Abstract
As sigma receptors are highly expressed on various cancer cells, radiolabeled sigma receptor ligands have been developed as imaging and therapeutic probes for cancer. Previously, we synthesized and evaluated a radioiodinated vesamicol derivative, 2-(4-[125I](4-iodophenyl)piperidine)cyclohexanol ((+)-[125I]pIV), and a radioiodinated aza-vesamicol derivative, trans-2-(4-(3-[125I](4-iodophenyl)propyl)piperazin-1-yl)cyclohexan-1-ol ([125I]2), as sigma-1 receptor-targeting probes. In order to obtain sigma receptor-targeting probes with superior biodistribution characteristics, we firstly synthesized twelve bromine-containing aza-vesamicol derivatives and evaluated their affinity for sigma receptors. One such derivative exhibited high selectivity for the sigma-1 receptor and another exhibited high affinity for both the sigma-1 and sigma-2 receptors. Thus, their halogen-substituted iodine- and radioiodine-containing compounds were prepared. The 125I-labeled compounds exhibited high uptake in tumor and lower uptake in non-target tissues than the two previously developed and evaluated 125I-labeled sigma receptor-targeting probes, [125I]pIV and [125I]2. Therefore, these novel radioiodine-labeled compounds should be promising as sigma receptor-targeting probes.
[125I]15 and [125I]17 exhibited high uptake in the tumor and low uptake in non-target tissues, indicating that [123/131I]15 and [123/131I]17 are promising compounds to be used as imaging and therapeutic probes for sigma receptor-positive tumors.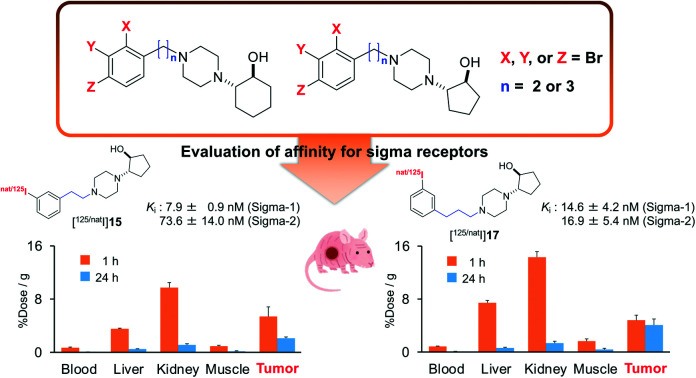
Introduction
Two subtypes of sigma receptors exist, and the sigma-1 receptor is one of them.2,3 The sigma-1 receptors are mainly located on the membrane of the endoplasmic reticulum (ER); they are composed of 223 amino acids, and their molecular weight is about 24 kDa. The sigma-1 receptor acts as a ligand-gated molecular chaperone protein modulating the ER-mitochondria Ca2+ transfer, protein folding, and ER stress pathways.4 Initially, studies on the sigma-1 receptor focused on its relationship with brain functions, such as signal transduction, pain modulation, memory, recognition, and emotions. Indeed, research on the sigma-1 receptor has been aimed at uncovering information on neurodegenerative diseases like Alzheimer's disease (AD), Parkinson's disease, amyotrophic lateral sclerosis (ALS), and multiple sclerosis (MS).5–7 For example, in preclinical models, sigma-1 receptor agonists have been reported to be effective protectants in these neurodegenerative diseases.8,9
On the other hand, the sigma-1 receptor has attracted attention in the oncology research field, because it is known to be highly expressed in various cancer cells.10 Moreover, antagonists of the sigma-1 receptor have been reported to inhibit the proliferation of cancer cells.11,12 Therefore, radiolabeled sigma-1 receptor ligands have been developed as imaging probes to diagnose not only neurodegenerative diseases but also cancer.13,14
The X-ray crystallography-derived crystal structures of the sigma-1 receptor bound to ligands have been reported; the relevant crystallographic data demonstrated that the electrostatic interaction between a protonated amino group of the ligand and the carboxylate group of Glu172 in the sigma-1 receptor is essential for receptor-ligand binding.15,16 Virtual screening experiments relying on docking studies indicated that compounds with an alkyl amino group and hydrophobic sites behave like high affinity ligands for the sigma-1 receptor.17 Notably, evidence also suggested the distances between the amino group nitrogen and hydrophobic sites to be an important factor determining the magnitude of the affinity of the ligand for the sigma-1 receptor.18
In 2017, the sigma-2 receptor was identified as TMEM97, an ER-resident transmembrane protein that regulates the lysosomal cholesterol transporter NPC1, whose loss causes Niemann–Pick disease type C1, a fatal lysosomal storage disorder.19,20 As the sigma-2 receptor is also overexpressed in many cancer cells, and this receptor has been reported to be a possible cancer cell biomarker,21 the sigma-2 receptor can be an attractive target for cancer imaging and therapy.22,23 Therefore, radiolabeled ligands of the sigma-2 receptor have been developed as imaging probes.24 Moreover, some radioligands exhibiting high affinity for both sigma receptor subtypes (sigma-1 and sigma-2 receptors) have been reported, given the fact that their use could enhance the extent of tumor-targeting compared to radioligands that are selective for a single subtype of the sigma receptors.25,26
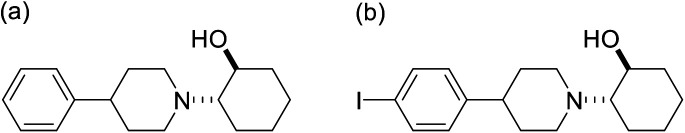
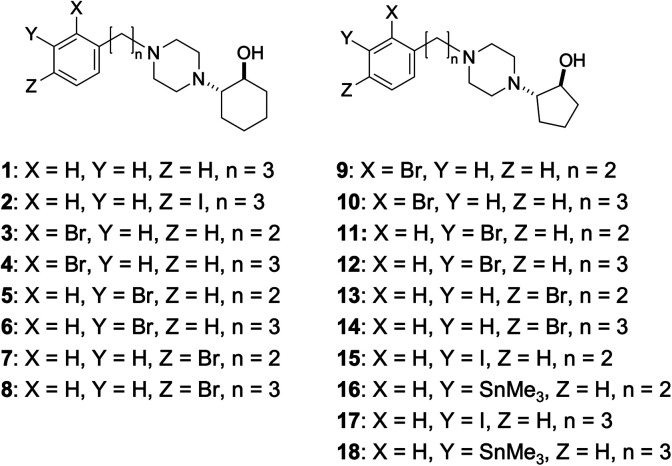
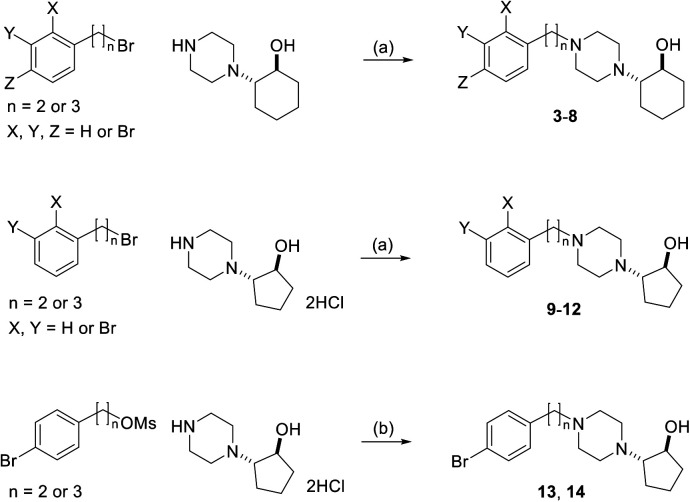
2-(4-Phenylpiperidino)cyclohexanol (vesamicol, Fig. 1a) has been studied as a ligand for the vesicular acetylcholine transporter as well as the sigma-1 receptor.27,28 We synthesized and evaluated radiohalogen-labeled vesamicol analogs as probes targeting the sigma-1 receptor.29–31 Our investigations of the probe-receptor interactions demonstrated that a compound with an iodine atom in the para position of the phenyl group in the vesamicol structure, 2-(4-(4-iodophenyl)piperidine)cyclohexanol (pIV, Fig. 1b), has over 10-fold higher affinity for the sigma-1 receptor than the lead compound, unmodified vesamicol.29 We hypothesized that insertion of an appropriate spacer between the halogenated phenyl group and the piperidine ring could increase the compounds' affinity for the sigma-1 receptor, because the halogenated phenyl group and the piperidine ring correspond to the above-mentioned hydrophobic site and alkyl amino group, respectively. Thus, compounds characterized by different lengths of the alkyl chain linking the phenyl group and the alkyl amino group were synthesized and evaluated.1 In the study, for the sake of synthetic accessibility, a piperazine ring was used as the alkyl amino group, instead of its piperidine counterpart. As a result, since trans-2-(4-(3-phenylpropyl)piperazin-1-yl)cyclohexan-1-ol (1, Fig. 2) exhibited high affinity for sigma-1 receptors, trans-2-(4-(3-[125I](4-iodophenyl)propyl)piperazin-1-yl)cyclohexan-1-ol ([125I]2, Fig. 2) was prepared and evaluated. Specifically, [125I]2 exhibited a superior biodistribution result, namely high tumor uptake and relatively rapid clearance from normal (non-cancerous) tissues. Although only iodinated aza-vesamicol derivatives whereby an iodine atom had been introduced in the para position of the phenyl group were investigated in a study conducted by our group, it remains unclear whether a compound comprising an iodine atom in the para position is the best option in the present study. Moreover, we recently reported that replacing the cyclohexanol in 2-(4-(2-iodophenyl)piperidine)cyclohexanol, a vesamicol analog containing an iodine atom, with cyclopentanol to obtain 2-(4-(2-iodophenyl)piperidine)cyclopentanol increased the compound's selectivity toward the sigma-1 receptor.32 Thus, in the present study, as a first step in the development of sigma-1 receptor-targeting radioligands for cancer theranostics, we designed twelve aza-vesamicol derivatives (3–14, Fig. 2) containing an ethylene or a propylene linker between the phenyl group and the piperazinylcyclohexanol or piperazinylcyclopentanol, as well as a bromine substituent in the ortho-, meta-, or para-position of the phenyl group. In spite of the fact that we aimed to develop radioiodine-labeled probes, based on the availability of the starting materials and the similarity between iodine and bromine,25,26 we first generated vesamicol derivatives comprising the bromine atom (3–14); indeed, we synthesized compounds by coupling trans-2-(1-piperazinyl)cyclohexanol or trans-2-(1-piperazinyl)cyclopentanol (20, Scheme 1) with bromophenylalkyl bromide or bromophenylalkyl methanesulfonate (Scheme 2).33,34
Fig. 1. Structures of (a) vesamicol and (b) pIV.
Fig. 2. Structures of compounds 1–18.
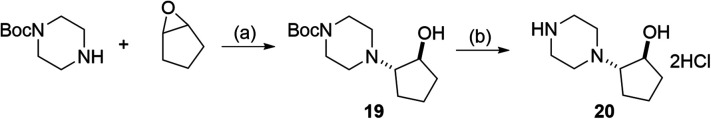
Scheme 1. Preparation of 19 and 20. (a) MeOH, 120 °C. (b) 4 M HCl in AcOEt, rt.
Scheme 2. Preparation of compounds 3–14. (a) K2CO3, MeCN, reflux; (b) N,N-diisopropylethylamine, NaI, DMF, 60–100 °C.
The binding affinities of 3–14 for the sigma receptors (sigma-1 and sigma-2) were determined by conducting competitive binding assays. Among them, one derivative (11) exhibited high selectivity for the sigma-1 receptor and another (12) exhibited high affinity for both the sigma-1 and sigma-2 receptors. Thus, their halogen-substituted iodine- and radioiodine-containing compounds 15, [125I]15, 17, and [125I]17 were synthesized (Schemes 3 and 4) and evaluated in vitro and in vivo. In this study, although we are interested in developing 123I (t1/2 = 12.3 h) labeled probes for single photon emission computed tomography (SPECT) imaging, 124I (t1/2 = 4.18 d) labeled probes for positron emission tomography (PET) imaging, or 131I (t1/2 = 8.02 d) labeled probes for radionuclide therapy, 125I (t1/2 = 59.4 d) was used as an alternative radionuclide to be used in preliminary basic research except SPECT experiments.
Scheme 3. Preparation of 15 and 17. (a) CuI, NaI, N,N′-dimethylethylenediamine, 1,4-dioxane, reflux.
Scheme 4. Preparation of [125I]15 and [125I]17. (a) (Me3Sn)2, Pd(PPh3)4, toluene, reflux; (b) [125I]NaI, 10% acetic acid, chloramine-T, rt.
Experimental
Materials and methods
DTG, (+)-pentazocine, and haloperidol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Other reagents were of reagent grade and used as received. [3H]1,3-o-Di-tolylguanidine ([3H]DTG) (1.1 TBq mmol−1), (+)-[3H]pentazocine (1.0 TBq mmol−1), and [125I]sodium iodide (644 GBq mg−1) were purchased from PerkinElmer (Waltham, MA, USA). [123I]Sodium iodide (71 MBq ng−1) was purchased from PDRadiopharma Inc. (Tokyo, Japan). Proton nuclear magnetic resonance (1H-NMR) spectra were obtained with JEOL JNM-ECS400 or JNM-ECA600 (JEOL Ltd, Tokyo, Japan). Electrospray ionization mass spectra (ESI-MS) were obtained with JEOL JMS-T100TD (JEOL Ltd). TLC analyses were performed with silica plates (Art 5553; Merck, Darmstadt, Germany). Radioactivity was determined with an auto well gamma counter (ARC-7010; Hitachi, Ltd., Tokyo, Japan).
1-Bromo-2-(2-bromoethyl)benzene,35 1-bromo-2-(2-bromopropyl)benzene,361-bromo-3-(2-bromoethyl)benzene,37 1-bromo-3-(3-bromopropyl)benzene,38 1-bromo-4-(2-bromoethyl)benzene,39 4-bromophenethyl methanesulfonate,40 and 3-(4-bromophenyl)propyl methanesulfonate,41trans-2-(1-piperazinyl)cyclohexanol,1 and tert-butyl piperazine-1-carboxylate42 were synthesized according to previous reports.
Synthesis of trans-2-(4-(2-(2-bromophenyl)ethyl)piperazin-1-yl)cyclohexan-1-ol (3)
2-(2-Bromoethyl)-1-bromobenzene (50.0 mg, 0.19 mmol) and trans-2-(1-piperazinyl)cyclohexanol (35.0 mg, 0.19 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (26.3 mg, 0.19 mmol) was added to the solution. The mixture was stirred under reflux for 54 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (1/0–20/1) as eluent. The obtained material was further purified by reversed phase (RP)-HPLC on Cosmosil 5C18-MS-II column (10 × 150 mm; Nacalai Tesque, Kyoto, Japan) at a flow rate of 4 mL min−1 with a gradient mobile phase of 70% methanol in water with 0.05% triethylamine (TEA) to 100% methanol with 0.05% TEA for 20 min (gradient system 1). The fraction containing 3 was determined by mass spectrometry, and was collected. The solvent was removed by lyophilization to provide 3 as a colorless solid (11.3 mg, 16%).
1H NMR (600 MHz, CDCl3): δ 1.12–1.30 (4H, m), 1.66–1.88 (3H, m), 2.10–2.28 (2H, m), 2.30–2.90 (8H, m), 2.60 (2H, t, J = 8.3 Hz), 2.95 (2H, t, J = 8.4 Hz), 3.37–3.39 (1H, m), 3.98 (1H, br s), 7.05–7.10 (1H, m), 7.17–7.33 (2H, m), 7.52 (1H, d, J = 8.3 Hz).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1367.
Synthesis of trans-2-(4-(3-(2-bromophenyl)propyl)piperazin-1-yl)cyclohexan-1-ol (4)
3-(2-Bromophenyl)propyl bromide (52.8 mg, 0.19 mmol) and trans-2-(1-piperazinyl)cyclohexanol (35.0 mg, 0.19 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (26.3 mg, 0.19 mmol) was added to the solution. The mixture was stirred under reflux for 24 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (40/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 4 as a colorless solid (7.2 mg, 10%).
1H NMR (400 MHz, CDCl3): δ 1.14–1.26 (4H, m), 1.70–1.71 (2H, m), 1.77–1.86 (5H, m), 2.11–2.23 (2H, m), 2.40–2.74 (6H, m), 2.42 (2H, t, J = 7.4 Hz), 2.73–2.77 (2H, m), 2.74 (2H, t, J = 7.8 Hz), 3.34–3.38 (1H, m), 3.97 (1H, br s), 7.03–7.07 (1H, m) 7.22–7.24 (1H, t, J = 8.0), 7.51 (1H, d, J = 7.6).
HRMS (ESI) m/z calcd for C19H30BrN2O (M + H)+, 383.1521; found 383.1505.
Synthesis of trans-2-(4-(2-(3-bromophenyl)ethyl)piperazin-1-yl)cyclohexan-1-ol (5)
1-Bromo-3-(2-bromoethyl)benzene (50.0 mg, 0.19 mmol) and trans-2-(1-piperazinyl)cyclohexanol (35.0 mg, 0.19 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (26.3 mg, 0.19 mmol) was added to the solution. After heating under reflux for 68 h, potassium carbonate was removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 5 as a colorless solid (15.2 mg, 22%).
1H NMR (400 MHz, CDCl3): δ 1.10–1.33 (4H, m), 1.64–1.89 (3H, m), 2.06–2.31 (2H, m), 2.47–2.88 (12H, m), 3.33–3.43 (1H, m), 3.97 (1H, br s), 7.10–7.19 (2H, m), 7.30–7.38 (2H, m).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1367.
Synthesis of trans-2-(4-(3-(3-bromophenyl)propyl)piperazin-1-yl)cyclohexan-1-ol (6)
3-(3-Bromophenyl)propyl bromide (50.0 mg, 0.18 mmol) and trans-2-(1-piperazinyl)cyclohexanol (33.2 mg, 0.18 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (24.9 mg, 0.18 mmol) was added to the solution. After heating under reflux for 24 h, potassium carbonate was removed by filtration, and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform–methanol (1/0–20/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 6 as a colorless solid (9.6 mg, 14%).
1H NMR (400 MHz, CDCl3): δ 1.09–1.31 (4H, m), 1.65–1.87 (5H, m), 2.08–2.24 (2H, m), 2.31–2.64 (6H, m), 2.35 (2H, t, J = 7.6 Hz), 2.60 (2H, t, J = 7.8 Hz), 2.76 (2H, t, J = 6.9 Hz), 3.31–3.40 (1H, m), 3.97 (1H, br s), 7.08–7.17 (2H, m), 7.29–7.36 (2H, m).
HRMS (ESI) m/z calcd for C19H30BrN2O (M + H)+, 383.1521; found 383.1534.
Synthesis of trans-2-(4-(2-(4-bromophenyl)ethyl)piperazin-1-yl)cyclohexan-1-ol (7)
1-(2-Bromoethyl)-4-bromobenzene (100.0 mg, 0.38 mmol) and trans-2-(1-piperazinyl)cyclohexanol (69.8 mg, 0.38 mmol) were dissolved in 4.0 mL of acetonitrile and then potassium carbonate (104.5 mg, 0.76 mmol) was added to the solution. After heating under reflux for 5 h, potassium carbonate was removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (1/0–50/1) as eluent. The obtained material was further purified by RP-HPLC performed using a Cosmosil 5C18-MS-II column (10 × 150 mm) at a flow rate of 4 mL min−1 with a gradient mobile phase of 80% methanol in water with 0.05% TEA to 100% methanol with 0.05% TEA for 20 min. The solvent was removed by lyophilization to provide 7 as a colorless solid (9.7 mg, 7%).
1H NMR (400 MHz, CDCl3): δ 1.13–1.28 (4H, m), 1.68–1.85 (3H, m), 2.09–2.25 (2H, m), 2.43–2.81 (12H, m), 3.33–3.40 (1H, m), 3.96 (1H, br s), 7.07 (2H, d, J = 8.4 Hz), 7.40 (2H, d, J = 8.4 Hz).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1360.
Synthesis of trans-2-(4-(3-(4-bromophenyl)propyl)piperazin-1-yl)cyclohexan-1-ol (8)
Compound 8 was synthesized in our previous study.1
Synthesis of trans-tert-butyl 4-(2-hydroxycyclopentyl)piperazine-1-carboxylate (19)
tert-Butyl piperazine-1-carboxylate (210 mg, 1.13 mmol) was dissolved in 1 mL of methanol. To the solution, 1,2-epoxycyclopentane (190.5 mg, 2.26 mmol) in 0.8 mL of methanol was slowly added dropwise. The reaction mixture was stirred at 120 °C for 47 h in a sealed tube. After the mixture was concentrated in vacuo, the residue was purified by chromatography on silica gel using chloroform/methanol (1/0–40/1) as eluent to afford 19 as a colorless oil (164.1 mg, 54%).
1H NMR (CDCl3): δ 1.48 (s, 9H), 1.57–1.97 (m, 6H), 2.50–2.57 (m, 5H), 3.44–3.48 (br, 4H), 4.11 (q, 1H, J = 4.3 Hz).
HRMS (ESI) m/z calcd for C14H27N2O3 (M + H)+, 271.2027; found 271.2022.
Synthesis of trans-2-(piperazin-1-yl)cyclopentan-1-ol dihydrochloride (20)
Compound 19 (164.1 mg, 0.61 mmol) was dissolved in 4 M HCl in ethyl acetate (1.0 mL) and the mixture was stirred at room temperature. After 7 h, the reaction mixture was concentrated in vacuo and the residue was suspended in ethyl acetate. The solid in the suspension was collected by filtration and washed with ethyl acetate to afford 20 as a colorless solid (97.7 mg, 66%).
1H NMR (400 MHz, D2O): δ 1.60–1.81 (4H, m), 2.02–2.11 (1H, m), 2.18–2.28 (1H, br s), 3.31–3.37 (1H, br s), 3.40–3.80 (8H, m), 4.22–4.28 (1H, m).
HRMS (ESI) m/z calcd for C18H38ClN4O2 (2M + HCl + H)+, 377.2674; found 377.2683.
Synthesis of trans-2-(4-(2-(2-bromophenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol (9)
2-(2-Bromophenyl)ethyl bromide (50.0 mg, 0.19 mmol) and 20 (32.4 mg, 0.133 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (26.3 mg, 0.19 mmol) was added to the solution. The mixture was stirred under reflux for 26 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (1/0–40/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 9 as a colorless solid (5.7 mg, 8%).
1H NMR (400 MHz, CDCl3): δ 1.58–1.79 (4H, m), 1.94–2.11 (2H, m), 2.66–2.73 (2H, m), 2.79–3.00 (11H, m), 4.42 (1H, q, J = 5.8 Hz) 7.05–7.12 (1H, m), 7.18–7.26 (2H, m), 7.53 (1H, d, J = 8.2 Hz).
HRMS (ESI) m/z calcd for C17H26BrN2O (M + H)+, 355.1208; found 355.1195.
Synthesis of trans-2-(4-(3-(2-bromophenyl)propyl)piperazin-1-yl)cyclopentan-1-ol (10)
3-(2-Bromophenyl)propyl bromide (50.0 mg, 0.18 mmol) and 20 (30.6 mg, 0.126 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (24.9 mg, 0.18 mmol) was added to the solution. The mixture was stirred under reflux for 42 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (1/0–20/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 10 as a colorless solid (11.7 mg, 18%).
1H NMR (400 MHz, CDCl3): δ 1.56–1.77 (4H, m), 1.80–1.89 (2H, m), 1.90–2.07 (2H, m), 2.48 (2H, t, J = 7.6 Hz), 2.61–2.81 (9H, m), 2.77 (2H, t, J = 7.8 Hz), 4.29 (1H, q, J = 6.5 Hz), 7.03–7.10 (1H, m), 7.21–7.27 (2H, m), 7.53 (1H, d, J = 7.3 Hz).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1343.
Synthesis of trans-2-(4-(2-(3-bromophenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol (11)
2-(2-Bromophenyl)ethyl bromide (50.0 mg, 0.19 mmol) and 20 (32.4 mg, 0.133 mmol), were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (26.3 mg, 0.19 mmol) was added to the solution. The mixture was stirred under reflux for 24 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform–methanol (1/0–40/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 11 as a colorless solid (28.2 mg, 42%).
1H NMR (400 MHz, CDCl3): δ 1.42–1.78 (4H, m), 1.87–2.05 (2H, m), 2.48–2.82 (13H, m), 4.14 (1H, q, J = 6.0 Hz), 7.11–7.18 (2H, m), 7.31–7.35 (1H, m), 7.36 (1H, s).
HRMS (ESI) m/z calcd for C17H26BrN2O (M + H)+, 355.1208; found 355.1200.
Synthesis of trans-2-(4-(3-(3-bromophenyl)propyl)piperazin-1-yl)cyclopentan-1-ol (12)
3-(3-Bromophenyl)propyl bromide (50.0 mg, 0.18 mmol) and 20 (30.7 mg, 0.126 mmol) were dissolved in 1.9 mL of acetonitrile and then potassium carbonate (24.9 mg, 0.18 mmol) was added to the solution. The mixture was stirred under reflux for 50 h. After cooling to room temperature, insoluble materials were removed by filtration and the filtrate was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–20/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 12 as a colorless solid (16.4 mg, 25%).
1H NMR (400 MHz, CDCl3): δ 1.42–2.02 (8H, m), 2.36 (2H, t, J = 7.6 Hz), 2.41–2.78 (8H, m), 2.51 (1H, q, J = 7.5 Hz), 2.61 (2H, t, J = 7.8 Hz), 4.14 (1H, q, J = 6.1 Hz), 7.08–7.20 (2H, m), 7.29–7.33 (1H, m), 7.34 (1H, s).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1378.
Synthesis of trans-2-(4-(2-(4-bromophenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol (13)
2-(4-Bromophenyl)ethyl methanesulfonate (50.0 mg, 0.18 mmol), 20 (65.1 mg, 0.27 mmol), NaI (27 mg, 0.18 mmol), and DIPEA (185 μL, 1.08 mmol) in DMF (1.0 mL) was stirred at 80 °C for 8 h. Then the temperature was elevated to 100 °C and stirred for 14 h. After cooling to room temperature, saturated aqueous NaHCO3 was added and the mixture was extracted with AcOEt/hexane (1/1). The organic phase was separated, washed with saturated aqueous NaHCO3 and brine, dried over Na2SO3, filtered and concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–20/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 13 as a colorless solid (10.0 mg, 16%).
1H NMR (600 MHz, CDCl3): δ 1.45–1.75 (4H, m), 1.89–2.01 (2H, m), 2.51–2.79 (9H, m), 2.57 (2H, t, J = 8.1 Hz), 2.76 (2H, t, J = 8.1 Hz), 4.15 (1H, q, J = 5.8 Hz), 7.08 (2H, d, J = 8.6 Hz), 7.40 (2H, d, J = 8.2 Hz).
HRMS (ESI) m/z calcd for C17H26BrN2O (M + H)+, 355.1208; found 355.1196.
Synthesis of trans-2-(4-(3-(4-bromophenyl)propyl)piperazin-1-yl)cyclopentan-1-ol (14)
A mixture of 3-(4-bromophenyl)propyl methanesulfonate (240 mg, 0.82 mmol), 20 (298 mg, 1.23 mmol), NaI (123 mg, 0.82 mmol), and DIPEA (842 μL, 4.92 mmol) in DMF (4.0 mL) was stirred at 50 °C for 3 h. Then the temperature was elevated to 80 °C and stirred for 5.5 h. After cooling to room temperature, saturated aqueous NaHCO3 was added and the mixture was extracted with AcOEt/hexane (1/1). The organic phase was separated, washed with saturated aqueous NaHCO3 and brine, dried over Na2SO3, filtered and concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–10/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 1). The solvent was removed by lyophilization to provide 14 as a colorless solid (21.1 mg, 7.0%).
1H NMR (600 MHz, CDCl3): δ 1.43–1.75 (4H, m), 1.75–1.82 (2H, m), 1.87–2.00 (2H, m), 2.35 (2H, t, J = 7.6 Hz), 2.38–2.82 (8H, m), 2.51 (1H, q, J = 7.6 Hz), 2.58 (2H, t, J = 7.7 Hz), 4.13 (1H, q, J = 6.2 Hz), 7.05 (2H, d, J = 8.3 Hz), 7.39 (2H, d, J = 8.2 Hz).
HRMS (ESI) m/z calcd for C18H28BrN2O (M + H)+, 369.1365; found 369.1346.
Synthesis of trans-2-(4-(2-(3-iodophenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol (15)
Compound 11 (50.0 mg, 0.14 mmol), CuI (27.0 mg, 0.14 mmol), NaI (212.8 mg, 1.42 mmol), and N,N′-dimethylethylenediamine (30.6 μL, 0.28 mmol) were dissolved in 500 μL of 1,4-dioxane. After heating under reflux for 103 h, the mixture was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–20/1) as eluent. The obtained material was further purified by RP-HPLC performed using a Cosmosil 5C18-MS-II column (10 × 150 mm) at a flow rate of 4 mL min−1 with a gradient mobile phase of 60% methanol in water with 0.05% TEA to 100% methanol with 0.05% TEA for 20 min (gradient system 2). The solvent was removed by lyophilization to provide 15 as a colorless solid (7.9 mg, 14%).
1H NMR (600 MHz, CDCl3): δ 1.44–1.53 (1H, m), 1.54–1.74 (3H, m), 1.88–2.01 (2H, m), 2.44–2.82 (9H, m), 2.57 (2H, t, J = 8.1 Hz), 2.74 (2H, t, J = 8.4 Hz), 4.13 (1H, q, J = 6.2 Hz), 7.01 (1H, t, J = 7.5 Hz), 7.17 (1H, d, J = 7.8 Hz), 7.53 (1H, d, J = 7.8 Hz), 7.57 (1H, s).
HRMS (ESI) m/z calcd for C17H26IN2O (M + H)+, 401.1090; found 401.1089.
Synthesis of trans-2-(4-(2-(3-(trimethylstannyl)phenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol (16)
Compound 11 (10.0 mg, 0.03 mmol), hexamethylditin (65.8 μL, 0.32 mmol), and Pd(PPh3)4 (8.7 mg, 0.01 mmol) were dissolved in 1 mL of toluene. After heating under reflux for 27 h, the mixture was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–30/1) as eluent to obtain 16 as a colorless solid (3.3 mg, 27%).
1H NMR (400 MHz, CDCl3): δ 0.28 (9H, t, J = 27.2 Hz), 1.56–1.78 (4H, m), 1.90–2.07 (2H, m), 2.56–2.95 (13H, m), 4.25–4.35 (1H, m), 7.15 (1H, d, J = 8.0 Hz), 7.21–7.40 (3H, m).
LRMS (ESI†) m/z calcd for C20H35N2OSn (M + H)+, 439.2; found 439.1.
Synthesis of trans-2-(4-(3-(3-iodophenyl)propyl)piperazin-1-yl)cyclopentan-1-ol (17)
Compound 12 (100.0 mg, 0.27 mmol), CuI (47.5 mg, 0.25 mmol), NaI (404.5 mg, 2.70 mmol), N,N′-dimethylethylenediamine (59.0 μL, 0.55 mmol) were dissolved in 800 μL of 1,4-dioxane. After heating under reflux for 54 h, the mixture was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–20/1) as eluent. The obtained material was further purified by RP-HPLC (gradient system 2). The solvent was removed by lyophilization to provide 17 as a colorless solid (11.6 mg, 10%).
1H NMR (600 MHz, CDCl3): δ 1.44–1.73 (4H, m), 1.78 (quin, 2H, J = 7.6 Hz), 1.86–2.00 (2H, m) 2.34 (2H, t, J = 7.5 Hz), 2.37–2.80 (9H, m), 2.57 (2H, t, J = 7.5 Hz), 4.12 (1H, q, J = 6.0 Hz), 7.00 (1H, t, J = 7.8 Hz), 7.14 (1H, d, J = 7.2 Hz) 7.51 (1H, d, J = 7.8 Hz), 7.55 (1H, s).
HRMS (ESI) m/z calcd for C18H28IN2O (M + H)+, 415.1246; found 415.1265.
Synthesis of trans-2-(4-(3-(3-(trimethylstannyl)phenyl)propyl)piperazin-1-yl)cyclopentan-1-ol (18)
Compound 12 (5.0 mg, 0.014 mmol), hexamethylditin (7.1 μL, 0.034 mmol), and Pd(PPh3)4 (0.9 mg, 0.8 μmol) were dissolved in 2 mL of toluene. After heating under reflux for 54 h, the mixture was concentrated in vacuo. The residue was purified by chromatography on silica gel using chloroform/methanol (100/1–30/1) as the eluent to obtain 18 as a colorless solid (0.2 mg, 6%).
1H NMR (400 MHz, CDCl3): δ 0.27 (9H, t, J = 27.0 Hz), 1.40–1.76 (4H, m), 1.79–2.05 (4H, m), 2.30–3.00 (13H, m), 4.14 (1H, q, J = 6.2 Hz), 7.10–7.21 (2H, m), 7.27–7.34 (2H, m).
LRMS (ESI†) m/z calcd for C21H37N2OSn (M + H)+, 453.2; found 453.1.
Preparation of 125I-labeled compounds, [125I]trans-2-(4-(2-(3-iodophenyl)ethyl)piperazin-1-yl)cyclopentan-1-ol ([125I]15) and [125I]trans-2-(4-(3-(3-iodophenyl)propyl)piperazin-1-yl)cyclopentan-1-ol ([125I]17)
Radiolabeled compounds [125I]15 and [125I]17 were prepared by a standard iodination reaction of the corresponding trimethylstannyl precursor.8 Briefly, 1 μL of [125I]NaI (2 MBq) solution, 100 μL of 16 or 18 (400 μg mL−1 in methanol), 15 μL of 10% acetic acid, and 20 μL of chloramine-T aqueous solution (4 mg mL−1) were added to the reaction vial. After standing the reaction mixture at room temperature for 10 min, the reaction was quenched by adding NaHSO3 aqueous solution. The reaction mixtures were purified by RP-HPLC performed using a Cosmosil 5C18-MS-II column (4.6 × 150 mm) at a flow rate of 1 mL min−1 with a gradient mobile phase of 60% methanol in water with 0.05% TEA to 90% methanol in water with 0.05% TEA for 20 min.
In vitro competitive binding assay
Rat brain and liver membranes for binding experiments were prepared from rat brains without cerebellum and rat liver in male Sprague-Dawley rats (200 g, Japan SLC, Inc., Hamamatsu, Japan), respectively, using a method described previously.43,44
A sigma-1 receptor binding assay was performed using the following method. Rat cerebral membranes (465–1193 μg protein) were incubated with 5 nM (+)-[3H]pentazocine and various concentrations of 3–15, 17, and haloperidol as a reference compound (from 10−10 to 10−5 M) in 0.5 ml of 50 mM Tris-HCl (pH 7.8) for 90 min at 37 °C. The incubated samples were quickly diluted with 5 mL of ice-cold Tris-HCl (pH 7.8) buffer followed by rapid filtration through Whatman Grade GF/B glass fiber filters (GE Healthcare UK Ltd, Amersham, UK) presoaked in 0.5% polyethylenimine using a cell harvester (Brandel, Gaithersburg, MD). Filters were washed three times with 5 mL of ice-cold buffer. Nonspecific binding was determined in the presence of 10 μM (+)-pentazocine. Radioactivity retained on the filters was measured with a liquid scintillation counter (LSC-5100; Hitachi, Ltd.).
A sigma-2 receptor binding assay was performed using the following method. Rat liver membranes (123–179 μg protein) were incubated with 5 nM [3H]DTG and various concentrations of 3–15, 17, and haloperidol (from 10−10 to 10−5 M) in 0.5 ml of 50 mM Tris-HCl (pH 7.8) for 90 min at 37 °C in the presence of 1 μM (+)-pentazocine to mask sigma-1 sites. Nonspecific binding was determined in the presence of 10 μM DTG and 1 μM (+)-pentazocine. The incubated samples were treated in the same manner as described for the sigma-1 receptor binding assays.
In vitro stability experiments
The stability experiments of [125I]15 and [125I]17 in phosphate buffer (PB) and murine plasma were performed as described previously with a slight modification.45 Briefly, [125I]15 and [125I]17 (22–82 kBq) in 0.1 M PB pH 7.4 (100 μL) were incubated at 37 °C for 24 h. At 24 h after incubation, the purities of radiotracers were analyzed by RP-HPLC.
Meanwhile, for stability assay in murine plasma, [125I]15 and [125I]17 (74–93 kBq) in freshly prepared murine plasma were incubated at 37 °C for 24 h. After incubation at 37 °C for 1 and 24 h, an equivalent amount of ice-cold acetonitrile was added. After centrifugation at 1700 g at 4 °C for 15 min, the supernatant was filtered through a 0.45 μm filter and analyzed using RP-HPLC to determine the radiochemical purities.
Determination of partition coefficient
The partition coefficients of [125I]15 and [125I]17 were measured as described previously.46 The partition coefficient was determined by calculating the ratio of radioactivity in 1-octanol to that in the 0.1 M phosphate buffer (pH 7.4), and expressed as a common logarithm (log P).
Cellular uptake experiments
DU-145 prostate cancer cell line was purchased from ATCC (Manassas, VA, USA). Cells were grown in cell culture dishes in RPMI 1640 medium with phenol red, 10% heat-inactivated fetal bovine serum (FBS), 100 μg mL−1 glutamine, 100 units per mL penicillin, and 100 μg mL−1 streptomycin at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
Cellular uptake experiments using a DU-145 cell line were performed as described previously.47 Briefly, the cells were seeded and pre-incubated for 24 h, and incubated in the culture medium containing [125I]15 or [125I]17 (3.7 kBq per well) for different time intervals (15, 30, 60, and 120 min). For a blocking study, [125I]15 or [125I]17 was incubated with 10 μM of haloperidol as a blocking agent. After incubation, the cells were washed twice with ice-cold PBS and were lysed by 1 M NaOH. The solutions were then collected and the radioactivity was determined and corrected for background radiation. The radioactivity of each sample was normalized for the protein level, which was determined using a Protein Assay Bicinchoninate Kit (Nacalai Tesque).
Biodistribution experiments of [125I]15 and [125I]17 in normal mice and tumor-bearing mice
The animal experimental protocols used were approved by the Committee on Animal Experimentation of Kanazawa University (AP-163723). Experiments with animals were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of Kanazawa University. The animals were housed with free access to food and water at 23 °C with a 12 h alternating light/dark schedule.
In the case of normal mice, 100 μL of [125I]15 or [125I]17 (18–33 kBq) in saline was intravenously administered into 6 weeks male ddY mice (28–30 g, Japan SLC, Inc.). The mice were sacrificed at 10, 30 min, 1, 4, 8, and 24 h postinjection. In the case of tumor-bearing mice, approximately 5 × 106 DU-145 cells were injected subcutaneously into the right dorsum of 4-week-old male BALB/c nude mice (15–19 g, Japan SLC, Inc.). After 14–21 days postinoculation, mice were intravenously administered 100 μL of [125I]15 or [125I]17 (37 kBq). At 1 and 24 h postinjection, the mice were sacrificed. Tissues of interest were removed and weighed, and radioactivity counts were determined with an auto well gamma counter and corrected for background radiation.
SPECT/CT imaging
[123I]15 and [123I]17 were prepared with 68 ± 4% (n = 3) and 47 ± 6% (n = 3) radiochemical yields, respectively, by the same method as the corresponding 125I-labeled compounds. After HPLC purification, SPECT/CT imaging experiments of [123I]15 and [123I]17 in the DU-145 tumor-bearing mice were performed using a small animal SPECT system (VECTor/CT, MIlabs, Houten, the Netherlands). SPECT scanning was performed at 24 h postinjection of [123I]15 (50 MBq) and [123I]17 (38 MBq), respectively.
Data were acquired in list mode and photopeak windows were set after the acquisition. The energy window of 143–175 keV was employed. Data were reconstructed using pixel-based order-subsets expectation maximization, with correction for attenuation on computed tomography, in 16 subsets and 6 iterations. The voxel size was 0.8 × 0.8 × 0.8 mm. The obtained SPECT/CT images were analyzed using an image-processing application (AMIDE Imaging software, version 1.0.4).
Results and discussion
In vitro competitive binding assay
Table 1 lists Ki values, which were utilized as indexes of the compounds' affinity for the sigma receptors. On the whole, the bromine atom-containing vesamicol derivatives 3–14 exhibited high affinity for the sigma-1 receptor. With the exception of 9 and 10, the Ki values measured for 3–14 for the sigma-1 receptor were lower than 20 nM. Moreover, the selectivity of 9, 11, and 13, which comprise an ethylene linker (n = 2), for the sigma-1 receptor was higher than that of other compounds; in fact, according to a previous published report, the substitution of cyclohexanol with cyclopentanol in vesamicol analogs increased the compounds' selectivity for the sigma-1 receptor.32 Additionally, although the cause for this observation is unclear, 12 and 14, which comprise a propylene linker (n = 3), showed high affinity for both the sigma-1 and sigma-2 receptors.
Affinities (nM) of synthesized vesamicol derivatives and haloperidol for sigma receptors.
| Sigma-1 (Ki) | Sigma-2 (Ki) | |
|---|---|---|
| 1 | 5.8 ± 0.9a | 50.4 ± 13.0a |
| 2 | 8.7 ± 1.0a | 22.3 ± 3.5a |
| 3 | 10.2 ± 2.3 | 41.8 ± 2.4 |
| 4 | 19.4 ± 1.5 | 32.5 ± 4.3 |
| 5 | 17.2 ± 7.6 | 25.1 ± 2.2 |
| 6 | 7.7 ± 1.4 | 22.2 ± 2.8 |
| 7 | 9.8 ± 5.7 | 44.5 ± 55.3 |
| 8 | 4.4 ± 0.7 | 26.0 ± 7.2 |
| 9 | 21.5 ± 6.4 | 371.1 ± 125.1 |
| 10 | 37.1 ± 10.7 | 116.1 ± 15.0 |
| 11 | 12.4 ± 2.5 | 137.9 ± 59.9 |
| 12 | 4.8 ± 1.4 | 8.1 ± 0.6 |
| 13 | 9.9 ± 0.8 | 148.3 ± 12.3 |
| 14 | 8.1 ± 0.9 | 12.6 ± 0.6 |
| 15 | 7.9 ± 0.9 | 73.6 ± 14.0 |
| 17 | 14.6 ± 4.2 | 16.9 ± 5.4 |
| Haloperidol | 13.9 ± 3.7 | 114.3 ± 18.7 |
Data were reported previously.1
In this study, we selected 11 as a ligand with high selectivity for the sigma-1 receptor and 12 as a ligand with high affinity for both the sigma-1 and sigma-2 receptors; we then synthesized the corresponding iodinated variants of 11 and 12, which we dubbed 15 and 17 (Fig. 2), via a copper-catalyzed iodine–bromine exchange reaction conducted on 11 and 12, respectively (Scheme 4). Notably, compounds 15 and 17 displayed similar affinities for the sigma receptors as the corresponding brominated compounds 11 and 12 (Table 1). These results were consistent with those of the previous studies wherein iodine- and bromine-containing vesamicol derivatives exhibited comparable binding affinities for sigma receptors.48,49
Preparation of radioiodine-labeled compounds [125I]15 and [125I]17
The radioiodinated compounds [125I]15 and [125I]17 were prepared with radiochemical yields of 62 ± 14% (n = 10) and 60 ± 17% (n = 8), respectively, implementing the standard electrophilic iododestannylation of the corresponding trimethylstannyl precursors 16 and 18, which were synthesized starting from the corresponding brominated compounds 11 and 12, respectively (Scheme 4). After purification by RP-HPLC, the obtained [125I]15 and [125I]17 exhibited radiochemical purities exceeding 97%. The identities of [125I]15 and [125I]17 were confirmed by comparing their HPLC retention times with those of the corresponding nonradioactive iodinated reference compounds 15 and 17, respectively (Fig. S1†).
In vitro stability experiments
The results of in vitro stability experiments of [125I]15 and [125I]17 are shown in Table 2. After incubation of [125I]15 and [125I]17 in 0.1 M PB (pH 7.4) for 24 h at 37 °C, over 95% of [125I]15 and [125I]17 remained intact.
In vitro stability of [125I]15 and [125I]17 in PB pH 7.4 and murine plasma.
| PB | Murine plasma | ||
|---|---|---|---|
| 24 h | 1 h | 24 h | |
| [125I]15 | 96.9 ± 0.2% | 95.6 ± 0.3% | 81.3 ± 1.0% |
| [125I]17 | 96.3 ± 0.2% | 96.5 ± 1.2% | 94.8 ± 0.5% |
Meanwhile, after incubation of [125I]15 and [125I]17 in murine plasma for 1 h at 37 °C, over 95% of [125I]15 and [125I]17 remained intact also. These results indicate that [125I]15 and [125I]17 are highly stable in vitro.
Partition coefficient
The octanol water partition coefficients of [125I]15 and [125I]17 were determined by the shake-flask method to evaluate these compounds' lipophilicity. The log P values of [125I]15 and [125I]17 were found to be 2.08 ± 0.02 and 2.50 ± 0.02, respectively [mean ± standard deviation (SD) for four samples]. The difference between these two values is derived from the different lengths of the alkyl chains linking the benzene ring to the piperazine ring. These results do not contradict the retention times determined in RP-HPLC analyses (Fig. S1†).
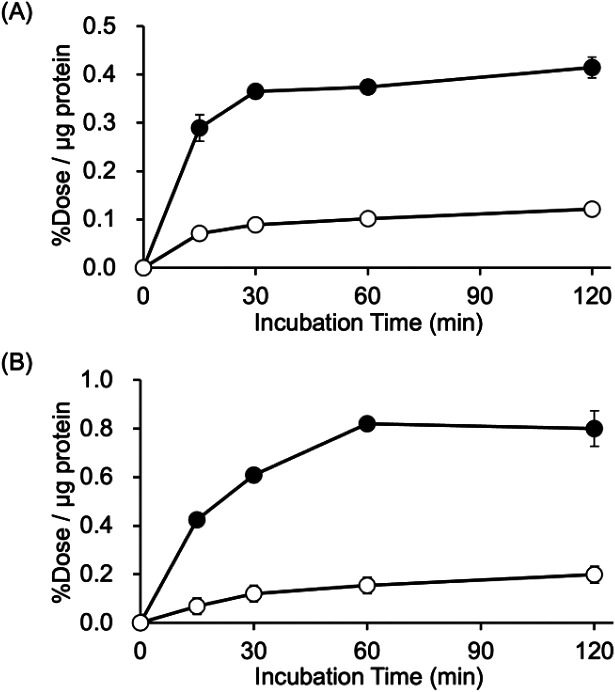
In vitro cellular uptake experiments
Experiments conducted to determine the uptake of [125I]15 and [125I]17 into DU-145 cells (a human prostate cancer cell line) were performed. The results indicated the rapid uptake of large amounts of [125I]15 and [125I]17 in DU-145 cells (Fig. 3). Moreover, the uptake of [125I]17 in DU-145 cells was higher than that of [125I]15. This observation may be due to the fact that DU-145 cells express not only the sigma-1 receptor but also the sigma-2 receptor.50 The uptake of [125I]15 and [125I]17 was observed to be significantly inhibited in the presence of an excess amount of haloperidol, a ligand of the sigma receptors. However, the inhibition of the uptake of [125I]15 and [125I]17 by haloperidol was not complete. The results suggested that a degree of nonspecific binding or uptake of [125I]15 and [125I]17 to DU-145 cells could take place.
Fig. 3. Cellular uptake study. (A) Cellular uptake study of [125I]15 (closed circles) and [125I]15 with 10 μM of haloperidol (open circles). (B) Cellular uptake study of [125I]17 (closed circles) and [125I]17 with 10 μM of haloperidol (open circles). All results are shown as mean ± SD.
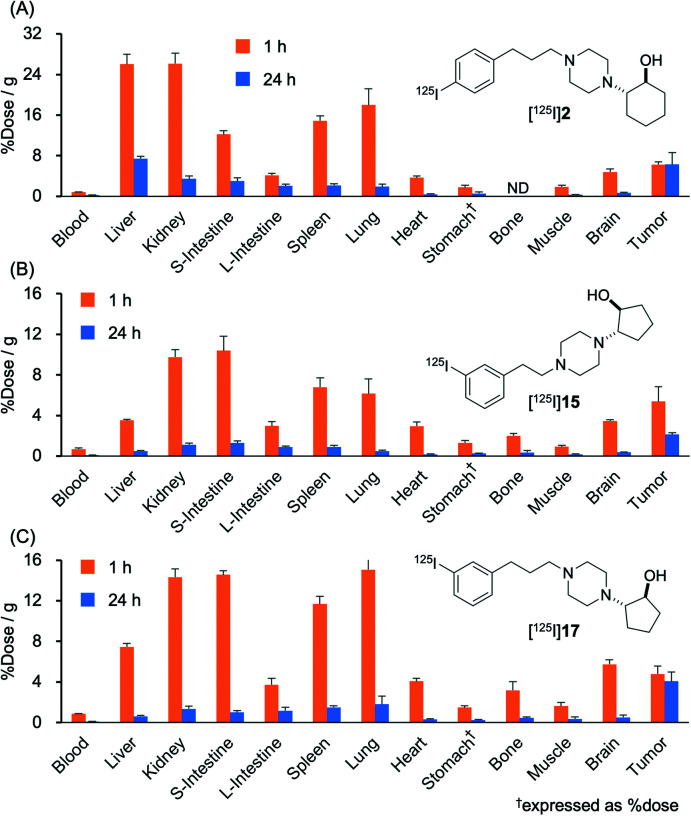
Biodistribution experiments
The results of the biodistribution of [125I]15 and [125I]17 in normal mice are shown in Tables S1 and S2.† The results of the biodistribution of [125I]2,1 [125I]15, and [125I]17 in DU-145 tumor-bearing mice are shown in Fig. 4 and Tables S3–S5.† These results showed a similar biodistribution tendency in normal tissues between normal mice and tumor-bearing mice. In normal mice, the blood clearance of radiotracers was so fast until 10 min; however, it was slow from 10 min to 8 h postinjection. Moreover, the accumulation of radioactivity in the neck gradually increased after 4 h. These results indicate that deiodination might occur after the accumulation of radiotracers in normal tissues, return to the blood circulation, and accumulate in the thyroid.
Fig. 4. Biodistribution experiments. Biodistribution of radioactivity at 1 and 24 h after intravenous injection of (A) [125I]2, (B) [125I]15, and (C) [125I]17 in DU-145 tumor-bearing mice. All results are shown as mean ± SD. †Expressed as % injected dose. ND means no data. Data of (A) [125I]2 was reported in a previous study.1.
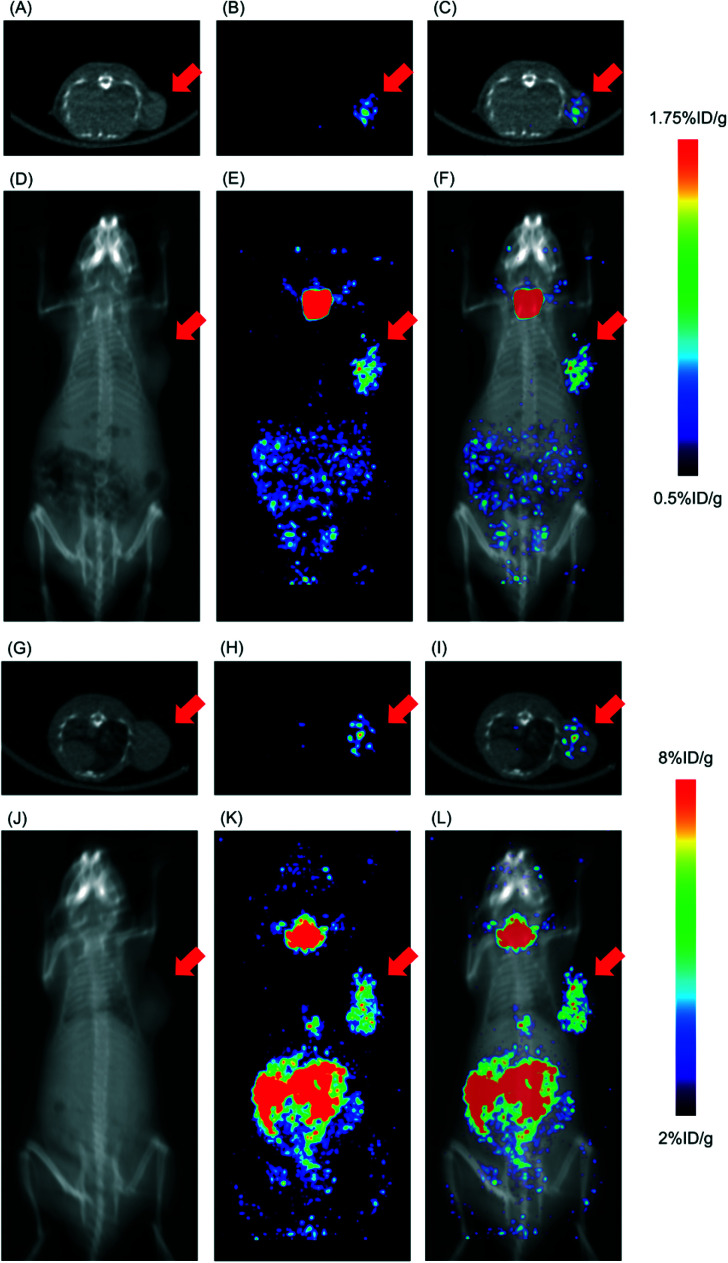
In tumor-bearing mice, a high tumor uptake was observed for both [125I]15 and [125I]17. Although the uptake of the mentioned compounds was not higher than that of [125I]2 and (+)-[125I]pIV, which had been evaluated as excellent tumor-targeting sigma receptor ligands in studies conducted previously by our group,1,46 accumulation of radioactivity in nonspecific tissues, such as the liver, kidney, and lung, was lower for [125I]15 and [125I]17 than for [125I]2 and (+)-[125I]pIV. Thus, the values for the tumor/non-target tissues accumulation ratios calculated for [125I]15 and [125I]17 were higher than those calculated for [125I]2 and (+)-[125I]pIV. In our previous study, we employed the 131I-labeled sigma receptor ligand (+)-[131I]pIV in targeted radionuclide therapy. Notably, (+)-[131I]pIV significantly inhibited the tumor growth in DU-145 tumor-bearing mice.46 As [125I]15 and [125I]17 were characterized by higher values for the tumor/non-target tissue ratios than (+)-[125I]pIV, the radioiodinated 15 and 17 are promising compounds not only for diagnostic imaging but also for radionuclide therapy applied to sigma receptor-positive tumors.
In the case of [125I]17, the tumor tissue exhibited longer retention of radioactivity than it was the case for [125I]15. In fact, at 24 h postinjection, the radioactivity due to [125I]15 decreased in the tumor tissue, although the decrease was less substantial than in other tissues. At 1 h postinjection, [125I]15 exhibited lower accumulation in almost all non-target tissues than [125I]17. We hypothesized that the observed difference in biodistribution between [125I]15 and [125I]17 was partially due to the difference in lipophilicity between these two compounds. In previous studies conducted by our group, a decrease in lipophilicity of a sigma-1 receptor ligand was observed to be associated with a decrease in the extent of accumulation in non-target tissues.46,47 Moreover, the high affinity of [125I]17 toward sigma-2 receptors may contribute to the observed difference in biodistribution.
On the other hand, vesamicol analogs have asymmetric carbons and were evaluated after optical resolution, such as pIV, in previous studies.12,23 However, in the present study, the racemic mixtures were utilized for [125I]15 and [125I]17, because the optical resolution of the relevant enantiomers could not be performed by the same method applied previously to vesamicol analogs. As (+)-vesamicol analogs have been reported to display higher affinity for the sigma-1 receptor than the corresponding (−)-vesamicol analogs, one of the enantiomers of [125I]15 and [125I]17 could have superior sigma receptor-binding characteristics to the others. Therefore, in the near future, we plan to investigate a method for their optical resolution and evaluate each enantiopure compound.
SPECT/CT imaging
SPECT/CT images of [123I]15 and [123I]17 are displayed in Fig. 5. The time point of SPECT imaging was set at 24 h postinjection of radiotracers, referring to the data from the biodistribution experiments. The SPECT images visualized the accumulation of [123I]15 and [123I]17 in the tumor. In addition, except for tumor accumulation, the accumulation of radioactivity in the abdominal organs, probably mainly the pancreas, and in the thyroid gland was observed. These are consistent with the results of the biodistribution experiments. Meanwhile, the high thyroid accumulation could be reduced by administering a saturated solution of potassium iodide, namely thyroid blockade,51 although it was not performed in this study.
Fig. 5. Representative SPECT/CT images. (A) an axial CT image, (B) an axial SPECT image, (C) an axial SPECT/CT fusion image, (D) a coronal maximal intensity projection (MIP) CT image, (E) a coronal MIP SPECT image, and (F) a coronal MIP SPECT/CT fusion image of a DU-145 tumor-bearing mouse at 24 h postinjection of [123I]15. (G) an axial CT image, (H) an axial SPECT image, (I) an axial SPECT/CT fusion image, (J) a coronal MIP CT image, (K) a coronal MIP SPECT image, and (L) a coronal MIP SPECT/CT fusion image of a DU-145 tumor-bearing mouse at 24 h postinjection of [123I]17. Red arrows indicate the site where DU-145 cells were inoculated.
Conclusions
We firstly synthesized twelve bromine-containing aza-vesamicol derivatives (3–14) comprising an ethylene or a propylene linker between the phenyl group and a piperazinylcyclohexanol or a piperazinylcyclopentanol moiety. Based on the results of the competitive binding assays, 11 and 12 were selected as sigma-1 receptor selective and sigma receptor non-selective ligands, respectively; moreover, radioiodine-labeled derivatives of these compounds were prepared, namely [125I]15 and [125I]17. Notably, [125I]15 and [125I]17 exhibited high uptake in the tumor in conjunction with lower uptake in non-target tissues than previously developed and tested 125I-labeled vesamicol derivatives, indicating that [123/131I]15 and [123/131I]17 are promising compounds to be used as imaging and therapeutic probes for sigma receptor-positive tumors.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported in part by Takeda Science Foundation and Hokkoku Foundation for Cancer Research.
Electronic supplementary information (ESI) available: HPLC chromatograms, biodistribution data, and NMR spectrum. See DOI: https://doi.org/10.1039/d2md00099g
Notes and references
- Ogawa K. Masuda R. Mishiro K. Wang M. Kozaka T. Shiba K. Kinuya S. Odani A. Bioorg. Med. Chem. 2019;27:1990–1996. doi: 10.1016/j.bmc.2019.03.054. [DOI] [PubMed] [Google Scholar]
- Quirion R. Bowen W. D. Itzhak Y. Junien J. L. Musacchio J. M. Rothman R. B. Su T. P. Tam S. W. Taylor D. P. Trends Pharmacol. Sci. 1992;13:85–86. doi: 10.1016/0165-6147(92)90030-A. [DOI] [PubMed] [Google Scholar]
- Schmidt H. R. Kruse A. C. Trends Pharmacol. Sci. 2019;40:636–654. doi: 10.1016/j.tips.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. J. Pharmacol. Sci. 2015;127:2–5. doi: 10.1016/j.jphs.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Miki Y. Mori F. Kon T. Tanji K. Toyoshima Y. Yoshida M. Sasaki H. Kakita A. Takahashi H. Wakabayashi K. Neuropathology. 2014;34:148–158. doi: 10.1111/neup.12080. [DOI] [PubMed] [Google Scholar]
- Mavlyutov T. A. Epstein M. L. Verbny Y. I. Huerta M. S. Zaitoun I. Ziskind-Conhaim L. Ruoho A. E. Neuroscience. 2013;240:129–134. doi: 10.1016/j.neuroscience.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng T. Y. Tsai S. A. Su T. P. J. Biomed. Sci. 2017;24:74. doi: 10.1186/s12929-017-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu A. Gradus T. Altman T. Maimon R. Saraf Avraham N. Geva M. Hayden M. Perlson E. Cell Death Dis. 2019;10(3):210. doi: 10.1038/s41419-019-1451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T. Expert Opin. Drug Discovery. 2021;16:373–389. doi: 10.1080/17460441.2021.1838483. [DOI] [PubMed] [Google Scholar]
- Vilner B. J. John C. S. Bowen W. D. Cancer Res. 1995;55:408–413. [PubMed] [Google Scholar]
- Crottès D. Rapetti-Mauss R. Alcaraz-Perez F. Tichet M. Gariano G. Martial S. Guizouarn H. Pellissier B. Loubat A. Popa A. Paquet A. Presta M. Tartare-Deckert S. Cayuela M. L. Martin P. Borgese F. Soriani O. Cancer Res. 2016;76:607. doi: 10.1158/0008-5472.CAN-15-1465. [DOI] [PubMed] [Google Scholar]
- Fallica A. N. Pittalà V. Modica M. N. Salerno L. Romeo G. Marrazzo A. Helal M. A. Intagliata S. J. Med. Chem. 2021;64:7926–7962. doi: 10.1021/acs.jmedchem.0c02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangangari K. K. Váradi A. Majumdar S. Larson S. M. Pasternak G. W. Pillarsetty N. K. Mol. Imaging Biol. 2020;22:358–366. doi: 10.1007/s11307-019-01369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agha H. McCurdy C. R. RSC Med. Chem. 2020;12:154–177. doi: 10.1039/D0MD00186D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. R. Zheng S. Gurpinar E. Koehl A. Manglik A. Kruse A. C. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. R. Betz R. M. Dror R. O. Kruse A. C. Nat. Struct. Mol. Biol. 2018;25:981–987. doi: 10.1038/s41594-018-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D. A. Schmidt H. R. Skiba M. A. Mandler M. D. Anderson J. R. Sliz P. Kruse A. C. ACS Med. Chem. Lett. 2020;11:1555–1561. doi: 10.1021/acsmedchemlett.9b00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis M. O. Karoutzou O. Foscolos A. S. Papanastasiou I. Molecules. 2017:22. doi: 10.3390/molecules22091408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon A. Schmidt H. R. Wood M. D. Sahn J. J. Martin S. F. Kruse A. C. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7160–7165. doi: 10.1073/pnas.1705154114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D. Wahlster L. Bartz F. Werenbeck-Ueding J. Praggastis M. Zhang J. Joggerst-Thomalla B. Theiss S. Grimm D. Ory D. S. Runz H. Hum. Mol. Genet. 2016;25:3588–3599. doi: 10.1093/hmg/ddw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S. Lu H. L. Zhang L. J. Wu Z. Med. Res. Rev. 2014;34:532–566. doi: 10.1002/med.21297. [DOI] [PubMed] [Google Scholar]
- Chen A. F. Ma W. H. Xie X. Y. Huang Y. S. Curr. Med. Chem. 2021;28:4172–4189. doi: 10.2174/0929867327666200902172615. [DOI] [PubMed] [Google Scholar]
- Zeng C. Riad A. Mach R. H. Cancers. 2020;12:1877. doi: 10.3390/cancers12071877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaghaghi Z. Alvandi M. Ghanbarimasir Z. Farzipour S. Emami S. Bioorg. Chem. 2021;115:105163. doi: 10.1016/j.bioorg.2021.105163. [DOI] [PubMed] [Google Scholar]
- Colabufo N. A. Abate C. Contino M. Inglese C. Niso M. Berardi F. Perrone R. Bioorg. Med. Chem. Lett. 2008;18:1990–1993. doi: 10.1016/j.bmcl.2008.01.109. [DOI] [PubMed] [Google Scholar]
- Yang D. Comeau A. Bowen W. D. Mach R. H. Ross B. D. Hong H. Van Dort M. E. Mol. Pharmaceutics. 2017;14:770–780. doi: 10.1021/acs.molpharmaceut.6b01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efange S. M. Khare A. B. Mach R. H. Parsons S. M. J. Med. Chem. 1999;42:2862–2869. doi: 10.1021/jm980560x. [DOI] [PubMed] [Google Scholar]
- Shiba K. Ogawa K. Ishiwata K. Yajima K. Mori H. Bioorg. Med. Chem. 2006;14:2620–2626. doi: 10.1016/j.bmc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Shiba K. Ogawa K. Mori H. Bioorg. Med. Chem. 2005;13:1095–1099. doi: 10.1016/j.bmc.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Ogawa K. Mizuno Y. Washiyama K. Shiba K. Takahashi N. Kozaka T. Watanabe S. Shinohara A. Odani A. Nucl. Med. Biol. 2015;42:875–879. doi: 10.1016/j.nucmedbio.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Ogawa K. Masuda R. Shiba K. Bunseki Kagaku. 2017;66:403–411. doi: 10.2116/bunsekikagaku.66.403. [DOI] [Google Scholar]
- Shigeno T. Kozaka T. Kitamura Y. Ogawa K. Taki J. Kinuya S. Shiba K. Ann. Nucl. Med. 2021;35:167–175. doi: 10.1007/s12149-020-01552-w. [DOI] [PubMed] [Google Scholar]
- Ogawa K. Takeda T. Yokokawa M. Yu J. Makino A. Kiyono Y. Shiba K. Kinuya S. Odani A. Chem. Pharm. Bull. 2018;66:651–659. doi: 10.1248/cpb.c18-00081. [DOI] [PubMed] [Google Scholar]
- Mishiro K. Nishii R. Sawazaki I. Sofuku T. Fuchigami T. Sudo H. Effendi N. Makino A. Kiyono Y. Shiba K. Taki J. Kinuya S. Ogawa K. J. Med. Chem. 2022;65:1835–1847. doi: 10.1021/acs.jmedchem.1c01211. [DOI] [PubMed] [Google Scholar]
- Bilodeau F. Brochu M.-C. Guimond N. Thesen K. H. Forgione P. J. Org. Chem. 2010;75:1550–1560. doi: 10.1021/jo9022793. [DOI] [PubMed] [Google Scholar]
- Virelli M. Moroni E. Colombo G. Fiengo L. Porta A. Ackermann L. Zanoni G. Chem. – Eur. J. 2018;24:16516–16520. doi: 10.1002/chem.201804244. [DOI] [PubMed] [Google Scholar]
- Cui H.-B. Xie L.-Z. Wan N.-W. He Q. Li Z. Chen Y.-Z. Green Chem. 2019;21:4324–4328. doi: 10.1039/C9GC01802F. [DOI] [Google Scholar]
- Yamashita Y. Tanaka K.-I. Yamakawa N. Asano T. Kanda Y. Takafuji A. Kawahara M. Takenaga M. Fukunishi Y. Mizushima T. Bioorg. Med. Chem. 2019;27:3339–3346. doi: 10.1016/j.bmc.2019.06.016. [DOI] [PubMed] [Google Scholar]
- Tassone J. P. Mawhinney R. C. Spivak G. J. J. Organomet. Chem. 2015;776:153–156. doi: 10.1016/j.jorganchem.2014.10.048. [DOI] [Google Scholar]
- Tam A. Soellner M. B. Raines R. T. J. Am. Chem. Soc. 2007;129:11421–11430. doi: 10.1021/ja073204p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C. Ferey V. Daumas M. Bernardelli P. Guérinot A. Cossy J. Org. Lett. 2020;22:6021–6025. doi: 10.1021/acs.orglett.0c02115. [DOI] [PubMed] [Google Scholar]
- Gobbo P. Gunawardene P. Luo W. Workentin M. S. Synlett. 2015;26:1169–1174. doi: 10.1055/s-0034-1380191. [DOI] [Google Scholar]
- Shiba K. Mori H. Matsuda H. Tsuji S. Kuji I. Sumiya H. Kinuya K. Tonami N. Hisada K. Sumiyosi T. Nucl. Med. Biol. 1995;22:205–210. doi: 10.1016/0969-8051(94)00093-Y. [DOI] [PubMed] [Google Scholar]
- Shiba K. Yano T. Sato W. Mori H. Tonami N. Life Sci. 2002;71:1591–1598. doi: 10.1016/S0024-3205(02)01919-7. [DOI] [PubMed] [Google Scholar]
- Effendi N. Mishiro K. Shiba K. Kinuya S. Ogawa K. Molecules. 2020:26. doi: 10.3390/molecules26010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K. Shiba K. Akhter N. Yoshimoto M. Washiyama K. Kinuya S. Kawai K. Mori H. Cancer Sci. 2009;100:2188–2192. doi: 10.1111/j.1349-7006.2009.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K. Kanbara H. Shiba K. Kitamura Y. Kozaka T. Kiwada T. Odani A. EJNMMI Res. 2012;2:54. doi: 10.1186/2191-219X-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K. Kanbara H. Kiyono Y. Kitamura Y. Kiwada T. Kozaka T. Kitamura M. Mori T. Shiba K. Odani A. Nucl. Med. Biol. 2013;40:445–450. doi: 10.1016/j.nucmedbio.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Ogawa K. Masuda R. Mizuno Y. Makino A. Kozaka T. Kitamura Y. Kiyono Y. Shiba K. Odani A. Nucl. Med. Biol. 2018;61:28–35. doi: 10.1016/j.nucmedbio.2018.03.005. [DOI] [PubMed] [Google Scholar]
- John C. S. Vilner B. J. Geyer B. C. Moody T. Bowen W. D. Cancer Res. 1999;59:4578–4583. [PubMed] [Google Scholar]
- Friedman N. C. Hassan A. Grady E. Matsuoka D. T. Jacobson A. F. J. Nucl. Med. 2014;55:211–215. doi: 10.2967/jnumed.113.124826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.