Abstract
Glycoconjugates are biologically significant molecules as they tend to serve a wide range of intra- and extra-cellular processes depending on their size and complexity. The secondary metabolites of the plant Myristica fragrans, eugenol and isoeugenol, have shown antifungal activities (IC50 1900 μM). Therefore, we envisioned that glycoconjugates based on these two scaffolds could prove to be potent antifungal agents. Triazole-containing compounds have shown prominent activities as antifungal agents. Based on this, we opined that a Cu(i) catalyzed click reaction could serve as the bridging tool between a eugenol/isoeugenol moiety and sugars to synthesize eugenol/isoeugenol based glycoconjugates. In our present work, we have coupled propargylated eugenol/isoeugenol and azido sugar to furnish eugenol/isoeugenol based glycoconjugates. In another approach, we have carried out hydroxylation of the double bond of eugenol and subsequent azidation of a primary alcohol followed by intramolecular coupling reactions leading to various other analogues. All the synthesized compounds were assayed against an opportunistic pathogenic fungus, Aspergillus fumigatus. Among the synthesized compounds, two analogues have exhibited significant antifungal activities with IC50 values of 5.42 and 9.39 μM, respectively. The study suggested that these two analogues inhibit cell wall-associated melanin hydrophobicity along with the number of conidia. The synthesized compounds were found to be non-cytotoxic to an untransformed cell line.
Design and synthesis of eugenol and isoeugenol based glycoconjugates and other analogous have been achieved using click chemistry. Two out of all the synthesized compounds exhibited significant antifungal activities against A. fumigatus.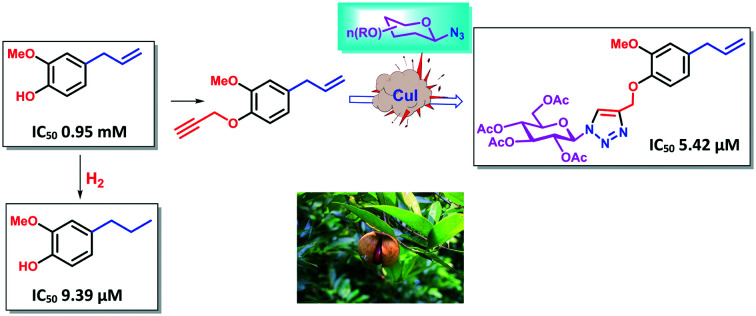
Introduction
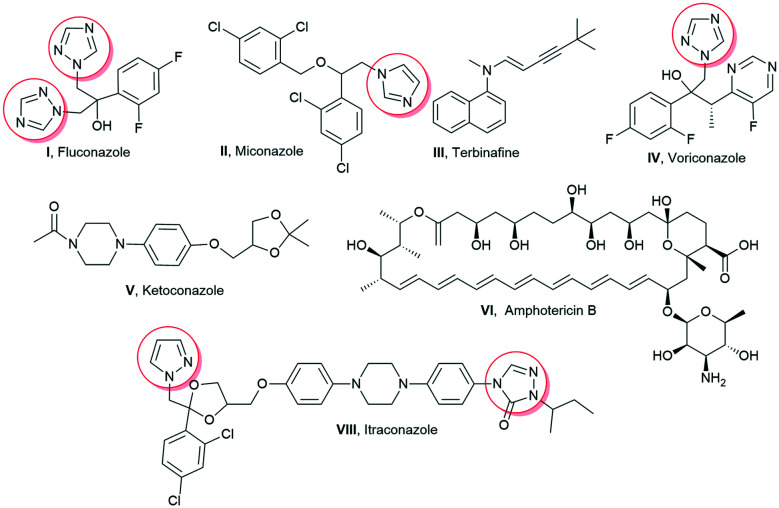
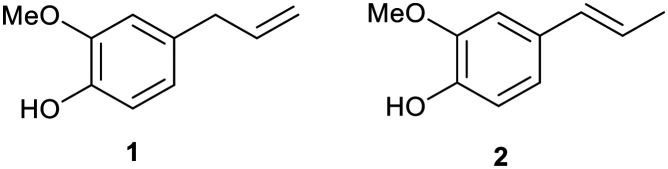
Aspergillus infections pose a significant threat to immune-compromised, organ transplant, neutropenic, and cancer patients and especially in patients having underlying lung diseases including asthma, tuberculosis, and chronic obstructive pulmonary disease. Multiple forms of aspergillosis have been estimated to affect about 1–4 million people worldwide, with more than 90 percent mortality rate.1 Recent studies have reported the incidence of invasive pulmonary aspergillosis in 19.6–33.3% of COVID-19 patients.2,3 The higher mortality rate of Aspergillus infections has been attributed to the virulence of the pathogen, delay in specific diagnosis, variable drug bioavailability, toxicity, and development of resistance towards drugs.4Aspergillus fumigatus is the most common Aspergillus species implicated in respiratory infections.4 It exhibits polygenic factors contributing to its virulence which include its small conidial size, biofilm formation, melanin layer, cell surface adhesion molecules, nutrient uptake that contributes towards protection during oxidative stress, and immunological inertness against the host killing mechanism.5 As the majority of virulence factors are associated with the fungal cell surface, it is a favourable target for the development of antifungal drugs. Subsequently, the increased incidence of fungal infections is also attributed to the availability of less toxic and efficient antifungal drugs. In the same regard, the matter of concern is the limited spectrum of efficacious antifungal drugs, which are not completely effective for the eradication of fungi. Amphotericin B (Amp B), itraconazole, fluconazole, voriconazole, ketoconazole, terbinafine, miconazole, etc. are available for the treatment of a wide array of fungal infections (Fig. 1). The majority of these antifungal drugs target cell membrane biosynthesis either at the final stage or by binding to the intermediary compound of its biosynthesis.6 There are a few other antifungal drugs targeting fungal cell wall components, nucleic acids, and microtubule biosynthesis. The toxicity associated with current antifungal drugs and the emergence of resistance have enthused an urgent need to improve existing antimicrobial scaffolds to develop novel antifungals. Hence, these antifungals have also been structurally modified to enhance their therapeutic index. Eugenol 1 and isoeugenol 2 are naturally occurring phenolic monoterpenes belonging to the phenylpropanoid group and have been traditionally isolated from Eugenia caryophyllata, Myristica fragrans, and Syzygium aromaticum (Fig. 2).7
Fig. 1. Clinically used antifungal drugs.
Fig. 2. Chemical structures of eugenol 1 and isoeugenol 2.
Isoeugenol 2 is produced by a range of plants such as the Petunia flower, Clarkia breweri, and Ocimum basilicum.7 Eugenol and isoeugenol have demonstrated analgesic, antioxidant, anti-inflammatory, anti-carcinogenic, anti-mutagenic, and even insect repellent properties.8 Besides, both of these compounds have been reported to exhibit antimicrobial activity.9 Eugenol 1 and isoeugenol 2 interfere with microbial membrane functions (membrane binding and permeability alteration) or suppress virulence factors including toxins, enzymes, the melanin biosynthesis pathway, hydrophobins, and formation of biofilms.10–12 They also induce the generation of H2O2 and increase the free Ca2+ concentration in the cytoplasm. Structural modifications in available drugs as well as naturally active lead moieties are conducted to enhance the drug efficacy and reduce their side effects. Alkylation of drug scaffolds has resulted in derivatives with promising antifungal activity. For example, 2,4-difluoro-2-(1H-1,2,4-triazol-1-yl) acetophenone compounds with linear C5–C8 alkyl chains13 and n-alkylated ebsulfur derivatives with a linear C5 alkyl chain displayed enhanced antifungal activity against Aspergillus species.14 In addition, aminoglycosides (kanamycin B and tobramycin) with linear C12 and C14 alkyl chains displayed significant antifungal activities. It has also been reported that the conjugation of a carbohydrate scaffold to naturally occurring bioactive molecules has led to enhanced bioactivity and manifestation of superior biological properties such as solubility, stability, and bioactivity compared to the parent molecule.15–17 Therefore, glycoconjugates of bioactive natural products can be ideal candidates for efficacious therapeutic agents. The hypervalent nature of sugar assists the drug molecule to reach target cells, thus playing a major role in the drug delivery system. In addition, the incorporation of the 1,2,4-triazole ring with therapeutically active compounds aids in the development of new antifungal agents.18,19
Results and discussion
We report herein the design and synthesis of novel glycoconjugates of eugenol 1 and isoeugenol 2 incorporating the 1,2,4-triazole ring and other derivatives and evaluated the antifungal activities of all the synthesised derivatives against A. fumigatus specifically targeting various cell wall associated virulence factors.
Synthesis and characterization of the synthesized compounds
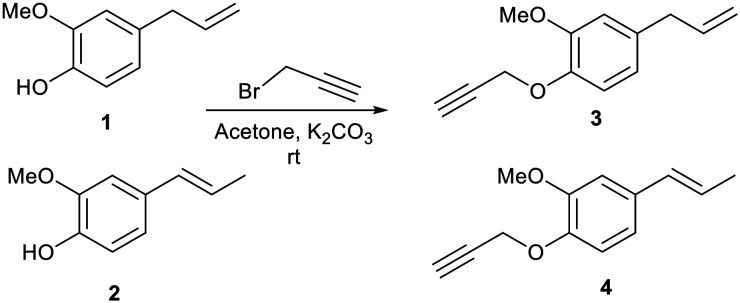
When we treated eugenol 1 and isoeugenol 2 with propargyl bromide in acetone in the presence of potassium carbonate (K2CO3) as base at room temperature, we obtained propargylated products 3 and 4, respectively (Scheme 1).
Scheme 1. Synthesis of propargylated derivatives of eugenol 1 and isoeugenol 2.
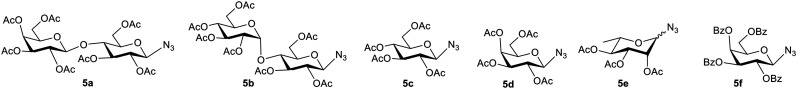
Azido sugars 5a–f (Fig. 3) were synthesized as per reported literature methods and their spectral data were found to be consistent with the reported literature data.15,20,21 The stereochemistries of the anomeric configurations of the azido sugars 5a–f were assigned as the β-configurations on the basis of comparison with reported data20 as well as their coupling constants in the 1H NMR (nuclear magnetic resonance) spectra. In general, the anomeric proton of the azido sugars appeared as a doublet integrated to one proton at 4.61–4.98 ppm in the 1H NMR spectra with a coupling constant in the range of 8.5–8.8 Hz, which unequivocally proves that the azide groups are β-oriented.15,20
Fig. 3. Structures of the azido sugars (5a–f) used for the synthesis of glycoconjugates.
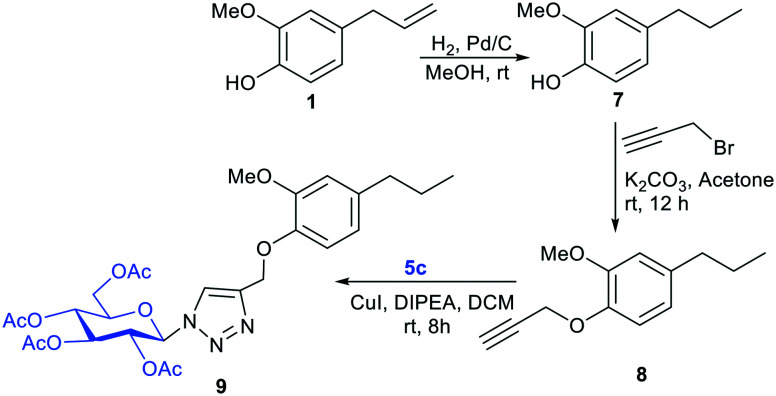
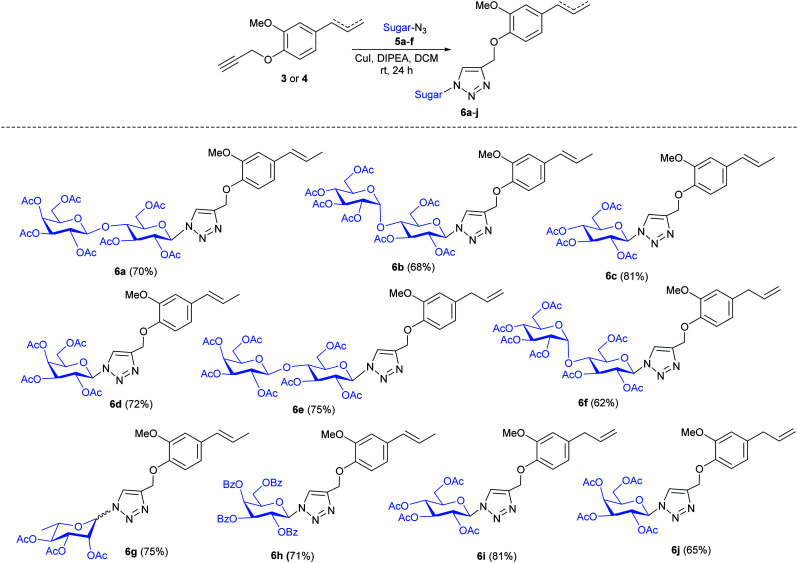
After synthesizing propargylated eugenol 3, isoeugenol 4, and azido sugars 5a–f, synthesis of eugenol and isoeugenol derived glycoconjugates was initiated utilizing click chemistry (Table 1).21–24 Propargylated eugenol 3, isoeugenol 4, and azido sugars 5a–f on reaction in the presence of catalytic amounts of copper iodide (CuI) and N,N-diisopropylethylamine (DIPEA) as base in dichloromethane (DCM) afforded glycoconjugates (6a–j) in good to very good yields. Slightly better yields of glycoconjugates were obtained in the case of reactions with lactose azide (6a and 6e) than those with maltose azide (6b and 6f). Azides of monosaccharides also furnished glycoconjugates (6c, d, and g–j) in good yields (Table 1). Azido glucose 5c furnished glycoconjugates (6c and 6i) in higher yields (81%, respectively) than glycoconjugates (6d and 6j) furnished by azido galactose 5d. Benzoate protected azido galactose (5f) also furnished glycoconjugate 6h in good yield. Compound 6i returned an IC50 value of 5.42 μM against A. fumigatus.
Substrate scopea,b.
|
Reaction conditions: propargylated eugenol/isoeugenol 3 or 4 (30 mg, 0.1485 mmol, 1 equiv.), azido sugars 5a–f (0.1782 mmol, 1.2 equiv.), DCM (3 ml), CuI (15.55 mg, 0.0816 mmol, 0.55 equiv.), and DIPEA (26 μl, 0.1485 mmol, 1.0 equiv.).
Isolated yield.
Furthermore, to explore the role of the exo-methylene double bond present in eugenol 1 in the antifungal activity, we reduced the exo-methylene double bond by using Pd–C/H2 to yield the reduced product, euganol 7 (ref. 25) (Scheme 2). The free –OH group of compound 7 was propargylated with propargyl bromide to afford compound 8. We then coupled compound 8 with 5c to provide compound 9. Among compounds 7, 8, and 9, compound 7 gave an IC50 value of 9.39 μM against A. fumigatus.
Scheme 2. Reduction of the double bond present in eugenol 1 and subsequent glyconjugation.
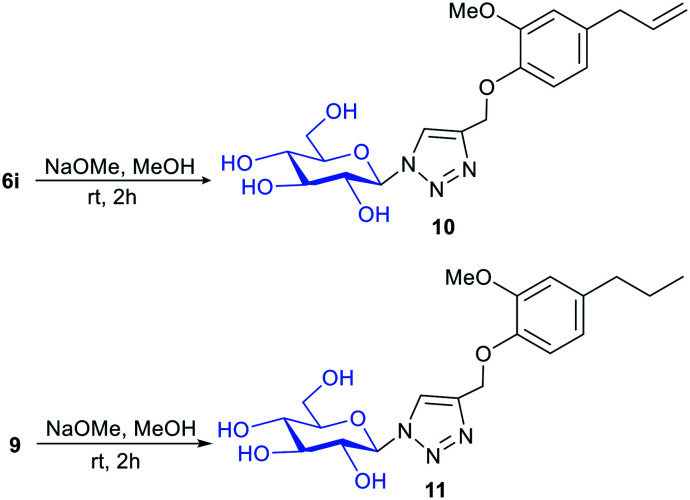
We further deprotected the acetyl groups present in compounds 6i and 9 in order to find out the role of the protecting group of sugar (Scheme 3).26 Compounds 10 and 11 were obtained after deprotection of acetyl groups present in compounds 6i and 9. However, the IC50 values of compounds 10 and 11 were found to be greater than 25 μM.
Scheme 3. Deacetylation of compounds 6i and 9.
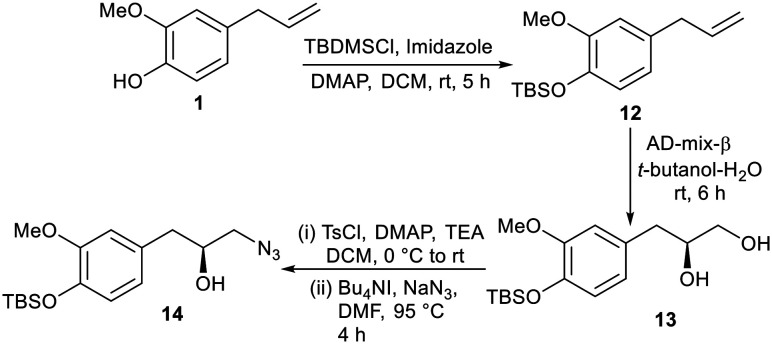
In order to synthesize more analogues other than glycoconjugates, the free –OH group of eugenol 1 was protected as OTBS by treating 1 with tert-butyldimethylsilyl chloride (TBDMS-Cl) in the presence of a combination of imidazole and 4-dimethyl-aminopyridine (DMAP) as bases in DCM to furnish compound 12. Compound 12 was subjected to dihydroxylation27 using AD-mix-β in a water–tert butanol solvent system to obtain compound 13 in which the primary hydroxyl group was treated with tosyl chloride only to facilitate azidation upon treatment with sodium azide (NaN3) in dimethylformamide (DMF) in the presence of a catalytic amount of tetrabutylammonium iodide (TBAI) to furnish compound 14 (Scheme 4).
Scheme 4. Dihydroxylation followed by azidation of eugenol 1.
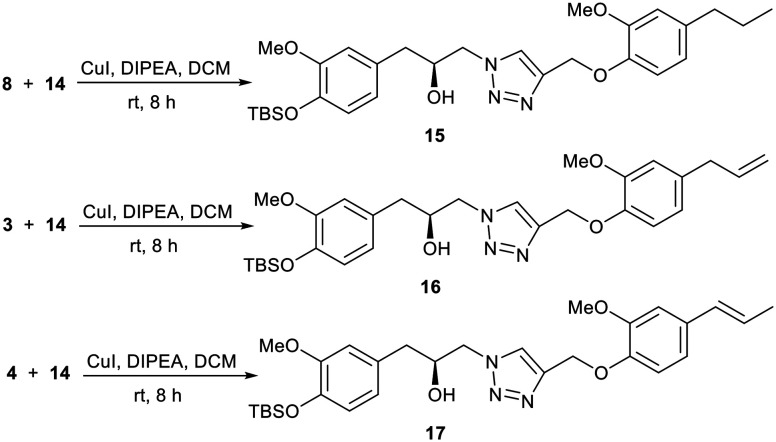
It is well established that often dimers of biologically active molecules exhibit higher activity than the monomeric unit.28,29 With compound 14 at hand, we envisaged to synthesize dimeric compounds. In this regard, we treated compound 14 with compounds 8, 3, and 4via a click reaction to afford compounds 15, 16, and 17, respectively (Scheme 5).
Scheme 5. Synthesis of dimeric compounds.
Biological evaluation of all the synthesized compounds
Eugenol 1, isoeugenol 2, and all the synthesized molecules (3, 4, 6a–j, and 7–17) were assayed for their anti-fungal activities against A. fumigatus. Preliminary data are furnished in Table 2.
IC50 values of compounds 3, 4, 6a–j, and 7–17 against A. fumigatus.
| Compound codes | IC50 values | Compound codes | IC50 values |
|---|---|---|---|
| 1 | 1900 μM | 6j | 10.86 μM |
| 2 | 1900 μM | 7 | 9.39 μM |
| 3 | >25 μM | 8 | >25 μM |
| 4 | >25 μM | 9 | > 25 μM |
| 6a | 14.47 μM | 10 | >25 μM |
| 6b | 14.47 μM | 11 | >25 μM |
| 6c | 21.73 μM | 12 | >25 μM |
| 6d | 10.86 μM | 13 | >25 μM |
| 6e | 14.47 μM | 14 | >25 μM |
| 6f | 14.47 μM | 15 | >25 μM |
| 6g | 12.08 μM | 16 | >25 μM |
| 6h | 15.18 μM | 17 | >25 μM |
| 6i | 5. 42 μM | Amp B | 1.08 μM |
As shown in Table 2, eugenol 1 demonstrated an IC50 value of 1900 μM against A. fumigatus. Conjugation of the glucose moiety with eugenol along with the azole group (compound 6i) increases the activity, as exhibited by the IC50 value of 5.42 μM. It is pertinent to mention here that the other sugars did not yield any notable activities. Galactose, which is the epimer of glucose having a different stereochemistry at C-4 than glucose, was also found to be less effective when linked to eugenol 1 which led us to believe that glucose, having all the OH bonds in equatorial positions, could be playing a pivotal role in terms of cellular interaction; however, when we coupled glucose with isoeugenol (compound 6c) we did not observe any significant activity. These observations envisaged us to look for the SAR associated with the position of the double bond as well and its presence. The non-activity of the isoeugenol–glucose conjugate 6c can be explained on the basis of non-terminal positioning of the double bond, whereas the eugenol–glucose conjugate 6i was found to be active with the terminal double bond. When we reduced the double bond of eugenol 1, the reduced eugenol 7 was found to exhibit an IC50 value of 9.39 μM. Inspired by this revelation we coupled compound 7 with glucose azide 5c. However, we did not observe any activity. We also assayed the dimeric compounds (15–17) with each of them returning an IC50 value greater than 25 μM.
The antifungal potency of the synthesized compounds of eugenol and isoeugenol (compounds 3, 4, 6a–j, and 7–17) against A. fumigatus was calculated using the micro-broth dilution assay as per the Clinical and Laboratory Standards Institute (CLSI) protocol.30 Amongst the synthesised compounds, only 6i and 7 inhibited the conidial as well as mycelial growth of A. fumigatus at significantly low MIC values i.e. 10.86 μM and 15.54 μM, respectively. The remaining compounds showed MIC >20 μM and, therefore, they were not considered for further experiments. The MIC and IC50 of both parent molecules (eugenol 1 and isoeugenol 2) were 3800 μM and 1900 μM, respectively. The MIC values of the drug control Amp B and itraconazole were 0.216 μM and 0.552 μM, respectively. The positive control showed A. fumigatus conidia with a characteristic greenish-grey color, whereas in compound-treated colonies white pigment-less conidia were visualized at IC50. The compounds exhibited MIC values ranging from 10–20 μM compared with the MIC value of 3800 μM of the parent molecules, indicating the higher antifungal potency of the compounds (more than 100-fold).
The crucial parameter for developing any antifungal molecule is its selective toxicity for fungal cells over mammalian cells. L-132 was referred to as an excellent cell line for cytotoxicity analysis.31 It consists of normal epithelial cells derived from human embryonic lungs. Hence, the compounds (6i and 7) were tested against the normal lung epithelial cell line L-132 along with the FDA-approved antifungal agent Amp B as a positive control. Cytotoxicity screening revealed that compounds were non-toxic on the untransformed cell line (compound 6i up to 43.46 μM and compound 7 up to 152.32 μM).
The concentration range selected for cytotoxicity covered the antifungal activity against A. fumigatus. The CC50 (cytotoxic concentration-50) values for compounds 6i and 7 were calculated as 43.46 μM and 150.5 μM. The reported sub-lethal cytotoxicity of the antifungal drug Amp B was in the range of 5.4–10.82 μM.32 The selectivity index (SI) values for 6i and 7 were calculated as 8.01 and 16.02, respectively. The compounds with a higher SI are considered to be promising drug candidates as the concentration of the compounds required to induce antimicrobial activity is lower than that that which induces cytotoxicity in host cells. The SI of eugenol was reported to be 0.2 against non-filamentous fungi.33 By comparison, the SI values of the compounds tested were higher than the reported value for eugenol. Therefore, our results indicate that the compounds are safe and can be further investigated as promising antifungal molecules. The magnitude of the biological activity of an active molecule, in terms of specific interactions with the molecular target, along with drug-related side effects is strongly influenced by its pharmacokinetics properties. The compounds (6i and 7) were screened through Lipinski's rule of five for oral bioavailability, health effects, maximum passive adsorption, and central nervous system (CNS) activity.
In silico prediction of the physico-chemical and pharmacokinetic properties of the two synthesised compounds (6i and 7) is summarized in Table 3. Compound 7 had a lower molecular weight of 164.12 g mol−1 possibly enhancing its absorption. Log−1P values of the two compounds (6i and 7) were less than 5. The in silico analysis of the compounds revealed that compound 7 conformed with all the listed parameters whereas compound 6i violated four physico-chemical properties. It is worth mentioning here that compound 7 complied with all the parameters of Lipinski's rule for oral bioavailability (30–70%). Compound 6i, on the other hand, violated three parameters according to Lipinski's rule, which might suggest poor oral bioavailability (<30%). Further, we studied the topological polar surface area (TPSA) of the active compounds 6i and 7. The TPSA value was less than 150 for 7, but 6i has a little higher value of TPSA. As can be seen in Table 3, compound 7 has TPSA values under the threshold of 140 Å, which might suggest good intestinal absorption and better CNS penetration, but not compound 6i. The synthesized compounds (6i and 7) meet the criteria of drug likeness with scores of −0.81 and −0.55 for 7 and 6i, respectively.
In silico prediction of the physico-chemical and pharmacokinetic properties of compounds 6i and 7.
| S. no. | Property | Criterion | In silico analysis | |
|---|---|---|---|---|
| Compound 6i | Compound 7 | |||
| 1. | Molecular weight (g mol−1) | <500 | 575.21 | 164.12 |
| 2. | Hydrogen donor | <5 | 0 | 0 |
| 3. | Hydrogen acceptor | <10 | 13 | 1 |
| 4. | Log P (lipophilicity) | <5 | 4.17 | 4.32 |
| 5. | Rotational bonds | <10.0 | 15 | 3 |
| 6. | Topological polar surface area (TPSA) | <140 Å2 | 163.6 Å 2 | 9.23 Å2 |
| 7. | Log S (solubility) | — | −4.54 | −3.58 |
| 8. | Oral bioavailability | — | <30% | 30–70% |
| 9. | Drug likeness score | — | −0.55 | −0.81 |
| Lipinski violations | 2 | 0 | ||
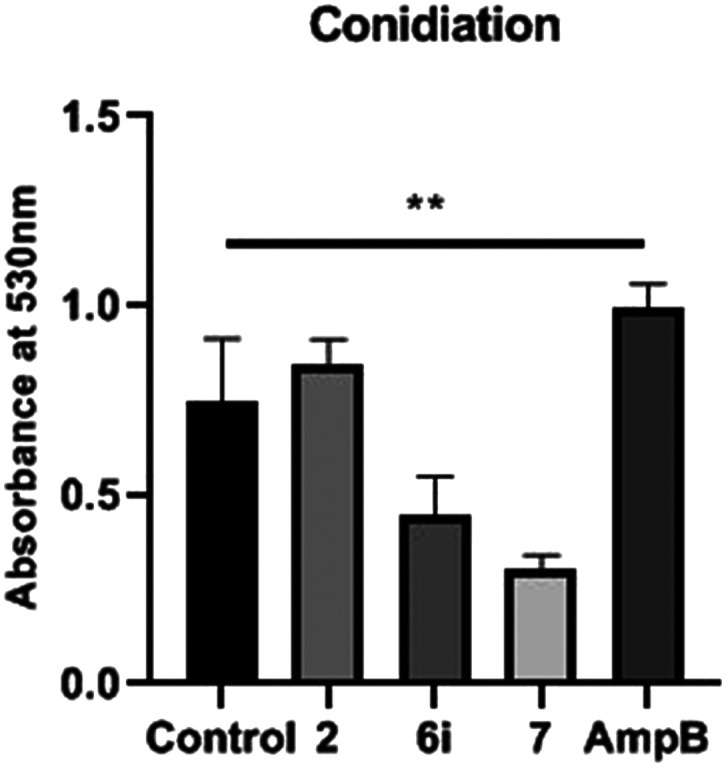
In order to determine the conidia formation in the presence of compound treatment, A. fumigatus was cultured at IC50. Consequently, the reduction in absorbance at 530 nm was significant which corresponds to the decrease in conidia formation on treatment with compounds 6i and 7; whereas in the case of eugenol and Amp B, there was an increase in conidia production compared to the positive control (Fig. 4; p < 0.005). No effect on hyphae formation was observed in any of the samples except the Amp B treated sample. The key regulator of the conidiation pathway, which has been well studied in Aspergillus nidulans as well as in A. fumigatus, is the transcription factor BrlA. It was observed that the ΔbrlA mutant was unable to form conidia, and it depicted increased hyphal growth34 and exhibited widespread transcriptional dysregulation of genes linked to conidiation, growth, and virulence. Molecular analysis is further warranted to gain insight into the target exposure of compounds.
Fig. 4. Effect of isoeugenol and the compounds (6i and 7) on the production of A. fumigatus conidia. Control – untreated A. fumigatus; p < 0.005 statistically.
Further, we carried out studies directed at the formation of DHN-melanin, a cell wall-associated parameter in A. fumigatus, which imparts a greenish-grey colour to the conidia. The formation of DHN-melanin is a fungal protective mechanism against diverse environmental factors such as UV radiation, oxidising agents, and extremes of temperatures. Since the compound-treated A. fumigatus conidia were pigment-less (albino), the melanin was extracted from the compound-treated as well as untreated control A. fumigatus conidia. Similar results were reported in our previously published paper showing a significant decrease in DHN-melanin in A. fumigatus in the presence of isoeugenol.12
A significant reduction in the melanin content of 6i and 7 treated conidia with reference to eugenol and the positive control was observed. According to Kumar et al.,35 the absorption spectra showed characteristic absorption peaks in the UV region ranging from 265–290 nm, but not in the visible region. The overall characteristic absorption peak was observed at 275 nm (see ESI† Fig. S1). The optical densities at 275 nm were 1.126, 0.399, 0.295, and 0.176 in the positive control and compound 2, 6i, and 7 treated conidia, respectively. Targeting the melanin biosynthesis process in A. fumigatus could be a potential strategy for antimicrobial drug development. The white colour of A. fumigatus conidia appears due to mutations in the pksP/alb1 gene, encoding a polyketide synthase required for conidial pigmentation. Pigment-less conidia were found to be less virulent than wild type strains of A. fumigatus in murine models of disseminated aspergillosis, possibly due to an increased susceptibility to phagocytosis and reactive oxygen species (ROS).36 In addition, the defect in the melanin biosynthesis pathway could contribute to the marked loss of adherence properties of the conidia.
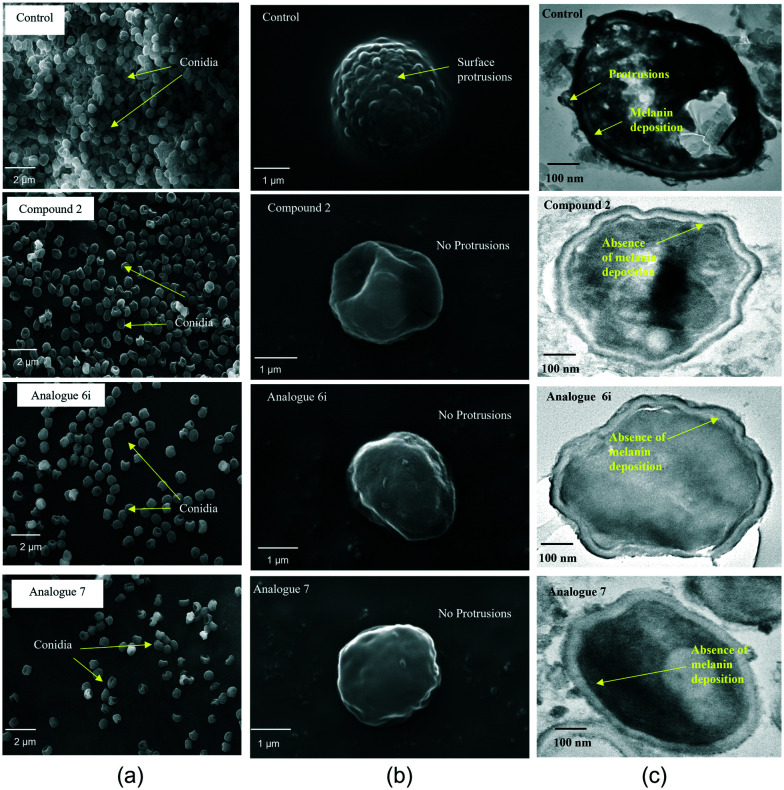
The cell wall of A. fumigatus conidia consists of a hydrophobin layer, a dense melanin layer, and a plasma membrane.37 According to Aimanianda et al.,38 conidia are normally round, but oval shapes were observed in rodlet mutants, which is responsible for hydrophobicity. Consequently, the conidial cell surface morphology was analysed and compared at IC50via scanning electron microscopy (SEM). Compound-treated conidia revealed a smooth conidial surface with the absence of protrusions and a decrease in the number of conidia (Fig. 5a), whereas untreated conidia possess a dense number of conidia with an echinulate surface (Fig. 5b). Correspondingly, transmission electron microscopy (TEM) analysis supported the presence of protrusions in untreated conidia as observed in SEM, whereas compound-treated conidial sections revealed a protrusionless outer surface with a visible clear inner surface indicating the absence of a melanin layer (Fig. 5c). Besides, there was accumulation of cytoplasmic content in wild type conidia, which was reduced in compound-treated conidia probably due to the alteration in the membrane, leading to leakage of cellular contents.
Fig. 5. SEM and TEM visualisations of the conidial surface of A. fumigatus: (a) reduction in the number of conidia in the compound-treated sample compared to the control; (b) compound-treated single conidium revealed loss of protrusions and a smooth surface compared to the echinulate conidial surface in the untreated control; (c) compound-treated single conidium showing loss of protrusions as well as the electron dense melanin layer of the conidial cell wall compared to control conidia showing protrusions and melanin deposition.
Fungal biofilms are very intricate structures, which provide a protective environment for a pathogen to thrive in hostile surroundings.39 This complex structure helps in developing resistance to antimicrobial agents. The signature proof of biofilm formation of a pathogen is the extracellular matrix (ECM) that is produced by the biofilm within 24 h of its colonisation in vitro as well as in vivo, in the case of A. fumigatus. In the A. fumigatus biofilm, ECM is a key component for colonisation by gluing together mycelial threads and is composed of polysaccharides (glucans and galactomannan), some hydrophobic proteins, and melanin.40 In contrast to other species, it lacks β-(1,3) glucan and chitin.
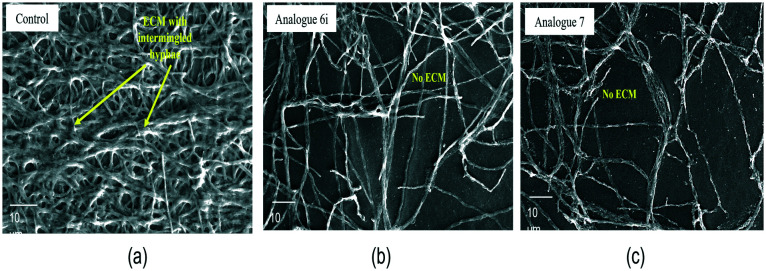
In the present study, the eradication of A. fumigatus biofilms was assessed under exposure to the synthesised compounds of eugenol. Compounds 6i and 7 eradicated preformed fungal biofilms effectively at concentration ranges of 69.53–86.92 μM and 243.60–304.5 μM, respectively. At the same concentrations, the surface morphology of fungal biofilms showed the complete absence of ECM on the hyphae. Meanwhile, in the present study, the biofilm eradication concentration was higher than that of the planktonic culture and greater than the CC50 of the compounds. Despite that, the compounds effectively worked to eliminate the formed ECM, which linked the hyphae to cause infection and were responsible for its stability as well. Under SEM analysis, the positive control of the A. fumigatus biofilm surface displayed a highly coordinated network of hyphal structures with dense ECM accumulation (Fig. 6a), whereas in compound-treated ones only hyphae were observed (Fig. 6b and c).
Fig. 6. SEM of the A. fumigatus biofilm surface. (a) A. fumigatus biofilm; the yellow arrow indicates ECM embedded in the hyphal network. (b) at IC50 of compound 6iA. fumigatus without ECM. (c) at IC50 of compound 7A. fumigatus without ECM. Magnification at 1K×; scale – 10 μm.
Conclusions
In summary, we have accomplished the design and synthesis of eugenol and isoeugenol derived glycoconjugates and investigated their antifungal activity against A. fumigatus. A small library of diverse molecules have been synthesised with various sugar moieties attached to the eugenol and isoeugenol core via click chemistry. Other analogues were also synthesized by functionalization of the olefinic group present in eugenol. It is pertinent to mention here that the glycoconjugates of eugenol were found to exhibit significant antifungal activities amongst all the synthesized compounds. It is presumed that these studies regarding the antifungal activity of eugenol and isoeugenol and related compounds could lead the way for designing future drug-like candidates.
Author contributions
L. Goswami and SP performed the synthesis of compounds and the click chemistry part; L. Gupta performed all experiments on antifungal activities of the synthesised compounds; MV critically reviewed the manuscript and corrected it; AKB and PV conceptualised the idea and critically analysed all the experiments as well as corrected the manuscript. All the authors searched the literature, recreated the figures, and drafted the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors would like to thank the Department of Science and Technology-Science and Engineering Research Board (DST-SERB) (EMR/2016/005752) New Delhi, Govt. of India for the financial support. LG and SP are grateful to UGC, New Delhi, India and CSIR, New Delhi, India for financial support in the form of UGC-SRF and CSIR-SRF, respectively. PV would like to thank Amity University Uttar Pradesh for providing the infrastructure and facilities for extensive research work.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2md00138a
Notes and references
- Kosmidis C. Denning D. W. Thorax. 2015;70:270–277. doi: 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- Lai C.-C. Yu W.-L. J. Microbiol., Immunol. Infect. 2021;54:46–53. doi: 10.1016/j.jmii.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A. M. Palmer M. D. Fraser M. Patterson Z. Mann C. Oliver D. Linton C. J. Gough M. Brown P. Dzietczyk A. Hedley M. McLachlan S. King J. Johnson E. M. J. Clin. Microbiol. 2020;59:e02136-20. doi: 10.1128/JCM.02136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. Bowyer P. Sabino R. Med. Mycol. 2019;57(Supplement_2):S145–S154. doi: 10.1093/mmy/myy149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft C. A. Culibrk L. Moore M. M. Tebbutt S. J. Front. Microbiol. 2016;7:472. doi: 10.3389/fmicb.2016.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzoni L. de Paula A. C. A. Silva E. Marcos C. M. Assato P. A. de Melo W. C. M. A. de Oliveira H. C. Costa-Orlandi C. B. Mendes-Giannini M. J. S. Fusco-Almeida A. M. Front. Microbiol. 2017;8:36. doi: 10.3389/fmicb.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T. Fridman E. Gang D. R. Vassão D. G. Jackson B. L. Kish C. M. Orlova I. Spassova S. M. Lewis N. G. Noel J. P. Baiga T. J. Dudareva N. Pichersky E. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10128–10133. doi: 10.1073/pnas.0603732103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung G. Im S. T. Kim Y. H. Jung S. J. Rhyu M.-R. Oh S. B. Neuroscience. 2014;261:153–160. doi: 10.1016/j.neuroscience.2013.12.047. [DOI] [PubMed] [Google Scholar]
- Pinto E. Vale-Silva L. Cavaleiro C. Salgueiro L. J. Med. Microbiol. 2009;58:1454–1462. doi: 10.1099/jmm.0.010538-0. [DOI] [PubMed] [Google Scholar]
- Marchese A. Barbieri R. Coppo E. Orhan I. E. Daglia M. Nabavi S. F. Izadi M. Abdollahi M. Nabavi S. M. Ajami M. Microbiology. 2017;43:668–689. doi: 10.1080/1040841X.2017.1295225. [DOI] [PubMed] [Google Scholar]
- Hoda S. Vermani M. Joshi R. K. Shankar J. Vijayaraghavan P. BMC Complementary Med. Ther. 2020;20:67. doi: 10.1186/s12906-020-2859-z. doi: 10.1186/s12906-020-2859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta L. Sen P. Bhattacharya A. K. Vijayaraghavan P. Arch. Microbiol. 2022;204:214. doi: 10.1007/s00203-022-02817-w. doi: 10.1007/s00203-022-02817-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S. K. Garzan A. Tsodikova S. G. Eur. J. Med. Chem. 2017;133:309–318. doi: 10.1016/j.ejmech.2017.03.075. [DOI] [PubMed] [Google Scholar]
- Ngo H. X. Shrestha S. K. Tsodikova S. G. ChemMedChem. 2016;11:1507–1516. doi: 10.1002/cmdc.201600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotammagari T. K. Paul S. Barik G. K. Santra M. K. Bhattacharya A. K. Org. Biomol. Chem. 2020;18:2252–2263. doi: 10.1039/D0OB00216J. [DOI] [PubMed] [Google Scholar]; , and references cited therein
- in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart and M. E. Etzler, Cold Spring Harbor Laboratory Press, 2nd edn, 2009 [PubMed] [Google Scholar]
- Szeja W. Grynkiewicz G. Rusin A. Curr. Org. Chem. 2017;21:218–235. doi: 10.2174/1385272820666160928120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D. Jain D. K. J. Adv. Pharm. Technol. Res. 2015;6:141–146. doi: 10.4103/2231-4040.161515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla B. S. Mahalinga M. Karthikeyan M. S. Poojary B. Akberali P. M. Kumari N. S. Eur. J. Med. Chem. 2005;40:1173–1178. doi: 10.1016/j.ejmech.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Goswami L. Paul S. Kotammagari T. K. Bhattacharya A. K. New J. Chem. 2019;43:4017–4021. doi: 10.1039/C8NJ05737K. [DOI] [Google Scholar]
- Kuijpers B. H. M. Groothuys S. Keereweer A. R. Quaedflieg P. J. L. M. Blaauw R. H. van Delft F. L. Rutjes F. P. J. T. Org. Lett. 2004;6:3123–3126. doi: 10.1021/ol048841o. [DOI] [PubMed] [Google Scholar]
- Huisgen R. Angew. Chem., Int. Ed. Engl. 1963;2:565–632. doi: 10.1002/anie.196305651. [DOI] [Google Scholar]
- Huisgen R. Angew. Chem., Int. Ed. Engl. 1963;2:633–696. doi: 10.1002/anie.196306331. [DOI] [Google Scholar]
- Rostovtsev V. V. Green L. G. Fokin V. V. Sharpless K. B. Angew. Chem., Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Bomon J. Bal M. Achar T. K. Sergeyev S. Wu X. Wambacq B. Lemière F. Sels B. F. Maes B. U. W. Green Chem. 2021;23:1995–2009. doi: 10.1039/D0GC04268D. [DOI] [Google Scholar]
- Bednarski M. Danishefsky S. J. Am. Chem. Soc. 1986;108:7060–7067. doi: 10.1021/ja00282a035. [DOI] [Google Scholar]
- Aratikatla E. K. Valkute T. R. Puri S. K. Srivastava K. Bhattacharya A. K. Eur. J. Med. Chem. 2017;138:1089–1105. doi: 10.1016/j.ejmech.2017.07.052. [DOI] [PubMed] [Google Scholar]
- Paquin A. Moreno C. R. Bérubé G. Molecules. 2021;26:2340. doi: 10.3390/molecules26082340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çapcı A. Herrmann L. Sampath Kumar H. M. Fröhlich T. Tsogoeva S. B. Med. Res. Rev. 2021;41:2927–2970. doi: 10.1002/med.21814. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI], Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, Clinical and Laboratory Standard Institute, Wayne, PA, 2nd edn, 2008 [Google Scholar]
- Kasper M. Roehlecke C. Witt M. Fehrenbach H. Hofer A. Miyata T. Weigert C. Funk R. H. W. Schleicher E. D. Am. J. Respir. Cell Mol. Biol. 2000;23:485–491. doi: 10.1165/ajrcmb.23.4.4117. [DOI] [PubMed] [Google Scholar]
- Harmsen S. McLaren A. C. Pauken C. McLemore R. Clin. Orthop. Relat. Res. 2011;469:3016–3021. doi: 10.1007/s11999-011-1890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho L. I. S. Alvarenga D. J. do Carmo L. C. F. de Oliveira L. G. Silva N. C. Dias A. L. T. Coelho L. F. L. de Souza T. B. Dias D. F. Carvalho D. T. J. Chem. 2017:e5207439. [Google Scholar]
- Stewart J. I. P. Fava V. M. Kerkaert J. D. Subramanian A. S. Gravelat F. N. Lehoux M. Howell P. L. Cramer R. A. Sheppard D. C. MBio. 2020;11:e03202–e03219. doi: 10.1128/mBio.03202-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C. G. Mongolla P. Pombala S. Kamle A. Joseph J. Lett. Appl. Microbiol. 2011;53:350–358. doi: 10.1111/j.1472-765X.2011.03116.x. [DOI] [PubMed] [Google Scholar]
- Sugareva V. Härtl A. Brock M. Hübner K. Rohde M. Heinekamp T. Brakhage A. A. Arch. Microbiol. 2006;186:345–355. doi: 10.1007/s00203-006-0144-2. [DOI] [PubMed] [Google Scholar]
- Latgé J. P. Beauvais A. Chamilos G. Annu. Rev. Microbiol. 2017;71:99–116. doi: 10.1146/annurev-micro-030117-020406. [DOI] [PubMed] [Google Scholar]
- Aimanianda V. Bayry J. Bozza S. Kniemeyer O. Perruccio K. Elluru S. R. Clavaud C. Paris S. Brakhage A. A. Kaveri S. V. Romani L. Latgé J.-P. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- Melloul E. Luiggi S. Anaïs L. Arné P. Costa J.-M. Fihman V. Briard B. Dannaoui E. Guillot J. Decousser J.-W. Beauvais A. Botterel F. PLoS One. 2016;11:e0166325. doi: 10.1371/journal.pone.0166325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N. A. R. Latge J.-P. Munro C. A. Microbiol. Spectrum. 2017;5 doi: 10.1128/microbiolspec.FUNK-0035-2016. doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.