedt, Synne Jenum, Marthe Jøntvedt Jørgensen, Elisabeth Toverud Landaas, Sarah Nur, Vidar Ormaasen, Frank Olav Pettersen, Else Quist-Paulsen, Dag Henrik Reikvam, Kjerstin Røstad, Linda Skeie, Anne Katrine Steffensen, Birgitte Stiksrud, Berit Gravrok, Vegard Skogen, Garth Daryl Tylden, Jan Terje Andersen, Anette Kolderup, Trine Kåsine, Fridtjof Lund-Johansen, Inge Christoffer Olsen, Karoline Hansen Skåra, Trung Tran, Cathrine Fladeby, Liv Hesstvedt, Mona Holberg-Petersen, Synne Jenum, Simreen Kaur Johal, Dag Henrik Reikvam, Kjerstin Røstad, Anne Katrine Steffensen, Birgitte Stiksrud, Eline Brenno Vaage, Erik Egeland Christensen, Marthe Jøntvedt Jørgensen, Sarah Nur, Vidar Ormaasen, Frank Olav Pettersen, Saad Aballi, Jorunn Brynhildsen, Waleed Ghanima, Anne Marie Halstensen, Åse Berg, Bjørn Blomberg, Reidar Kvåle, Nina Langeland, Kristin Greve Isdahl Mohn, Olav Dalgard, Ragnhild Eiken, Richard Alexander Molvik, Carl Magnus Ystrøm, Gernot Ernst, Lars Thoresen, Lise Tuset Gustad, Lars Mølgaard Saxhaug, Nina Vibeche Skei, Raisa Hannula, Mette Haugli, Roy Bjørkholt Olsen, Hedda Hoel, Dag Arne Lihaug Hoff, Asgeir Johannessen, Bjørn Åsheim-Hansen, Bård Reikvam Kittang, Lan Ai Kieu Le, Ravinea Manotheepan, Lena Bugge Nordberg, Hans Schmidt Rasmussen, Grethe-Elisabeth Stenvik, Ruth Foseide Thorkildsen, Leif Erik Vinge, Pawel Mielnik, Vegard Skogen, Hilde Skudal, and Birgitte Tholin
Notes
Study group members. The Norwegian SARS-CoV-2 study group investigators are: Cathrine Austad, Mette Bogen, Anne Hermann, Hanne Opsand, Trude Steinsvik, Bjørn Martin Woll (Vestre Viken Hospital Trust); Erik Egeland Christensen, Kristin Eftestøl, Liv Hesstvedt, Synne Jenum, Marthe Jøntvedt Jørgensen, Elisabeth Toverud Landaas, Sarah Nur, Vidar Ormaasen, Frank Olav Pettersen, Else Quist-Paulsen, Dag Henrik Reikvam, Kjerstin Røstad, Linda Skeie, Anne Katrine Steffensen, Birgitte Stiksrud (Oslo University Hospital); Berit Gravrok, Vegard Skogen, Garth Daryl Tylden (University Hospital of North Norway). The NOR-SOLIDARITY consortium members are: Jan Terje Andersen, Anette Kolderup, Trine Kåsine, Fridtjof Lund-Johansen, Inge Christoffer Olsen, Karoline Hansen Skåra, Trung Tran, Cathrine Fladeby, Liv Hesstvedt, Mona Holberg-Petersen, Synne Jenum, Simreen Kaur Johal, Dag Henrik Reikvam, Kjerstin Røstad, Anne Katrine Steffensen, Birgitte Stiksrud, Eline Brenno Vaage, Erik Egeland Christensen, Marthe Jøntvedt Jørgensen, Sarah Nur, Vidar Ormaasen, Frank Olav Pettersen (Oslo University Hospital); Saad Aballi, Jorunn Brynhildsen, Waleed Ghanima, Anne Marie Halstensen (Østfold Hospital Trust); Åse Berg (Stavanger University Hospital); Bjørn Blomberg, Reidar Kvåle, Nina Langeland, Kristin Greve Isdahl Mohn (Haukeland University Hospital); Olav Dalgard (Akershus University Hospital); Ragnhild Eiken, Richard Alexander Molvik, Carl Magnus Ystrøm (Innlandet Hospital Trust); Gernot Ernst, Lars Thoresen (Vestre Viken Hospital Trust); Lise Tuset Gustad, Lars Mølgaard Saxhaug, Nina Vibeche Skei (Nord-Trøndelag Hospital Trust); Raisa Hannula (Trondheim University Hospital); Mette Haugli, Roy Bjørkholt Olsen (Sørlandet Hospital Trust); Hedda Hoel (Lovisenberg Diaconal Hospital); Dag Arne Lihaug Hoff (Ålesund Hospital); Asgeir Johannessen, Bjørn Åsheim-Hansen (Vestfold Hospital Trust); Bård Reikvam Kittang (Haraldsplass Deaconess Hospital); Lan Ai Kieu Le (Haugesund Hospital); Ravinea Manotheepan, Lena Bugge Nordberg, Hans Schmidt Rasmussen, Grethe-Elisabeth Stenvik, Ruth Foseide Thorkildsen, Leif Erik Vinge (Diakonhjemmet Hospital); Pawel Mielnik (Førde Hospital); Vegard Skogen (University Hospital of North Norway); Hilde Skudal (Telemark Hospital Trust); Birgitte Tholin (Molde Hospital).
Author contributions. T. U., T. B. D., B. H., S. L. M., P. A., and A. T. were responsible for the study conception and execution of the present substudy. A. B. D., A. M. D. R., K. N. H., A. M., A. K. F., P. A., and M. T. were responsible for the management, coordination, research activity planning, and execution of the NOR-Solidarity trial. J. C. H., A. M. D. R., L. H., A. B. K., A. T., A. L., K. T., A. R. H., K. E. M., S. G. D., B. F., F. M., and S. B. were responsible for the management, coordination, research activity, planning, and execution of the Norwegian SARS-CoV-2 study. T. V. L. and O. H. S. were responsible for the 3-month follow-up protocol for pulmonary function testing. A. M. D. R., T. B. D., A. B. D., B. H., P. A., B. K. G., and J. C. H. coordinated the collection and storage of the biobank material. T. B. D., T. U., A. E. M., M. K., A. L., S. L. M., and B. H. were responsible for the biochemical analyses and in vitro studies. All authors revised and approved the final version of the manuscript.
Disclaimer. The funders had no role in the study design, data collection, analysis, or decision to publish this article.
Financial support. This work was supported by the National Program of Clinical Therapy Research in the Specialist Health Services, Norway (grant number 2020201 to NOR-SOLIDARITY trial); Oslo University Hospital; the Research Council of Norway (grant number 312780 to Norwegian SARS-CoV-2 study); a philanthropic donation from Vivaldi Invest A/S, owned by Jon Stephenson von Tetzchner (to Norwegian SARS-CoV-2 study); the South-Eastern Norway Regional Health Authority (grant number 2021071); and the European Union Horizon 2020 Research and Innovation Program (grant number 71029 to European Virus Archive GLOBAL project). Funding to pay the Open Access publication charges for this article was provided by the University of Oslo.
Data availability
Anonymized patient-level data for participants in this study, statistical analysis plan, and statistical coding can be made available after the approval of the institutional review board. Requests should be made to the corresponding author.
References
- 1. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020; 369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Masood KI, Yameen M, Ashraf J, et al. Upregulated type I interferon responses in asymptomatic COVID-19 infection are associated with improved clinical outcome. Sci Rep 2021; 11:22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lucas C, Wong P, Klein J, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020; 584:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 2020; 27:992–1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020; 130:2620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 2020; 11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol 2020; 20:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moderbacher C R, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adamo S, Chevrier S, Cervia C, et al. Profound dysregulation of T cell homeostasis and function in patients with severe COVID-19. Allergy 2021; 76:2866–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller G, Hopken UE, Lipp M. The impact of CCR7 and CXCR5 on lymphoid organ development and systemic immunity. Immunol Rev 2003; 195:117–35. [DOI] [PubMed] [Google Scholar]
- 11. Damas JK, Smith C, Oie E, et al. Enhanced expression of the homeostatic chemokines CCL19 and CCL21 in clinical and experimental atherosclerosis: possible pathogenic role in plaque destabilization. Arterioscler Thromb Vasc Biol 2007; 27:614–20. [DOI] [PubMed] [Google Scholar]
- 12. Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM II, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum 2011; 63:914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuwabara T, Ishikawa F, Yasuda T, et al. CCR7 ligands are required for development of experimental autoimmune encephalomyelitis through generating IL-23-dependent Th17 cells. J Immunol 2009; 183:2513–21. [DOI] [PubMed] [Google Scholar]
- 14. Schreiber T, Ehlers S, Aly S, et al. Selectin ligand-independent priming and maintenance of T cell immunity during airborne tuberculosis. J Immunol 2006; 176:1131–40. [DOI] [PubMed] [Google Scholar]
- 15. Halvorsen B, Dahl TB, Smedbakken LM, et al. Increased levels of CCR7 ligands in carotid atherosclerosis: different effects in macrophages and smooth muscle cells. Cardiovasc Res 2014; 102:148–56. [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann-Vold AM, Hesselstrand R, Fretheim H, et al. CCL21 as a potential serum biomarker for pulmonary arterial hypertension in systemic sclerosis. Arthritis Rheumatol 2018; 70:1644–53. [DOI] [PubMed] [Google Scholar]
- 17. Su CM, Wang L, Yoo D. Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci Rep 2021; 11:13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barratt-Due A, Olsen IC, Nezvalova-Henriksen K, et al. Evaluation of the effects of remdesivir and hydroxychloroquine on viral clearance in COVID-19: a randomized trial. Ann Intern Med 2021; 174:1261–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. WHO Solidarity Trial Consortium, Pan H, Peto R, et al. Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. N Engl J Med 2021; 384:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. ISARIC Clinical Characterisation Group, Garcia-Gallo E, Merson L, et al. ISARIC-COVID-19 dataset: a prospective, standardized, global dataset of patients hospitalized with COVID-19. Sci Data 2022; 9:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Helsedirektoratet . Covid-19—antall innlagte pasienter på sykehus.https://www.helsedirektoratet.no/statistikk/antall-innlagte-pasienter-pa-sykehus-med-pavist-covid-19. Accessed 15 March 2022.
- 22. Veneti L, Valcarcel Salamanca B, Seppala E, et al. No difference in risk of hospitalization between reported cases of the SARS-CoV-2 Delta variant and Alpha variant in Norway. Int J Infect Dis 2022; 115:178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lind A, Barlinn R, Landaas ET, et al. Rapid SARS-CoV-2 variant monitoring using PCR confirmed by whole genome sequencing in a high-volume diagnostic laboratory. J Clin Virol 2021; 141:104906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lerum TV, Maltzahn NN, Aukrust P, et al. Persistent pulmonary pathology after COVID-19 is associated with high viral load, weak antibody response, and high levels of matrix metalloproteinase-9. Sci Rep 2021; 11:23205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boumaza A, Gay L, Mezouar S, et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J Infect Dis 2021; 224:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delorey TM, Ziegler CGK, Heimberg G, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021; 595:107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desai N, Neyaz A, Szabolcs A, et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun 2020; 11:6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun 2020; 11:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arunachalam PS, Wimmers F, Mok CKP, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020; 369:1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill SE, Dos Santos CC, O’Gorman DB, et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med Exp 2020; 8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng HY, Xu M, Yang CX, et al. Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct Target Ther 2020; 5:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thair SA, He YD, Hasin-Brumshtein Y, et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 2021; 24:101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Overmyer KA, Shishkova E, Miller IJ, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst 2021; 12:23–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McClain MT, Constantine FJ, Henao R, et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat Commun 2021; 12:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ng DL, Granados AC, Santos YA, et al. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci Adv 2021; 7:eabe5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan YH, Fong SW, Poh CM, et al. Asymptomatic COVID-19: disease tolerance with efficient anti-viral immunity against SARS-CoV-2. EMBO Mol Med 2021; 13:e14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020; 26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mothes R, Pascual-Reguant A, Koehler R, et al. Local CCL18 and CCL21 expand lung fibrovascular niches and recruit lymphocytes, leading to tertiary lymphoid structure formation in prolonged COVID-19. medRxiv [preprint], 27 March 2022. doi: 10.1101/2022.03.24.22272768. [DOI] [Google Scholar]
- 39. Sulaiman I, Chung M, Angel L, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol 2021; 6:1245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glasner DR, Puerta-Guardo H, Beatty PR, Harris E. The good, the bad, and the shocking: the multiple roles of dengue virus nonstructural protein 1 in protection and pathogenesis. Annu Rev Virol 2018; 5:227–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glasner DR, Ratnasiri K, Puerta-Guardo H, Espinosa DA, Beatty PR, Harris E. Dengue virus NS1 cytokine-independent vascular leak is dependent on endothelial glycocalyx components. PLoS Pathog 2017; 13:e1006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar A, Prasoon P, Kumari C, et al. SARS-CoV-2-specific virulence factors in COVID-19. J Med Virol 2021; 93:1343–50. [DOI] [PubMed] [Google Scholar]
- 43. Rauti R, Shahoha M, Leichtmann-Bardoogo Y, et al. Effect of SARS-CoV-2 proteins on vascular permeability. Elife 2021; 10:e69314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konno Y, Kimura I, Uriu K, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 2020; 32:108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li JY, Liao CH, Wang Q, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 2020; 286:198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heine A, Held SA, Daecke SN, et al. The JAK-inhibitor ruxolitinib impairs dendritic cell function in vitro and in vivo. Blood 2013; 122:1192–202. [DOI] [PubMed] [Google Scholar]
- 47. Rivas-Caicedo A, Soldevila G, Fortoul TI, Castell-Rodriguez A, Flores-Romo L, Garcia-Zepeda EA. Jak3 is involved in dendritic cell maturation and CCR7-dependent migration. PLoS One 2009; 4:e7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized patient-level data for participants in this study, statistical analysis plan, and statistical coding can be made available after the approval of the institutional review board. Requests should be made to the corresponding author.