Abstract
The peptidoglycan (murein) of Helicobacter pylori has been investigated by high-performance liquid chromatography and mass spectrometric techniques. Murein from H. pylori corresponded to the A1γ chemotype, but the muropeptide elution patterns were substantially different from the one for Escherichia coli in that the former produced high proportions of muropeptides with a pentapeptide side chain (about 60 mol%), with Gly residues as the C-terminal amino acid (5 to 10 mol%), and with (1→6)anhydro-N-acetylmuramic acid (13 to 18 mol%). H. pylori murein also lacks murein-bound lipoprotein, trimeric muropeptides, and (l-d) cross-linked muropeptides. Cessation of growth and transition to coccoid shape triggered an increase in N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu (approximately 20 mol%), apparently at the expense of monomeric muropeptides with tri- and tetrapeptide side chains. Muropeptides with (1→6)anhydro-muramic acid and with Gly were also more abundant in resting cells.
Helicobacter pylori colonizes the human stomach and establishes a chronic infection associated with an inflammatory response of the gastric epithelium. A subpopulation of infected individuals develop peptic ulcer disease (3, 10, 28). In addition, H. pylori has been recognized as a risk factor for gastric adenocarcinoma (12, 34). There appears to be no substantial reservoir of H. pylori aside from the human stomach (6).
H. pylori cells growing actively in vitro are curved rods which, after prolonged incubation, evolve into metabolically active but nonculturable coccoid cells (2, 4, 33). In the stomach mostly spiral-shaped bacteria are found, but coccoid cells have been observed in the more severely damaged regions of the gastric mucosa (8, 21). The recent isolation of H. pylori from the feces of adults and children implicates a fecal-oral transmission (13). The coccoid cells may be a persistent form in which H. pylori can exist in the environment (6). If the coccoid form could replicate after ingestion by humans, as recently shown for a murine animal model, they could be instrumental for H. pylori infection (1, 7, 47). Under the electron microscope coccoid cells appear as U-shaped bacilli with both ends joined by a membranous structure (2, 8). The morphological transition is the result of a global physiological change involving drastic modifications in metabolic activity (2, 9, 31, 32, 44).
Paradoxically, the involvement of the cell wall, the primary bacterial morphogenetic element (20), in this transition has not been studied in detail. Structural modifications of the cell wall could play a role in H. pylori pathogenesis, in particular if cell wall fragments were released. Liberation of peptidoglycan (murein) fragments may trigger inflammatory and arthropathic processes (17, 22, 24, 41, 45, 48, 49). Furthermore, murein fragments are capable of highly specific interactions with particular host cell types, as exemplified by Bordetella pertussis tracheal cytotoxin [N-acetylglucosaminyl-(1→6)-anhydro-N-acetylmuramyl–l-Ala–d-Glu–(γ)-meso-diaminopimelyl–d-Ala] (16, 25, 26), which leads to destruction of infected ciliated tracheal cells in whooping cough (18, 19). An identical toxin, released by Neisseria gonorrhoeae, promotes ciliated-cell-specific damage in the fallopian tube mucosa (29, 30).
Gram-negative bacteria are presumed to have structurally homogeneous cell walls (43). However, application of high-resolution techniques demonstrated numerous variations of the common basic structure (11, 38). The structural variations could be in part responsible for the specific biological activities of cell wall fragments from different bacteria. Therefore, a detailed investigation of the structural evolution of the H. pylori cell wall throughout the morphological transition from spiral to coccoid has been undertaken.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For studies of spiral-shaped bacteria H. pylori NCTC 11637 was grown overnight in brucella broth (Difco, Detroit, Mich.) supplemented with 0.8 μg of amphotericin B ml−1, 5 μg of trimethoprim ml−1, and 10% fetal bovine serum at 37°C in an atmosphere of 10% CO2 obtained with CampyPak Plus envelopes (Becton Dickinson Microbiology Systems). To obtain coccoid bacteria, cultures were left for 15 days under the same conditions. The identity of H. pylori was confirmed by characteristic urease and catalase activities. Purity of cultures and bacterial morphology were routinely confirmed by optical microscopy of Gram-stained samples. Escherichia coli MC6RP1 (37) was grown in Luria-Bertani medium at 37°C (23).
Murein preparation, HPLC analysis, and purification of muropeptides.
Bacterial cultures were slowly dropped onto an equal volume of a boiling solution of 8% sodium dodecyl sulfate under strong magnetic stirring and further processed for high-performance liquid chromatography (HPLC) analysis as described previously (40). Muramidase (Cellosyl; Hoechst, Frankfurt am Main, Germany)-digested samples were analyzed by HPLC according to the method of Glauner (14) on a Hypersil ODS18 reverse-phase column (250 mm by 4 mm; 3-μm particle size; Teknochroma, Barcelona, Spain). Muropeptides were detected by monitoring the A204 and, when required, collected individually at the UV detector outlet. Purified muropeptides were vacuum dried, resuspended in MilliQ water (Millipore), and desalted by HPLC as described previously (40).
Amino acid analysis.
Samples for amino acid analysis were subjected to acid hydrolysis in 6 N HCl for 12 h at 105°C, vacuum dried, resuspended in an appropriate volume of MilliQ water, and further processed for ortho-phthaldialdehyde pre-column derivatization and HPLC analysis as described previously (40).
Galactosylation of purified sacculi.
Sacculi (100 to 200 μg) were sedimented (at 100,000 × g for 10 min) in a TL-100 ultracentrifuge with a TL-100.3 rotor (Beckman Instruments Inc., Palo Alto, Calif.), resuspended in an equal volume of 20 mM MnCl2–100 mM morpholinepropanesulfonic acid (MOPS), pH 7.4, and subjected to galactosylation of the terminal N-acetylglucosamine residues with UDP-galactose and cow milk galactosyl transferase (Sigma, St. Louis, Mo.) (40). Galactosylated sacculi were further processed for muramidase digestion and HPLC analysis as described above.
MALDI-MS.
Positive- and negative-ion MALDI mass spectrometry (MALDI-MS) analyses were performed in the linear mode on a Kompact MALDI IV time-of-flight instrument (Shimadzu Kratos Analytical, Manchester, United Kingdom) equipped with a delayed-extraction device and a nitrogen laser. Mass spectra were obtained by signal averaging of 50 consecutive laser shots. The thin-layer sample preparation technique was applied by using a saturated matrix solution of α-cyano-4-hydroxycinnamic acid in acetone. Matrix solution (0.5 μl) was deposited into a target well and followed by crystallization under atmospheric pressure at room temperature. Each lyophilized peptidoglycan fraction was redisolved in 10 μl of water and diluted (1:10) if required. Sample solution (0.5 μl) was deposited onto the crystallized matrix layer. After unforced solvent evaporation took place, the dried samples were washed on the MALDI target wells with 2 μl of cold water to remove low-mass, water-soluble impurities.
RESULTS
HPLC analysis of H. pylori murein.
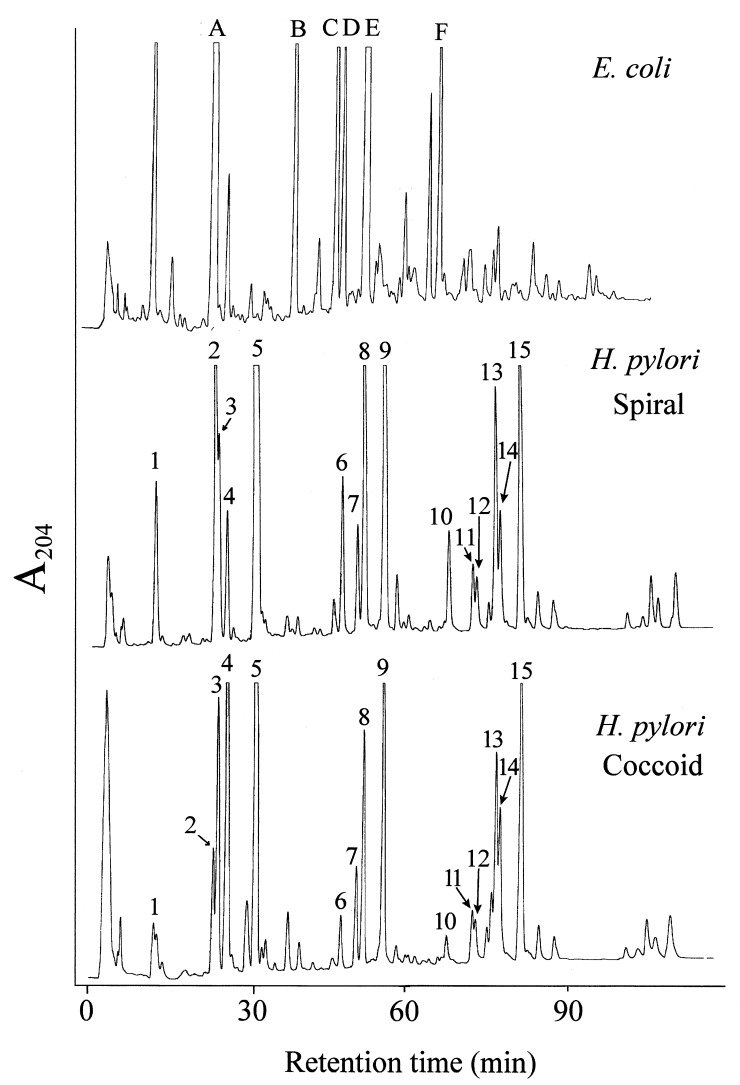
Murein samples (200 to 300 μg) from spiral H. pylori cells, 15 day-old coccoid H. pylori cells, and E. coli MC6RP1 cells were muramidase-digested, and the solubilized fractions were further processed for amino acid and muropeptide HPLC analyses. The results indicated that H. pylori and E. coli mureins are made up of the same amino acids; meso-diaminopimelic acid (mDAP), Glu, and Ala. More than 99% of the total mDAP in the samples was recovered in the soluble fractions, indicating an essentially complete solubilization of murein. The HPLC muropeptide elution profiles for H. pylori and E. coli murein samples were very distinct (Fig. 1), indicating substantial differences in muropeptide composition. Several of the major H. pylori muropeptides were not present in significant amounts in E. coli (peaks 3, 5, 7, 9, and 10 to 15 in Fig. 1), whereas in H. pylori no peaks were detectable at the positions corresponding to the E. coli lipoprotein-bound, (ld)-mDap-mDap cross-linked, and trimeric major muropeptides (peaks B, C, and F, respectively, in Fig. 1) (27).
FIG. 1.
HPLC elution patterns of murein samples purified from spiral and coccoid H. pylori cells. Muropeptide mixtures were analyzed as described in the text, and the A204 of the eluent was monitored. A murein sample from E. coli was analyzed under identical conditions for comparative purposes. Numbers in H. pylori panels identify corresponding peaks in both spiral and coccoid cell samples. Muropeptides shown in the E. coli panel correspond to the basic structure N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–(γ)-mDap–R1R2, where R1 and R2 are substituents at the l-carboxy and d-amino groups of mDap, respectively. R1 and R2 for the muropeptides shown are as follows: A, R1 = →d-Ala, R2 = −H; B, R1 = →Lys-Arg, R2 = −H (Braun’s lipoprotein anchoring muropeptide); C, R1 = N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–(d-Ala)mDap←, R2 = −H; D, R1 = N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–mDap–d-Ala←, R2 = −H; E, R1 = N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–(d-Ala)mDap–d-Ala←, R2 = −H; F, R1 = N - acetylglucosaminyl - N - acetylmuramyl–l - Ala–d - Glu–(d - Ala)mDap–d - Ala ←, R2 = N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–mDap–d-Ala→.
No qualitative differences were observed between the elution profiles for spiral- and coccoid-cell mureins. However, important quantitative variations were evident, in particular in a group of three peaks with retention times corresponding to the monomeric muropeptides N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–(γ)-mDap–d-Ala, N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu–(γ)-mDap–d-Ala–Gly, and N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu of E. coli (peaks 2, 3, and 4 in Fig. 1) (27).
Identification of individual muropeptides from spiral and coccoid cells by MALDI-MS.
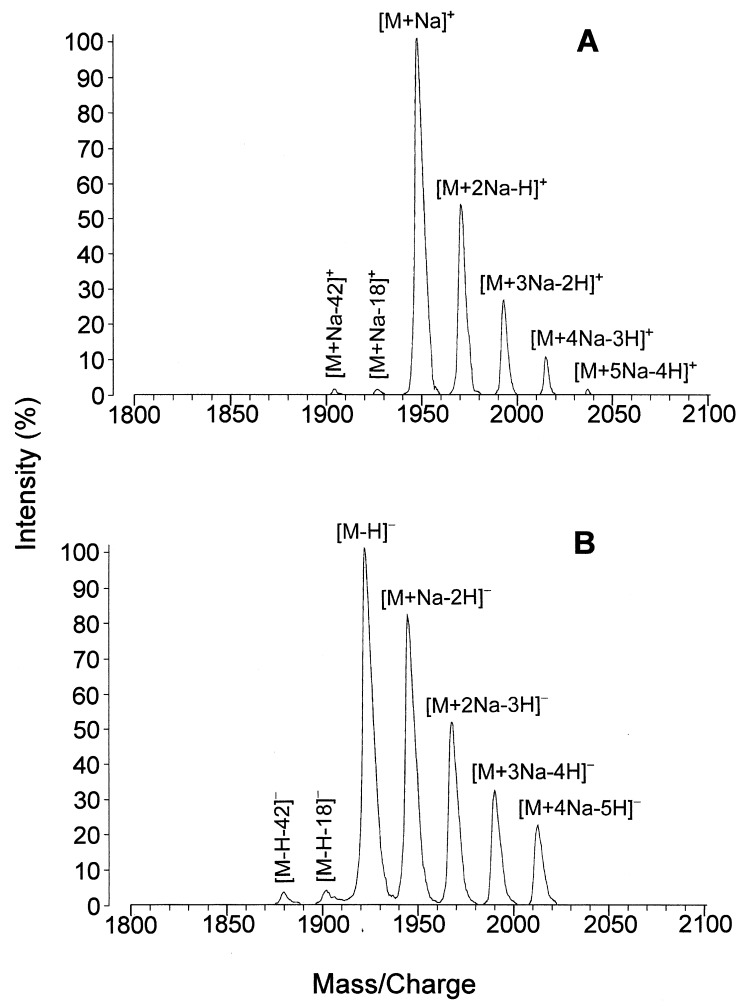
The substantial differences observed in muropeptide elution patterns made a direct identification of H. pylori muropeptides imperative. This was performed by positive- and negative-ion MALDI-MS of the individual muropeptides purified from murein of both spiral and coccoid cells, as in previous instances (36, 39). The sodiated molecules of the major murein monomers and dimers could be detected as dominating ions in the positive-ion, linear, delayed-extraction mode (Fig. 2A). More complex sodium adduct ions—[M+2Na-H]+, [M+3Na-2H]+, [M+4Na-3H]+, and [M+5Na-4H]+—were formed due to incomplete desalting. Sensitivity was significantly higher in the negative-ion mode than in the positive-ion mode. The negative-ion mass spectra exhibited ions of the type [M-H]− as the most abundant peaks (Fig. 2B), although ions of the type [M+nNa-(n+1)H]− (n = 1 to 4) were detected, too. Furthermore, in both ion modes the loss of a neutral water molecule (18 mass units) and an acetyl group from the base peak was detected with a significant abundance (Fig. 2). To deduce the putative muropeptide structures, experimental relative molecular mass values were compared to values calculated for murein fragments made up of the amino acids detected in the murein and the amino sugars N-acetylglucosamine and N-acetylmuramic acid or its (1→6)anhydro derivative, found in the glycan strand-terminating muropeptides (anhydro-muropeptides) (15, 27, 38, 40).
FIG. 2.
Molecular ion region of the positive- and negative-ion MALDI mass spectrum of fraction 7 from murein of spiral H. pylori cells. (A) The positive-ion mass spectrum exhibited complex adduct ions at m/z 1945.0 (calculated m/z, 1945.9), 1966.9, 1989.0, 2011.1, and 2033.0, corresponding to [M+Na]+, [M+2Na-H]+, [M+3Na-2H]+, [M+4Na-3H]+, and [M+5Na-4H]+ ions, respectively. (B) In the negative-ion mass spectrum, a similar complex adduct ion pattern ([M+Na-2H]−, [M+2Na-3H]−, [M+3Na-4H]−, and [M+4Na-5H]−) was observed, but with the deprotonated molecule [M-H]− (found m/z, 1921.0; calculated, m/z 1921.9) as the base peak.
More than 80% of the total material in H. pylori chromatograms was identified by MALDI-MS. The results confirmed the same basic structure for H. pylori and E. coli mureins (chemotype A1γ), with N-acetylglucosaminyl-N-acetylmuramyl-l-Ala-d-Glu-(γ)-mDap-d-Ala-d-Ala as the basic monomeric subunit (Table 1) (43). Components eluting at equal retention times upon HPLC separation corresponded to identical muropeptides in H. pylori and E. coli. The more prominent peaks in H. pylori murein corresponded to muropeptides with a pentapeptide side chain terminated with either d-Ala (5, 9, 10, and 15 in Fig. 1 and Table 1) or Gly (3 and 7 in Fig. 1 and Table 1). Muropeptides with (1→6)anhydro-muramic acid residues were also among the dominant peaks (10 to 15 in Fig. 1).
TABLE 1.
Analysis of murein purified from H. pylori cells at different stages of the spiral-coccoid morphological transition
Relative molecular mass values for NaBH4-reduced muropeptides purified from spiral- and coccoid-cell murein. Measurements of the relative molecular mass for the sodiated molecular ions [M+Na]+ (not shown) confirmed the results presented in the table. Spiral cells were taken from a 5-h-old culture; coccoid cells were taken from a 15-day-old culture.
Calculated as described by Glauner et al. (20).
More than 95% of the cells had the indicated morphology.
Numbers correspond with peaks in Fig. 1. In the lower part of the table, muropeptides are grouped according to structural similarity. Monomers, monomeric muropeptides (1 to 5 and 10); dimers, cross-linked muropeptides (6 to 9 plus 11 to 15); anhydro, muropeptides with (1→6)anhydro-muramic acid (10 to 15); dipeptide, muropeptides with dipeptide side chain (4); d-Ala–d–Ala, muropeptides with a pentapeptidic side chain terminated in d-Ala–d-Ala (5, 9, 10, and 15); d-Ala–Gly, muropeptides with a pentapeptidic side chain terminated in d-Ala–Gly (3 and 7).
Muropeptides best fitting the relative molecular masses measured by MALDI-MS. Abbreviations: NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid; (an)NAM, (1→6)anhydro-N-acetylmuramic acid.
Culture in the exponential phase of growth.
Culture in the early (4 h) stationary phase of growth.
Muropeptides 11 and 12 and muropeptides 13 and 14 are position isomers for the (1→6)anhydro-N-acetylmuramic acid residue relative to the peptide bridge. The alternative positions for (an)NAM and NAM residues are shown in brackets.
Presumably an unresolved mixture of two position isomers as for muropeptides 11 and 12 and muropeptides 13 and 14. The alternative positions for (an)NAM and NAM residues are shown in brackets.
Evolution of muropeptide composition throughout the spiral-coccoid morphological transition of H. pylori.
Murein purified from H. pylori cultures of increasing age was subjected to HPLC analysis, and the relative abundance of muropeptides for each sample was calculated according to the method of Glauner et al. (15). Cell morphology in each culture was checked by optical microscopy. After the second day of incubation coccoid cells started to accumulate, and by the fourth day no spiral cells could be observed. The muropeptide composition for each sample is shown in Table 1.
The prevalence, in both spiral and coccoid cells, of muropeptides with a pentapeptide side chain was remarkable. More than 50% of total muropeptides retained the canonical d-Ala–d-Ala dipeptide, and an additional 5 to 10% had d-Ala–Gly as the terminal dipeptide. The proportion of Gly-containing muropeptides doubled rapidly when cells went into the stationary phase of growth, while the proportion of d-Ala-terminated ones decreased moderately (Table 1).
Anhydro-muropeptides were exceptionally abundant (13 to 18 mol%) and hyper-cross-linked (about 90% cross-linkage) in H. pylori murein. The abundance of muropeptides with anhydro-muramic acid and pentapeptide side chains led to the accumulation of two muropeptides (anhydro-disaccharide pentapeptide and the cross-linked dimer disaccharide tetrapeptide-disaccharide pentapeptide with one anhydro-muramic acid residue [peaks 10 and 15 in Fig. 1]) which, to our knowledge, had not been detected in other bacteria. Shape transition had a notable influence on the proportion of anhydro-muropeptides, which increased sharply by about 30% (from 14 to 18 mol%) once active growth stopped (Table 1).
Cross-linkage was similar in spiral and coccoid cells (around 30%), although it was slightly lower in actively growing than in resting cells (Table 1). Cross-linked anhydro-muropeptides accounted for about one-half of the total cross-linkage, and their contribution was apparently independent of the morphological transition.
Galactosylation of H. pylori sacculi.
Incubation of sacculi with milk galactosyl transferase and UDP-galactose under appropriate conditions results in the specific galactosylation of the N-acetylglucosamine residues at the terminus of glycan strands (40, 42), allowing the identification of N-acetylglucosaminyl terminal muropeptides. A sample of purified sacculi from 15-day-old coccoid cells was divided into two identical aliquots. One aliquot was subjected to galactosylation, while the second was spared as a reference. After galactosylation both aliquots were muramidase digested and analyzed by HPLC. The proportion of galactosylated muropeptides was estimated from the reduction in peak size. Dimeric anhydro-muropeptides were the most susceptible to galactosylation. After the reaction, 45% of the anhydro-dimers were galactosylated, whereas only 27% of nonanhydro-dimers and 3% of monomers were modified. Therefore, about one-half of the cross-linked anhydro-muropeptides contain a glycan strand-terminating N-acetylglucosamine residue.
DISCUSSION
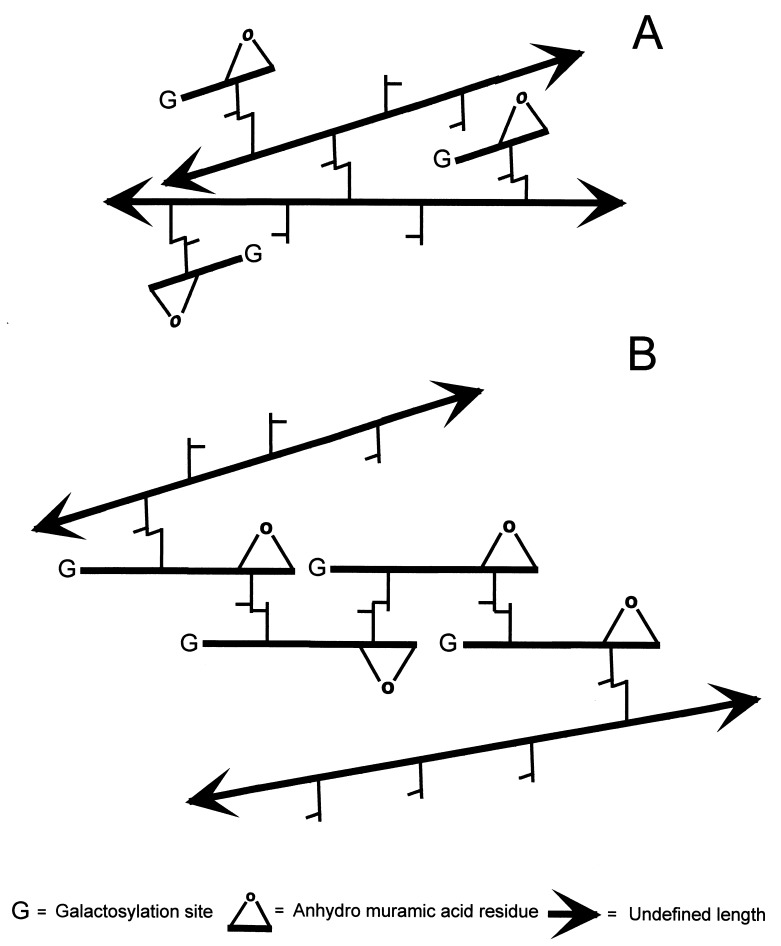
High-resolution analysis confirmed that H. pylori murein was of the A1γ chemotype (43) but had a unique muropeptide composition. Compared to other gram-negative bacteria, cross-linkage of H. pylori murein was similar in extension but simpler, as it was exclusively mediated by (dd)-d-Ala→mDAP cross-linked dimers. The elevated proportion of glycan chain-terminating anhydro-muropeptides implied a short mean length for glycan strands (5 to 7 disaccharide units). Therefore, very short (1 to 3 disaccharides) strands must be abundant. The very high cross-linkage of anhydro-muropeptides (80 to 88%) and murein galactosylation results suggested the presence of unit-length cross-linked muropeptides in sizable amounts and/or an extensive head-to-tail cross-linking of short strands. Unit-length cross-linked muropeptides cannot interconnect adjacent peptidoglycan strands and therefore cannot contribute to the strength of the sacculus. On the contrary, a number of very short strands cross-linked head to tail could eventually connect distant, long strands and therefore become an integral part of the stress-bearing structure (Fig. 3). The abundance of muropeptides with d-Ala- and Gly-terminated pentapeptide side chains could be due to the absence of ld- and dd-carboxypeptidases, as reported for Caulobacter crescentus (27). Misincorporation of Gly instead of d-Ala by d-Ala–d-Ala ligases has been postulated as the origin of Gly-containing muropeptides in E. coli (15). Sacculi from H. pylori were apparently devoid of covalently bound lipoproteins, which play an important role, anchoring the outer membrane to the sacculus in other gram-negative bacteria (5). Muropeptide analysis therefore indicated that H. pylori has a relatively simple murein compared to those of other gram-negative bacteria.
FIG. 3.
Alternative models accounting for the high proportion in cross-linked anhydro-muropeptides susceptible to galactosylation. (A) Single anhydro-disaccharide units are cross-linked to nearby long glycan strands. Muropeptides with this configuration do not contribute to the physical strength of the sacculus. (B) Short strands (two disaccharide units) are head-to-tail cross-linked to each other, connecting longer strands. In this configuration very short chains could effectively contribute to the strength of the sacculus. Muramidase digestion would release equal amounts of galactosylated, anhydro-cross-linked dimers in both instances. The models shown in both panels could coexist.
Analysis of murein from cells undergoing morphological transition revealed substantial variation in muropeptide proportions. The accumulation of dipeptide monomers and a concomitant reduction in tri- and tetrapeptide monomers constituted the most dramatic modification (Table 1). The results suggest that activation of a (γ)-glutamyl-diaminopimelate endopeptidase leads to massive conversion of tri- and tetrapeptide monomers into dipeptide monomers, as previously observed in sporulating Bacillus sphaericus (46). Thus, the accumulation of disaccharide-dipeptides appears to be a result of convergent evolution between the distantly related bacteria H. pylori and B. sphaericus in the genesis of resistant forms, i.e., coccoid cells and endospores, respectively. The proportions of anhydro-muropeptides and Gly-terminated muropeptides also increased significantly in coccoid cells with respect to spiral ones, as did cross-linkage to a lesser extent.
The changes observed above speak of an important modification of the sacculus associated with the morphological transition. Nevertheless, time course analysis showed that variations in cross-linkage and anhydro-muropeptides were more likely linked to the transition in the state of growth, as shown for E. coli (35), than to the transition in morphology. In both cases the values after 4 h in the stationary phase remained essentially constant for up to 15 days. In contrast, the plateau value for accumulation of dipeptide monomers was only reached when most cells (>95%) were coccoid, indicating a connection with the change in shape. Muropeptide composition was essentially stable from the time coccoid cells became predominant (day 4) and remained so for at least 11 more days.
In summary, H. pylori murein has a unique muropeptide composition and undergoes substantial modifications, requiring the activation of specific enzymes, when cells stop active growth and become committed to morphological transition.
ACKNOWLEDGMENTS
We thank J. C. Quintela for his helpful advice and J. de la Rosa for technical assistance.
This work was supported by grant PM97-0148-C02-01 Programa Sectorial de Promoción del Conocimiento, Ministerio de Educación y Cultura, Spain; grant 08.2/0029/1997 from the Consejería de Educación y Cultura, Comunidad de Madrid, Spain; an institutional grant from the Fundación Ramón Areces to M. A. de Pedro; and grant P11183 from the Austrian Fonds zur Förderung der wissenchaftlichen Forschung to G. Allmaier. Interchange between Austrian and Spanish laboratories was funded by Acción Integrada Austria-España grant HU1997-0032 to G. Allmaier and M. A. de Pedro. K. Costa was supported by a fellowship from the Gulbenkian Foundation (PGDBM) and program PRAXIS XXI (BD/9807/96). L. Engstrand and P. Falk were supported by grants from the Swedish Medical Research Council, the Swedish Cancer Society, and the Swedish Foundation for Strategic Research.
REFERENCES
- 1.Aleljung P, Nilsson H O, Wang X, Nyberg P, Morner T, Warsame I, Wadstrom T. Gastrointestinal colonisation of BALB/cA mice by Helicobacter pylorimonitored by heparin magnetic separation. FEMS Immunol Med Microbiol. 1996;13:303–309. doi: 10.1111/j.1574-695X.1996.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 2.Benaïssa M, Babin P, Quellard N, Pezennec L, Cenatiempo Y, Fauchère J L. Changes in Helicobacter pyloriultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun. 1996;64:2331–2335. doi: 10.1128/iai.64.6.2331-2335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser M J. Helicobacter pyloriand the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 4.Bode G, Mauch F, Malfertheiner P. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol Infect. 1993;111:483–490. doi: 10.1017/s0950268800057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publisher; 1994. pp. 319–341. [Google Scholar]
- 6.Cave D R. How is Helicobacter pylori transmitted? Gastroenterology. 1997;113:S9–14. doi: 10.1016/s0016-5085(97)80004-2. [DOI] [PubMed] [Google Scholar]
- 7.Cellini L, Allocati N, Angelucci D, Iezzi T, Di Campli E, Marzio L, Dainelli B. Coccoid Helicobacter pylorinot culturable in vitro reverts in mice. Microbiol Immunol. 1994;38:843–850. doi: 10.1111/j.1348-0421.1994.tb02136.x. [DOI] [PubMed] [Google Scholar]
- 8.Chan W Y, Hui P K, Leung K M, Chow J, Kwok F, Ng C S. Coccoid forms of Helicobacter pyloriin the human stomach. Am J Clin Pathol. 1994;102:503–507. doi: 10.1093/ajcp/102.4.503. [DOI] [PubMed] [Google Scholar]
- 9.Cole S P, Cirillo D, Kagnoff M F, Guiney D G, Eckmann L. Coccoid and spiral Helicobacter pyloridiffer in their abilities to adhere to gastric epithelial cells and induce interleukin-8 secretion. Infect Immun. 1997;65:843–846. doi: 10.1128/iai.65.2.843-846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folkening W J, Nogami W, Martin S A, Rosenthal R S. Structure of Bordetella pertussispeptidoglycan. J Bacteriol. 1987;169:4223–4227. doi: 10.1128/jb.169.9.4223-4227.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forman D, Newell D G, Fullerton F, Yarnell J W, Stacey A R, Wald N, Sitas F. Association between infection with Helicobacter pyloriand risk of gastric cancer: evidence from a prospective investigation. BMJ. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox J G. Non-human reservoirs of Helicobacter pylori. Aliment Pharmacol Ther. 1995;9(Suppl. 2):93–103. [PubMed] [Google Scholar]
- 14.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 15.Glauner B, Holtje J V, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 16.Goldman W E, Cookson B T. Structure and functions of the Bordetellatracheal cytotoxin. Tokai J Exp Clin Med. 1988;13(Suppl.):187–191. [PubMed] [Google Scholar]
- 17.Hazenberg M P, Klasen I S, Kool J, Ruseler-van Embden J G, Severijnen A J. Are intestinal bacteria involved in the etiology of rheumatoid arthritis? APMIS. 1992;100:1–9. doi: 10.1111/j.1699-0463.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 18.Heiss L N, Flak T A, Lancaster J R J, McDaniel M L, Goldman W E. Nitric oxide mediates Bordetella pertussistracheal cytotoxin damage to the respiratory epithelium. Infect Agents Dis. 1993;2:173–177. [PubMed] [Google Scholar]
- 19.Heiss L N, Moser S A, Unanue E R, Goldman W E. Interleukin-1 is linked to the respiratory epithelial cytopathology of pertussis. Infect Immun. 1993;61:3123–3128. doi: 10.1128/iai.61.8.3123-3128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höltje J V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janas B, Czkwianianc E, Bak-Romaniszyn L, Bartel H, Tosik D, Planeta-Malecka I. Electron microscopic study of association between coccoid forms of Helicobacter pyloriand gastric epithelial cells. Am J Gastroenterol. 1995;90:1829–1833. [PubMed] [Google Scholar]
- 22.Kohashi O, Pearson C M, Watanabe Y, Kotani S, Koga T. Structural requirements for arthritogenicity of peptidoglycans from Staphylococcus aureus and Lactobacillus plantarumand analogous synthetic compounds. J Immunol. 1976;116:1635–1639. [PubMed] [Google Scholar]
- 23.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 24.Lichtman S N, Bachmann S, Munoz S R, Schwab J H, Bender D E, Sartor R B, Lemasters J J. Bacterial cell wall polymers (peptidoglycan-polysaccharide) cause reactivation of arthritis. Infect Immun. 1993;61:4645–4653. doi: 10.1128/iai.61.11.4645-4653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luker K E, Collier J L, Kolodziej E W, Marshall G R, Goldman W E. Bordetella pertussistracheal cytotoxin and other muramyl peptides: distinct structure-activity relationships for respiratory epithelial cytopathology. Proc Natl Acad Sci USA. 1993;90:2365–2369. doi: 10.1073/pnas.90.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luker K E, Tyler A N, Marshall G R, Goldman W E. Tracheal cytotoxin structural requirements for respiratory epithelial damage in pertussis. Mol Microbiol. 1995;16:733–743. doi: 10.1111/j.1365-2958.1995.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 27.Markiewicz Z, Glauner B, Schwarz U. Murein structure and lack of dd- and ld-carboxypeptidase activities in Caulobacter crescentus. J Bacteriol. 1983;156:649–655. doi: 10.1128/jb.156.2.649-655.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 29.Martin S A, Rosenthal R S, Biemann K. Fast atom bombardment mass spectrometry and tandem mass spectrometry of biologically active peptidoglycan monomers from Neisseria gonorrhoeae. J Biol Chem. 1987;262:7514–7522. [PubMed] [Google Scholar]
- 30.Melly M A, McGee Z A, Rosenthal R S. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeaeto damage human fallopian-tube mucosa. J Infect Dis. 1984;149:378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- 31.Mizoguchi H, Fujioka T, Kishi K, Nishizono A, Kodama R, Nasu M. Diversity in protein synthesis and viability of Helicobacter pyloricoccoid forms in response to various stimuli. Infect Immun. 1998;66:5555–5560. doi: 10.1128/iai.66.11.5555-5560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narikawa S, Kawai S, Aoshima H, Kawamata O, Kawaguchi R, Hikiji K, Kato M, Iino S, Mizushima Y. Comparison of the nucleic acids of helical and coccoid forms of Helicobacter pylori. Clin Diagn Lab Immunol. 1997;4:285–290. doi: 10.1128/cdli.4.3.285-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilius M, Strohle A, Bode G, Malfertheiner P. Coccoid like forms (CLF) of Helicobacter pylori. Enzyme activity and antigenicity. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;280:259–272. doi: 10.1016/s0934-8840(11)80964-3. [DOI] [PubMed] [Google Scholar]
- 34.Parsonnet J, Hansen S, Rodríguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H. Helicobacter pyloriinfection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 35.Pisabarro A G, de Pedro M A, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coliassociated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pittenauer E, Schmid E R, Allmaier G, Pfanzagl B, Loffelhardt W, Fernandez C Q, de Pedro M A, Stanek W. Structural characterization of the cyanelle peptidoglycan of Cyanophora paradoxa by 252Cf plasma desorption mass spectrometry and fast atom bombardment/tandem mass spectrometry. Biol Mass Spectrom. 1993;22:524–536. doi: 10.1002/bms.1200220906. [DOI] [PubMed] [Google Scholar]
- 37.Prats R, de Pedro M A. Normal growth and division of Escherichia coliwith a reduced amount of murein. J Bacteriol. 1989;171:3740–3745. doi: 10.1128/jb.171.7.3740-3745.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quintela J C, Caparros M, de Pedro M A. Variability of peptidoglycan structural parameters in gram-negative bacteria. FEMS Microbiol Lett. 1995;125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- 39.Quintela J C, de Pedro M A, Zollner P, Allmaier G, Garcia-del Portillo P F. Peptidoglycan structure of Salmonella typhimuriumgrowing within cultured mammalian cells. Mol Microbiol. 1997;23:693–704. doi: 10.1046/j.1365-2958.1997.2561621.x. [DOI] [PubMed] [Google Scholar]
- 40.Quintela J C, Pittenauer E, Allmaier G, Aran V, de Pedro M A. Structure of peptidoglycan from Thermus thermophilusHB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietschel E T, Schletter J, Weidemann B, El-Samalouti V, Mattern T, Zahringer U, Seydel U, Brade H, Flad H D, Kusumoto S, Gupta D, Dziarski R, Ulmer A J. Lipopolysaccharide and peptidoglycan: CD14-dependent bacterial inducers of inflammation. Microb Drug Resist. 1998;4:37–44. doi: 10.1089/mdr.1998.4.37. [DOI] [PubMed] [Google Scholar]
- 42.Schindler M, Mirelman D, Schwarz U. Quantitative determination of N-acetylglucosamine residues at the non-reducing ends of peptidoglycan chains by enzymic attachment of [14C]-d-galactose. Eur J Biochem. 1976;71:131–134. doi: 10.1111/j.1432-1033.1976.tb11098.x. [DOI] [PubMed] [Google Scholar]
- 43.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorberg M, Nilsson M, Hanberger H, Nilsson L E. Morphologic conversion of Helicobacter pylorifrom bacillary to coccoid form. Eur J Clin Microbiol Infect Dis. 1996;15:216–219. doi: 10.1007/BF01591357. [DOI] [PubMed] [Google Scholar]
- 45.Takada H, Tsujimoto M, Kato K, Kotani S, Kusumoto S, Inage M, Shiba T, Yano I, Kawata S, Yokogawa K. Macrophage activation by bacterial cell walls and related synthetic compounds. Infect Immun. 1979;25:48–53. doi: 10.1128/iai.25.1.48-53.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vacheron M J, Guinand M, Francon A, Michel G. Characterisation of a new endopeptidase from sporulating Bacillus sphaericus which is specific for the gamma-d-glutamyl-l-lysine and gamma-d-glutamyl-(l)meso-diaminopimelate linkages of peptidoglycan substrates. Eur J Biochem. 1979;100:189–196. doi: 10.1111/j.1432-1033.1979.tb02048.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Sturegard E, Rupar R, Nilsson H O, Aleljung P A, Carlen B, Willen R, Wadstrom T. Infection of BALB/c A mice by spiral and coccoid forms of Helicobacter pylori. J Med Microbiol. 1997;46:657–663. doi: 10.1099/00222615-46-8-657. [DOI] [PubMed] [Google Scholar]
- 48.Weidemann B, Schletter J, Dziarski R, Kusumoto S, Stelter F, Rietschel E T, Flad H D, Ulmer A J. Specific binding of soluble peptidoglycan and muramyldipeptide to CD14 on human monocytes. Infect Immun. 1997;65:858–864. doi: 10.1128/iai.65.3.858-864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells A F, Hightower J A, Parks C, Kufoy E, Fox A. Systemic injection of group A streptococcal peptidoglycan-polysaccharide complexes elicits persistent neutrophilia and monocytosis associated with polyarthritis in rats. Infect Immun. 1989;57:351–358. doi: 10.1128/iai.57.2.351-358.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]