Abstract
The N-terminal domain of the OmpA protein from Escherichia coli, consisting of 170 amino acid residues, is embedded in the outer membrane, in the form of an antiparallel β-barrel whose eight transmembrane β-strands are connected by three short periplasmic turns and four relatively large surface-exposed hydrophilic loops. This protein domain serves as a paradigm for the study of membrane assembly of integral β-structured membrane proteins. In order to dissect the structural and functional roles of the surface-exposed loops, they were shortened separately and in all possible combinations. All 16 loop deletion mutants assembled into the outer membrane with high efficiency and adopted the wild-type membrane topology. This systematic approach proves the absence of topogenic signals (e.g., in the form of loop sizes or charge distributions) in these loops. The shortening of surface-exposed loops did not reduce the thermal stability of the protein. However, none of the mutant proteins, with the exception of the variant with the fourth loop shortened, served as a receptor for the OmpA-specific bacteriophage K3. Furthermore, all loops were necessary for the OmpA protein to function in the stabilization of mating aggregates during F conjugation. An OmpA deletion variant with all four loops shortened, consisting of only 135 amino acid residues, constitutes the smallest β-structured integral membrane protein known to date. These results represent a further step toward the development of artificial outer membrane proteins.
The two-domain outer membrane protein OmpA of Escherichia coli is a well-established model for the study of membrane assembly in vivo (8, 17, 18) as well as in vitro (16, 40). The three-dimensional structure of its N-terminal membrane-embedded domain, which assembles into the outer membrane as efficiently as the full-length protein (36), has been solved at atomic resolution (31). This 170-residue protein domain resides in the outer membrane in the form of an eight-stranded antiparallel β-barrel (Fig. 1), a finding that is reminiscent of the porins which are also built up from antiparallel β-strands that are connected by relatively large and hydrophilic surface-exposed loops and short periplasmic turns (5, 44). However, the truncated OmpA derivative has the advantage over the porins and the full-length OmpA of being a monomeric one-domain protein and of having a much smaller membrane moiety. For these reasons, this OmpA domain was chosen as a paradigm for the study of membrane assembly of integral β-structured membrane proteins. Systematic studies demonstrated that circularly permuted variants (20) and even split variants (21) of this protein domain could assemble into the outer membrane with high efficiency. Recently, the role of individual transmembrane β-strands was probed by a randomization mutagenesis approach (22). In this context, a systematic study of the role of the large surface-exposed loops was now performed.
FIG. 1.
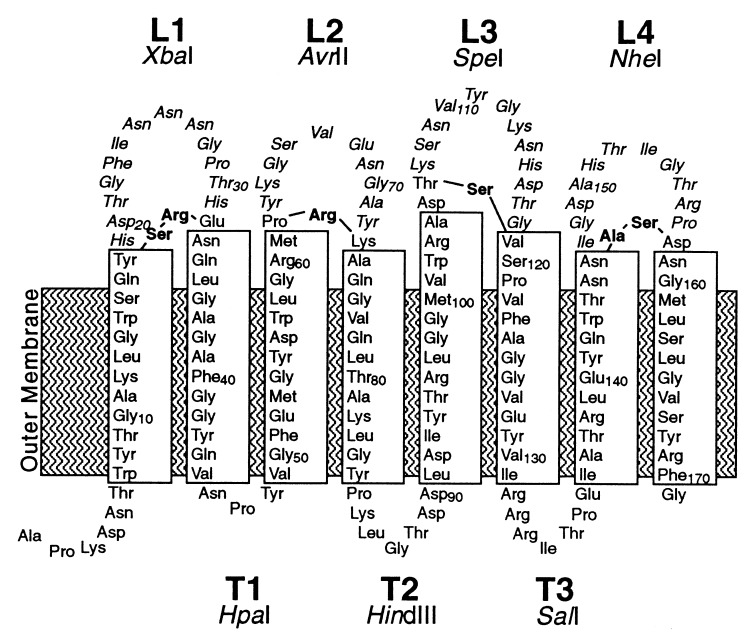
Two-dimensional model of the arrangement of the N-terminal β-barrel domain of OmpA in the outer membrane, based on the prediction by Vogel and Jähnig (42). Transmembrane β-strands are boxed. The surface-exposed loops and periplasmic turns have been labeled L1 to L4 and T1 to T3, respectively. Amino acid residues are numbered according to their position in the wild-type sequence. In this study, an OmpA variant with two modifications at its periplasmic turns has been used. At the second turn, Ile-87 was replaced by Lys-Leu-Gly. At turn T3, the peptide Arg-Arg-Arg-Ile was introduced between Ile-131 and Thr-132, and Ala-130 was converted to Val (Materials and Methods and Table 1). Residues that have been removed upon loop deletion mutagenesis are shown in italics; amino acids that have been introduced in their places are shown in boldface. Unique restriction sites which are associated with alterations at the encoded loops and turns are indicated.
Among the surface-exposed loops, some are very important for the function of outer membrane proteins. They are involved in the recognition of many ligands, e.g., small-molecule nutrients such as iron-siderophore complexes or sugars (13, 15), toxic agents such as bacteriophages or colicins (3, 14, 30), and probably eukaryotic target cells for bacterial pathogens (2, 24, 33). However, it is an open question whether these surface-exposed loops also play an important structural role, e.g., by determining the membrane topology of the polypeptide chain. The conservation of their relatively large and hydrophilic character is at the very least striking. This is surprising insofar as they must somehow cross the hydrophobic lipid bilayer. Whereas charge distributions of the extramembranous regions are crucial for the membrane topology of cytoplasmic membrane proteins (10, 27), it is unknown whether the extramembranous regions of outer membrane proteins play a similar role. Although previous studies in which surface-exposed loops were shortened (1, 11, 15, 35) suggested that these loops do not contain topogenic information, they did not rule out the existence of possibly redundant topogenic signals in the remaining loops. Never had all loops been shortened simultaneously, and a LamB mutant with three shortened loops still contained more than half of its surface-exposed residues (15). Also, charge distributions were never exchanged between surface-exposed and periplasmic protein segments. Here, I shortened all four surface-exposed loops of OmpA separately and in all possible combinations (thus removing 83% of all surface-exposed loop residues) and compared the 16 resulting OmpA variants with respect to structure and function.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophages.
If not indicated otherwise, an ompA mutant of strain UH300 (not synthesizing the protein [18]) was used. Synthesis of full-length OmpA mutants, resulting from read-through of an amber stop codon (see below), was achieved in the ompA supF strain UH203 (7). Some protease digestion experiments were performed with the deep-rough mutant BB12 of E. coli B (34). Strains XL1-Blue, carrying a Tn10-tagged F′ plasmid specifying resistance to tetracycline (Stratagene, La Jolla, Calif.), and SOAR1 (a spontaneously isolated ompA derivative of the streptomycin-resistant strain UL4Sm [19, 23]) served as donor and recipient in F-conjugation experiments. For labeling experiments, strains were transformed with plasmid pGP1-2 (41), which allows temperature-induced expression of the T7 RNA polymerase and, hence, of genes under the control of a T7-specific promoter.
All plasmids encoding OmpA loop deletion variants were derived from the pBluescript KS(+) (Stratagene) derivative pKSO171, which carries a promoterless ompA allele with an amber stop codon corresponding to amino acid residue 172 of the mature OmpA protein downstream of a T7-specific promoter (36). In order to facilitate the easy combination of individual loop deletion mutations, the ompA allele of plasmid pKSO171 was modified to contain unique restriction sites at the regions encoding the three periplasmic turns: an HpaI site at the first turn, a HindIII site at the second turn, and a SalI site at the third turn (Fig. 1). The resulting plasmid, pKSO171ΔHdΔSal-HS, was constructed as follows. (i) A plasmid harboring an ompA gene variant containing four SalI linkers at the region corresponding to the third turn (Table 1, turn T3S4) (36) was digested with SalI and religated, resulting in a variant with only one SalI linker insertion (Table 1, turn T3S1). It should be noted that during the original SalI linker insertion mutagenesis codon 130 (GCG) was changed to GTG, thus substituting Val-130 for Ala (Table 1, turn T3B0) (36). (ii) This single SalI linker insertion was combined with a HindIII linker insertion at the region corresponding to the second turn (ompA-T2-1.5) (36). DNA sequencing revealed that the published amino acid sequence listed two nonexisting residues; the corrected sequence is given in Table 1 (turn T2H1). (iii) Both linker insertions were then transferred into a pKSO171 derivative whose HindIII and SalI sites of the multiple cloning site had been destroyed before by filling in and religation. Notably, amino acids Asn-46–Pro-47 of the first turn are encoded by a unique HpaI site in the wild-type ompA allele.
TABLE 1.
Nucleotide and derived amino acid sequences of the regions corresponding to periplasmic turns and surface-exposed loops
| OmpA turn and loop variants (reference) | DNA and derived amino acid sequencesa | Relevant restriction site(s) |
|---|---|---|
| Periplasmic turns | ||
| T2, wild type | Gly Tyr Pro Ile Thr | BstEII |
| 84 85 86 87 88 | ||
| GGT TAC CCA ATC ACT | ||
| T2H1 (36) | Gly Tyr Pro Lys Leu Gly Thr | HindIII |
| 84 85 86 88 | ||
| GGT TAC CCC AAG CTT GGG ACT | ||
| T2M1 (this study) | Gly Tyr Pro Lys Leu Arg Arg Thr Arg Ala Ser Gln Leu Gly Thr | BsmBI, MluI, BsmBI |
| 84 85 86 88 | ||
| GGT TAC CCC AAG CTG AGA CGC ACG CGT GCG TCT CAG CTT GGG ACT | ||
| T2S0 (this study) | Gly Tyr Pro Gly Thr | SmaI |
| 84 85 86 88 | ||
| GGT TAC CCC GGG ACT | ||
| T3, wild type | Ala Ile Thr | None |
| 130 131 132 | ||
| GCG ATC ACT | ||
| T3B0 (36) | Val Ile Thr | BclI |
| 131 132 | ||
| GTG ATC ACT | ||
| T3S4 (36) | Val Ile Arg Arg Arg Val Asp Ala Ser Thr Arg Arg Arg Ile Thr | SalI (4 times) |
| 131 132 | ||
| GTG ATC CGT CGA CGC GTC GAC GCG TCG ACG CGT CGA CGG ATC ACT | ||
| T3S1 (this study) | Val Ile Arg Arg Arg Ile Thr | SalI |
| 131 132 | ||
| GTG ATC CGT CGA CGG ATC ACT | ||
| T3B1 (this study) | Val Ile Arg Arg Ala Gly Cys Ala Gln Pro Ala Arg Arg Ile Thr | BspMI, BssHII, BspMI |
| 131 132 | ||
| GTG ATC GCT CGA GCA GGT TGC GCG CAA CCT GCT CGA CGG ATC ACT | ||
| Surface-exposed loops | ||
| ΔL1 (this study) | Tyr Ser Arg Glu Asn | XbaI |
| 18 32 33 | ||
| TAC TCT AGA GAA AAC | ||
| ΔL2 (this study) | Met Pro Arg Lys Ala | AvrII |
| 61 62 73 74 | ||
| ATG CCT AGG AAA GCT | ||
| ΔL3 (this study) | Ala Asp Thr Ser Val | SpeI |
| 104 105 106 119 | ||
| GCA GAC ACT AGT GTT | ||
| ΔL4 (this study) | Asn Ala Ser Asp Asn | NheI |
| 146 158 159 | ||
| AAC GCT AGC GAC AAC |
Introduced linker sequences of turn variants and introduced nucleotides of loop deletion mutants are shown in boldface; relevant restriction sites are underlined. Wild-type amino acid residues are numbered according to the mature wild-type protein.
Bacteriophage K3 is a T-even-type OmpA-specific coliphage (30).
OmpA loop deletion mutagenesis.
To perform OmpA loop deletion mutagenesis, a PCR-based megaprimer method was employed (38). The primers (purchased from MWG Biotech, Ebersberg, Germany) were about 60 nucleotides in length and had at least 20 complementary nucleotides for first-round amplification (25 cycles, Pfu polymerase [Stratagene]) and at least 30 complementary nucleotides for second-round amplification (32 cycles, Taq polymerase). After cloning of the PCR-amplified DNA and restriction fragment exchange to minimize the probability of second-site mutations, all loop mutations were confirmed by DNA sequencing. The resulting sequences are listed in Table 1. All 16 possible combinations of different OmpA loop deletion mutations were constructed by using the three unique restriction sites at the regions corresponding to the periplasmic turns (see above), which separate the four surface-exposed loops.
Excision linker insertion mutagenesis.
Excision linkers (29) introduce some extra base pairs (e.g., codons) which, in coding sequences, will lead to the addition of some extra amino acid residues in the encoded protein. Second, these linkers and the adjacent regions in the target sequence can be removed by digestion with a type IIS restriction endonuclease, thus creating small deletions. To obtain linker insertions at the regions corresponding to the last two periplasmic turns, plasmids were digested with HindIII or SalI, respectively. For annealing, self-complementary nonphosphorylated oligonucleotides with compatible overhangs were dissolved in 10 mM Tris-HCl (pH 8.5), boiled for 10 min, and cooled to room temperature over a period of 20 min. Oligonucleotides T2-ex (5′-AGCTGAGACGCACGCGTGCGTCTC) and T3-ex (5′-TCGAGCAGGTTGCGCGCAACCTGC) were used for enlargement of the second and third turns, respectively (T2M1 and T3B1 [Table 1]). Fifteen picomoles of the oligonucleotides was ligated with 0.15 pmol of linearized plasmid DNA. Prior to transformation of competent cells, the ligation mixture was digested with HindIII or SalI, respectively, boiled for 10 min, and cooled to 4°C over a period of 30 min. This procedure should lead to the dissociation of the noncovalently joined strand of the ligated double-stranded linker DNA, followed by reannealing of the covalently joined strands of the linkers at both termini of the plasmid and concomitant ring closure. Clones with the desired linker insertions were screened by restriction analysis, making use of the facts that linker insertion results in the removal of the original plasmid-encoded site and that the introduction of more than one linker would lead to the introduction of an additional restriction site at the boundaries of two adjacent linkers (T2-ex, PvuII; T3-ex, XhoI).
Radioactive labeling of proteins and protease digestion experiments.
Radioactive labeling of OmpA variants, conversion of cells to spheroplasts, and protease digestion experiments have been performed essentially as previously described by Koebnik (21). Strains carrying plasmid pGP1-2 (41) served as a host. If not indicated otherwise, cells were subjected to protease digestion (0.1 mg of protease per ml) for 15 min at 30°C. Thermal stability of proteins was estimated as described previously (21). Proteins were analyzed by sodium dodecyl sulfate (SDS)–16% polyacrylamide gel electrophoresis (PAGE). Gels were autoradiographed, and protein bands were quantitated with a laser densitometer (UltroScan XL; Pharmacia LKB, Bromma, Sweden). Peaks were integrated with the Pharmacia LKB GelScan XL 2.1 program.
F conjugation and phage sensitivity assays.
F conjugation in liquid medium (L broth) was carried out as described elsewhere (26). In brief, exponentially growing donor and recipient cells were mixed at a ratio of 1:10 and were incubated for 30 min with shaking at 37°C. Then, mixtures were cooled on ice and vortexed for 10 s, and dilutions were plated on selective medium. Both streptomycin (20 μg/ml) and ampicillin (80 μg/ml) counterselected against the donor; tetracycline (16 μg/ml) counterselected against the recipient.
Phage receptor activity of OmpA variants was assayed as described previously (20).
RESULTS AND DISCUSSION
Construction and expression of OmpA loop deletion mutants.
For simultaneous replacement of all surface-exposed loops of the OmpA β-barrel domain by much shorter loops, a tailor-made ompA allele with unique restriction sites at the DNA regions corresponding to the three periplasmic turns was constructed (Materials and Methods). This procedure resulted in the introduction of two and four additional amino acids at the second and third turns (turns T2H1 and T3S1 [Table 1]), respectively, and this wild-type-like OmpA variant was dubbed OmpA-T2H1-T3S1.
The design of loop deletion mutations was based on the available topological model (42) and guided by the following three considerations. First, wild-type surface loops, which were predicted to be 12 to 14 residues long, should be replaced by new loops, consisting of only three, preferably turn-promoting, amino acids (12). Second, all four new loops should be encoded by DNA sequences that contain unique and compatible restriction endonuclease recognition sites in the same reading frame, thus giving the possibility of using the new plasmids as versatile surface exposure expression vectors. Third, these four cleavage sites should be arranged such that the original adjacent residues were kept whenever possible. According to these criteria, four OmpA loop deletion mutants were constructed (Materials and Methods); the resulting mutations are summarized in Fig. 1 and Table 1. These single deletions were then combined in all possible combinations, such that a series of 16 ompA alleles was obtained, encoding OmpA variants with none (i.e., wild type) to all four surface-exposed loops shortened. The deletion variants were designated by their shortened loops, i.e., OmpAΔL234 means that loops L2, L3, and L4 were shortened while loop L1 was retained.
By use of the bacteriophage T7 RNA polymerase-promoter system (41), the deletion variants were labeled radioactively. SDS-PAGE demonstrated that all 16 OmpA variants were expressed and processed with efficiency similar to that of the wild-type loop variant (data not shown and Table 2). No degradation products accumulated during the 15-min chase, indicating their stability in vivo. Furthermore, all 16 OmpA variants were heat modifiable (examples are given in Fig. 2 and 3), a behavior that corresponds to that of wild-type OmpA, thus strongly suggesting their folding into a β-structure conformation and incorporation into the outer membrane (39). It should be noted that the C-terminally truncated OmpA behaves in a manner opposite to that of the full-length protein with respect to its heat modifiability, i.e., the heat-denatured molecules migrate faster than the nondenatured molecules (36). Hence, all 16 OmpA loop deletion derivatives with different combinations of wild-type and shortened surface-exposed loops were stably expressed and efficiently assembled into the outer membrane of E. coli, regardless of the presence or absence of large hydrophilic extramembranous loops.
TABLE 2.
Molecular masses and subtilisin susceptibilities of OmpA loop deletion variants in spheroplasts
| OmpA variant | Molecular mass (kDa) | Digestion classa | Mapped cleavage siteb | Efficiency of digestion (%)c |
|---|---|---|---|---|
| Wild-type loops | ||||
| OmpA-T2H1-T3S1 | 19.5 | NC | ||
| One loop shortened | ||||
| OmpAΔL1 | 18.4 | NC | ||
| OmpAΔL2 | 18.6 | I | Loop L1 | 20 |
| OmpAΔL3 | 18.3 | I | Loop L1 | 30 |
| OmpAΔL4 | 18.6 | I | Loop L1 | 35 |
| Two loops shortened | ||||
| OmpAΔL12 | 17.4 | NC | ||
| OmpAΔL13 | 17.1 | II | Loop L4 | 20 |
| OmpAΔL14 | 17.4 | III | Loop L3 | 20 |
| OmpAΔL23 | 17.4 | I | Loop L1 | 45 |
| OmpAΔL24 | 17.6 | I | Loop L1 | 70 |
| OmpAΔL34 | 17.3 | I | Loop L1 | 70 |
| Three loops shortened | ||||
| OmpAΔL123 | 16.2 | II | Loop L4 | 25 |
| OmpAΔL124 | 16.5 | III | Loop L3 | 20 |
| OmpAΔL134 | 16.2 | NC | ||
| OmpAΔL234 | 16.4 | I | Loop L1 | 75 |
| Four loops shortened | ||||
| OmpAΔL1234 | 15.3 | NC |
According to the size differences between the undigested OmpA variants and their largest cleavage products upon proteolytic digestion (see also Fig. 2 and 3), three digestion classes could be identified.
See the text for the mapping of cleavage sites.
The amounts of cleaved molecules after digestion for 15 min at 30°C are shown. NC, no cleavage observed.
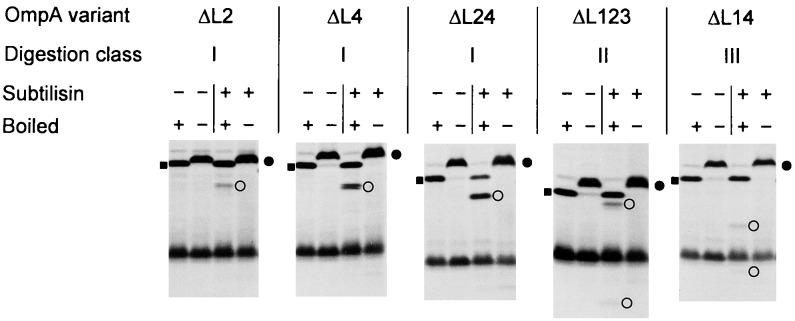
FIG. 2.
SDS-PAGE analysis of OmpA loop deletion variants after subtilisin digestion. After conversion of cells to spheroplasts, radioactively labeled and chased variants were subjected to proteolytic digestion. Heat-modifiable forms are marked by solid circles, and undigested forms that had been boiled prior to the electrophoresis for 10 min are indicated by solid squares. OmpA-related cleavage products are indicated by open circles.
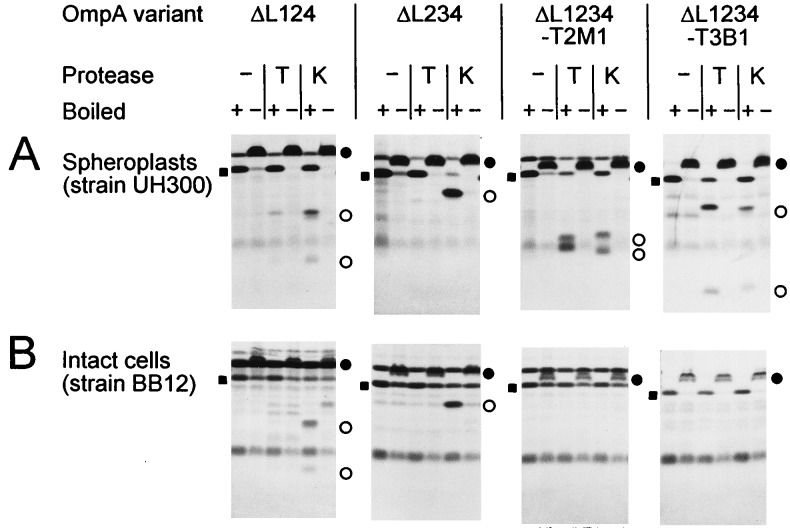
FIG. 3.
SDS-PAGE analysis of OmpA loop deletion variants after proteolytic digestion of spheroplasts (A) or intact cells (B), as described in the legend to Fig. 2. Cells of strain BB12 were digested for 30 min. −, no protease added; T, trypsin added; K, proteinase K added. For definitions of symbols, see the legend to Fig. 2.
Membrane topology of OmpA loop deletion mutants.
In order to study the membrane topology of OmpA loop deletion variants, protease protection experiments were performed. In general, surface-exposed loops of outer membrane proteins are expected to be folded tightly in order to prevent their proteolytic digestion by proteases that are present in their natural environment. If some parts of the molecule are misfolded, they should become totally degraded by proteases. In contrast, when a protein is only locally distorted then it is expected to be cleaved only in that region of the molecule (1, 4, 19, 36). After radioactive labeling of the OmpA derivatives, cells were converted to spheroplasts and subjected to subtilisin digestion. SDS-PAGE demonstrated that the heat-modifiable forms of most loop deletion variants were cleaved to one or two distinct smaller species (Table 2; examples are given in Fig. 2). However, the fact that these fragments were recovered intact indicates that they were protected from further degradation by being in the bilayer.
Comparative proteolytic digestion of membrane-assembled molecules in spheroplasts versus intact cells gives information about the orientation of a protein in the membrane. Sites that are cleaved in both cases are located at the cell surface, whereas exclusive cleavage in spheroplasts indicates that these sites are probably located on the periplasmic side. Digestion of intact cells synthesizing variants OmpAΔL123, OmpAΔL124, OmpAΔL134, and OmpAΔL234 led to the same cleavage patterns as were observed upon digestion of spheroplasts (e.g., ΔL124 and ΔL234 [Fig. 3]), provided that the deep-rough mutant strain BB12 was used for expression, thus demonstrating the surface location of the cleavage sites. Interestingly, use of a standard K-12 strain did not yield any cleavage (data not shown). Obviously, only after destroying the interactions between protein and lipopolysaccharide molecules either by treatment with EDTA (25), which is used during conversion of cells to spheroplasts, or by truncating the lipopolysaccharide molecules genetically (e.g., in strain BB12) do proteases gain access to potential cleavage sites.
The apparent molecular masses of the cleavage products were estimated by SDS-PAGE. This way, three digestion classes, I, II, and III, could be identified (Table 2), and the corresponding cleavage sites could be mapped to one of the remaining wild-type loops at about amino acid positions Asp-20 (loop L1), Asn-159 (loop L4), and Lys-107 (loop L3), respectively (numbering is according to the wild-type OmpA sequence). Class I comprised all variants with wild-type loop L1 and a deletion in loop L2, L3, or L4 (OmpAΔL2, OmpAΔL3, OmpAΔL4, OmpAΔL23, OmpAΔ24, OmpAΔL34, and OmpAΔL234). In these cases, about 2.3 kDa was removed by the subtilisin protease. Proteins of class II were cleaved into two visible (labeled) fragments, a very small one and a fragment which was only slightly reduced in size (by about 1.3 kDa); this class included all variants with an intact loop L4 and deletions in loops L1 and L3 (OmpAΔL13 and OmpAΔL123). Finally, class III, whose members were cleaved into a small fragment and a fragment which was about 6.5 kDa smaller than the undigested molecules, comprised all variants with a retained loop L3 and deletions in loops L1 and L4 (OmpAΔL14 and OmpAΔL124). In all cases where two cleavage products were observed, their sizes added up to the size of the undigested molecules. Apparently, cleavage at these loops resulted from lack of protection by the now-truncated neighboring loops.
In this context, another point of interest concerns the different protease sensitivities among the loop deletion variants. Under the conditions used for subtilisin digestion of spheroplasts, reactions were not complete (Table 2). Obviously, proteases can act better not only when the interaction with lipopolysaccharide molecules is weakened but also when the network of intramolecular interactions is destroyed by shortening of more than one loop. Proteins of digestion class I illustrate this point very well (Fig. 2). In this series, variants OmpAΔL234, OmpAΔL34, and OmpAΔL24 were most efficiently digested, followed by intermediately digested variants OmpAΔL23, OmpAΔL4, and OmpAΔL3 and rather slowly digested variant OmpAΔL2. This means that a protease cleavage site in loop L1 was unmasked by the shortening of one neighboring loop (ΔL2 or ΔL4) and that this cleavage site was further unmasked by simultaneous shortening of both loops (ΔL24).
Mapping of class I cleavage to the first loop was also consistent with the detection of only one fragment since the extreme N-terminal sequence did not contain a methionine residue that could have been labeled. In addition to subtilisin, trypsin and proteinase K were tested, leading to essentially the same results (examples are given in Fig. 3). However, the first loop, which does not contain any potential trypsin cleavage site, could not be cleaved by trypsin, thus giving further support for the notion that cleavage by subtilisin or proteinase K occurred in this region (e.g., OmpAΔL234 [Fig. 3]).
As additional controls, derivatives with small diagnostic peptide insertions at two of the periplasmic turns in an OmpAΔL1234 surface loop background were constructed by the introduction of excision linkers. The resulting sequences at the second (turn T2M1) and third (turn T3B1) turns are listed in Table 1. The same linker was also introduced at the third periplasmic turn of the wild-type-loop OmpA variant (OmpA-T2H1-T3B1). That such an addition of 10 or 12 extra residues to turns T2 and T3 can lead to cleavage at these sites upon proteolytic digestion of spheroplasts has been shown previously (36). When these linker insertion variants were digested, three different proteases (trypsin, proteinase K, and subtilisin) gave essentially the same results (Fig. 3). Whereas with spheroplasts proteolytic cleavage at the insertion sites was observed, these insertions were fully protected from cleavage during digestion of intact cells, even in a deep-rough mutant strain (Fig. 3), thus indicating that the diagnostic peptides were indeed exposed at the periplasmic face of the outer membrane. By converting the excision linker insertion at the second turn to a small deletion with the type IIS restriction endonuclease BsmBI, an even smaller variant consisting of only 135 amino acid residues was constructed (OmpAΔL1234-T2S0 [Table 1]). As judged by its heat modifiability and protease protection, this variant was also efficiently assembled into the outer membrane (data not shown). This is the smallest β-structured membrane protein known to date.
In summary, comparative proteolytic digestion demonstrated that all deletion variants assembled with wild-type membrane topology and orientation. Thus, since the periplasmic turns also do not possess topogenic information (21, 36), this information must therefore be entirely encoded in the transmembrane β-strands. This finding is at variance with proteins of the cytoplasmic membrane whose topology is, according to the positive-inside rule (43), mainly governed by the adjacent charges next to the membrane-spanning segments. Wild-type OmpA has a net charge of −3 at its periplasmic turns and of +1 at its surface-exposed loops. At the other extreme, variants OmpAΔL34 and OmpAΔL134 have an opposite charge distribution (+1 at the periplasmic turns and −1 at the surface-exposed loops); yet these variants assembled in a wild-type manner. Therefore, the membrane topology of outer membrane proteins is determined by another, as yet unknown mechanism.
Thermal stability of OmpA loop deletion mutants.
For comparison of the thermal stabilities of OmpA loop deletion variants, the half-life of the conversion of the heat-modifiable form to the completely denatured form at 72°C was estimated (Fig. 4). For this purpose, full-length variants of the OmpA derivatives (including the C-terminal periplasmic domain) were synthesized in an amber mutant strain, thus suppressing the stop codon at amino acid position 172 (36).
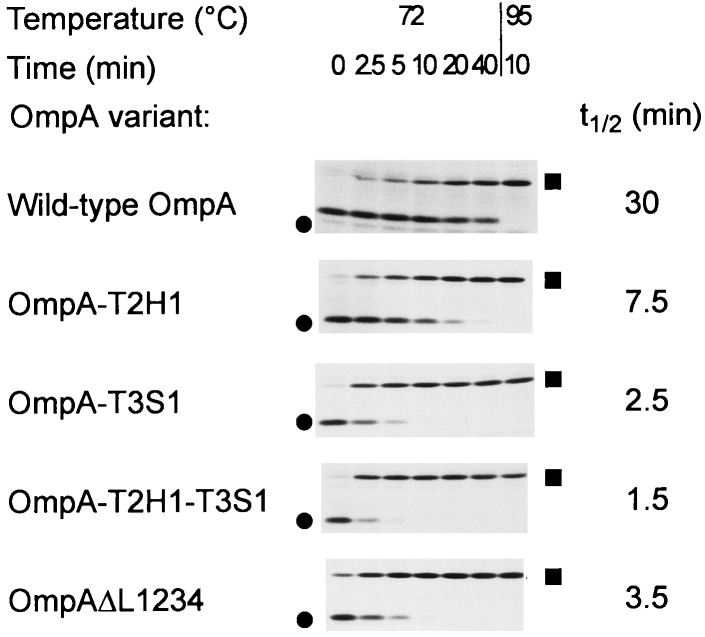
FIG. 4.
SDS-PAGE analysis of thermal stability of OmpA loop deletion variants. OmpA mutants were radioactively labeled in the supF strain UH203 and chased for 30 min. Proteins were solubilized in SDS-PAGE sample buffer containing 2% (wt/vol) SDS and were incubated at 72°C for the time indicated prior to electrophoresis. Heat-modifiable forms are marked by circles, and denatured forms are marked by squares.
Whereas wild-type OmpA had a half-life of conversion of about 30 min, all analyzed mutants of the loop deletion series (OmpAΔL1, OmpAΔL2, OmpAΔL3, OmpAΔL4, and OmpAΔL1234) as well as the wild-type-like OmpA-T2H1-T3S1 variant with all four surface loops retained had a half-life of 5 min or less (e.g., OmpAΔL1234 [Fig. 4]). The only differences between the authentic wild-type protein and the wild-type-like variant of the deletion series are found at the last two periplasmic turns. In order to dissect the roles of both turns, variants with alterations at only the second (OmpA-T2H1) or third (OmpA-T3S1) turn were constructed in an otherwise wild-type surface loop background. Their stabilities were intermediate, and both changes combined (OmpA-T2H1-T3S1) led to the least stable variant (OmpA-T2H1-T3S1 [Fig. 4]). Direct comparison of pairs with or without all surface-exposed loops in the same turn background revealed that the loop deletion variants were even slightly more stable (OmpAΔL1234 versus OmpA-T2H1-T3S1 [Fig. 4]; OmpAΔL1234-T3B1 versus OmpA-T2H1-T3B1). Hence, the thermal stabilities of all loop variants were very similar, and yet they were significantly reduced in comparison to that of the wild-type protein, an observation that could be attributed to the alterations at the periplasmic turns.
This apparent lack of contribution of the surface loops to the thermal stability of the protein was unexpected and suggests that these loops might be highly mobile but yet protease resistant as long as the neighboring loops are intact. This finding is corroborated by the X-ray structural analysis, which failed to locate the residues of three loops in the electron density map (31). Only loop L3 could be modeled; however, this loop is involved in a crystal contact around Lys-113 and might therefore be accidentally frozen. Hence, main chain-main chain interactions between adjacent β-strands and the packing of side chains inside the β-barrel domain are, in addition to the periplasmic turns, the main contributors to thermal stability, which leads to the so-called heat modifiability of the proteins.
Functional characterization of OmpA loop deletion mutants.
An ompA mutant host strain (not expressing the chromosomal ompA gene) transformed with the appropriate plasmids was assayed for its sensitivity to OmpA-specific bacteriophage K3, a well-characterized T-even-type phage (37). Only strains expressing the wild-type-like OmpA-T2H1-T3S1 or OmpAΔL4 variants were fully sensitive to phage K3, thus giving further evidence that these two proteins were correctly assembled into the outer membrane. Although they were also correctly assembled (see above), all other loop deletion variants did not allow plaque formation. These data are entirely consistent with previous studies which demonstrated that amino acid substitutions in loops L2 and L3 caused resistance to phage K3 whereas substitutions in the fourth loop that mediated resistance to other OmpA-specific bacteriophages did not influence the efficiency of plating of phage K3 (30). Hence, except for the fourth loop, all other loops were necessary for the OmpA protein to function as a receptor for phage K3.
For efficient F conjugation in liquid media, expression of the OmpA protein by the recipient cell is required. In this function, the wild-type protein can be replaced by its β-barrel domain (23). Host strain SOAR1, expressing different plasmid-encoded OmpA variants, served as a recipient; strain XL1-Blue (harboring F′::Tn10) served as a donor. For conjugation, a mixture of equal amounts of all 16 loop deletion variants was used. Cells were plated in parallel on plates allowing growth of only the exconjugants and on plates which also allowed growth of the recipient cells. Colonies from both plate types were pooled separately (about 500 recipient clones and 750 exconjugant clones), and their plasmid DNAs were isolated. By use of two unique restriction enzymes (NruI and SphI), plasmids encoding OmpA variants with a different number of loops shortened could be distinguished. Agarose gel electrophoresis demonstrated that the recipient pool represented, as expected, an equal mixture of all 16 loop deletion variants (data not shown). However, only full-length variants were identified in the pool of exconjugants. This competition experiment demonstrated that loop shortening is not compatible with the function of OmpA in F conjugation. Furthermore, this assay underscores the power of competitive F conjugation in enrichment for wild-type cells among an excess of nonfunctional ompA mutants. In a similar experiment, it was found that ompA wild-type cells could be enriched among a thousandfold surplus of ompA mutants (23). This method will be very useful in further experiments involving random libraries of OmpA mutants.
Finally, studies on the role of the surface-exposed loops during the invasion of brain microvascular endothelial cells by pathogenic E. coli (33) are in progress.
Conclusion and outlook.
This study demonstrates, for the first time, that an engineered outer membrane protein depleted of any large hydrophilic extramembranous loops still maintains the potential to fold into a structure which is very similar to that of the wild-type protein. Hence, not only do surface-exposed loops of outer membrane proteins tolerate small insertions (4, 9, 19, 28) or deletions (1, 11, 15, 35) in one or a few loops, but they can also tolerate the essentially complete removal of all loops at the same time. This way, this study excludes the possibility that some redundant topogenic information might be scattered throughout several loops. Although my work is limited to the effects of loop shortening on membrane assembly of the outer membrane protein OmpA, it is not unreasonable to suggest that such loops do not play an active role in the assembly of other, if not all, integral β-structured membrane proteins.
Moreover, these findings represent a significant step toward the development of artificial outer membrane proteins. It may simplify such a task because it now appears possible to start with only a minimum β-barrel core domain. It is not necessary to put too much effort into the design of the turns or loops connecting the β-strands. Such artificial membrane proteins may have several biomedical or biotechnological applications, for instance, as a framework for the docking of binding or catalytic sites useful for vaccine development, immunoassays, or liposome-mediated drug delivery (for a review, see reference 32). The use of an artificial membrane-embedded core as a protein fusion partner for producing two-dimensional crystals or in the development of biosensors (6) might represent another application. The design principles make the OmpAΔL1234 variant an excellent starting point for such an endeavor.
ACKNOWLEDGMENTS
I wish to thank Ulf Henning for his interest in this research and for his generous support. Helpful discussions with Michèle Loewen, Kaspar Locher, and Patrick Van Gelder during preparation of the manuscript are highly appreciated.
Part of this investigation was supported by grant Ko1686/1-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Agterberg M, Adriaanse H, Tijhaar E, Resink A, Tommassen J. Role of the cell-surface-exposed regions of outer membrane protein PhoE of Escherichia coliK12 in the biogenesis of the protein. Eur J Biochem. 1989;185:365–370. doi: 10.1111/j.1432-1033.1989.tb15124.x. [DOI] [PubMed] [Google Scholar]
- 2.Beer K B, Miller V L. Amino acid substitutions in naturally occurring variants of Ail result in altered invasion activity. J Bacteriol. 1992;174:1360–1369. doi: 10.1128/jb.174.4.1360-1369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charbit A, Clement J M, Hofnung M. Further sequence analysis of the phage lambda receptor site. Possible implications for the organization of the LamB protein in Escherichia coliK12. J Mol Biol. 1984;175:395–401. doi: 10.1016/0022-2836(84)90355-3. [DOI] [PubMed] [Google Scholar]
- 4.Charbit A, Ronco J, Michel V, Werts C, Hofnung M. Permissive sites and topology of an outer membrane protein with a reporter epitope. J Bacteriol. 1991;173:262–275. doi: 10.1128/jb.173.1.262-275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coliporins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich C, Schmitt L, Tampe R. Molecular organization of histidine-tagged biomolecules at self-assembled lipid interfaces using a novel class of chelator lipids. Proc Natl Acad Sci USA. 1995;92:9014–9018. doi: 10.1073/pnas.92.20.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freudl R, Braun G, Hindennach I, Henning U. Lethal mutations in the structural gene of an outer membrane protein (OmpA) of Escherichia coliK12. Mol Gen Genet. 1985;201:76–81. doi: 10.1007/BF00397989. [DOI] [PubMed] [Google Scholar]
- 8.Freudl R, Schwarz H, Stierhof Y D, Gamon K, Hindennach I, Henning U. An outer membrane protein (OmpA) of Escherichia coliK-12 undergoes a conformational change during export. J Biol Chem. 1986;261:11355–11361. [PubMed] [Google Scholar]
- 9.Freudl R. Insertion of peptides into cell-surface-exposed areas of the Escherichia coliOmpA protein does not interfere with export and membrane assembly. Gene. 1989;82:229–236. doi: 10.1016/0378-1119(89)90048-6. [DOI] [PubMed] [Google Scholar]
- 10.Gafvelin G, von Heijne G. Topological frustration in multispanning E. coliinner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Jeanteur D, Pattus F, Hancock R E. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosaporin OprD. Mol Microbiol. 1995;16:931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson E G, Thornton J M. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Killmann H, Videnov G, Jung G, Schwarz H, Braun V. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and φ80 and colicin M bind to the gating loop of FhuA. J Bacteriol. 1995;177:694–698. doi: 10.1128/jb.177.3.694-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klebba P E, Hofnung M, Charbit A. A model of maltodextrin transport through the sugar-specific porin, LamB, based on deletion analysis. EMBO J. 1994;13:4670–4675. doi: 10.1002/j.1460-2075.1994.tb06790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinschmidt J H, Tamm L K. Folding intermediates of a β-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- 17.Klose M, MacIntyre S, Schwarz H, Henning U. The influence of amino substitutions within the mature part of an Escherichia coliouter membrane protein (OmpA) on assembly of the polypeptide into its membrane. J Biol Chem. 1988;263:13297–13302. [PubMed] [Google Scholar]
- 18.Klose M, Störiko A, Stierhof Y D, Hindennach I, Mutschler B, Henning U. Membrane assembly of the outer membrane protein OmpA of Escherichia coli. J Biol Chem. 1993;268:25664–25670. [PubMed] [Google Scholar]
- 19.Koebnik R, Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coliK-12. J Bacteriol. 1993;175:826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koebnik R, Krämer L. Membrane assembly of circularly permuted variants of the E. coliouter membrane protein OmpA. J Mol Biol. 1995;250:617–626. doi: 10.1006/jmbi.1995.0403. [DOI] [PubMed] [Google Scholar]
- 21.Koebnik R. In vivo membrane assembly of split variants of the E. coliouter membrane protein OmpA. EMBO J. 1996;15:3529–3537. [PMC free article] [PubMed] [Google Scholar]
- 22.Koebnik R. Membrane assembly of the E. coliouter membrane protein OmpA: exploring sequence constraints on transmembrane β-strands. J Mol Biol. 1999;285:1801–1810. doi: 10.1006/jmbi.1998.2405. [DOI] [PubMed] [Google Scholar]
- 23.Koebnik, R. Unpublished results.
- 24.Kupsch E M, Knepper B, Kuroki T, Heuer I, Meyer T F. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeaefor human leukocytes and epithelial cells. EMBO J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leive L. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun. 1965;21:290–296. doi: 10.1016/0006-291x(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 26.Manoil C, Rosenbusch J P. Conjugation-deficient mutants of Escherichia colidistinguish classes of functions of the outer membrane OmpA protein. Mol Gen Genet. 1982;187:148–156. doi: 10.1007/BF00384398. [DOI] [PubMed] [Google Scholar]
- 27.McGovern K, Ehrmann M, Beckwith J. Decoding signals for membrane protein assembly using alkaline phosphatase fusions. EMBO J. 1991;10:2773–2782. doi: 10.1002/j.1460-2075.1991.tb07826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merck K B, de Cock H, Verheij H M, Tommassen J. Topology of the outer membrane phospholipase A of Salmonella typhimurium. J Bacteriol. 1997;179:3443–3450. doi: 10.1128/jb.179.11.3443-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mormeneo S, Knott R, Perlman D. Precise nucleotide sequence modifications with bidirectionally cleaving class-IIS excision linkers. Gene. 1987;61:21–30. doi: 10.1016/0378-1119(87)90361-1. [DOI] [PubMed] [Google Scholar]
- 30.Morona R, Klose M, Henning U. Escherichia coliK-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol. 1984;159:570–578. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pautsch A, Schulz G E. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 32.Popot J L, Saraste M. Engineering membrane proteins. Curr Opin Biotechnol. 1995;6:394–402. doi: 10.1016/0958-1669(95)80068-9. [DOI] [PubMed] [Google Scholar]
- 33.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S H, Kim K S. Outer membrane protein A of Escherichia colicontributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prehm P, Jann B, Jann K, Schmidt G, Stirm S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coliB. J Mol Biol. 1976;101:277–281. doi: 10.1016/0022-2836(76)90377-6. [DOI] [PubMed] [Google Scholar]
- 35.Rehm B H A, Hancock R E W. Membrane topology of the outer membrane protein OprH from Pseudomonas aeruginosa: PCR-mediated site-directed insertion and deletion mutagenesis. J Bacteriol. 1996;178:3346–3349. doi: 10.1128/jb.178.11.3346-3349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ried G, Koebnik R, Hindennach I, Mutschler B, Henning U. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coliK-12. Mol Gen Genet. 1994;243:127–135. doi: 10.1007/BF00280309. [DOI] [PubMed] [Google Scholar]
- 37.Riede I, Drexler K, Eschbach M L, Henning U. DNA sequence of genes 38 encoding a receptor recognizing protein of bacteriophages T2, K3 and of K3 host range mutants. J Mol Biol. 1987;194:31–39. doi: 10.1016/0022-2836(87)90713-3. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 39.Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–6069. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surrey T, Jähnig F. Kinetics of folding and membrane insertion of a beta-barrel membrane protein. J Biol Chem. 1995;270:28199–28203. doi: 10.1074/jbc.270.47.28199. [DOI] [PubMed] [Google Scholar]
- 41.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel H, Jähnig F. Models for the structure of outer-membrane proteins of Escherichia coliderived from Raman spectroscopy and prediction methods. J Mol Biol. 1986;190:191–199. doi: 10.1016/0022-2836(86)90292-5. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]