Abstract
Detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is essential for diagnosis, treatment, and infection control. Polymerase chain reaction (PCR) fails to distinguish acute from resolved infections, as RNA is frequently detected after infectiousness. We hypothesized that nucleocapsid in blood marks acute infection with the potential to enhance isolation and treatment strategies. In a retrospective serosurvey of inpatient and outpatient encounters, we categorized samples along an infection timeline using timing of SARS-CoV-2 testing and symptomatology. Among 1860 specimens from 1607 patients, the highest levels and frequency of antigenemia were observed in samples from acute SARS-CoV-2 infection. Antigenemia was higher in seronegative individuals and in those with severe disease. In our analysis, antigenemia exhibited 85.8% sensitivity and 98.6% specificity as a biomarker for acute coronavirus disease 2019 (COVID-19). Thus, antigenemia sensitively and specifically marks acute SARS-CoV-2 infection. Further study is warranted to determine whether antigenemia may aid individualized assessment of active COVID-19.
Keywords: nucleocapsid, antigenemia, COVID-19, SARS-CoV-2
We evaluate SARS-CoV-2 nucleocapsid antigenemia as a marker of acute COVID-19 in a large, diverse serosurvey and investigate outliers to better understand the phenomenon. Findings suggest antigenemia is a biomarker of acute SARS-CoV-2 infection with potential diagnostic utility in multiple contexts.
Although it is the standard of care for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), reverse transcription polymerase chain reaction (RT-PCR) remains an imperfect diagnostic marker for coronavirus disease 2019 (COVID-19) because SARS-CoV-2 RNA commonly persists beyond the period of acute infection [1–3]. Accordingly, Centers for Disease Control and Prevention (CDC) guidelines do not recommend retesting most individuals by RT-PCR within 90 days following diagnosis. Instead, isolation guidelines are based on time from symptom onset [4, 5]. This creates a dilemma when screening tests detect SARS-CoV-2 RNA in a patient without well-defined onset or resolution of COVID-19–like illness. Alternative molecular markers for acute infection are not widely available [6] and low-sensitivity respiratory antigen testing may be effectively applied at a population level [7, 8], but there remains a need for more sensitive and specific diagnostics to provide individualized guidance.
The presence of viral nucleocapsid protein in peripheral blood (antigenemia) has been demonstrated in SARS-CoV-1 and SARS-CoV-2 infection [9–21]. A blood-based antigen biomarker may have inherent advantages over upper respiratory tract antigen testing, or biomarkers such as RT-PCR cycle threshold (Ct) value and subgenomic RNA (sgRNA), because specimen quality and quantity can be standardized. Reports of antigenemia test performance as a diagnostic biomarker are inconsistent, likely due to varying assay composition and inconsistent reference standards as many studies compare against respiratory RT-PCR as a gold standard and fail to account for the persistence of RNA beyond acute infection.
In this study, we present evidence from a large serosurvey of adults in inpatient and outpatient settings to explore the hypothesis that nucleocapsid antigenemia is a sensitive and specific marker of acute infection as defined by a clinical timeline. Specifically, each blood sample was categorized through rigorous review of clinical history and respiratory SARS-CoV-2 testing in a schema that assumes a typical course of COVID-19 for all subjects. Our study is novel among others evaluating nucleocapsid antigenemia in that we were able to capture blood samples from each stage of infection to evaluate the performance of antigenemia testing for staging acuity, which we define based on onset of symptoms and timing of respiratory SARS-CoV-2 positivity. Furthermore, our evaluation of outliers in each staging category uncovered evidence of reinfection as well as persistence of antigenemia in immunocompromised individuals. Overall, we find a strong association between acute infection and nucleocapsid antigenemia, which also correlates with serostatus and disease severity. Together our findings suggest antigenemia may clarify disease timing and provide needed insight in many clinical settings.
METHODS
Clinical Specimens
We collected a convenience sample of residual plasma, serum, and whole blood specimens from the clinical chemistry laboratory of Emory Medical Laboratories 1 day per week between 11 January 2021 and 12 March 2021. These specimens were originally collected for routine clinical testing from inpatient (medical/surgical wards, intensive care, obstetrics) and outpatient settings (clinics, emergency department, infusion centers, ambulatory surgery). Samples were transferred to a −80°C repository after clinical testing was completed, but prior to being discarded. More than 1 blood sample from the same patient was permitted with a minimum time of 5 days between samples. This study was approved and granted complete Health Insurance Portability and Accountability Act (HIPAA) and consent waiver by the Emory University Institutional Review Board (STUDY00000510). This study was approved by the Institutional Review Board of Emory University for the use of residual clinical specimens. Written informed consent by the patients was not required.
Nucleocapsid Assay
Nucleocapsid antigenemia was quantified on the Quanterix HD-X platform. Residual serum and plasma samples were thawed once after storage at −80°C and diluted 1 to 3 in assay sample diluent. Diluted samples were then run using the ultrasensitive SIMOA SARS-CoV-2 N Protein Antigen assay (Quanterix) on the automated Quanterix HD-X platform (Quanterix), which has a validated limit of detection of 0.099 pg/mL in respiratory and saliva samples. Analytical validation in serum and plasma samples is reported by Quanterix for research use outside of their Emergency Use Authorization. Samples with antigen levels too high for the linear range of the assay were further diluted 1 to 20 and retested. Final antigen concentrations were determined by interpolation after sigmoidal fitting of duplicate calibration curves run on each test plate.
Serological Testing
In-house developed single-dilution serological screening assays for SARS-CoV-2 receptor binding domain (RBD) and nucleocapsid antibodies were used to establish serological status at the time of antigenemia testing. Antibody class-specific RBD serologies were performed as previously described [22–24]. Nucleocapsid antibody testing was performed using an in-house developed enzyme-linked immunosorbent assay (ELISA; Supplementary material).
Medical Record Review
Patient medical record number was recorded at the time of specimen collection. The Emory Healthcare Clinical Data Warehouse (CDW) was queried for SARS-CoV-2 nucleic acid amplification tests (NAAT) and antigen tests, clinical notes, International Classification of Diseases-Tenth Revision (ICD-10) codes, laboratory values, and, if applicable, requirement for mechanical ventilation and date of death. All Ct values were obtained directly from reports produced by the manufacturer’s software (Supplementary material).
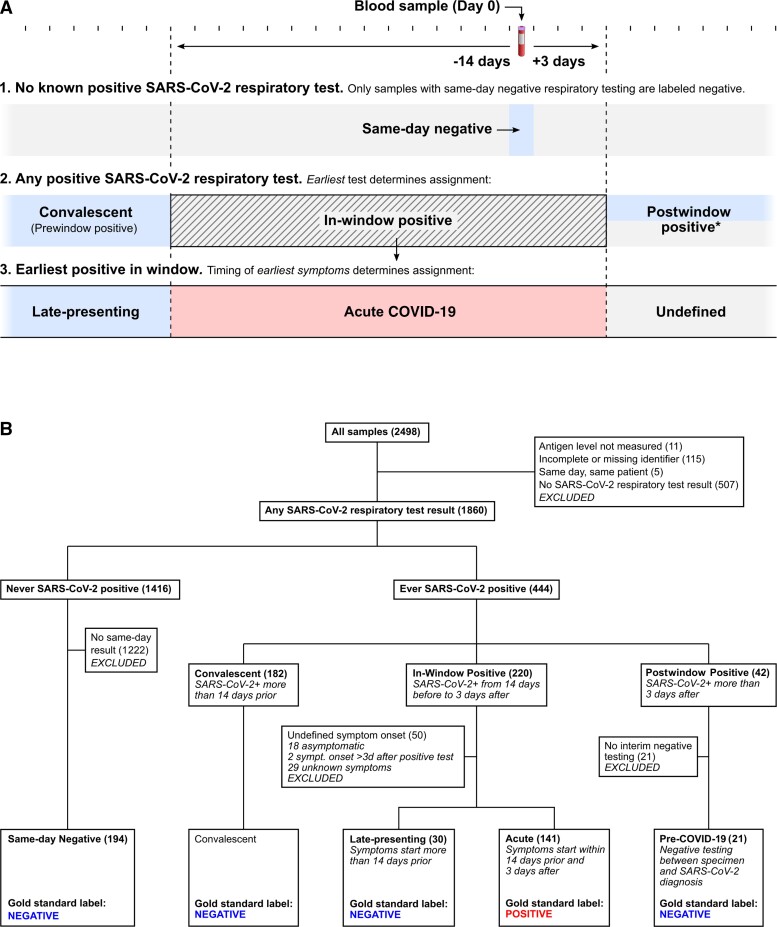
A COVID-19 status label (positive or negative) and a category (convalescent, late-presenting, acute, pre-COVID-19, and same-day negative) were assigned to each blood sample based on that patient’s (1) SARS-CoV-2 respiratory testing (including NAAT or antigen), (2) date of earliest positive test, and (3) date of symptom onset (Figure 1).
Figure 1.
A, Schematic of the process for COVID-19 status assignment. Samples from patients with no record of positive SARS-CoV-2 respiratory testing were only considered negative if corresponding negative respiratory testing occurred on the same day. Due to the lack of a gold standard for active SARS-CoV-2 infection, samples from individuals with history of positive SARS-CoV-2 testing were labeled based on earliest known positive SARS-CoV-2 respiratory test and time since symptom onset. *Samples with postwindow-positive SARS-CoV-2 testing were labeled negative if a negative SARA-CoV-2 test was available following the sample but before the positive test. Otherwise the sample was labeled unknown. B, Flow chart of categorization and labeling process indicating number of samples assigned to each group. Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Chart review began with automated review of SARS-CoV-2 respiratory results available in the medical record. Blood samples from a patient with a positive test more than 14 days prior to sample collection were labeled convalescent and no further review of the medical record for categorization purposes was performed. History and physical clinical notes dated within 14 days before or after the date of the blood sample were then reviewed, if available, for all patients not labeled convalescent. Date of COVID-19–like symptom onset (including fever, fatigue, malaise, myalgia, headache, dyspnea, cough, wheezing, anosmia, ageusia, congestion, rhinorrhea, or diarrhea) and earliest positive SARS-CoV-2 testing was recorded if these had been described in the history narrative or clinician’s assessment and plan.
The original medical records were then reviewed for all patients (other than those labeled convalescent) with a positive SARS-CoV-2 test who did not yet have date of symptom onset recorded in our data set. The entire medical record was available during this stage, but the reviewer was blinded to antigenemia status, which was not considered in labeling of COVID-19 status or category assignment.
Given that reinfection with SARS-CoV-2 was rare at the time of this study [25], our approach assumed that no reinfection events were captured in our sample set, which spanned 3 months. Patients without any record of SARS-CoV-2 testing were excluded from analysis. Further detail is provided in Supplementary material.
Data Analysis
Data obtained during specimen collection were stored in Microsoft Excel. CDW reports were provided in comma-separated values (csv) format. All data were then imported into MATLAB (The MathWorks, Inc) for analysis. Wilcoxon rank-sum test was used for comparisons.
RESULTS
Specimens and COVID-19 Status Assignments
In total, 2498 serum and plasma samples were targeted for evaluation during the study period (Figure 1B). Eleven samples were not evaluated for antigenemia due to preanalytical factors such as insufficient sample volume. Thus, 2487 samples were available for quantification of antigenemia, of which 255 (10.2%) exhibited detectable nucleocapsid; 115 of 2487 were excluded due to lack of patient identifiers and 5 additional samples were excluded as they had been collected on the same day as another blood sample from a single patient. Clinical data were examined for the remaining 2367 samples from 2101 unique patients (Table 1). Of the 2367 samples, 507 were excluded because of no record of SARS-CoV-2 testing, and 11 of these 507 (2.1%) had detectable antigenemia. The remaining 1860 samples from 1607 patients had SARS-CoV-2 testing records to guide categorization and were classified as described in Figure 1B and Table 2.
Table 1.
Summary of Patient Characteristics by COVID-19 Status
| Characteristic | SARS-CoV-2 Infection Status | ||
|---|---|---|---|
| Positive | Negative | Undefined | |
| n | 130 | 385 | 1622 |
| Age, y, mean (IQR) | 60.6 (52.2–73.0) | 54.2 (39.2–69.6) | 55.0 (39.8–70.2) |
| Female, % | 47.7 | 57.1 | 55.5 |
| Vaccinated, % | 1.5 | 4.7 | 9.9 |
| Race, % | |||
| African American or black | 78.5 | 72.0 | 60.7 |
| American Indian or Alaskan Native | 0.0 | 0.0 | 0.2 |
| Asian | 0.8 | 1.0 | 1.6 |
| Caucasian or white | 13.1 | 20.0 | 29.0 |
| Native Hawaiian or other Pacific Islander | 0.0 | 0.3 | 0.2 |
| Multiple | 0.0 | 0.5 | 0.4 |
| Unknown, unavailable or unreported | 7.7 | 6.2 | 7.9 |
| Ethnicity | |||
| Non-Hispanic or Latino | 83.9 | 86.0 | 84.4 |
| Unreported, unknown, unavailable | 13.9 | 7.8 | 11.9 |
| Hispanic or Latino | 0.8 | 5.5 | 3.0 |
| Not recorded | 1.5 | 0.8 | 0.7 |
| Antigenemia, %a | 85.8 | 10.1 | 3.9 |
| Settinga | |||
| Inpatient | 70.9 | 42.9 | 39.3 |
| ER or CDU | 29.1 | 27.2 | 14.1 |
| Outpatient | 0.0 | 26.7 | 45.3 |
| Peripartum | 0.0 | 3.3 | 1.1 |
Abbreviations: CDU, clinical decision unit; ER, emergency room.
Reflects all included samples (including multiple samples for a unique patient).
Table 2.
Categories Determined by Chart Review for Samples and Patients Included in the Analysis
| Category | Samples | Unique Patients |
|---|---|---|
| Never SARS-CoV-2 positive | 1416 | 1249 |
| Same-day negative test | 194 | 194 |
| Ever SARS-CoV-2 positivea | 444 | 360 |
| Convalescent | 182 | 153 |
| Late presenting | 30 | 30 |
| Acute | 141 | 130 |
| Sampled 3 or more days prior to diagnosis | 42 | 34 |
| Negative interim testing | 21 | 16 |
Includes in-hospital nucleic acid amplification tests (NAAT) as well as community NAAT or antigen testing if reported in the clinical narrative.
Diagnostic Performance of Antigenemia for Acute COVID-19
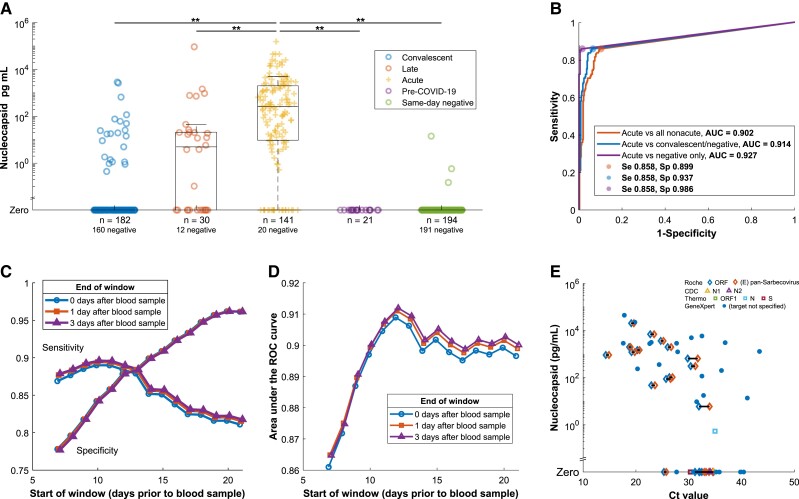
Nucleocapsid antigenemia was present at higher frequency and with a higher median concentration in acute COVID-19 samples compared to samples categorized as late presenting, convalescent, pre-COVID-19, or same-day negative (P < .001 for all comparisons; Figure 2A). Receiver operating characteristic (ROC) analysis demonstrated area under the curve (AUC) of 0.902 in distinguishing samples from patients experiencing acute infection from all nonacute categories, and sensitivity and specificity were 85.2% and 89.9%, respectively (Figure 2B). Test characteristics with censoring of the potentially ambiguous late-presenting group showed AUC 0.914, sensitivity 85.8%, and specificity 93.7%, while the most stringent comparison (censoring of the convalescent and late-presenting groups) demonstrated AUC 0.972, sensitivity 85.8%, and specificity 98.6%. Sensitivity improved to 93.9% when the comparison was only made among seronegative individuals (Supplementary Figure 1).
Figure 2.
A, Prevalence of antigenemia and serum or plasma nucleocapsid levels for blood samples by category. Unexpected results (presence of nucleocapsid in the convalescent and same-day negative groups, absence of nucleocapsid in the acute group) are examined in Supplementary Tables 2–5. B, ROC curve for diagnostic performance of detectable antigenemia with reference to a −14/+3 day window for acute infection. The additional curves progressively exclude ambiguous categories. C, Impact on sensitivity and specificity of varying the window period, which defines the reference standard for acute COVID-19. D, AUC for the same varied window periods. E, Antigenemia compared to RT-PCR Ct value for those specimens with a Ct value available from the clinical laboratory on the same day. Symbols correspond to assay and gene target with horizontal line linking Ct values for different targets detected in the same sample. This includes data from 4 assays on 3 thermocycler platforms described in further detail in the Supplementary material. Abbreviations: AUC, area under the curve; CDC, Centers for Disease Control and Prevention; COVID-19, coronavirus disease 2019; Ct, cycle threshold; N, nucleocapsid; ORF, open reading frame; ROC, receiver operating characteristic; RT-PCR, reverse transcription polymerase chain reaction; S, spike; Se, sensitivity; Sp, specificity.
Test characteristics were also examined when adjusting the reference standard by varying parameters of the acuity window. Sensitivity decreased as the window start period increased beyond −11 days (Figure 2C). Meanwhile, specificity consistently increased as the period of the acuity window was lengthened. Maximum AUC was observed with a window period opening at −12 days (AUC = 0.912 with window close at +3 days) with minimal effect of varying the postsampling period from 0 to +3 days (Figure 2D).
Ct values from positive nasopharyngeal RT-PCR were available from the same day as a blood sample for 49 specimens. Only 6 of 17 samples with corresponding to Ct values greater than 33 had antigenemia and 4 of 6 of these were from the GeneXpert assay (Figure 2E). All except for 2 samples with corresponding Ct values less than 30 exhibited antigenemia.
Temporal Trends in Antigenemia
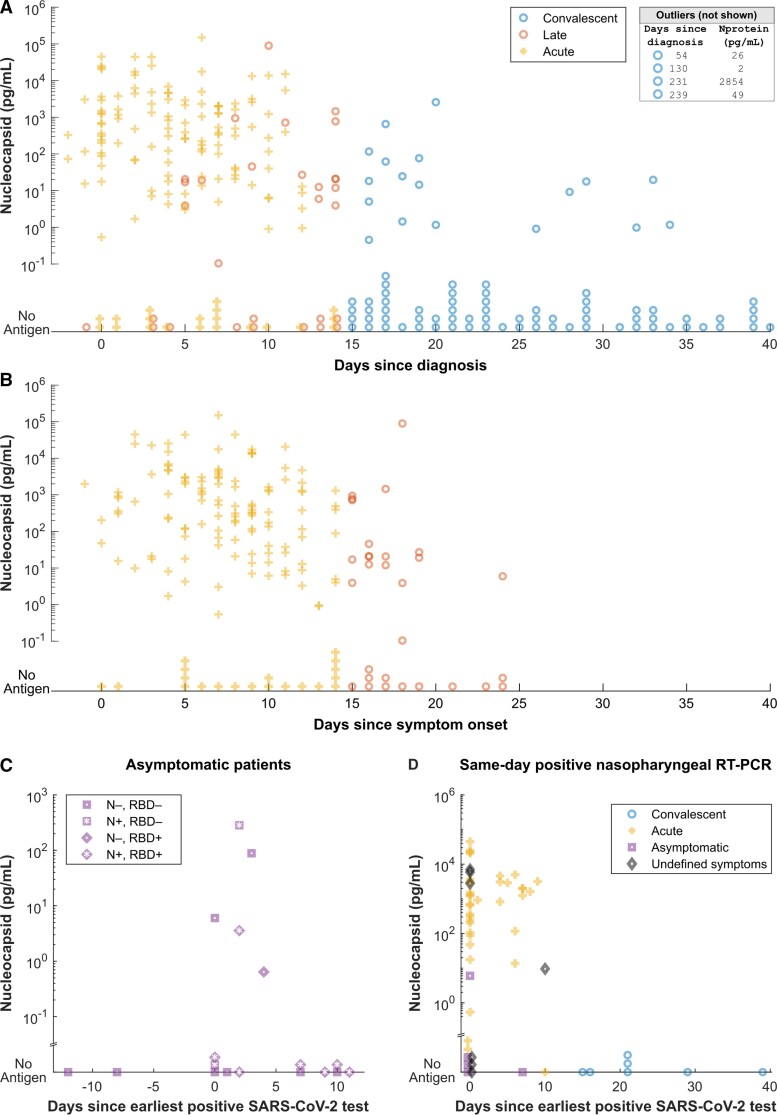
We analyzed the dynamics of antigen level over time in samples from the acute, late-presenting, and convalescent groups. The frequency of detectable nucleocapsid and antigen concentration decreased over time following diagnosis and reported symptom onset (Figure 3A and 3B). Eighteen samples were identified from patients who were asymptomatic at the time of COVID-19 diagnosis, 5 (27.7%) of which had detectable nucleocapsid antigenemia. Nucleocapsid antigen was detected more frequently (50.0%) in the subset of samples available from asymptomatic patients within 3 days of their diagnosis (Figure 3C). Among 55 samples from individuals with positive respiratory RT-PCR testing on the same day, 7 convalescent samples did not exhibit antigenemia (Figure 3D and Supplementary Table 1) and acute infections primarily exhibited high antigenemia. Among this subset of patients, no antigenemia was observed more than 14 days after the earliest known positive test (Figure 3D).
Figure 3.
A and B, Serum or plasma nucleocapsid plotted against time since diagnosis (A) and symptom onset (B). Samples without antigen detected are shown stacked on the horizontal axis. Four samples with antigenemia beyond 41 days are listed in the box and 93 samples without antigenemia between 41 and 351 days after earliest diagnosis are not shown. C, Serum or plasma nucleocapsid in patients whose COVID-19 course was described as asymptomatic in clinical records. The x-axis reflects time between first known positive respiratory test and the day the blood sample used in our analysis was collected. Shape and shading of each symbol classify serological status of asymptomatic patients. D, Serum or plasma nucleocapsid for individuals with positive nasopharyngeal RT-PCR on the same day as blood sample collection. Shape and shading of each symbol classify clinical status of patients. Abbreviations: COVID-19, coronavirus disease 2019; N, nucleocapsid; RBD, receptor-binding domain; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Examination of Outliers
We reviewed medical records for individuals with unexpected presence or absence of antigenemia based on staging category. Twenty-one of 182 (11.5%) in the convalescent group had antigenemia (Supplementary Table 2 and Supplementary Figure 2); 15 of these 21 (71.4%) convalescent samples of interest had clinical evidence that might explain persistent antigen positivity. Among these, 2 individuals had clinical history consistent with reinfection by SARS-CoV-2, 2 were highly immunocompromised, and 11 samples (median time from diagnosis 20 days; interquartile range, 16.5–28.5 days) had severe COVID-19 marked by need for high-flow oxygen, intubation, or death. End-stage renal disease or dialysis was more common among samples in the convalescent group with antigenemia compared to those without antigenemia (fraction = 0.41; 95% confidence interval [CI], .20–.61] vs 0.13; 95% CI, .07–.18) whereas other comorbidities were not significantly different (Supplementary Figure 3). Three individuals had negative respiratory SARS-CoV-2 testing and antigenemia on the same day, none of which had evidence of COVID-19–related symptoms (Supplementary Table 3). Eighteen samples had antigenemia after more than 14 days of symptoms, of which 14 were seropositive for both N IgG and RBG IgG, 2 seropositive for RBD IgG only, and 2 were seronegative for both. Thirteen had nucleocapsid level less than 46 pg/mL while the other 5 exceeded 700 pg/mL, including both N- and RBD-seronegative patients and N negative/RBD positive sample (Supplementary Table 4, Supplementary Figure 4 and Supplementary Figure 5). Twenty individuals with samples categorized in the acute COVID-19 group did not have antigenemia; 10 of these were collected 10 or more days after symptom onset (Supplementary Table 5 and Supplementary Figure 6).
Antigenemia Trends by Antibody Serostatus
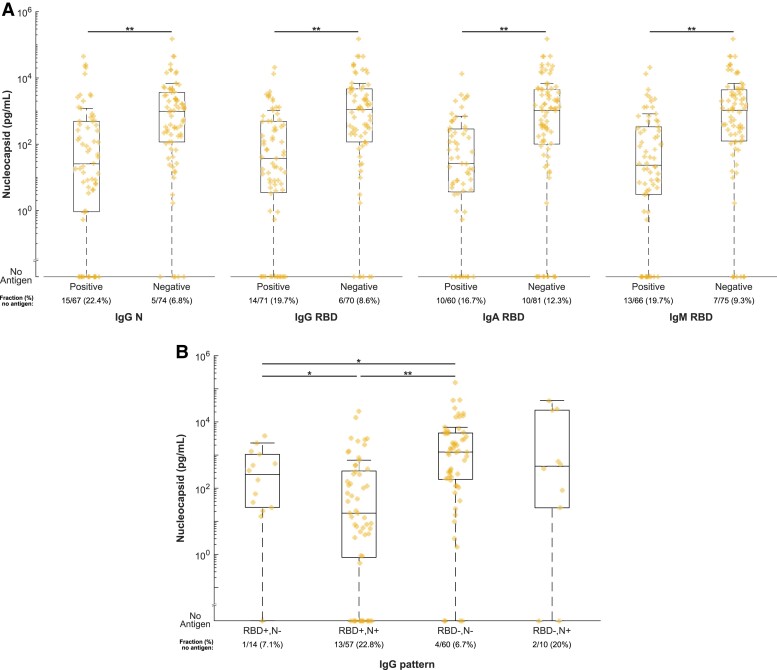
Distribution of nucleocapsid levels in the acute COVID-19 group were significantly different with higher median values in seronegative samples compared to seropositive samples for nucleocapsid IgG, RBD IgG, RBD IgA, and RBD IgM (P < .001 for each comparison; Figure 4A). Seropositive samples were also more likely to have undetectable antigenemia. Similar trends were seen in the late-presenting group except for the comparison based on IgM, which was not significant (Supplementary Figure 7). In addition, we compared antigenemia levels in groups defined by patterns of serostatus consistent with vaccination (RBD positive/N negative) as well as natural infection (RBD positive/N positive). Antigenemia levels were significantly lower in acutely ill patients with serological evidence of natural infection or vaccination compared to seronegative individuals (Figure 4B).
Figure 4.
A, Comparison of serum or plasma nucleocapsid levels in individuals with and without SARS-CoV-2–specific antibodies. Samples were tested by in-house developed serological tests for nucleocapsid- and receptor binding domain-specific IgG as well as receptor binding domain-specific IgA and IgM. Levels of nucleocapsid are plotted and compared in samples stratified by seropositivity for each antibody type. B, Comparison of nucleocapsid levels by serostatus pattern. RBD positive/N negative is most consistent with prior vaccination. RBD positive /N positive is most consistent with natural infection with or without prior vaccination. Abbreviations: Ig, immunoglobulin; N, nucleocapsid; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
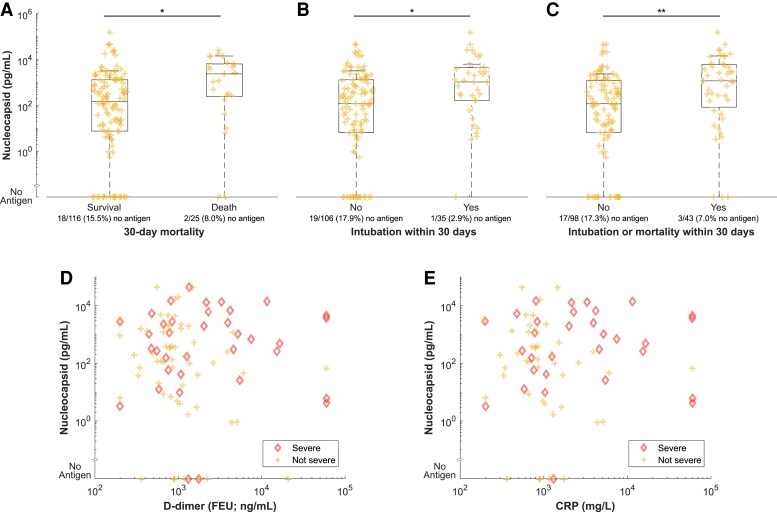
Association of Antigenemia With COVID-19 Severity
In the acute COVID-19 group, distribution of nucleocapsid antigen was significantly different and median value was higher in samples from patients who died or required intubation within 30 days of sampling compared to those who survived or did not require intubation (Figure 5A–C). This observation held true for comparison based on the composite of intubation or mortality. Levels of nucleocapsid antigenemia were not significantly associated with elevated D-dimer (cutoff 500 ng/mL) but were associated with elevated C-reactive protein (P = .002 in comparison based on 40 mg/L cutoff; Figure 5D and 5E).
Figure 5.
Comparison of serum or plasma nucleocapsid levels by (A–C) severity and (D and E) inflammatory biomarkers. B and C, Intubation includes intubation within 30 days before or after the blood sample was collected. D and E, Individuals with severe COVID-19 as defined by the composite of 30-day intubation or mortality are highlighted. Abbreviations: COVID-19, coronavirus disease 2019; CRP, C-reactive protein; FEU, fibrinogen equivalent unit.
DISCUSSION
This analysis of blood samples from routine clinical specimens collected during the ongoing COVID-19 pandemic demonstrates the following. First, antigenemia is a sensitive and specific marker for acute SARS-CoV-2 infection, as defined by timing after clinical diagnosis and symptom onset. Second, nucleocapsid is elevated in samples without evidence of antinucleocapsid (IgG) and antispike (IgG, IgM, and IgA) seroconversion. Third, antigenemia is associated with disease severity.
Evolving CDC isolation guidance during the COVID-19 pandemic reflects the difficulty of objectively defining resolution of SARS-CoV-2 infection. Underlying this is the persistence of RNA targets beyond the period of acute infection in immunocompetent individuals [1–3]. Meanwhile, persistence of replication-competent virus for months has been demonstrated by viral culture in immunocompromised hosts [25–29]. This creates a diagnostic dilemma when RT-PCR is persistently positive for weeks after diagnosis, when reinfection with SARS-CoV-2 is a consideration, or when encountering positive SARS-CoV-2 RT-PCR test results in an asymptomatic individual without history of prior objective diagnosis or prior COVID-19–like illness. Our data suggest that nucleocapsid antigenemia occurs most frequently and at higher levels in the acute stages of SARS-CoV-2 infection as defined by the timing of molecular diagnosis and symptomatology in a large serosurvey representing diverse patient encounters. While prospective studies with comparison presence of viable virus are needed to more rigorously determine the clinical utility of antigenemia testing on an individualized level, our findings are consistent with a model in which antigenemia marks acute infection.
Further, our data compels interest in whether antigenemia may provide direct evidence of active viral replication, with potential to aid in evaluation of infectiousness or guide therapeutics at an individualized level. For example, antiviral agents are not likely to benefit a patient without active SARS-CoV-2 replication. Clinical trial data therefore may be confounded by failure to stratify patients according to such a marker, as late presenters after cessation of viral replication would likely fail to show benefit or may even suffer harm from investigatory antiviral agents. In fact, recent evidence emphasizes the greatest benefit of antivirals early in infection [30]. In showing its association with acute SARS-CoV-2 infection and characterizing outliers (eg, convalescent patients with persistent antigenemia), our data suggest that nucleocapsid should be further investigated as a marker of viral activity, infectiousness, and a predictor of therapeutic response.
While other studies have reported similar findings, strengths and novelty of our study include a diverse cohort that is among the largest in which nucleocapsid antigenemia has been quantified to date and rigorous assignment of COVID-19 status through medical record review. Prior studies restricting the definition of a positive case to no more than 2 weeks after symptom onset report sensitivities between 90.9% and 97.5% and specificities between 94.2% and 100% [15–20] (Supplementary Table 6), and our data are consistent with these findings. Of further interest, our data revealed detectable antigen in 11 (2.1%) of the blood samples obtained in the primary serosurvey even though these patients were never screened with nasopharyngeal RT-PCR testing in our health care system. These represent likely infectious patients who may have had a missed SARS-CoV-2 diagnosis and suggest a potential role for antigenemia screening in a population for whom blood is already being sampled to complement existing infection control measures.
We also detected antigenemia in a small number of patients with subclinical SARS-CoV-2 infection. Individuals who test positive for SARS-CoV-2 without antecedent or subsequent COVID-19–like symptoms either represent shedding of replication-competent virus during subclinical disease or persistent RNA shedding following subclinical disease. While we corroborate previous findings that levels of antigenemia are associated with disease severity [19, 20], the presence of antigenemia in 5 asymptomatic individuals with SARS-CoV-2 demonstrates that antigenemia can also be present in subclinical infection. Despite the difficulty associated with identifying these cases, further investigation of the prevalence of antigenemia in acute asymptomatic infection is needed to clarify its role in screening broad populations.
This study is limited by use of a convenience sampling approach and retrospective data collection. Symptom onset as recorded in the medical record can be subjective and influenced by recall bias. Because of the ubiquity of community-based testing, SARS-CoV-2 diagnosis was documented prior to evaluation in our health care system for a subset of these patients and was only known to us when documented in the clinical narrative in addition to being subject to biases and imprecision. In addition, nucleocapsid-specific immunoglobulin may interfere with quantitation of antigenemia in individuals who have seroconverted although it is currently unknown whether total (Ig-bound and unbound) antigen or free (unbound only) antigen is a more meaningful clinical indicator. The primary analysis relies on the assumption that each subject is immunocompetent, that immunocompetent hosts have similar duration of acute COVID-19, and that there are no other confounding factors which may result in prolonged antigenemia. Recognizing these limitations, we performed a post hoc investigation of outlier cases, which facilitated hypothesis generation regarding reasons for prolonged antigenemia such as reduced renal function, prolonged critical illness, and immune compromise (Supplementary Tables 1–5 and 7). Several studies have demonstrated high specificity of antigenemia by evaluation of prepandemic samples [15, 17, 20], suggesting many false positives in our study are likely to have active infection beyond the parameters for acute infection defined in our reference standard schema. This will be further clarified as more robust comparisons to viral culture, sgRNA, RT-PCR Ct value, and respiratory antigen testing can be achieved.
Together our data demonstrate that nucleocapsid antigenemia is a sensitive and specific biomarker of acute COVID-19, wherein COVID-19 status is defined by time since earliest positive testing and symptom onset. We propose that nucleocapsid antigenemia is a candidate biomarker for active viral replication—recognizing that the available evidence points to this being an individualized process that cannot be broadly defined based on a generic timeline. Further prospective studies with rigorous documentation of clinical course and correlation with viral culture and other potential biomarkers of viral replication are needed.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Hans P Verkerke, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Gregory L Damhorst, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA.
Daniel S Graciaa, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Kaleb McLendon, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
William O’Sick, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Chad Robichaux, Emory Healthcare, Atlanta, Georgia, USA.
Narayanaiah Cheedarla, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Sindhu Potlapalli, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Shang-Chuen Wu, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Kristin R V Harrington, Department of Epidemiology, Emory University Rollins School of Public Health, Atlanta, Georgia, USA.
Andrew Webster, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Colleen Kraft, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Christina A Rostad, Department of Pediatrics and Center for Childhood Infections and Vaccines, Emory University School of Medicine and Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
Jesse J Waggoner, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA; Emory Healthcare, Atlanta, Georgia, USA; Department of Pediatrics and Center for Childhood Infections and Vaccines, Emory University School of Medicine and Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
Neel R Gandhi, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Department of Epidemiology, Emory University Rollins School of Public Health, Atlanta, Georgia, USA.
Jeannette Guarner, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Sara C Auld, Department of Epidemiology, Emory University Rollins School of Public Health, Atlanta, Georgia, USA; Division of Pulmonary, Allergy, Critical Care and Sleep Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Andrew Neish, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
John D Roback, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Wilbur A Lam, The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA; Department of Pediatrics and Center for Childhood Infections and Vaccines, Emory University School of Medicine and Children’s Healthcare of Atlanta, Atlanta, Georgia, USA; Aflac Cancer and Blood Disorders Center at Children’s Healthcare of Atlanta, Atlanta, Georgia, USA; Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA.
N Sarita Shah, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Department of Epidemiology, Emory University Rollins School of Public Health, Atlanta, Georgia, USA.
Sean R Stowell, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Notes
Acknowledgments. The authors thank Lisa Cole, Cecillitha J Williams, and Mark Meyers for assistance with specimen collection and Heather Jones for assistance with obtaining Ct values.
Financial support. This work was supported by the Woodruff Health Sciences Center COVID-19 Center for Urgent Research Engagement; and the National Institutes of Health (grant numbers U54 CA260563-01 Immune Regulation of COVID-19 Infection in Cancer and Autoimmunity to J. D. R., R01 HL138656 COVID-19 supplement to S. R. S., R01 AI138646 supplement to N. S. S., K24 AI114444 to N. R. G., U54 EB027690 03, and UL1TR002378 ).
References
- 1. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2:e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yan D, Zhang X, Chen C, et al. Characteristics of viral shedding time in SARS-CoV-2 infections: a systematic review and meta-analysis. Front Public Health 2021; 9:652842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drain PK. Rapid diagnostic testing for SARS-CoV-2. N Engl J of Med 2022; 386:264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention . COVID-19 quarantine and isolation. https://www.cdc.gov/coronavirus/2019-ncov/your-health/quarantine-isolation.html. Accessed 29 November 2021.
- 5. Centers for Disease Control and Prevention . CDC updates and shortens recommended isolation and quarantine period for general population. CDC Newsroom Releases; 2021. https://www.cdc.gov/media/releases/2021/s1227-isolation-quarantine-guidance.html. Accessed 29 November 2021. [Google Scholar]
- 6. Binnicker MJ. Can testing predict SARS-CoV-2 infectivity? The potential for certain methods to be a surrogate for replication-competent virus. J Clin Microbiol 2021; 59:e0046921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J of Med 2020; 383:e120. [DOI] [PubMed] [Google Scholar]
- 8. Chin ET, Huynh BQ, Chapman LAC, Murrill M, Basu S, Lo NC. Frequency of routine testing for coronavirus disease 2019 (COVID-19) in high-risk healthcare environments to reduce outbreaks. Clin Infect Dis 2021; 73:e3127–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Che XY, Hao W, Wang Y, et al. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis 2004; 10:1947–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li YH, Li J, Liu XE, et al. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J Virol Methods 2005; 130:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li T, Wang L, Wang H, et al. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front Cell Infect Microbiol 2020; 10:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lebedin YS, Lyang OV, Galstyan AG, Panteleeva AV, Belousov VV, Rebrikov DV. Serum SARS-CoV-2 nucleocapsid antigen detection is essential for primary diagnostics of SARS-CoV-2-associated pneumonia. medRxiv. 25September2020, preprint: not peer reviewed. doi: 10.1101/2020.09.24.20200303. [DOI] [Google Scholar]
- 13. Su B, Yin J, Lin X, et al. Quantification of SARS-CoV-2 antigen levels in the blood of patients with COVID-19. Sci China Life Sci 2021; 64:1193–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogata AF, Maley AM, Wu C, et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin Chem 2020; 66:1562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hingrat QL, Visseaux B, Laouenan C, et al. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect 2020; 27:789.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahava M, Kurkela S, Kuivanen S, et al. Detection of SARS-CoV-2 nucleocapsid antigen from serum can aid in timing of COVID-19 infection. J Virol Methods 2022; 302:114469. doi: 10.1016/j.jviromet.2022.114469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shan D, Johnson JM, Fernandes SC, et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat Commun 2021; 12:1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Ong CM, Yun C, et al. Diagnostic value of nucleocapsid protein in blood for SARS-CoV-2 infection. Clin Chem 2021; 68:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H, Hogan CA, Verghese M, et al. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin Chem 2021; 68:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Favresse J, Bayart JL, David C, et al. Serum SARS-CoV-2 antigens for the determination of COVID-19 severity. medRxiv. 21November2021, preprint: not peer reviewed. doi: 10.1101/2021.11.18.21266478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thudium RF, Stoico MP, Hogdall E, et al. Early laboratory diagnosis of COVID-19 by antigen detection in blood samples of the SARS-CoV-2 nucleocapsid protein. J Clin Microbiol 2021; 59:e0100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verkerke H, Horwath M, Saeedi B, et al. Comparison of antibody class-specific SARS-CoV-2 serologies for the diagnosis of acute COVID-19. J Clin Microbiol 2021; 59:e02026-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verkerke H, Saeedi BJ, Boyer D, et al. Are we forgetting about IgA? A re-examination of coronavirus disease 2019 convalescent plasma. Transfusion 2021; 61:1740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen JWL, Verkerke H, Owens J, et al. Serum pooling for rapid expansion of anti-SARS-CoV-2 antibody testing capacity. Transfus Clin Biol 2021; 28:51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aydillo T, Gonzalez-Reiche AS, Aslam S, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 2020; 383:2586–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hensley MK, Bain WG, Jacobs J, et al. Intractable COVID-19 and prolonged SARS-CoV-2 replication in a CAR-T-cell therapy recipient: a case study. Clin Infect Dis 2021; 73:e815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021; 371:1139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gottlieb RL, Vaca CE, Paredes R, et al. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N Engl J of Med 2021; 386:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.