Abstract
A λ-EMBL3 genomic library of Bacillus stearothermophilus T-6 was screened for hemicellulolytic activities, and five independent clones exhibiting β-xylosidase activity were isolated. The clones overlap each other and together represent a 23.5-kb chromosomal segment. The segment contains a cluster of xylan utilization genes, which are organized in at least three transcriptional units. These include the gene for the extracellular xylanase, xylanase T-6; part of an operon coding for an intracellular xylanase and a β-xylosidase; and a putative 15.5-kb-long transcriptional unit, consisting of 12 genes involved in the utilization of α-d-glucuronic acid (GlcUA). The first four genes in the potential GlcUA operon (orf1, -2, -3, and -4) code for a putative sugar transport system with characteristic components of the binding-protein-dependent transport systems. The most likely natural substrate for this transport system is aldotetraouronic acid [2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose] (MeGlcUAXyl3). The following two genes code for an intracellular α-glucuronidase (aguA) and a β-xylosidase (xynB). Five more genes (kdgK, kdgA, uxaC, uxuA, and uxuB) encode proteins that are homologous to enzymes involved in galacturonate and glucuronate catabolism. The gene cluster also includes a potential regulatory gene, uxuR, the product of which resembles repressors of the GntR family. The apparent transcriptional start point of the cluster was determined by primer extension analysis and is located 349 bp from the initial ATG codon. The potential operator site is a perfect 12-bp inverted repeat located downstream from the promoter between nucleotides +170 and +181. Gel retardation assays indicated that UxuR binds specifically to this sequence and that this binding is efficiently prevented in vitro by MeGlcUAXyl3, the most likely molecular inducer.
The plant cell wall is composed of three major polymeric constituents: cellulose, hemicellulose, and lignin. The hemicelluloses are a series of heteropolysaccharides including glucans, mannans, arabinans, and xylans (8, 9). The latter are the most common hemicellulosic polysaccharides and are composed of β-(1-4)-linked d-xylopyranosyl units. Depending on the source, xylan structures vary from linear to highly branched heteropolysaccharides (66). In hardwoods, the xylan backbone is modified with 4-O-methyl-d-glucuronosyl residues attached to the C-2 of the xylose units and with acetyl esters at O-2 or O-3 positions of the d-xylosyl residues. In nonacetylated softwood xylans, the substituents are 4-O-methyl-d-glucuronosyl and l-arabinofuranosyl residues attached to the main chain by α-1,3-glucosidic linkages (8). Due to its complex structure, the complete degradation of xylan requires the concerted action of several hydrolytic enzymes which specifically cleave various linkages in this polymer (7, 13, 16). Endo-1,4-β-xylanase (EC 3.2.1.8) cleaves the xylan backbone, and β-xylosidase (EC 3.2.1.37) cleaves xylose monomers from the nonreducing end of xylo-oligomers. Removal of the side groups is catalyzed by α-glucuronidase (EC 3.2.1.139), α-l-arabinosidase (EC 3.2.155), and acetylesterase (EC 3.1.1.72).
Although over a hundred hemicellulolytic enzymes have been purified and characterized from both fungi and bacteria (64), little is known about their regulation at the molecular level. The regulation of degradative enzyme synthesis in Bacillus spp. is complex and mediated via several mechanisms, including induction-repression, catabolite repression, and transition phase regulation (17). To date, there is only limited information regarding the transport mechanisms by which xylan degradation products enter the cell. This is surprising, since in the past few years, there have been an increasing number of studies concerning microbial xylanolytic systems. In the yeast Cryptococcus albidus (and possibly in Cryptococcus flavus), xylobiose and xylotriose are taken up by an active transport system called the β-xyloside permease (35). A transport system which exhibits preferential use of xylo-oligosaccharides over xylose may operate in Clostridium thermocellum (69). The transport of xylobiose in Streptomyces lividans does not proceed via the phosphoenolpyruvate-sugar phosphotransferase system but depends on an ATP-binding protein (MsiK) involved in energy coupling of the sugar uptake system (30). In other organisms capable of utilizing xylan, the genes involved in transport of xylobiose or xylotriose have not been cloned or characterized.
Bacillus stearothermophilus T-6 was isolated based on its ability to secrete an extracellular, thermostable, alkaline-tolerant xylanase (33). This enzyme was used in large-scale biobleaching mill trials (41) and is of potential industrial interest. Strain T-6 also produces other thermostable hemicellulolytic enzymes, genes for some of which have been cloned and characterized elsewhere (19, 20, 65).
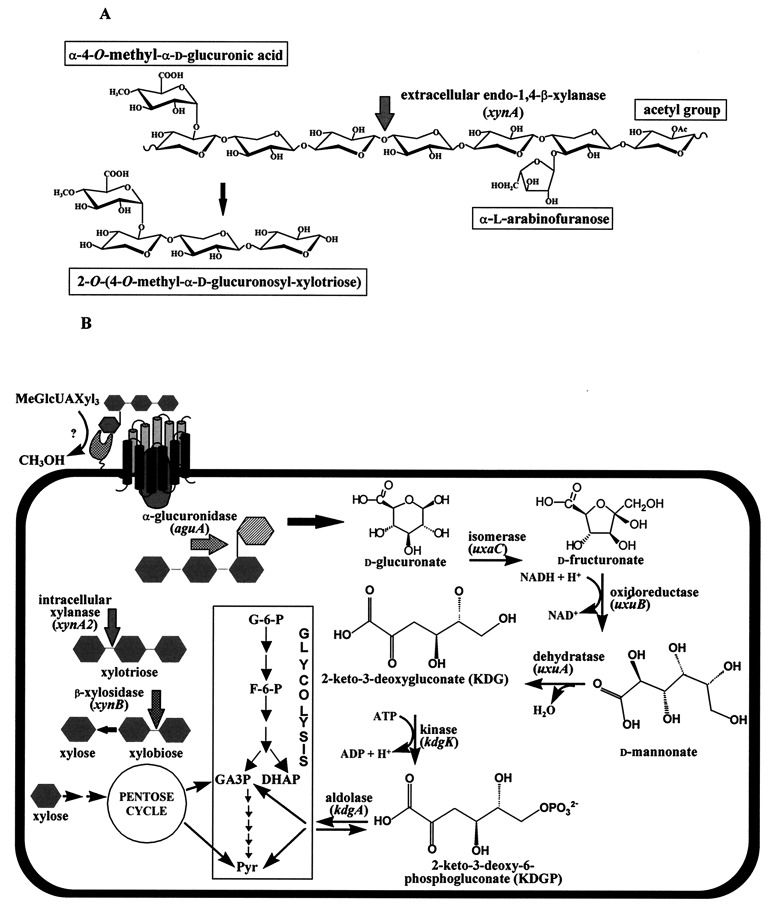
The degradation of xylan by B. stearothermophilus T-6 seems to follow the scheme in Fig. 1. An extracellular xylanase (xylanase T-6) cleaves the main backbone of xylan and generates xylobiose and short oligoxylose units (two to four sugars) with various branched substitutions. These units enter the cell by specialized permeases and are then further degraded to monomers by intracellular hemicellulases, including α-l-arabinofuranosidase (20), α-d-glucuronidase (65), and β-xylosidase (Fig. 1).
FIG. 1.
A proposed degradation pathway of MeGlcUAXyl3 in B. stearothermophilus T-6. (A) Xylan is composed of β-1,4-linked xylopyranose units which can be substituted with l-arabinofuranosyl, methyl-d-glucuronic acid, and acetyl side chains. The key enzyme in the degradation of xylan is an extracellular endo-1,4-β-xylanase (xynA). This enzyme releases short xylose units (xylobiose, xylotriose, and xylotetraose) which can be substituted with various side chains such as l-arabinose, d-glucuronic acid, or its 4-O-methyl ether, as in 2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose (MeGlcUAXyl3). (B) MeGlcUAXyl3 is demethylated and enters the cell via a specific transporting system. This system resembles the binding-protein-dependent transport systems in which a solute-binding lipoprotein interacts with integral membrane protein components that are involved in translocating the substrate across the membrane (22). Inside the cell, GlcUAXyl3 is cleaved by α-glucuronidase to yield xylotriose and d-glucuronic acid. The xylo-oligomers are hydrolyzed to xylose by intracellular xylanase and β-xylosidase. Xylose is converted into xylulose-5-phosphate, which can enter the pentose cycle. d-Glucuronic acid is converted into KDG by a three-step pathway catalyzed by uronate isomerase (uxaC), d-mannonate oxidoreductase (uxuB), and d-mannonate hydrolase (uxuA). KDG is then phosphorylated by KDG kinase (kdgK) to give KDGP, which is finally cleaved by KDGP aldolase (kdgA) to yield glyceraldehyde 3-phosphate (GA3P) and pyruvate. The latter two compounds can enter the Embden-Meyerhof-Parnas pathway.
In the present study, we describe the cloning and sequence analysis of a 23.5-kb chromosomal segment from B. stearothermophilus T-6. This segment contains a putative 15.5-kb-long operon involved in the utilization of α-d-glucuronic acid (GlcUA). Based on the identified genes, a novel transport system for branched xylo-oligosaccharides substituted with methyl d-glucuronic acid (MeGlcUA) is suggested, together with the biochemical pathway for the utilization of glucuronic acid. To date, the metabolism of d-glucuronic acid has not been described for B. stearothermophilus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. stearothermophilus T-6 (NCIMB 40222) was obtained following enrichment procedures for strains capable of producing alkaline-tolerant, extracellular, thermostable xylanases (33). Escherichia coli strains used were KW251 for the library construction in λ-EMBL3 (Promega, Madison, Wis.), XL-1 Blue for general cloning in pBluescript II KS(+) (Stratagene, La Jolla, Calif.) or pSL301 (Invitrogen, San Diego, Calif.), and JM109(DE3)(pLysS) (Promega) for expression via the T7 RNA polymerase expression system with pET11d or pET9d (Novagen, Madison, Wis.).
Growth conditions.
Growth medium for B. stearothermophilus was basic salt medium (BSM) supplemented with 0.5% glucose or xylose. BSM contained the following per liter: KH2PO4, 0.4 g; MgSO4 7H2O, 0.1 g; (NH4)2SO4, 2 g; MOPS (N-morpholinepropanesulfonic acid) buffer, 10.4 g; and trace element solution, 4 ml. Trace element solution contained the following per liter: CaCl2 7H2O, 0.92 g; FeSO4 7H2O, 1.51 g; MnSO4 4H2O, 0.148 g; ZnSO4 7H2O, 0.105 g; CuSO4 5H2O, 0.156 g. The solution was adjusted to pH 2.0 with sulfuric acid.
DNA and RNA isolation and manipulation.
B. stearothermophilus T-6 genomic DNA was isolated by the procedure of Marmur (44) as outlined by Johnson (32). Plasmid DNA was purified with the Qiagen plasmid kit (Qiagen Inc., Chatsworth, Calif.). DNA was manipulated by standard procedures (5, 58). Total RNA was isolated with the RNeasy kit (Qiagen) according to the protocol obtained from the supplier.
Construction of genomic libraries.
Genomic DNA was partially digested with Sau3A and then separated on a 0.7% agarose gel. DNA fragments of 13 to 20 kb were extracted from the gel with activated glass beads (Geneclean II kit; Bio 101, La Jolla, Calif.), ligated into EMBL3 BamHI arms, and packaged in phage particles with the EMBL3 BamHI arm cloning system together with the Packgene system (Promega).
DNA sequencing.
DNA sequencing was performed on both strands, either manually by the dideoxy chain termination method of Sanger et al. (59), with the T7 sequencing kit (Pharmacia, Uppsala, Sweden), or with an automated sequencer (Perkin-Elmer 377) at the sequencing unit in the Weizmann Institute, Rehovot, Israel.
Computer analysis.
Nucleotide and amino acid sequences were analyzed with MacVector (IBI, New Haven, Conn.) or the sequence analysis software package of the Genetics Computer Group, version 9 (University of Wisconsin, Madison). Sequence homologies were searched with FASTA (50) or BLAST (2) algorithms.
Transcriptional analyses.
All transcriptional analyses were performed on total RNA which was extracted from exponentially growing cells. Primer extension reactions were performed as described previously (47) with avian myeloblastosis virus reverse transcriptase (Promega), 40 μg of total RNA, and the primer PE_221420 (5′-GCATATTTCTCCCTCCAATCCC-3′), which was designed to correspond to the template strand of ORF1 at positions 331 to 352 (see Fig. 3). Northern (RNA) blot analysis was performed by the procedure described by Moran (47). Slot blot RNA analysis was carried out with the Manifold II slot blot system (Schleicher & Schuell, Inc., Keene, N.H.), according to the supplier’s protocol.
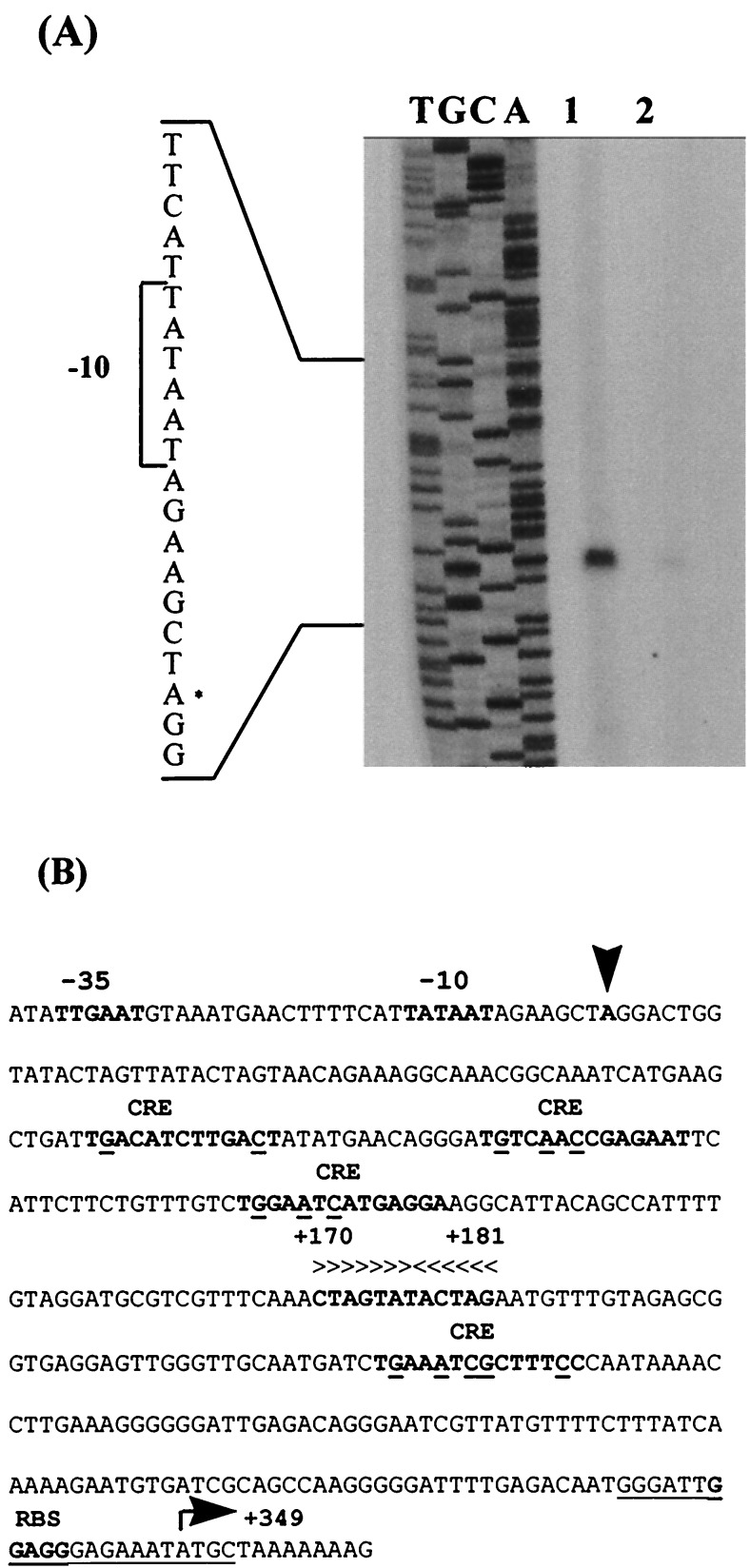
FIG. 3.
(A) Mapping the 5′ termini of the GlcUA cluster by primer extension analysis. Total RNA was isolated from mid-exponential-phase cultures of B. stearothermophilus T-6 grown in BSM supplemented with 0.5% xylose and 0.5% glucose (lane 1) or with 0.5% glucose as the sole carbon source (lane 2). Dideoxynucleotide sequence reactions were carried out with the same primer used for the reverse transcriptase reactions. The position of the transcriptional start point is indicated with an asterisk on the inferred nontemplate strand sequence. (B) Sequence data for the regulatory region. The transcriptional start point (+1) is indicated by a vertical arrowhead. The −35 and −10 regions, the proposed ribosome binding site (RBS), the initiating methionine codon, and the potential CREs are in boldface. The CRE sequence is TGT/AAANC|GNTNA/TCA, where underlined letters represent the most critical bases, N is any base, and the vertical line denotes an axis of symmetry (28, 68). The GlcUA operator is indicated by horizontal arrowheads above the inverted repeat. The sequence of the primer used for primer extension experiments is underlined.
Cloning and expression of the uxuR gene.
Based on the DNA sequence of the uxuR gene, two PCR primers that allow the in-frame cloning of the gene in the pET vectors were designed. The N-terminal primer (5′-GATCATCCATGGACTTTATCACTGCCA-3′) was made to contain an ATG translational start codon inside an NcoI restriction site (CCATGG). The C-terminal primer (5′-GATATTGGATCCTTATACAAAATAATGAGG-3′) contained a stop codon (TAG) and a BamHI restriction site (GGATCC) after the end of the gene. Following PCR amplification, the construct was cloned in vectors pET9d and pET11d (linearized with NcoI and BamHI), resulting in plasmids pET9d-uxuR and pET11d-uxuR, respectively. Expression of uxuR was carried out by growing 200-ml cultures of E. coli JM109(DE3)(pLysS) carrying pET11d-uxuR in terrific broth (58), supplemented with kanamycin (25 μg/ml) and carbenicillin (50 μg/ml) at 37°C. Induction by 4 mM isopropyl-β-d-thiogalactoside (IPTG) was carried out at a cell turbidity of 0.6 U of optical density at 600 nm. After 3 h of incubation, the cells were harvested, resuspended in 20 ml of solution A (50 mM Tris-Cl [pH 7.5], 100 mM KCl, 10% glycerol, 1 mM EDTA containing 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol), and disrupted by a single passage through a French press. Following centrifugation of the cell extract (14,000 × g for 15 min), the soluble fraction was used for gel retardation assays.
Mobility shift DNA-binding assay.
The DNA probe for the gel retardation assays was a 30-bp double-stranded DNA fragment containing the putative GlcUA operator (from positions +162 to +190). The double-stranded probe was made from two synthetic complementary oligonucleotides, 5′-TTGTTTCAAACTAGTATACTAGAATGTTTG-3′ and 5′-TTCAAACATTCTAGTATACTAGTTTGAAAC-3′. The two oligonucleotides were designed to have two noncomplementary T nucleotides at the 5′ end for end labeling with Klenow fragment in the presence of [α-32P]dATP or α-35S-dATP. The araD operator (21) was used as a nonspecific competitor DNA probe and was made from two synthetic complementary oligonucleotides, 5′-AAATAGAAAAATTGTACGTACAATAGTATAAT-3′ and 5′-AAATTATACTATTGTACGTACAATTTTTCTAT-3′. This probe was end labeled with γ-35S-dATP with T4 polynucleotide kinase.
The binding reaction mixture (30-μl total volume) contained 20 μl of solution A, 2 μg of salmon sperm DNA, 0.66 mM dithiothreitol, 33 μg of bovine serum albumin, 0.08 ng of labeled probe (about 50,000 cpm), and the indicated amount of protein. The binding mixture was incubated for 30 min at 45°C and then separated on a 6.6% nondenaturing polyacrylamide gel prepared in Tris-borate-EDTA buffer (58) and run for 1 to 2 h. Dried gels were subjected to autoradiography. Binding of UxuR to the GlcUA operator in the presence of various sugars was carried out under the same conditions described above but with 0.16 μg of crude protein extract and 10 mM indicated sugars. Aldotetraouronic acid (MeGlcUAXyl3) was obtained as described below, and a combined solution of aldotriouronic acid (MeGlcUAXyl2) (80%) and aldobiouronic acid (MeGlcUAXyl1) (20%) was obtained from Megazyme International (Dublin, Ireland). d-Glucuronic acid, d-xylose, d-glucose, l-arabinose, and xylobiose were obtained from Sigma (St. Louis, Mo.).
Preparation of 2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose.
To obtain the substrate 2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose (MeGlcUAXyl3), 4-O-methyl-d-glucurono-d-xylan (Sigma) was extensively digested with recombinant endo-β-1,4-xylanase and β-xylosidase. The two enzymes are from strain T-6 and were overexpressed in E. coli BL21(DE3) with the T7 RNA polymerase expression system. The resulting soluble products included mainly MeGlcUAXyl3, xylobiose, and xylose. These sugars were easily separated with a BioGel P-2 (Bio-Rad, Richmond, Calif.) gel filtration column (100 by 2 cm) running with H2O at room temperature. The acidic compound (MeGlcUAXyl3) was eluted in the void volume, presumably separated by partition principles. Final identification of the compound was made by thin-layer chromatography analysis and mass spectra using a TSQ-70B mass spectrometer under fast atom bombardment.
Nucleotide sequence accession number.
The 23,467-bp sequence of the uxu region has been assigned GenBank accession no. AF098273.
RESULTS
Cloning of the xylan utilization genes from B. stearothermophilus T-6.
Our general strategy for cloning xylan-utilizing genes was to screen genomic libraries for hemicellulolytic activities. A λ-EMBL3 genomic library of B. stearothermophilus T-6 was prepared, and individual phage lysates of infected E. coli host cells were tested for hemicellulolytic activities by using a mixture of p-nitrophenyl glycosides as substrates. Four independent clones (as shown by their insert size) exhibiting β-xylosidase activity were isolated. All of the four clones proved to have common regions among them and with the previously characterized 13-kb clone, λD1, which harbors the extracellular xylanase gene (xynA) (19). A compilation restriction map, together with Southern blot analysis, suggested that the combined inserts of the five clones represent a 23-kb segment of the strain T-6 chromosome (Fig. 2).
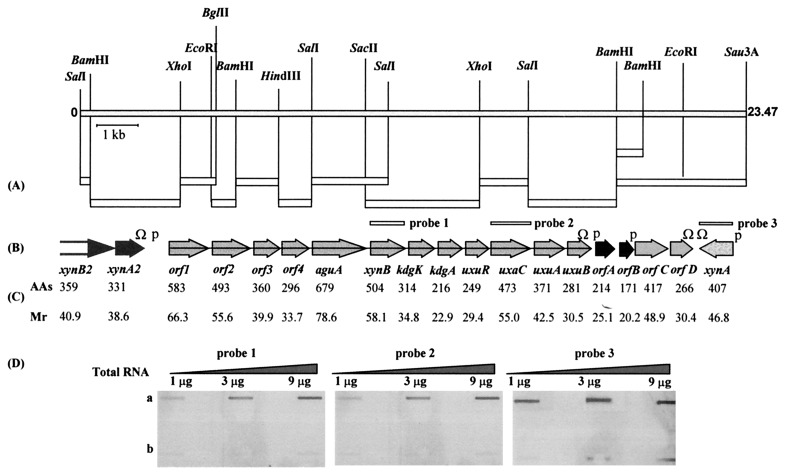
FIG. 2.
Genetic map of the 23.5-kb segment containing xylan and glucuronic acid utilization genes. (A) Fragments cloned directly from phage DNA into pKS or pSL301 are shown as open rectangles. (B) The positions of the 19 ORFs. The sequence of the N terminus of the xynB2 gene product is incomplete. The letter P indicates the proposed promoter regions, and Ω indicates rho-independent terminator-like transcription terminators. (C) The number of amino acids and the calculated molecular weight (in thousands) of the putative protein encoded by each gene are given below it. (D) Detection of xynB, uxaC, and xynA expression at the RNA level by slot blot RNA. Total RNA was isolated from mid-exponential-phase cultures of B. stearothermophilus T-6 grown in BSM supplemented with 0.5% xylose and 0.5% glucose (a) or with 0.5% glucose as the sole carbon source (b), applied to Schleicher & Schuell BA85 nitrocellulose, and annealed separately to three different 35S-labeled DNAs for xynB, uxaC, and xynA. The hybridization solution contained about 5,000 cpm of each probe per ml.
Sequence analysis.
To facilitate sequencing of the entire segment, fragments of the cloned DNAs were subcloned into appropriate vectors [pBluescript II KS(+) or pSL301] (Fig. 2A). The sequence data revealed 18 open reading frames (ORFs) and one partial ORF (xynB2) organized in at least three transcriptional units, in a region of 23,467 bp (Fig. 2B). These units include the previously characterized xynA gene (encoding xylanase T-6), a partial transcriptional unit containing the genes for an intracellular xylanase (xynA2) and β-xylosidase (xynB2), and a putative 15.5-kb-long unit consisting of 12 genes involved in d-glucuronic acid (GlcUA) utilization. In addition, there are four ORFs encoding proteins with unpredicted functions. The lengths of the ORFs and the calculated Mrs of the deduced gene products are given in Fig. 2C. The results of BLAST analysis of the various ORFs are summarized in Table 1 together with the putative ribosome-binding sites and initiation codons.
TABLE 1.
Homologies and Shine-Dalgarno analysis of the putative ORFs of the 23.5-kb chromosomal segment
| ORF | Representative homologous protein (sp./accession no.)a | Homologous gene product | Identity (%)/amino acid overlap | SD-initiation codonb |
|---|---|---|---|---|
| xynB2 | XylA (B. stearothermophilus 21/P45702) | β-Xylosidase | 94.2/359 | Incomplete in NH3 end |
| xynA2 | XynA (B. stearothermophilus 21/P45703) | Xylanase | 81.8/336 | GTTGGAGGAGGGGGAATGAAGatg |
| orf1 | ORF HO698 (Halobacterium sp. strain NRC-1/AF016485) | Unknown | 25.5/456 | AATGGGATTGGAGGGAGAAATatg |
| orf2 | LplA (B. subtilis/P37966) | Unknown | 21/381 | AGGGCATTTTTCGATACTGGGatg |
| orf3 | AraP (B. subtilis/X89810) | Integral membrane protein | 25.9/224 | AAGAAGTGAATGAATGGTATCatg |
| orf4 | MsmG (Streptococcus mutans/M77351) | Integral membrane protein | 25.3/217 | TTTTAAAGGGAGGGGAGGTCTatg |
| aguA | AguA (T. maritima/P96105) | α-Glucuronidase | 60.6/578 | GTAACAGGAGGAGAATTCGACatg |
| xynB | XynB (T. saccharolyticum/ P36906) | β-Xylosidase | 64.4/503 | CGGTGAATAGGGGAGAGGAGCatg |
| kdgK | KdgK (E. chrysanthemi/P45416) | KDG kinase | 26.5/223 | AAATGGTTTTGGAGGGACGAGatg |
| kdgA | KdgA (E. chrysanthemi/P38448) | KDGP aldolase | 30.1/196 | CGATAAAGGAGATGAACCTACatg |
| uxuR | UxuR (E. coli/P39161) | Regulatory protein | Partial homology | AAGAAAACGCATGATCATGGatg |
| uxaC | UxaC (E. coli/P42607) | Uronate isomerase | 48.1/420 | GCTTTGCGGGAGGGAGACAGTgtg |
| uxuA | UxuA (E. coli/P24215) | d-Mannonate hydrolase | 47.7/132 | TGAGTGGGGGATGAGATCAGAatg |
| uxuB | YjmF (B. subtilis/AF015825) | Putative d-mannonate oxidoreductase | 59.5/285 | AAGCAGGAAGGTGAGAAGACatg |
| orfA | YteU (B. subtilis/AF008220) | Unknown | 41.5/197 | ACAAGAAAAGGTGGATGAATTatg |
| orfB | Unknown | Unknown | No significant similarities | TCAGAGGAGGGGGAGGAGGACgtg |
| orfC | Rv2812 (Mycobacterium tuberculosis/Z81331) | Unknown | 34.2/374 | CGCGAAGGAGGGAGAACACGgtg |
| orfD | Rv2813 (M. tuberculosis/Z81331) | Unknown | 33.7/261 | TGAGACAAGGAGGAGATTCGAatg |
| xynA | XynA (Bacillus sp. strain C-125/P07528) | Extracellular xylanase | 58.7/375 | TAAAGAATGAGGGGGAATAGTatg |
The reported homologous proteins are those showing the highest score among proteins with an experimentally defined function. In the absence of relevant biochemical data, the most similar putative protein was reported.
Nucleotides which are complementary to the 3′ end of the 16S rRNA of B. subtilis (3′-UCUUUCCUCCACUAC-5′) are underlined. The ATG and GTG start codons are in lowercase letters. SD, Shine-Dalgarno sequence.
Glucuronic acid utilization gene cluster.
Within the 23.5-kb chromosomal segment, 12 genes are oriented in the same direction without any obvious transcriptional initiation signals or transcriptional terminators separating them. Thus, it is likely that they constitute a polycistronic 15.5-kb operon. Downstream from the last gene of the operon (uxuB), there is a palindromic sequence corresponding to an mRNA hairpin loop with a ΔG of −20 kcal/mol (PC/Gene; Intelligenetics, Inc., Geneva, Switzerland). The stem-loop is followed by a series of T nucleotides typical of a rho-independent terminator (10).
orf1, orf2, orf3, and orf4.
ORF1 has no similarity to other known proteins in the databases. The N terminus of ORF1 has features characteristic of signal peptides of bacterial lipoproteins and contains a putative cleavage site with a perfect match to the consensus cleavage site sequence for the lipoprotein signal peptidase II (Leu-Ala-Gly/Ala↓Cys) (67). Sequence analysis of orf1 revealed the presence of a termination codon (UAG) corresponding to amino acid position 71. Thus, it is likely that strain T-6 contains a stop mutation in the first gene of the operon. ORF2 has limited similarity to the lipoprotein LplA from Bacillus subtilis, but the protein does not contain the signal peptide sequence typical of bacterial lipoproteins. Hydropathy plots of the orf2 gene product, with the algorithms of Kyte and Doolittle (37), Eisenberg et al. (15), Klein et al. (34), and Rao and Argos (54), did not reveal transmembrane segments, and it was, therefore, classified as a peripheral protein. Hydropathy analyses for ORF3 and ORF4 showed a pattern of hydrophobic and hydrophilic regions, indicating that these proteins are likely to have membrane-spanning regions. Hydrophobic-moment analysis of ORF3 and ORF4 predicted six and five transmembrane helices, respectively. The two proteins showed homology to several integral cytoplasmic membrane proteins involved in sugar transport and contain a conserved hydrophilic segment with the consensus sequence EAA---G---------I-LP, typical of integral membrane proteins from binding-protein-dependent transport systems (12). On the basis of this signature, ORF3 and ORF4 can be assigned to the disaccharide subcluster of bacterial binding-protein-dependent permeases. This subcluster contains proteins involved in the transport of short (di- and tri-) oligosaccharides, including malto-oligosaccharides, raffinose-mellibiose, and lactose (60). It is possible that ORF1 to ORF4 constitute a four-component transport system for glucuronic acid attached to xylo-oligosaccharides.
xynB and aguA.
The xynB and aguA genes code for β-xylosidase and α-glucuronidase, respectively. Both genes were subcloned and expressed in E. coli, and their corresponding gene products were characterized biochemically (45, 65). The xynB gene product shows significant similarity to β-xylosidases from two thermophilic, anaerobic bacteria, Thermoanaerobacterium saccharolyticum B6A-RI and Caldocellum saccharolyticum (38, 42). The α-glucuronidase shows high similarity to the α-glucuronidase from the marine hyperthermophilic bacterium Thermotoga maritima MSB8 (DSM 3109) (57) and to the extracellular α-glucuronidases from Trichoderma reesei and Aspergillus tubingensis (14, 43). Neither ORF included any recognizable gram-positive bacterial signal peptide; thus, they are likely to be intracellular.
kdgK, kdgA, uxaC, uxuA, and uxuB.
Five genes in the cluster code for proteins that are homologous to enzymes involved in the catabolism of galacturonate and glucuronate (Table 1). The kdgK and kdgA gene products show high similarity to 2-keto-3-deoxygluconate (KDG) kinase and 2-keto-3-deoxy-6-phosphogluconate (KDGP) aldolase from Erwinia chrysanthemi. These enzymes are responsible for the utilization of KDG, a common degradation end product of pectic polymers. KdgK also shows some similarity to several bacterial kinases specific for fructose. The uxaC gene encodes a putative protein with high similarity to uronate isomerase from E. coli. UxuA exhibits partial similarity to d-mannonate hydrolase from E. coli. UxuB shows homology to several oxidoreductases, but even though it is likely to be a mannonate oxidoreductase, it does not have any homology with UxuB from E. coli. The uxuB gene product from strain T-6 has 59.5% (285-amino-acid overlap) identity with YjmF from B. subtilis, which was reported elsewhere as a putative mannonate oxidoreductase (55).
uxuR.
The uxuR gene is located immediately downstream of the kdgA gene and encodes a protein with weak overall similarity to several regulatory proteins. A search for sequence motifs with the program MOTIFS (Genetics Computer Group, Inc.) revealed, in the N-terminal region, a typical helix-turn-helix DNA-binding motif. This motif has similarity with regulatory proteins from the GntR family (23).
Transcriptional analyses of the GlcUA gene cluster.
To determine whether the 12 genes constitute a polycistronic operon, Northern blot and slot blot RNA analyses were performed. Total RNA was isolated from T-6 cultures grown in the presence or absence of xylose (a potential inducer) and was annealed separately to DNA probes for xynB and uxaC. The Northern blot analysis indicated that both probes hybridized to a high-molecular-weight RNA of over 15 kb in length (data not shown). Slot blot analysis revealed that the level of xynB and uxaC transcripts is about the same in cultures grown in the presence of xylose (Fig. 2D). No hybridization was detected with RNA from cultures grown without xylose, suggesting that xylose could be an inducer of the GlcUA gene cluster.
Mapping of the 5′ end of the glucuronic acid (GlcUA) locus.
By primer extension analysis, the apparent transcriptional start point of the GlcUA locus was assigned to an A nucleotide, 349 bases upstream from the initiation ATG codon of orf1 (Fig. 3A). The potential −35 region (TTGAAT), with four of six bases matching the ςA consensus, is separated by 17 bp from the potential −10 region (TATAAT), which matches the ςA consensus perfectly (48). A perfect 12-bp inverted repeat (5′-CTAGTATACTAG-3′), resembling operator sequences, is located downstream of the promoter region between nucleotides +170 and +181. Based on sequence homology, four potential operator sites for catabolite-responsive regulation (catabolite-responsive elements [CREs]) were located between the promoter and the ATG initiation codon. The CRE consensus sequence is TGWNANCGNTNWCA (W = A or T) (28, 68) (Fig. 3B).
Expression of the UxuR protein in E. coli.
The uxuR gene was cloned via PCR in T7 RNA polymerase expression vectors. Attempts to introduce pET9d-uxuR into the T7 RNA polymerase-containing E. coli JM109(DE3) or JM109(DE3)(pLysS) were unsuccessful; no viable transformants were obtained, suggesting that the UxuR protein is highly toxic to E. coli. In these strains, the basal level (uninduced) of T7 RNA polymerase can direct measurable expression of target genes (63). A second attempt to express the UxuR protein was made by using the pET11d vector. This vector contains the lacI gene and a 25-bp lac operator sequence downstream from the T7 promoter region. Binding of the lac repressor at this site effectively reduces transcription by T7 polymerase. Indeed, JM109(DE3)(pLysS) cells harboring the pET11d-uxuR vector were viable and permitted low-level expression of UxuR.
UxuR binds to the GlcUA operator.
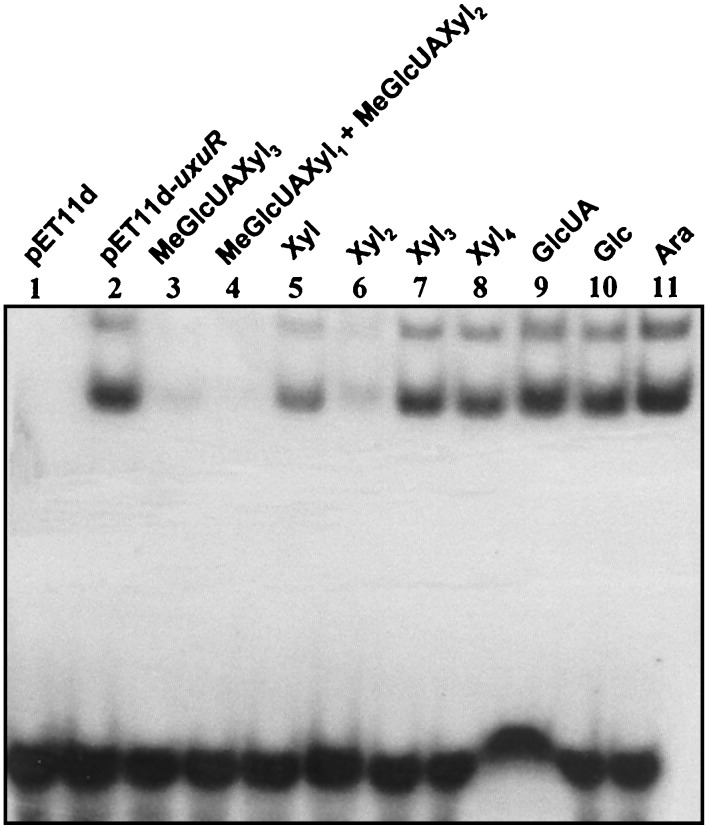
The ability of the UxuR protein to bind the potential GlcUA operator was tested in gel retardation assays. The electrophoretic mobility of the GlcUA DNA fragment was retarded when the fragment was incubated with a cell extract of E. coli expressing UxuR (Fig. 4A). Two bands with shifted mobilities were usually observed, indicating that two different complexes are formed. No DNA-protein complex was formed when the pET11d-containing extract was used (Fig. 4A). Addition of a 250-fold excess of an unlabeled DNA fragment containing the GlcUA operator eliminated the labeled-complex formation, suggesting specific interaction (Fig. 4B). The binding specificity was confirmed by showing that UxuR did not shift an unrelated 32-bp fragment containing a 14-bp invert repeat (the araD operator), and this fragment did not prevent formation of a complex of UxuR with its operator (Fig. 4C). To identify the molecular inducer of the GlcUA operon, the binding of UxuR to the GlcUA operator in the presence of various sugars was tested (Fig. 5). Binding was prevented in the presence of 10 mM aldotetraouronic acid (MeGlcUAXyl3), a solution of aldotriouronic acid (MeGlcUAXyl2) (80%) and aldobiouronic acid (MeGlcUAXyl1) (20%) and xylobiose. Xylose at 10 mM reduced the binding only partially, whereas xylotriose, xylotetraose, d-glucose, d-glucuronic acid, and l-arabinose showed no effect. Based on these results, MeGlcUAXyl3 seems to be the best inducer of the GlcUA operon in vitro.
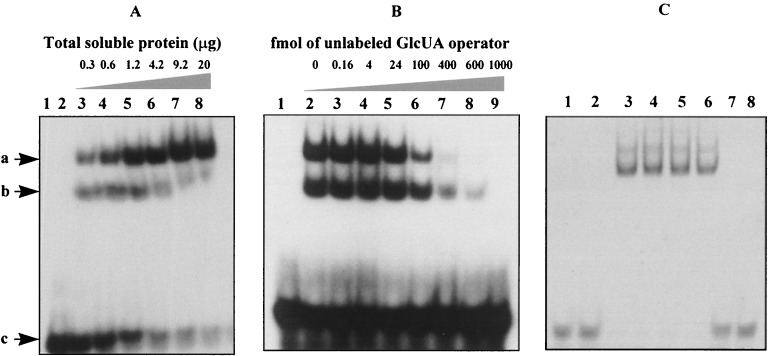
FIG. 4.
(A) Gel retardation of a 32P-labeled GlcUA operator fragment by crude extracts from E. coli cells producing UxuR. All lanes contained about 4 fmol (0.08 ng) of radioactively labeled DNA fragment containing the synthetic GlcUA operator. Lane 1 contained no extract. Lane 2 contained 20 μg of crude extract from cells carrying only the vector (pET11d). Lanes 3 to 8 contained different amounts of crude extracts from E. coli producing UxuR. The shifted bands are indicated by arrows: a, higher-mobility band; b, lower-mobility band; c, free DNA (not shifted). (B) Competition experiments using unlabeled synthetic GlcUA operator. Lane 1 contained 0.62 μg of crude extract from cells carrying only the vector (pET11d). Lanes 2 to 9 contained crude extracts from E. coli producing UxuR together with different amounts of the unlabeled synthetic GlcUA operator. (C) Competition experiments using synthetic araD operator. Lanes 1 to 6 contained the 35S-labeled synthetic GlcUA operator with no extract (lane 1); extract from cells carrying only the vector (pET11d) (lane 2); and extracts from E. coli producing UxuR together with 0, 500, 1,000, and 2,000 fmol of unlabeled synthetic araD operator, (lanes 3 to 6, respectively). Lanes 7 and 8 contained 4 fmol of 35S-labeled synthetic araD operator without (lane 7) or with (lane 8) 20 μg of crude extracts from E. coli producing UxuR.
FIG. 5.
Binding of UxuR to the GlcUA operator in the presence of various sugars. All lanes contained 0.08 ng of radioactively labeled DNA. Lane 1 contained no extract. Lane 2 contained 0.16 μg of crude extract from cells carrying only the vector (pET11d). Lanes 3 to 11 contained 0.16 μg of crude extracts from E. coli producing UxuR (pET11d-uxuR) together with a 10 mM concentration of the following sugars: aldotetraouronic acid [2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose (MeGlcUAXyl3)] (lane 3), a solution of aldobiouronic acid (MeGlcUAXyl1) (20%) and aldotriouronic acid (MeGlcUAXyl2) (80%) (lane 4), xylose (Xyl) (lane 5), xylobiose (Xyl2) (lane 6), xylotriose (Xyl3) (lane 7), xylotetraose (Xyl4) (lane 8), d-glucuronic acid (GlcUA) (lane 9), d-glucose (Glc) (lane 10), and l-arabinose (Ara) (lane 11).
DISCUSSION
The sequencing of a 23.5-kb segment of the B. stearothermophilus T-6 chromosome revealed a cluster of genes which play a role in the utilization of xylan and glucuronic acid. The segment contains a putative 15.5-kb operon, consisting of 12 genes, involved in the utilization of d-glucuronic acid (GlcUA).
Transport system of MeGlcUAXyl3.
The first four genes in the GlcUA cluster (orf1, -2, -3, and -4) appear to encode a binding-protein-dependent transport system (26). Such systems normally include two identical or similar integral membrane proteins; an extracytoplasmic, ligand-specific, binding protein (1, 3); and two identical or similar ATP-binding proteins (4). ORF1 appears to correspond to the extracellular substrate-binding protein, anchored to the cytoplasmic membrane via an amino-lipid group (22, 51). If so, it is presumably inactive in strain T-6, since orf1 contains a stop codon at position 71. In this regard, when strain T-6 is grown on 4-O-methyl-d-glucurono-d-xylan, MeGlcUAXyl3 remains in the culture fluid whereas xylose and xylobiose are consumed. To determine the role of ORF1 and the transport system, we are now in the process of isolating T-6 mutants capable of utilizing MeGlcUAXyl3 as a sole carbon source. ORF1 may also function as a methylesterase removing the methoxyl group from methylglucuronic acid. In E. chrysanthemi 3939, pectin methylesterase genes (pem) are grouped into several clusters, and one of these genes (pemB) has been characterized as encoding an outer membrane lipoprotein (61).
The deduced amino acid sequences of ORF3 and ORF4 reveal several features of the type often associated with integral membrane proteins of the binding-protein-dependent transport system. ORF2 does not contain the typical ATP-binding domain which is found in all binding-protein-dependent transport systems described so far (25). Thus, either there is an additional ATPase gene located elsewhere on the chromosome or this binding-protein-dependent transport system is ATP independent. We note that both membrane components (ORF3 and ORF4) contain the conserved EAAX3GX9IXLP motif which is thought to be involved in the interaction with ATP-binding subunits (60).
The most likely substrate for the putative transport system is aldotetraouronic acid [2-O-(4-O-methyl-α-d-glucupyranosyluronic acid)-d-xylotriose] (MeGlcUAXyl3). This conclusion is based on the following observations. (i) MeGlcUAXyl3 (together with xylose and xylobiose) is found in the supernatant of T-6 cultures grown on 4-O-methyl-d-glucurono-d-xylan. In addition, these compounds are the main end products of 4-O-methyl-d-glucurono-d-xylan following total hydrolysis with recombinant xylanase T-6. (ii) Strain T-6 possesses three intracellular enzymes that participate in the breakdown of MeGlcUAXyl3. Two of the enzymes, α-glucuronidase and β-xylosidase, are part of the GlcUA gene cluster. The purified α-glucuronidase was shown elsewhere to hydrolyze MeGlcUAXyl3 to 4-O-methyl-d-glucuronic acid and xylotriose (65). A gene for an intracellular xylanase, xynA2 (required for the hydrolysis of xylotriose), is located just upstream of the GlcUA cluster. The XynA2 protein has 81.8% identity (336-amino-acid overlap) with a xylanase (xynA gene) from B. stearothermophilus 21 (6) that, to the best of our judgment, lacks a signal sequence and has erroneously been described as an extracellular xylanase. (iii) In gel retardation assays, MeGlcUAXyl3 was particularly effective in preventing the binding of UxuR to a synthetic operator.
In B. subtilis, galacturonate and glucuronate enter the cells via the hexuronate transporter encoded by a single gene (exuT) (46).
Metabolism of glucuronic acid.
Five genes of the cluster are apparently involved in the metabolism of glucuronic acid (Fig. 1). Based on sequence homologies for these genes, it appears that in strain T-6, d-glucuronic acid is metabolized via a pathway similar to that of E. chrysanthemi, E. coli, and B. subtilis (29, 52, 56). In these organisms, d-glucuronic acid is converted to KDG by a three-step pathway catalyzed by uronate isomerase (uxaC), d-mannonate oxidoreductase (uxuB), and d-mannonate hydrolase (uxuA). KDG is then phosphorylated by KDG kinase (kdgK) to give KDGP, which is finally cleaved by KDGP aldolase (kdgA) to yield glyceraldehyde 3-phosphate and pyruvate. The latter two compounds can enter the Embden-Meyerhof-Parnas pathway (Fig. 1).
A schematic view of the catabolism of MeGlcUAXyl3 by B. stearothermophilus T-6 is shown in Fig. 1. MeGlcUAXyl3 is demethylated and enters the cell through a specialized transport system. Inside the cell, GlcUAXyl3 is hydrolyzed to d-glucuronic acid and xylotriose by the intracellular α-d-glucuronidase. Xylotriose is hydrolyzed to xylose and xylobiose; the latter is further hydrolyzed to xylose by a β-xylosidase (XynB). Upon isomerization and phosphorylation of xylose, xylulose-5-phosphate enters the pentose cycle (27, 31).
Regulatory sites of the glucuronic acid gene cluster.
The regulatory sites of the GlcUA operon were studied by primer extension and gel retardation analyses. The apparent transcriptional start point corresponds well with putative −10 and −35 regions for promoters recognized by the vegetative sigma factors of E. coli, B. subtilis, and B. stearothermophilus (40). The 5′ untranslated region preceding the first coding sequence is 349 nucleotides in length and, based on sequence analysis, contains several potential regulatory elements. A perfect 12-bp inverted repeat is located between nucleotides +170 and +181 with respect to the transcription start site. This sequence resembles an operator site for a DNA-binding protein; a DNA fragment containing this region was shown by the gel retardation assays to bind UxuR specifically.
Surprisingly, the operator site does not overlap the −45 to +20 region, as is the case for most known operators (11). In a compilation of 76 repressible promoters from E. coli (11), only in the purB gene was the operator located at a significant distance from the promoter. In that case, repression of the purB gene is by a transcriptional roadblock mechanism that inhibits transcript elongation (24). In the case of the B. subtilis hut operon, the hutOCR2 site, which is required for complete hut catabolite repression, is located over 200 bp downstream of the transcriptional start site (70). Further analysis of the regulatory region revealed four potential CREs. In other gram-positive bacteria, CREs are sites of negative regulation by the CcpA protein. Recently, we have cloned and sequenced the ccpA gene from strain T-6; the deduced amino sequence showed 68% identity to that of CcpA of B. subtilis (36).
Regulatory protein UxuR.
The UxuR protein appears to be highly toxic to E. coli, perhaps due to binding to essential elements on host DNA. A similar phenomenon was described by Ramesh et al. (53), when transcription activator C of bacteriophage Mu was overexpressed in a T7 expression system. Only by applying stringent control over basal expression levels of T7 RNA polymerase were we able to produce the uxuR gene product successfully in E. coli.
Gel retardation assays indicated that MeGlcUAXyl3 was the most effective compound tested in preventing the binding of UxuR to the synthetic operator. Xylobiose and MeGlcUAXyl1-2 were also effective, although xylotriose was not. Xylose had only a partial effect on the binding in vitro. However, xylose can induce the GlcUA operon in vivo based on the slot blot RNA analysis and the fact that α-glucuronidase activity can be detected in cultures grown in the presence of xylose alone. Thus, the mutation in orf1 probably prevents MeGlcUAXyl3 from entering the cells but does not block transcription (induced by xylose) completely. In A. tubingensis, the expression of the aguA gene is induced by xylose but not by glucuronic acid (14). It should be mentioned that strain T-6 grows very well on xylose and xylan, probably due to an effective transport system for xylose, whereas B. subtilis 168 grows on xylan but grows very poorly on xylose (39). In strain T-6, xylose is also the molecular inducer of the extracellular xylanase T-6 (xynA) (19) and of xylulose kinase (xylB) (62). Since the regulatory region of the xynA gene does not contain an operator sequence resembling that of the GlcUA operon, it is likely that at least two regulatory proteins are involved in the regulation of the xylose- and the glucuronic acid-utilizing genes. Although UxuR binds the GlcUA operator in vitro, its direct role as a repressor has to be verified. Since strain T-6 is not transformable, we will be using B. subtilis as a host for expressing the uxuR gene and measuring its effect on the expression of a GlcUA regulatory region-lacZ fusion.
The uxuR gene is part of the putative GlcUA operon, suggesting that the operon is autoregulated. The fact that the uxuR gene lacks the canonical GGAGG sequence in its ribosome binding site (Table 1) suggests that the translation of the gene is poor compared to that of the other genes in the operon (49). In the case of the gluconate (gnt) operon of B. subtilis, which also encodes its own transcriptional regulator, the gnt repressor appears to be posttranscriptionally repressed by an unidentified mechanism (18). The regulatory protein of the hexuronate utilization locus, ExuR, is also likely to regulate its own operon (46). This type of autoregulation has the advantage that the expression of the operon is controlled over a wide range of inducer concentrations. However, the significance of this phenomenon for the physiology of glucuronic acid and xylan utilization has yet to be determined.
ACKNOWLEDGMENTS
This research was supported by grants no. 93-171 and 96-178 (to Y.S. and A.L.S.) from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. Support was also provided by the Technion Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation, Germany.
REFERENCES
- 1.Adams M D, Wagner L M, Graddis T J, Landick R, Antonucci T K, Gibson A L, Oxender D L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport system of Escherichia coli. J Biol Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ames G F-L. Bacterial periplasmic transport systems: structure, mechanism and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 4.Ames G F-L, Mimura C S, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia colito human: traffic ATPases. FEMS Microbiol Rev. 1990;75:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 6.Baba T, Shinke R, Nanmori T. Identification and characterization of clustered genes for thermostable xylan-degrading enzymes, β-xylosidase and xylanase, of Bacillus stearothermophilus21. Appl Environ Microbiol. 1994;60:2252–2258. doi: 10.1128/aem.60.7.2252-2258.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286–290. [Google Scholar]
- 8.Biely P. Biochemical aspects of the production of microbial hemicellulases. In: Coughlan M P, Hazlewood G P, editors. Hemicellulose and hemicellulases. London, United Kingdom: Portland Press; 1993. pp. 29–52. [Google Scholar]
- 9.Browning B L. The composition and chemical reactions of wood. In: Browning B L, editor. The chemistry of wood. New York, N.Y: John Wiley and Sons, Inc.; 1963. pp. 58–101. [Google Scholar]
- 10.Carafa Y A, Brody E, Thermes C. Prediction of rho-independent Escherichia colitranscription terminators: a statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 11.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dassa E, Hofnung M. Sequence of gene malG in Escherichia coliK12: homologies between integral membrane components from binding-protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker R F H, Richards G M. Hemicellulases: their occurrence, purification, properties and mode of action. Adv Carbohydr Chem Biochem. 1985;32:277–352. doi: 10.1016/s0065-2318(08)60339-x. [DOI] [PubMed] [Google Scholar]
- 14.De Vries R P, Poulsen C H, Madrid S, Visser J. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J Bacteriol. 1998;180:243–249. doi: 10.1128/jb.180.2.243-249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson K E L, Blanchette R A, Ander P. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 17.Ferrari E, Jarnagin A S, Schmidt B F. Commercial production of extracellular enzymes. In: Sonenshein A L, Hoch J A, Losick R M, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 917–937. [Google Scholar]
- 18.Fujita Y, Fujita T, Miwa Y. Evidence for posttranscriptional regulation of synthesis of the Bacillus subtilisGnt repressor. FEBS Lett. 1990;267:71–74. doi: 10.1016/0014-5793(90)80290-y. [DOI] [PubMed] [Google Scholar]
- 19.Gat O, Lapidot A, Alchanati I, Regueros S, Shoham Y. Cloning and DNA sequence of the gene coding for Bacillus stearothermophilusT-6 xylanase. Appl Environ Microbiol. 1994;60:1889–1896. doi: 10.1128/aem.60.6.1889-1896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilead S, Shoham Y. Purification and characterization of an α-l-arabinofuranosidase from Bacillus stearothermophilusT-6. Appl Environ Microbiol. 1995;61:170–174. doi: 10.1128/aem.61.1.170-174.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilead-Gropper S. Ph.D. thesis. Haifa, Israel: Technion-Israel Institute of Technology; 1998. [Google Scholar]
- 22.Gilson E, Alloing G, Schmidt T, Claverys J-P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in Gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haydon D J, Guest J R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;79:291–296. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 24.He B, Zalkin H. Repression of Escherichia coli purBis by a transcriptional roadblock mechanism. J Bacteriol. 1992;174:7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 26.Higgins C F, Hyde S C, Mimmack M M, Gileadi U, Gill D R, Gallagher M P. Binding protein-dependent transport systems. J Bioenerg Biomembr. 1990;22:571–592. doi: 10.1007/BF00762962. [DOI] [PubMed] [Google Scholar]
- 27.Horecker B L. Pentose metabolism in bacteria. New York, N.Y: John Wiley & Sons, Inc.; 1962. [Google Scholar]
- 28.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for Gram-positive bacteria? Mol Microbiol. 1995;15:359–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 29.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Hexuronate catabolism in Erwinia chrysanthemi. J Bacteriol. 1987;169:1223–1231. doi: 10.1128/jb.169.3.1223-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurtubise Y, Shareck F, Kluepfel D, Morosoli R. A cellulase/xylanase-negative mutant of Streptomyces lividans1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding protein. Mol Microbiol. 1995;17:367–377. doi: 10.1111/j.1365-2958.1995.mmi_17020367.x. [DOI] [PubMed] [Google Scholar]
- 31.Jeffries T W. Utilization of xylose by bacteria and fungi. Adv Biochem Eng Biotechnol. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- 32.Johnson J L. Genetic characterization. In: Gerhardt P, Murray R G E, Costilow E W, Nester W A, Wood N R, Krieg R, Philips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 450–472. [Google Scholar]
- 33.Khasin A, Alchanati I, Shoham Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilusT-6. Appl Environ Microbiol. 1993;59:1725–1730. doi: 10.1128/aem.59.6.1725-1730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein P, Kanehisa M, Delisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 35.Kratky Z, Biely P. Inducible beta-xylosid permease as a constituent of the xylan-degrading enzyme system of the yeast Cryptococcus albidus. Eur J Biochem. 1980;112:367–373. doi: 10.1111/j.1432-1033.1980.tb07214.x. [DOI] [PubMed] [Google Scholar]
- 36.Kraus A, Kuster E, Wagner A, Hoffmann K, Hillen W. Identification of a co-repressor binding site in catabolite control protein CcpA. Mol Microbiol. 1998;30:955–963. doi: 10.1046/j.1365-2958.1998.01123.x. [DOI] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y E, Zeikus J G. Genetic organization, sequence and biochemical characterization of recombinant β-xylosidase from Thermoanaerobacterium saccharolyticumstrain B6A-RI. J Gen Microbiol. 1993;139:1235–1243. doi: 10.1099/00221287-139-6-1235. [DOI] [PubMed] [Google Scholar]
- 39.Linder C, Stukle J, Hecker M. Regulation of xylanolytic enzymes in Bacillus subtilis. Microbiology. 1994;140:753–757. doi: 10.1099/00221287-140-4-753. [DOI] [PubMed] [Google Scholar]
- 40.Lonetto M, Gribskov M, Gross C A. The ς70family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundgren K R, Bergkvist L, Hogman S, Joves H, Eriksson G, Bartfai T, van der Laan J, Rosenberg E, Shoham Y. TCF mill trail on softwood pulp with Korsnäs thermostable and alkaline stable xylanase T-6. FEMS Microbiol Rev. 1994;13:365–368. [Google Scholar]
- 42.Luthi E, Love D R, McAnulty J, Wallace C, Caughey P A, Saul D, Bergquist P L. Cloning, sequence analasis, and expression of genes encoding xylan-degrading enzymes from the thermophile “Caldocellum saccharolyticum.”. Appl Environ Microbiol. 1990;56:1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margolles-Clark E, Saloheimo M, Siika-Aho M, Penttila M. The α-glucuronidase-encoding gene of Trichoderma reesei. Gene. 1996;172:171–172. doi: 10.1016/0378-1119(96)00167-9. [DOI] [PubMed] [Google Scholar]
- 44.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 45.Mechaly A. Ph.D. thesis. Haifa, Israel: Technion-Israel Institute of Technology; 1998. [Google Scholar]
- 46.Mekjian K R, Bryan E M, Beall B W, Moran C P. Regulation of hexuronate utilization in Bacillus subtilis. J Bacteriol. 1999;181:426–433. doi: 10.1128/jb.181.2.426-433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran C P. Measuring gene expression in Bacillus. In: Harwood C R, Cutting S M, editors. Molecular biological methods for bacillus. Chichester, United Kingdom: John Wiley & Sons; 1990. pp. 267–293. [Google Scholar]
- 48.Moran C P, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 49.Mountain A. Gene expression for Bacillus subtilis. In: Harwood C R, editor. Bacillus. New York, N.Y: Plenum Press; 1989. pp. 73–114. [Google Scholar]
- 50.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtilisplays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 52.Pujic P, Dervyn R, Sorokin A, Ehrlich S D. The kdgRKAT operon of Bacillus subtilis: detection of the transcript and regulation by the kgdR and ccpAgenes. Microbiology. 1998;144:3111–3118. doi: 10.1099/00221287-144-11-3111. [DOI] [PubMed] [Google Scholar]
- 53.Ramesh V, De A, Nagaraja V. Overproduction and purification of C protein, the late gene transcription activator from phage Mu. Protein Expr Purif. 1994;5:379–384. doi: 10.1006/prep.1994.1055. [DOI] [PubMed] [Google Scholar]
- 54.Rao J K M, Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- 55.Rivolta C, Soldo B, Lazarevic V, Joris B, Mauel C, Karamata D. A 37.7 kb DNA fragment from Bacillus subtilischromosome containing a putative 12.3 kb operon involved in hexuronate catabolism and a perfectly symmetrical hypothetical catabolite-responsive element. Microbiology. 1998;144:877–884. doi: 10.1099/00221287-144-4-877. [DOI] [PubMed] [Google Scholar]
- 56.Robert-Baudouy J, Portalier R, Stoeber F. Regulation of hexuronate system genes in Escherichia coli K-12: multiple regulation of the uxu operon by exuR and uxuRgene products. J Bacteriol. 1981;145:211–220. doi: 10.1128/jb.145.1.211-220.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruile P, Winterhalter C, Liebl W. Isolation and analysis of a gene encoding α-glucuronidase, an enzyme with a novel primary structure involved in the breakdown of xylan. Mol Microbiol. 1997;23:267–279. doi: 10.1046/j.1365-2958.1997.2011568.x. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saurin W, Koster W, Dassa E. Bacterial binding protein-dependent permeases: characterization for distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 61.Shevchik V E, Condemine G, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Characterization of pectin methylesterase B, an outer membrane lipoprotein in Erwinia chrysanthemi3937. Mol Microbiol. 1996;19:455–466. doi: 10.1046/j.1365-2958.1996.389922.x. [DOI] [PubMed] [Google Scholar]
- 62.Shulami S. M.S. thesis. Haifa, Israel: Technion-Israel Institute of Technology; 1995. [Google Scholar]
- 63.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 64.Sunna A, Antranikian G. Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol. 1997;17:39–67. doi: 10.3109/07388559709146606. [DOI] [PubMed] [Google Scholar]
- 65.Teplitsky A, Shulami S, Moryles S, Zaide G, Shoham Y, Shoham G. Crystallization and preliminary X-ray analysis of α-d-glucuronidase from Bacillus stearothermophilusT-6. Acta Crystallogr Sect D. 1999;55:869–872. doi: 10.1107/s0907444998012918. [DOI] [PubMed] [Google Scholar]
- 66.Timell T E. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol. 1967;1:45–70. [Google Scholar]
- 67.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 68.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolic repression operator sequences in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiegel G, Mothershed C P, Puls J. Differences in xylan degradation by various noncellulolytic thermophilic anaerobes and Clostridium thermocellum. Appl Environ Microbiol. 1985;49:656–659. doi: 10.1128/aem.49.3.656-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wray L V, Pettengill J F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requirs a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]