Abstract
Five endopectate lyases from the phytopathogenic bacterium Erwinia chrysanthemi, PelA, PelB, PelD, PelI, and PelL, were analyzed with respect to their modes of action on polymeric and oligomeric substrates (degree of polymerization, 2 to 8). On polygalacturonate, PelB showed higher reaction rates than PelD, PelI, and PelA, whereas the reaction rates for PelL were extremely low. The product progression during polygalacturonate cleavage showed a typical depolymerization profile for each enzyme and demonstrated their endolytic character. PelA, PelI, and PelL released oligogalacturonates of different sizes, whereas PelD and PelB released mostly unsaturated dimer and unsaturated trimer, respectively. Upon prolonged incubation, all enzymes degraded the primary products further, to unsaturated dimer and trimer, except for PelL, which degraded the primary products to unsaturated tetramer and pentamer in addition to unsaturated dimer and trimer. The bond cleavage frequencies on oligogalacturonates revealed differences in the modes of action of these enzymes that were commensurate with the product progression profiles. The preferential products formed from the oligogalacturonates were unsaturated dimer for PelD, unsaturated trimer for PelB, and unsaturated tetramer for PelI and PelL. For PelA, preferential products were dependent on the sizes of the oligogalacturonates. Whereas PelB and PelD displayed their highest activities on hexagalacturonate and tetragalacturonate, respectively, PelA, PelI, and PelL were most active on the octamer, the largest substrate used. The bond cleavage frequencies and reaction rates were used to estimate the number of subsites of each enzyme.
Erwinia chrysanthemi is a plant pathogenic enterobacterium which causes soft-rot disease. This bacterium secretes various enzymes that permit the degradation of pectin, which is present in the middle lamella and primary plant cell walls. Pectin is a complex carbohydrate consisting of smooth homogalacturonan regions, where the backbone is partially methylesterified and/or acetylated (4), and of the so-called hairy regions, consisting of alternating galacturonate-rhamnose stretches. To the rhamnose residues, side chains like arabinans or galactans can be attached (17). To date, pectin methylesterase, pectin acetylesterase, pectate lyase, pectin lyase, and polygalacturonase activities have been detected in E. chrysanthemi (1, 6, 19). Among those activities, the pectate lyases appear to be the most abundant, and they have been shown to play an important role in soft-rot disease (2). The pectate lyases cleave internal glycosidic bonds in polygalacturonate by β-elimination, releasing both saturated and unsaturated oligogalacturonates. The pectate lyase activity of E. chrysanthemi results from the combined action of numerous pectate lyases, including five major enzymes, PelA, PelB, PelC, PelD, and PelE (23), and five minor enzymes, PelX (3, 21), PelL (10, 18), PelZ (14), PelI (20), and PelW (22). All pectate lyases are extracellular and have an endo mode of action, except for PelW and PelX, which are localized in the cytoplasm and the periplasm, respectively, and have an exolytic activity (21, 22).
With three-dimensional (3D) structures now available for three pectate lyases, namely, Plc and Ple from E. chrysanthemi EC16 (9, 24, 25) and Pel from Bacillus subtilis (13), we have addressed the mode of action of five pectate lyases to understand their specificity as determined by the nature and number of subsites. PelA, PelB, and PelD represent the acidic, neutral, and basic pectate lyases from family 1, respectively (5), and PelI and PelL are representatives of families 3 and 4, respectively (20). The enzymes were studied with respect to bond cleavage frequencies and reaction rates on oligogalacturonates with degrees of polymerization (n) of 2 to 8 and with respect to the reaction rates and the product progression on polygalacturonate. This provides a quantitative basis for their different modes of action and allows an estimation of the number of subsites.
MATERIALS AND METHODS
Abbreviations.
The abbreviations used are as follows: Pel, pectate lyase; (GalpA)n, oligogalacturonate with indicated degree of polymerization (n); u(GalpA)n, Δ4,5-unsaturated oligogalacturonate with indicated degree of polymerization (n); HPAEC, high-performance anion-exchange chromatography; PAD, pulsed amperometric detection.
Overproduction of the pectate lyases.
The bacterial strains, plasmids, and techniques used to overproduce and purify the five pectate lyases examined in the present study were described previously (18, 20, 23). Protein concentrations were determined by the Bio-Rad protein assay with bovine serum albumin as a standard.
Substrates.
The oligogalacturonates of defined length (n = 2 to 8) were prepared as described previously (7). Polygalacturonate (degree of polymerization, 150 [8]) was obtained from U.S. Biochemical Corp. (Cleveland, Ohio).
Enzymatic assays.
Activities on polygalacturonate and oligogalacturonates were determined spectrophotometrically by measuring the change in absorbance at 235 nm due to the formation of Δ4,5-unsaturated products (ɛ235 = 4,600 M−1 · cm−1 [11]). All assays were conducted at 37°C in 0.5 ml with 2.5 g of polygalacturonate liter−1 or 0.5 mM oligogalacturonate (n = 2 to 8) in 20 mM buffer–1 mM CaCl2. The buffers used were Tris-HCl (pH 8.0) for PelA, PelD, and PelL and 2-amino-methyl-propanol–HCl (pH 9.0) for PelB and PelI. These buffers were chosen after determination of the pH optima. One unit of pectate lyase activity is the amount of enzyme which liberates 1 μmol of product per min.
Mode of action.
For the determination of the bond cleavage frequencies on oligogalacturonates (n = 2 to 8) and for the analysis of the products released from polygalacturonate, enzyme reactions were performed under conditions identical to those described for the enzymatic assays: 2.5 g of polygalacturonate liter−1 or 0.5 mM oligogalacturonate (n = 2 to 8), 1 mM CaCl2, and 20 mM Tris-HCl (pH 8.0) (for PelA, PelD, and PelL) or 2-amino-methyl-propanol–HCl (pH 9.0) (for PelB and PelI).
To determine the bond cleavage frequencies, enzyme amounts were chosen to minimize the risk of secondary cleavage product formation. Reactions were stopped by lowering the pH to 3.5 to 4 by the addition of 0.2 volume of 1.2% (vol/vol) acetic acid. Reaction products were analyzed by HPAEC with PAD (Dionex Inc., Sunnyville, Calif.) and UV detection at 235 nm (Pye-Unicam LC-UV detector). The elution conditions used were the same as those outlined previously (12). The saturated products were quantitated from the PAD signal and the PAD response from a calibration mixture of oligogalacturonates (n = 1 to 8) at a concentration of 0.1 mM each. The unsaturated products formed were estimated from the peak areas of the UV profile. From the combined data, the bond cleavage frequencies were calculated.
The product formation during polygalacturonate cleavage was studied for up to 24 h. Normalized amounts of enzyme were added (1.80 U/ml for PelA, 2.13 U/ml for PelB, 1.87 U/ml for PelD, 2.08 U/ml for PelI, and 1.90 U/ml for PelL). At timed intervals, 50-μl samples were withdrawn and reactions were stopped by lowering the pH to 3.5 to 4 by the addition of 0.2 volume of 1.2% (vol/vol) acetic acid. Reaction products were analyzed by HPAEC as described above. The unsaturated products formed were quantitated from the UV profiles and a standard containing unsaturated dimer at 0.2 mM. For each enzyme, the percentage of substrate conversion after 24 h was estimated from the combined mass of the unsaturated products formed as quantified by HPAEC.
3D structure modelling.
In order to model the 3D structures of PelB (accession no. X67475) and PelD (accession no. AJ132101) based on the E. chrysanthemi EC16 Plc (PDB accession no. 2PEC) and Ple (PDB accession no. 1PCL) structures, respectively, the Modeler routine of the Homology module as provided by the program Insight II (version 97.0; Molecular Simulations Inc., San Diego, Calif.) was used. The quality of the models was estimated from Ramachandran plots.
RESULTS AND DISCUSSION
Reaction rates and product progression on polygalacturonate.
The five enzymes studied showed very different specific activities on polygalacturonate (Table 1). PelB and PelD were the most active, whereas PelI and PelA showed less activity. The activity of PelL was very low.
TABLE 1.
Specific activities of E. chrysanthemi 3937 endopectate lyases at their pH optima with polygalacturonate as the substratea
| Enzyme | pH | Sp act (U · mg−1) |
|---|---|---|
| PelA | 8.0 | 36 |
| PelB | 9.0 | 532 |
| PelD | 8.0 | 467 |
| PelI | 9.0 | 104 |
| PelL | 8.0 | 1.9 |
Conditions of this experiment included the following: 2.5 g of polygalacturonate liter−1, 1 mM CaCl2, and 20 mM Tris-HCl (pH 8.0) or 20 mM 2-amino-methyl-propanol–HCl (pH 9.0) at 37°C.
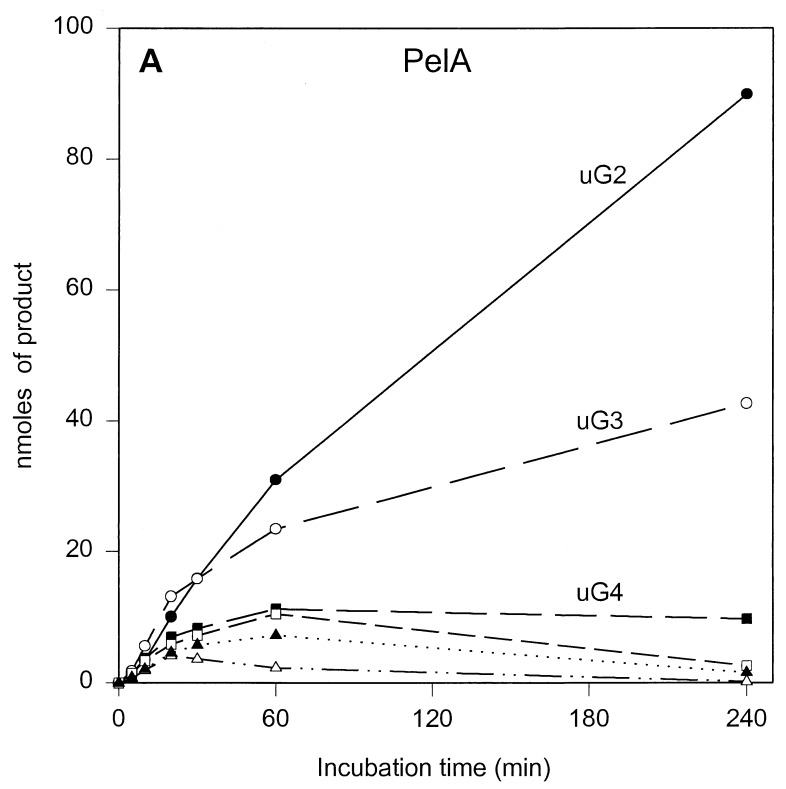
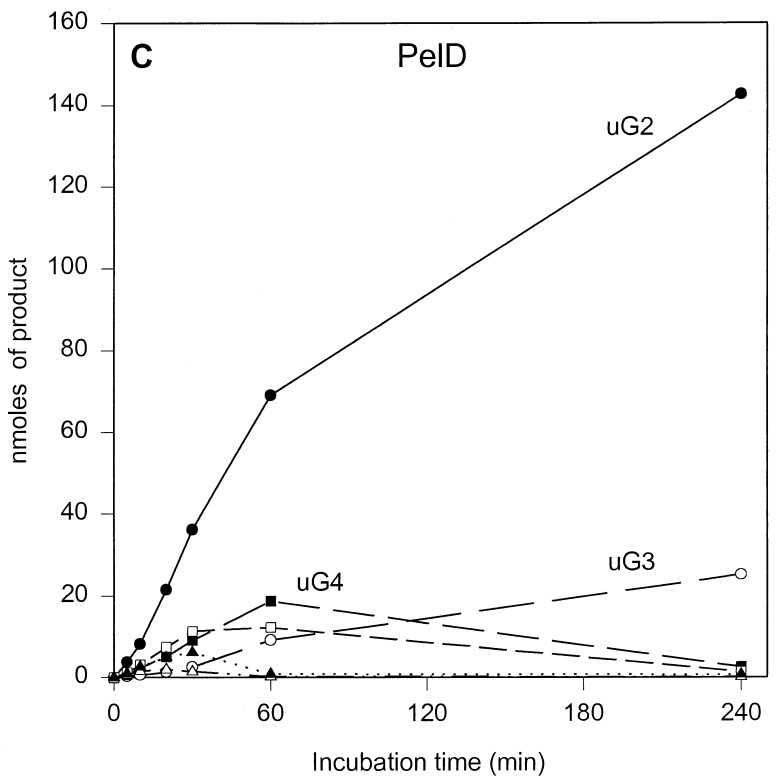
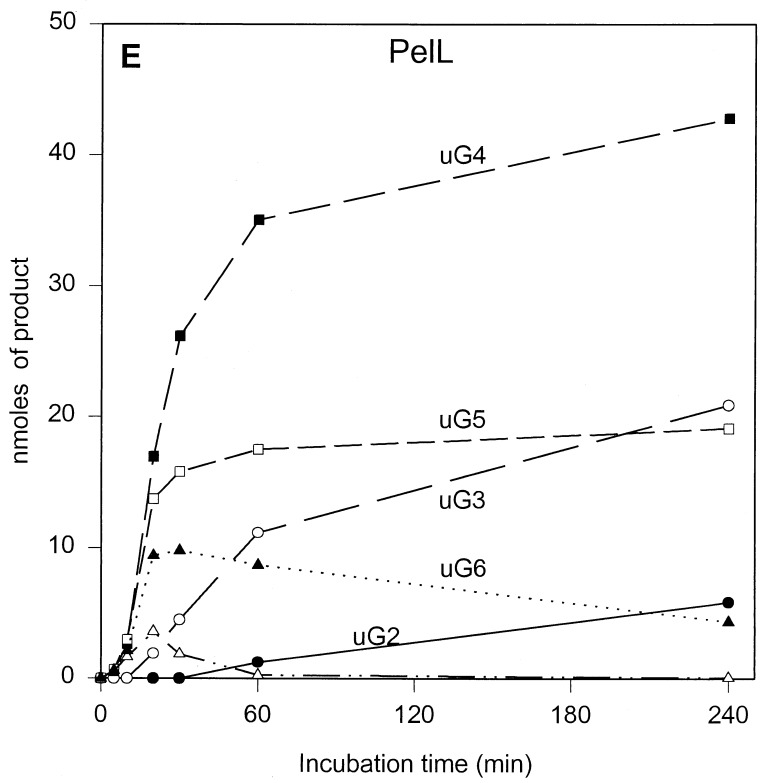
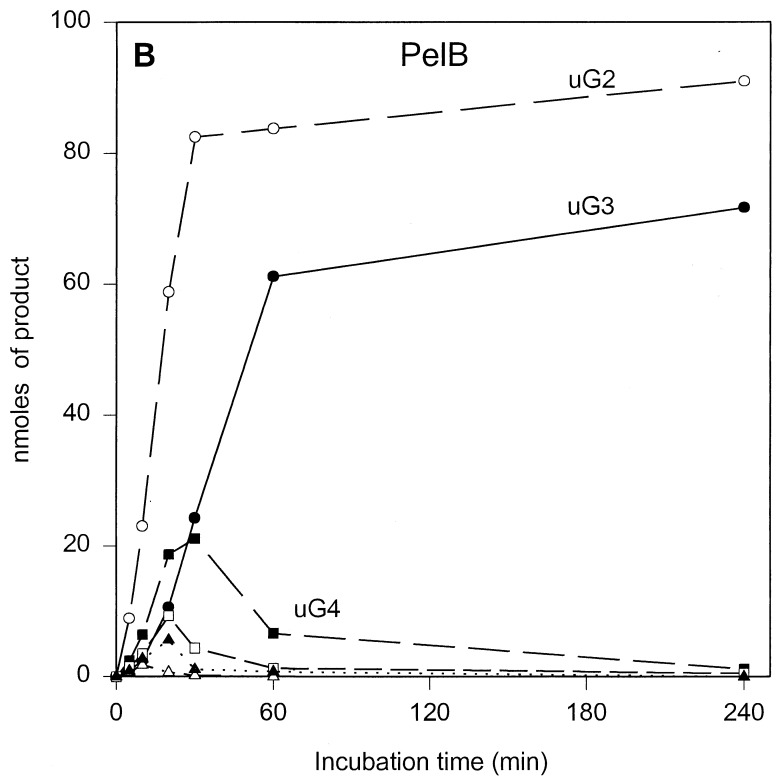
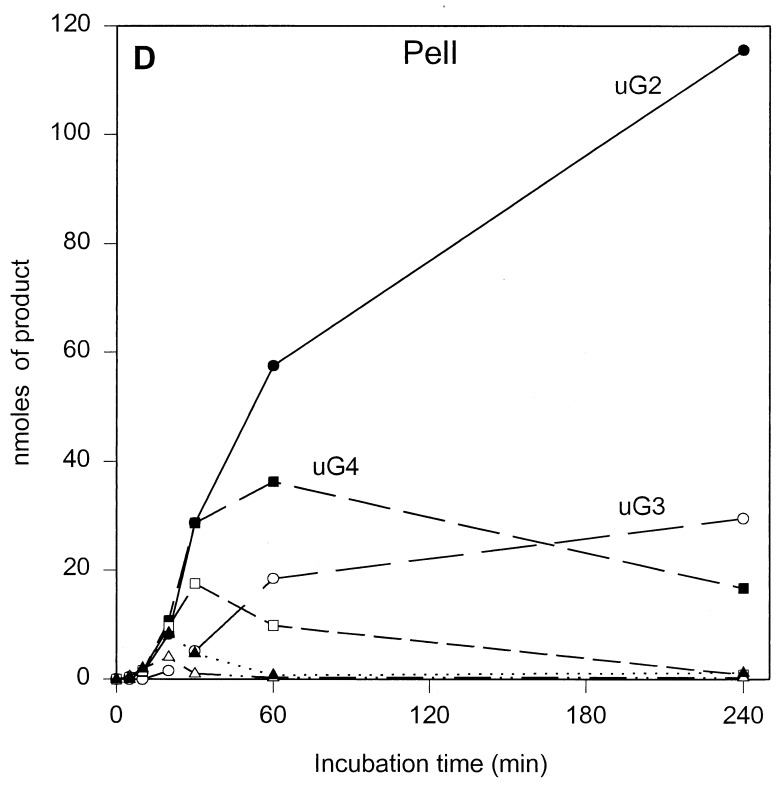
The results of the product progression data for each enzyme are presented in Fig. 1. For clarity, only the data for the period up to 4 h of incubation are shown. Only minor further changes were observed after 4 h. These involved mainly a small increase in u(GalpA)2, except in the case of PelL, which revealed a further increase in u(GalpA)2 and u(GalpA)3 at the expense of u(GalpA)5 and u(GalpA)6. After 24 h, 70 to 78% of the polygalacturonate initially present, depending on the enzyme, was converted (data not shown). For all five enzymes, the data showed the initial formation of oligogalacturonates larger than u(GalpA)5, which were gradually converted into smaller products, compatible with an endolytic mode of attack of the polymer substrate. The depolymerization profiles, which are a result of the number of subsites, the affinity of the subsite(s) for the substrate, and the intrinsic reaction rate, are different for all enzymes. For PelB and PelD, one major product accumulated quickly, namely, u(GalpA)3 and u(GalpA)2, respectively, indicating a strong preference for the substrate to bind in the particular mode resulting in those products. Furthermore, for PelB, a strong accumulation of u(GalpA)2 was also observed, although with a lag. This suggests that smaller products bind in a different preferred productive mode as compared to the larger products. For PelA, PelI, and PelL, less pronounced favored products were formed. PelA converted the initial larger products formed into u(GalpA)2 and u(GalpA)3. For PelI, a prominent transient increase in u(GalpA)4, which was degraded into u(GalpA)2 upon further incubation was recorded. PelL also showed accumulation of u(GalpA)4 and u(GalpA)5, but u(GalpA)4 was not further cleaved.
FIG. 1.
Unsaturated reaction products formed upon incubation of polygalacturonate with the E. chrysanthemi 3937 pectate lyases PelA (A), PelB (B), PelD (C), PelI (D), and PelL (E). Conditions are described in Materials and Methods. The following concentrations of enzymes were added: 1.80 U · ml−1 for PelA, 2.13 U · ml−1 for PelB, 1.87 U · ml−1 for PelD, 2.08 U · ml−1 for PelI, and 1.90 U · ml−1 for PelL. uGn, u(GalpA)n. Symbols: ●, u(GalpA)2; ○, u(GalpA)3; ■, u(GalpA)4; □, u(GalpA)5; ▴, u(GalpA)6; ▵, u(GalpA)7–10.
Previously, product progression profiles were reported for Pla, Plb, Plc, and Ple from E. chrysanthemi EC16 (15). Pla, Plc, and Ple are most closely related to PelA, PelB, and PelD of E. chrysanthemi 3937, respectively, with 29 amino acid substitutions for Pla, 27 amino acid substitutions for Plc, and 22 amino acid substitutions and 7 amino acid insertions for Ple. Surprisingly, despite these mutations, the product progression profiles for PelA and PelD closely resemble those for Pla and Ple (15). However, for PelB, a strong accumulation of u(GalpA)2, which was in contrast to the profile for Plc, was observed. The origin of these similarities and differences will be discussed below.
Reaction rates and bond cleavage frequencies on oligogalacturonates (n = 2 to 8).
In order to understand the origin of the differences between the individual enzymes in product progression on polygalacturonate and their mode of action, the turnover rates of oligogalacturonates (n = 2 to 8) and corresponding bond cleavage frequencies for each enzyme were studied.
None of these endo-acting enzymes showed degradation of (GalpA)2, which is also reflected in the product progression profiles where u(GalpA)2 accumulated as the smallest product. Another major product observed in the product progression profile was u(GalpA)3. The fact that this product accumulated must be attributed to the very low conversion rates of this substrate by all five enzymes, as revealed in studies using unsaturated oligomers as substrates (data not shown). This is also corroborated by the observed low reaction rates on (GalpA)3 for all five enzymes (Table 2). Thus, the enzymes require substrates with an n of >3 for effective catalysis.
TABLE 2.
Bond cleavage frequencies and specific activities of the E. chrysanthemi 3937 endopectate lyases PelA, PelI, PelL, PelD, and PelB acting on oligogalacturonates of defined lengthsa
| Enzyme | nb | Oligogalacturonatec | Activity (U · mg−1) |
|---|---|---|---|
| PelA | 3 | G — G — G | 0.6 |
| 100 | |||
| 4 | G — G — G — G | 17 | |
| 100 | |||
| 5 | G — G — G — G — G | 12 | |
| 23 77 | |||
| 6 | G — G — G — G — G — G | 15 | |
| 33 6 61 | |||
| 7 | G — G — G — G — G — G — G | 23 | |
| 70 9 2 19 | |||
| 8 | G — G — G — G — G — G — G — G | 25 | |
| 62 19 6 2 11 | |||
| PelI | 3 | G — G — G | 0.6 |
| 100 | |||
| 4 | G — G — G — G | 11 | |
| 62 38 | |||
| 5 | G — G — G — G — G | 87 | |
| 43 22 34 | |||
| 6 | G — G — G — G — G — G | 252 | |
| 74 23 3 | |||
| 7 | G — G — G — G — G — G — G | 504 | |
| 4 91 5 | |||
| 8 | G — G — G — G — G — G — G — G | 630 | |
| 2 33 61 4 | |||
| PelL | 3 | G — G — G | 0 |
| 100 | |||
| 4 | G — G — G — G | 0.02 | |
| 100 | |||
| 5 | G — G — G — G — G | 0.22 | |
| 28 72 | |||
| 6 | G — G — G — G — G — G | 2.2 | |
| 64 34 2 | |||
| 7 | G — G — G — G — G — G — G | 3.5 | |
| 4 91 5 | |||
| 8 | G — G — G — G — G — G — G — G | 3 | |
| 3 38 57 2 | |||
| PelD | 3 | G — G — G | 13 |
| 100 | |||
| 4 | G — G — G — G | 418 | |
| 100 | |||
| 5 | G — G — G — G — G | 310 | |
| 8 92 | |||
| 6 | G — G — G — G — G — G | 268 | |
| 3 3 94 | |||
| 7 | G — G — G — G — G — G — G | 250 | |
| 4 7 16 73 | |||
| 8 | G — G — G — G — G — G — G — G | 222 | |
| 1 14 10 75 | |||
| PelB | 3 | G — G — G | 40 |
| 100 | |||
| 4 | G — G — G — G | 2,400 | |
| 43 57 | |||
| 5 | G — G — G — G — G | 3,660 | |
| 69 31 | |||
| 6 | G — G — G — G — G — G | 4,070 | |
| 10 52 38 | |||
| 7 | G — G — G — G — G — G — G | 3,780 | |
| 2 6 62 30 | |||
| 8 | G — G — G — G — G — G — G — G | 3,160 | |
| 11 67 22 |
Conditions of this experiment included the following: 0.5 mM oligogalacturonate (n = 3 to 8), 1 mM CaCl2, and 20 mM Tris-HCl (pH 8.0) (for PelA, PelD, and PelL) or 20 mM amino-methyl-propanol–HCl (pH 9.0) (for PelB and PelI) at 37°C. Product analysis was done as described in Materials and Methods.
n, degree of polymerization.
Bond cleavage frequencies are given in percentages. The reducing ends of the oligogalacturonates are printed in bold type.
For PelA, two preferred cleavage positions were observed depending on the size of the oligogalacturonate. Exclusive cleavage at the second bond from the reducing end for (GalpA)3 and (GalpA)4 and preferential cleavage at this bond for (GalpA)5 and (GalpA)6 were found, whereas (GalpA)7 and (GalpA)8 were preferentially cleaved at the second bond from the nonreducing end. This shift of the preferred bond cleaved, together with the observation that the penultimate bond from both ends was preferred over more central bonds for oligogalacturonates with an n of >5, strongly indicates that the number of subsites with the highest affinity for homogalacturonan is four, stretching from positions −2 to +2. This is further substantiated by the lower cleavage rates for (GalpA)5 and (GalpA)6 relative to that for (GalpA)4. The increased reaction rate for (GalpA)7 and (GalpA)8 indicates that the actual number of subsites exceeds four.
Despite a 100-fold difference in reaction rates between PelI and PelL, both enzymes showed their highest activities on the longer oligogalacturonates (n ≥ 6) and displayed comparable bond cleavage frequencies, with a preference for the fourth glycosidic linkage from the reducing end. Subsequent conversion of the smaller oligomers that were formed proceeded for both enzymes at a much lower rate, particularly for PelL, and with different end products. For PelI, u(GalpA)4 was still a good substrate, explaining the decrease of this product in the progression profiles. Due to the relatively much lower activity of PelL on (GalpA)5 and (GalpA)4, these substrates are not further converted as long as oligogalacturonates with a degree of polymerization of >5 are present. For the smaller oligogalacturonates, the preferred cleavage was observed at the nonreducing end for PelI whereas PelL preferred the second glycosidic bond from the reducing end. Since the reaction rates for PelI and PelL increased with oligogalacturonates with an n up to 7 with increasing preference for the fourth glycosidic linkage from the reducing end, it can be concluded that for both enzymes the minimum number of subsites is seven, stretching from positions −3 to +4. It should be pointed out that in view of the very low activity of PelL, but its pronounced macerating activity (20), homogalacturonan may not be the natural substrate.
PelD was the most prominent enzyme with respect to the preferred bond cleaved. For oligogalacturonates with an n of 6, hardly any products other than u(GalpA)2 were detected, and for those with an n of 7 or 8, the second bond from the reducing end was still strongly preferred. Thus, the bond cleavage frequencies explain the product progression well. The strong preference of PelD for the second glycosidic linkage from the reducing end and the highest activity on (GalpA)4 strongly suggest that the array of four subsites stretches from positions −2 to +2.
For PelB, cleavage of oligogalacturonates with an n of >4 occurred predominantly at the third glycosidic bond from the reducing end whereas u(GalpA)2 was formed mainly from (GalpA)4 and (GalpA)3. The highest reaction rate was observed on (GalpA)6. In combination with the predominant cleavage of oligogalacturonates with an n of >4 at the third linkage from the reducing end, this indicates the presence of six subsites extending from positions −3 to +3.
As mentioned above, the product progression for PelB differed from that of Plc, the E. chrysanthemi EC16 counterpart, mainly with respect to u(GalpA)2 formation. A comparison of the data for PelB with those obtained for Plc under slightly different conditions (16) reveals only small changes with respect to bond cleavage frequencies and even a stronger preference for Plc to form u(GalpA)2 from (GalpA)4 (75%). However, the rate at which (GalpA)4 is cleaved by Plc relative to that of the larger oligogalacturonates is lower than the same cleavage ratio for PelB [(GalpA)4/(GalpA)6 cleavage ratios for Plc and PelB are 1 to 4.6 and 1 to 1.7, respectively]. This indeed can account for the observed difference in product progression.
Since PelB and Plc differ with respect to only 27 of 353 amino acids, the PelB 3D structure was modeled by using the Plc 3D structure (16). Most amino acid substitutions took place at a site that was remote from the substrate binding cleft (16) in the N-terminal part of the enzyme that forms an α-helix capping the β-helix and at the surface of the enzyme. Those mutations are, therefore, probably not involved in enzyme specificity. However, three amino acid substitutions (G161N, A167S, and S239G; the last residues refer to those of Plc) were found in the substrate binding cleft. At least one of the substitutions, S239G, may involve (in)direct contact with the substrate due to the change of the functionality of the side chain. In the modeled PelB structure, the G161N mutation results in a changed conformation of D160. In the Plc-substrate structure, D160 and D162 coordinate a Ca2+ ion that is in contact with the substrate (16). The A167S mutation most probably does not affect the enzymatic properties since its position below the Ca2+ ion present in both the substrate-free enzyme and the enzyme-substrate complex is not likely to influence the Ca2+ ion that is coordinated by the four conserved acidic residues D129, D131, E166, and D170. Thus, the observed changes in the modeled 3D structure of PelB, notably G161N and S239G, may well account for the different product progression profiles of PelB and Plc.
The limited number of amino acid differences observed for PelD and Ple also allowed the modeling of the PelD 3D structure based on the Ple 3D structure (9). None of the amino acid substitutions was found to occur in the substrate binding cleft as inferred from the Plc-substrate complex (16). The similar product progression profiles for PelD and Ple and those differing for PelB and Plc indicate that the naturally occurring mutations do not change the catalytic properties unless they occur in the substrate binding cleft. From this, it logically follows that the amino acid differences between PelA and Pla are most likely in positions remote from the substrate binding cleft.
For all of the enzymes studied here, the bond cleavage frequencies and substrate reaction rates on oligogalacturonates (n = 2 to 8) easily explain the product progression on the model substrate polygalacturonate. Still puzzling are the much higher activities of PelB and PelI on oligogalacturonates relative to those on polygalacturonate. A plausible explanation would be that the polymer substrate obviously extends beyond the array of subsites and that this hampers its effective binding.
In physiological terms, PelD, PelB, and PelI, being the most active enzymes and generating u(GalpA)2, u(GalpA)3, and u(GalpA)4, respectively, might serve as the major substrate supply for the bacterium. Both u(GalpA)2 and u(GalpA)3 may be internalized directly (6). PelX might cleave u(GalpA)4 into u(GalpA)2 (22).
It has been shown that PelD and PelI are very active in tissue maceration, that PelB and PelL are moderately active in this respect, and that PelA is virtually devoid of this activity (1, 10, 20). So far, we have not been able to establish a relationship between specific activity, mode of action, and macerating properties of the enzymes. Further elucidation of the substrate specificity of the pectate lyases must await the isolation of well-defined complex substrates and the elucidation of 3D structures of enzyme-substrate complexes.
ACKNOWLEDGMENT
This work was supported by E.C. grant AIR2-CT941345 to J.V. and J.R.-B.
REFERENCES
- 1.Barras F, Van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 2.Boccara M, Diolez A, Rouve M, Kotoujansky A. The role of individual pectate lyases of Erwinia chrysanthemi strain 3937 in pathogenicity on Saintpauliaplants. Physiol Mol Plant Pathol. 1988;33:95–104. [Google Scholar]
- 3.Brooks A D, He S Y, Gold S, Keen N T, Collmer A, Hutcheson S W. Molecular cloning of the structural gene for exopolygalacturonate lyase from Erwinia chrysanthemiEC16 and characterization of the enzyme product. J Bacteriol. 1990;172:6950–6958. doi: 10.1128/jb.172.12.6950-6958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vries J A, Hansen M, Søderberg J, Glahn P E, Pedersen J K. Distribution of methoxyl groups in pectins. Carbohydr Polym. 1986;6:165–176. [Google Scholar]
- 5.Henrissat B, Heffron S E, Yoder M D, Lietzke S E, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 7.Kester H C M, Visser J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem. 1990;12:150–160. [PubMed] [Google Scholar]
- 8.Kester H C M, Kusters-van Someren M A, Müller Y, Visser J. Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur J Biochem. 1996;240:738–746. doi: 10.1111/j.1432-1033.1996.0738h.x. [DOI] [PubMed] [Google Scholar]
- 9.Lietzke S E, Scavetta R D, Yoder M D, Jurnak F. The refined three-dimensional structure of pectate lyase E from Erwinia chrysanthemiat 2.2 Å resolution. Plant Physiol. 1996;111:73–92. doi: 10.1104/pp.111.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi3937. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 11.MacMillan G P, Vaughn R H. Purification and properties of a polygalacturonate-trans-eliminase produced by Clostridium multifermentans. Biochemistry. 1964;3:564–572. doi: 10.1021/bi00892a016. [DOI] [PubMed] [Google Scholar]
- 12.Pařenicová L, Benen J A E, Kester H C M, Visser J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur J Biochem. 1998;251:72–80. doi: 10.1046/j.1432-1327.1998.2510072.x. [DOI] [PubMed] [Google Scholar]
- 13.Pickersgill R, Jenkins J, Harris G, Nasser W, Robert-Baudouy J. The structure of Bacillus subtilispectate lyase in complex with calcium. Nat Struct Biol. 1994;1:717–723. doi: 10.1038/nsb1094-717. [DOI] [PubMed] [Google Scholar]
- 14.Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Regulation of pelZ, a gene of the pelB-pelC cluster encoding a new pectate lyase of Erwinia chrysanthemi3937. J Bacteriol. 1996;178:7187–7196. doi: 10.1128/jb.178.24.7187-7196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preston J F, Rice J D, Ingram L O, Keen N T. Differential depolymerization mechanisms of pectate lyases secreted by Erwinia chrysanthemiEC16. J Bacteriol. 1992;174:2039–2042. doi: 10.1128/jb.174.6.2039-2042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scavetta, R. D., S. R. Herron, A. T. Hotchkiss, N. Kita, N. T. Keen, J. A. E. Benen, H. C. M. Kester, J. Visser, and F. Jurnak. Structure of plant cell wall fragment complexed to PelC. Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 17.Schols H A, Posthumus M A, Voragen A G J. Structural features of hairy regions of pectins isolated from apple juice produced by the liquefaction process. Carbohydr Res. 1990;206:117–130. [Google Scholar]
- 18.Shevchik V E, Scott M, Mayans O, Jenkins J. Crystallization and preliminary X-ray analysis of a member of a new family of pectate lyases, PelL from Erwinia chrysanthemi. Acta Crystallogr Sect D. 1998;54:419–422. doi: 10.1107/s0907444997012043. [DOI] [PubMed] [Google Scholar]
- 19.Shevchik V E, Hugouvieux-Cotte-Pattat N. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi. Mol Microbiol. 1997;24:1285–1301. doi: 10.1046/j.1365-2958.1997.4331800.x. [DOI] [PubMed] [Google Scholar]
- 20.Shevchik V E, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Pectate lyase PelI of Erwinia chrysanthemi3937 belongs to a new family. J Bacteriol. 1997;179:7321–7330. doi: 10.1128/jb.179.23.7321-7330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shevchik V E, Kester H C M, Benen J A E, Visser J, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the exopolygalacturonate lyase, PelX, of Erwinia chrysanthemi3937. J Bacteriol. 1999;181:1652–1663. doi: 10.1128/jb.181.5.1652-1663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shevchik, V. E., G. Condemine, J. Robert-Baudouy, and N. Hugouvioux-Cotte-Pattat. The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of the pectin catabolism in Erwinia chrysanthemi 3937. J. Bacteriol, in press. [DOI] [PMC free article] [PubMed]
- 23.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemipectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoder M D, Keen N T, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 25.Yoder M D, Jurnak F. The refined three-dimensional structure of pectate lyase C from Erwinia chrysanthemiat 2.2 Angstrom resolution. Plant Physiol. 1995;107:349–364. doi: 10.1104/pp.107.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]