Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with a hyperinflammatory state and lymphocytopenia, a hallmark that appears as both signature and prognosis of disease severity outcome. Although cytokine storm and a sustained inflammatory state are commonly associated with immune cell depletion, it is still unclear whether direct SARS-CoV-2 infection of immune cells could also play a role in this scenario by harboring viral replication. We found that monocytes, as well as both B and T lymphocytes, were susceptible to SARS-CoV-2 infection in vitro, accumulating double-stranded RNA consistent with viral RNA replication and ultimately leading to expressive T cell apoptosis. In addition, flow cytometry and immunofluorescence analysis revealed that SARS-CoV-2 was frequently detected in monocytes and B lymphocytes from coronavirus disease 2019 (COVID-19) patients. The rates of SARS-CoV-2-infected monocytes in peripheral blood mononuclear cells from COVID-19 patients increased over time from symptom onset, with SARS-CoV-2-positive monocytes, B cells, and CD4+ T lymphocytes also detected in postmortem lung tissue. These results indicated that SARS-CoV-2 infection of blood-circulating leukocytes in COVID-19 patients might have important implications for disease pathogenesis and progression, immune dysfunction, and virus spread within the host.
Keywords: SARS-CoV-2, COVID-19, lymphocytopenia, peripheral blood mononuclear cell (PBMC), lymphocytes, monocytes, apoptosis
Introduction
In late 2019, a new coronavirus emerged as the cause of a severe acute respiratory syndrome named coronavirus disease 2019 (COVID-19). The causal agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is classified in the family Coronaviridae, genus Betacoronavirus, based on its similarity to SARS-CoV (Zhou et al., 2020a). Since its emergence, SARS-CoV-2 has become globally distributed, infecting >323 million people, resulting in 5.5 million deaths (https://covid19.who.int). The main clinical features of COVID-19 are fever, cough, dyspnea, anosmia, and dysgeusia, but some patients rapidly evolve to severe respiratory distress syndrome (Huang et al., 2020; Vargas-Gandica et al., 2020).
Previous studies identify an inflammatory cytokine storm and lymphocytopenia as hallmarks of severe COVID-19 cases, with extreme functional exhaustion of T lymphocytes (Diao et al., 2020; Huang et al., 2020; Zheng et al., 2020). Peripheral blood mononuclear cells (PBMCs) from COVID-19 patients show an upregulation of autophagy and apoptosis (Xiong et al., 2020), suggesting that dampening of the immune system by SARS-CoV-2 infection might impact disease outcomes. In addition, a decrease in circulating lymphocytes has been associated with poor COVID-19 outcomes (Huang et al., 2020). Still, it remains uncertain whether lymphocytopenia is caused by intense lymphocyte recruitment to infected sites or by direct viral infection of lymphocytes, with consequent cell death.
To initiate infection, the large glycoprotein spike (S) on the surface of SARS-CoV-2 binds to angiotensin-converting enzyme 2 (ACE2) on the target cell membrane (Hoffmann et al., 2020b). Upon ACE2 binding, the S protein can be cleaved by transmembrane protease serine 2 (TMRPSS2), exposing the fusion peptide and promoting membrane fusion. Fusion occurs at the plasma membrane or following endocytic uptake with endosomal cathepsins facilitating the cleavage of S protein at acidic pH (Hoffmann et al., 2020b).
Because ACE2 plays a critical role in viral entry, it is one of the main factors of cellular tropism (Letko et al., 2020; Hoffmann et al., 2020b). ACE2 expression analysis showed that the respiratory and gastrointestinal tract, kidney, and myocardium are ACE2-enriched (To and Lo, 2004; Qi et al., 2020; Zou et al., 2020). Interestingly, COVID-19 patients’ heart autopsies showed viral particles in cardiomyocytes and cardiac macrophages (Dolhnikoff et al., 2020; Tavazzi et al., 2020; Bailey et al., 2021). Single-cell RNA sequencing of bronchoalveolar lavage fluid (BALF) samples identified immune cells harboring SARS-CoV-2 RNA replication intermediates, as well as antigens in CD3+ cells in lung autopsies. Furthermore, SARS-CoV-2 antigens were found postmortem in the spleen and lymph nodes with pathological signs of damage (Xiang et al., 2021). Together, these pieces of evidence raise essential questions about whether these immune cells become infected in the inflamed tissues or before reaching secondary lymphoid tissues.
In the present study, we investigated the susceptibility and permissiveness of human PBMCs to SARS-CoV-2. We found that PBMCs are susceptible and permissive to SARS-CoV-2 infection, both in vitro and in COVID-19 patients. Furthermore, infection and cell death of PBMCs may reduce circulating lymphocytes in infected individuals, directly impacting disease outcomes and thus contributing to virus dissemination to other organs.
Results
SARS-CoV-2 infects and replicates in human PBMCs in vitro
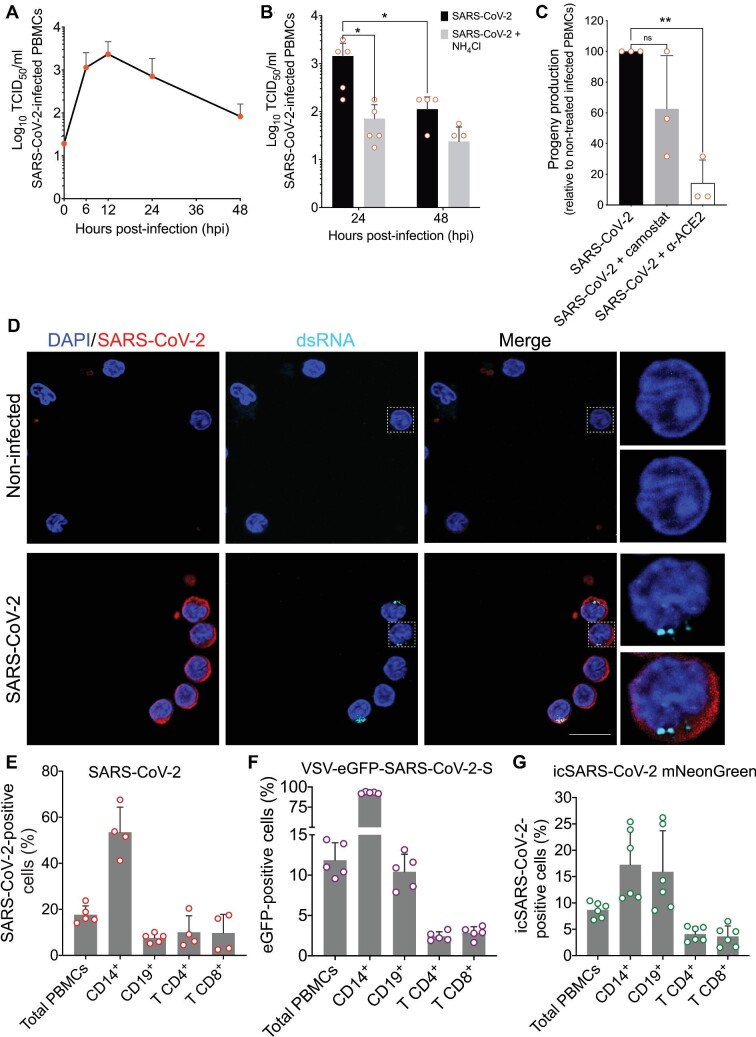
To determine whether primary cultures of human PBMCs could be infected with SARS-CoV-2, we obtained PBMCs from five healthy donors and inoculated them with virus at an multiplicity of infection (MOI) of 1. Following 1 h of incubation at 37°C, cells were washed extensively, incubated for 48 h, and the viral yield determined at periodic intervals using a 50% tissue culture infectious dose (TCID50) assay (Figure 1A). SARS-CoV-2 titers peaked between 6 and 12 hours post-infection (hpi), with a 100-fold increase from the initial input, followed by a steady decrease (Figure 1A). Thus, SARS-CoV-2 infection of PBMCs produced progeny capable of inducing a cytopathic effect on monolayers of Vero E6 cells. Pretreatment of PBMCs with ammonium chloride (NH4Cl), which elevates intracellular pH and hampers processes such as endosomal acidification, inhibited viral replication, as evidenced by a 10-fold reduction in SARS-CoV-2 progeny production after 24 h (P = 0.017) (Figure 1B). This result suggests that SARS-CoV-2 infection in human PBMCs depends on intracellular acidic pH (Figure 1B). To evaluate whether NH4Cl treatment impairs viral entry, we used a chimeric vesicular stomatitis virus (VSV), in which the native glycoprotein was substituted by S proteins from different coronaviruses. Treatment of Calu-3 cells with NH4Cl similarly reduced the infection by VSV expressing the enhanced green fluorescent reporter protein (VSV-eGFP) chimeric viruses expressing SARS-CoV-2-S or Middle East respiratory syndrome coronavirus S protein (MERS-CoV-S) (Supplementary Figure S1A and B). Consistent with high levels of TMPRSS2 expression in Calu-3 cells that can facilitate direct entry of SARS-CoV-2 at the plasma membrane, NH4Cl pretreatment did not prevent VSV-SARS-CoV-2 infection (Hoffmann et al., 2020a). As expected, NH4Cl inhibited the infection of Calu-3 by VSV, which requires endosome acidification to facilitate G-mediated membrane fusion. Infection of Calu-3 cells by a chimeric VSV expressing the fusion and hemagglutinin glycoproteins from the paramyxovirus parainfluenza 5 (PIV5) was unaffected by NH4Cl, consistent with PIV5 fusing directly at the plasma membrane.
Figure 1.
SARS-CoV-2 differentially infects subsets of human PBMCs in vitro. PBMCs from healthy donors were infected in vitro with SARS-CoV-2 Brazil/SPBR-02/2020 (MOI = 1) and cultured at 37°C. (A) Replication curve of SARS-CoV-2 with mean (and standard deviations, SDs) of virus titers (in TCID50/ml) in supernatants from cultures of PBMCs. (B) SARS-CoV-2 progeny titers in supernatants from infected PBMCs in culture for 24 and 48 hpi, with and without treatment with NH4Cl. (C) Effects on SARS-CoV-2 progeny production by blocking infection in PBMCs with antibody anti-ACE2 or camostat mesylate, an inhibitor of TMPRSS2. Titers were determined in supernatants at 24 hpi. Progeny titers after treatments were normalized to the progeny titer from untreated PBMCs. (D) Immunofluorescence for SARS-CoV-2 and dsRNA in PBMCs at 6 hpi with SARS-CoV-2. Cells were immunostained for SARS-CoV-2 (red) and dsRNA (cyan), and then analyzed by confocal microscopy. Magnification, 630× . Scale bar, 10 μm. (E) The total frequency and subpopulation's frequency of SARS-CoV-2-positive cells at 24 hpi with authentic SARS-CoV-2. PBMCs from healthy donors were infected, then at 24 hpi stained with mouse polyclonal anti-SARS-CoV-2, and analyzed by flow cytometry. (F) The total frequency and subpopulation's frequency of eGFP-positive cells at 8 hpi with the chimera VSV-eGFP-SARS-CoV-2-S virus. (G) The total frequency and subpopulation's frequency of neon green positive cells at 24 hpi with the infectious clone icSARS-CoV-2 mNeonGreen reporter virus. Individual values are plotted with circle signs; histograms depict mean ± SD. The percentage of each cell subtype was normalized against the total PBMC value. Statistical analysis was done by one-way or two-way analysis of variance (ANOVA). Tukey's or Holm–Sidak post-tests were applied when suitable. P-values < 0.05 were considered significant.
Even though ACE2 expression in human PBMCs is considered to be minimal (Dong et al., 2020; Song et al., 2020), we examined whether SARS-CoV-2 infection was sensitive to the blockade of ACE2 and TMPRSS2. Pretreatment of PBMCs with the TMPRSS2 inhibitor Camostat had no overall discernible effect on virus yields, using either wild-type or fluorescent SARS-CoV-2. In contrast, pre-incubation with an antibody blocking ACE2 reduced virus titers 5-fold over 24 hpi (P = 0.0016) (Figure 1C). Conversely, there was no significant reduction in virus yield when PBMCs were pre-treated with unrelated isotype control antibodies (Supplementary Figure S1C and D). Similar treatment also affected the entry of chimeric VSV-SARS-CoV-2-S, but not VSV-eGFP. Even though the amounts of eGFP+ cells were similar, the lower mean fluorescent intensity in infected PBMCs suggests that the viruses could still enter cells but through a less efficient pH-independent pathway. Taken together, the evidence indicates that the infection of PBMCs is not mediated exclusively by the interaction with ACE2 and in a pH-dependent manner (Supplementary Figure S1E and F).
Coronavirus replication produces several RNA-replicative intermediates and accumulates double-stranded RNA (dsRNA) in the cytoplasm of infected cells, thus making a reliable indicator of virus replicative activity. We, therefore, examined infected PBMCs for SARS-CoV-2 antigen and dsRNA using confocal microscopy. SARS-CoV-2 antigen-positive cells were also positive for dsRNA, and double-positive cells visualized at 6 hpi followed a pattern that roughly matched the accumulation of virus progeny (Figure 1D). The dsRNA staining was seen as puncta, consistent with the juxtanuclear staining of viral replication complexes seen for other coronaviruses (Deng et al., 2017). Together, these data demonstrate that human peripheral blood cells are susceptible to infection by SARS-CoV-2 in vitro.
The finding that human PBMCs can be infected with SARS-CoV-2 in vitro led us to examine which cell types were susceptible to infection. PBMCs from healthy donors were incubated with SARS-CoV-2 at MOI of 1, and the intracellular expression of SARS-CoV-2 antigens was assessed by flow cytometry with a polyclonal antibody (Figure 1E). At 24 hpi, 17.7% ± 3.8% of all PBMCs were SARS-CoV-2-positive (Figure 1E), with CD14+ monocytes representing an average of 53.5% of SARS-CoV-2-positive cells. In addition to monocytes, CD4+ (10%) and CD8+ (9%) T lymphocytes and CD19+ B cells (7.58%) were also positive for SARS-CoV-2 antigens. To examine whether this was unique to SARS-CoV-2, we used chimeric VSV containing the S genes of MERS-CoV, SARS-CoV, and SARS-CoV-2 (Fukuma et al., 2015; Hoffmann et al., 2020a, b; Zang et al., 2020). Approximately 30% of PBMCs were susceptible to VSV-eGFP-mediated infection, and this was reduced for VSV-SARS-CoV-2-S (12%), VSV-SARS-CoV-S (9%), and VSV-MERS-CoV-S (20%) (Figure 1F; Supplementary Figure S1G). These proportions are consistent with what has been reported for authentic SARS-CoV and MERS-CoV (Li et al., 2003; Yilla et al., 2005; Chu et al., 2016). Consistent with the cell types infected by SARS-CoV-2, VSV-SARS-CoV-2-S infects CD14+ monocytes (92%), CD4+ and CD8+ T lymphocytes (2.4% and 2.8%, respectively), and CD19+ B cells (11%) (Figure 1F). We also used the strategy based on a SARS-CoV-2 infectious clone (icSARS-CoV-2) engineered to express a replication-dependent fluorescent protein reporter (Figure 1G) (Xie et al., 2020). At 24 hpi (MOI = 1), 8.6% of the total PBMCs were positive for the replication reporter. From all the positive live cells, the most abundantly infected cells were CD14+ monocytes (17%), followed by CD19+ B cells (15.9%), CD4+ T lymphocytes (4.1%), and CD8+ T lymphocytes (3.6%). To show that the fluorescent protein detection was only possible if the virus was viable, a stock aliquot was ultraviolet (UV)-inactivated, and the lack of infectivity was confirmed by plaque assay (Supplementary Figure S1H). The UV-inactivated virus was used in the same amount as that of infectious viruses to infect PBMCs, and no fluorescence was detected by flow cytometry (Supplementary Figure S1I). Together, the data show that SARS-CoV-2 can infect and replicate in different subsets of peripheral blood cells.
SARS-CoV-2 infection of PBMCs in vitro induces apoptosis of T lymphocytes
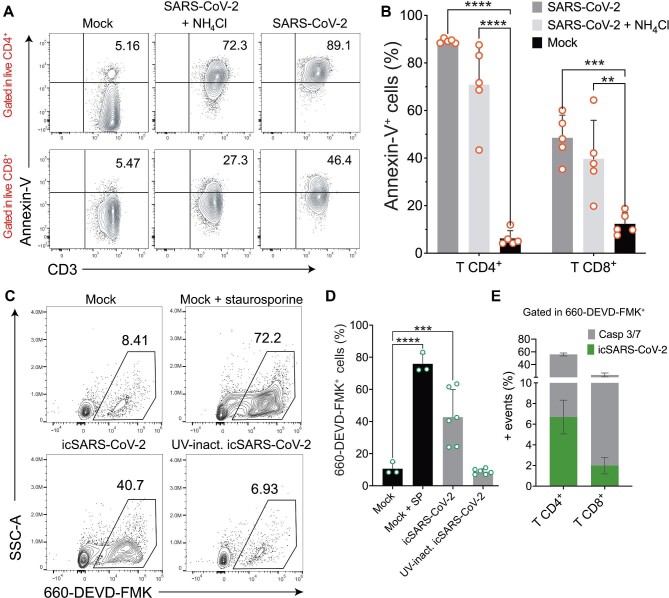
As CD4+ and CD8+ T cells were susceptible to SARS-CoV-2 infection, we next investigated whether this could cause cell death. For this analysis, we infected PBMCs from healthy donors and monitored the translocated phosphatidylserine (PS) to the outer leaflet of the plasma membrane by annexin-V staining at 24 hpi. Representative plots are depicted in Figure 2A. In mock-treated control cells, some basal annexin-V staining was detected (CD4+ mean 6.24%, CD8+ mean 12.36%) (Figure 2B). Infection with SARS-CoV-2 resulted in an increase in annexin-V staining in live CD4+ (89.1% ± 0.9%) and CD8+ T lymphocytes (48.5% ± 9.5%) (Figure 2B). Differences were still significant when cells were analyzed independently of live/dead staining. In the case of CD8+ T cells, the difference was even greater (59.64%, P = 0.0001) (Supplementary Figure S2A), indicating that a considerable percentage of dead CD8+ T cells were annexin-V-positive. A small reduction in annexin-V staining was observed in CD4+ and CD8+ T cells after NH4Cl treatment (70.88% ± 17.2% and 39.7% ± 16.1%, respectively). In addition to PS translocation, we evaluated the activation of caspase 3/7 as indicative of apoptosis. PBMCs from healthy donors were infected at MOI of 1 with icSARS-CoV-2 and evaluated at 24 hpi by flow cytometry. Representative plots are shown in Figure 2C. Quantitatively, 42.73% ± 17.18% of PBMCs were positive for active caspase 3/7 upon icSARS-CoV-2 infection, compared with 75.9% ± 8.3% of PBMCs induced with staurosporine, a known apoptosis inducer (Figure 2D). The rates of apoptotic cells in PBMCs exposed to UV-inactivated virus (8.7% ± 1.3%) were similar to the rates seen in mock-infected PBMCs (10.5% ± 3.8%) (Figure 2D). This indicates that a viable replicating virus is needed to induce apoptosis in these cells. Among PBMCs, T lymphocytes accounted for most of the caspase 3/7-positive cells. CD4+ T lymphocytes comprised 49.2% ± 2.3% of the cells with active caspase 3/7, followed by CD8+ T cells (21% ± 3%) (Figure 2E). The positivity rates for active caspase 3/7 were similarly low in B cells (1.8% ± 0.4%) and monocytes (1.1% ± 0.4%) (Supplementary Figure S2B). Only ∼6.7% of CD4+ and 2% of CD8+ T cells were simultaneously positive for active caspase 3/7 and icSARS-CoV-2, suggesting that apoptosis induction occurs independently of viral replication in these cells (Figure 2E). Taken together, the results indicate that SARS-CoV-2 induces apoptosis and increases cell death markers in circulating T lymphocytes.
Figure 2.
SARS-CoV-2 activates caspase 3/7 and increases the expression of cell death markers. (A) Representative flow cytometry plots of live CD4+ and CD8+ T cells positive for annexin-V staining in PBMCs at 24 hpi with SARS-CoV-2 (MOI = 1), in the presence or absence of NH4Cl. (B) Percentages of live lymphocytes positive for annexin-V and expressing PS at the cell surface after SARS-CoV-2 infection. (C) Representative plots of PBMCs presenting active caspase 3/7. Cells from healthy donors were infected with mock (in the absence or presence of the apoptosis inducer staurosporine), icSARS-CoV-2 (MOI=1), or UV-inactivated icSARS-CoV-2. At 24 hpi, cells were labelled with the fluorochrome-labelled inhibitors of caspases (FLICA) substrate 660-DEVD-FMK and analyzed by flow cytometry. (D) Frequencies of active caspase 3/7 in total PBMCs. SP, staurosporine. (E) Frequencies of T lymphocytes positive for caspase 3/7 (gray) and icSARS-CoV-2 (green) from the total PBMCs. Mean ± SD is indicated for all bar graphs. Significance was determined by two-way ANOVA and Bonferroni's post-test.
Circulating immune cells from COVID-19 patients are infected and harbor SARS-CoV-2 replication
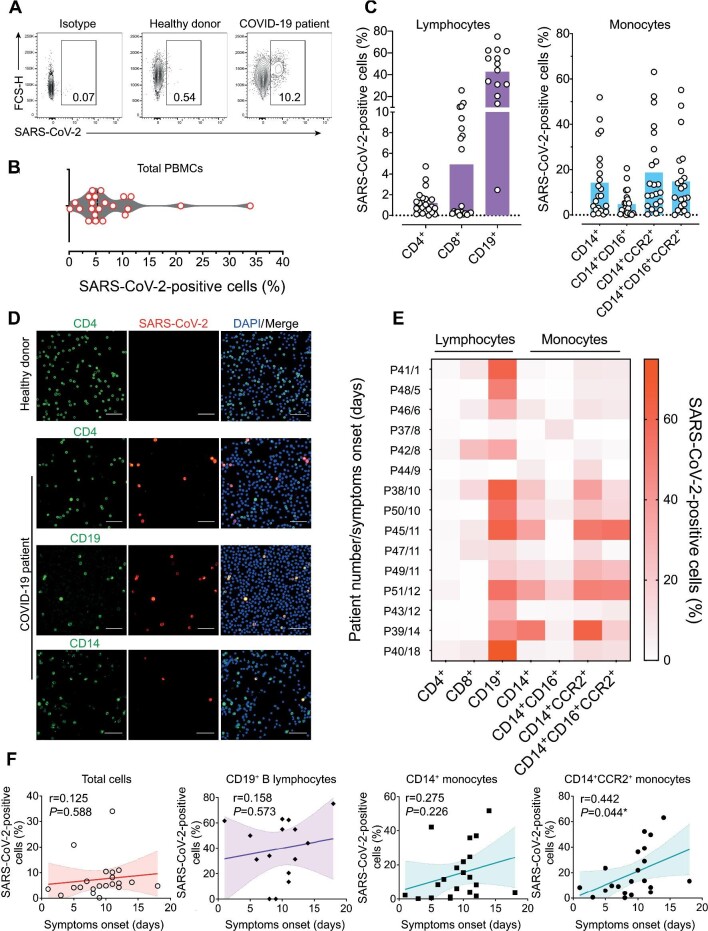
As PBMCs were robustly infected in vitro, we analyzed the PBMCs from severe COVID-19 patients. Patients’ clinical and demographic characteristics are listed in Supplementary Table S1. The PBMCs from 22 of the 29 patients enrolled (seven samples were discarded due to severe lymphocytopenia) and 12 healthy donors were analyzed by flow cytometry for SARS-CoV-2 antigen staining (Figure 3A). Gating strategies for lymphocytes and monocytes are shown in Supplementary Figure S3A and B, respectively. SARS-CoV-2 antigens were detected in 7.68% ± 1.56% of PBMCs from COVID-19 patients (Figure 3B). However, not all COVID-19 patients showed expressive staining for SARS-CoV-2 in PBMCs, and the rates of SARS-CoV-2-positive cells ranged from 0.16% to 33.9% (Figure 3B).
Figure 3.
Detection of SARS-CoV-2 in PBMCs from hospitalized COVID-19 patients. Twenty-two ICU-hospitalized COVID-19 patients presenting a moderate to severe condition and 12 age- and gender-matched healthy donors PCR negative for SARS-CoV-2 were enrolled. (A) Representative flow cytometry plots indicating SARS-CoV-2 positivity of PBMCs from COVID-19 patients in comparison with isotype control and healthy donors. (B) Violin plot showing the frequencies of SARS-CoV-2-infected cells from COVID-19 patients. (C) Percentages of SARS-CoV-2-infected cells considering different immunophenotypes in COVID-19 patients. (D) Immunofluorescence of PBMCs from COVID-19 patients labelling for SARS-CoV-2 (red), nuclei (blue), and immunophenotypes as CD4, CD19, or CD14 (green). Scale bar, 50 µm. (E) Heat map indicating SARS-CoV-2-positive cell frequencies for each immunophenotype, stratified by time from symptom onset (patient number/days after symptom onset). Data were plotted individually for each COVID-19 patient analyzed. (F) Correlation and linear regression analysis between time after symptom onset and frequencies of SARS-CoV-2-positive cells. Both P and r values are indicated in the graphs. The best-fitting line is displayed in colors, while the light-colored area represents the confidence interval. P-values <0.05 were considered significant.
We also monitored PMBCs from 15 of the 22 COVID-19 patients for SARS-CoV-2 RNA levels by real-time reverse transcription–polymerase chain reaction (RT–PCR). Virus genome was detected in 8 out of 15 patients’ PBMCs (53.3%), with a mean viral load of 3.8 × 104 copies per µg RNA (Supplementary Table S2). Immunophenotyping of cells from COVID-19 patients indicated that the highest proportion of SARS-CoV-2-positive cells was found in B lymphocytes (42.73% ± 4.3%). Infection of CD4+ and CD8+ T lymphocytes corresponded to 1.2% ± 1.19% and 4.93% ± 6.6% of the positive PBMCs, respectively. A high rate of CD14+ monocytes (14.19% ± 15.26%) was positive for SARS-CoV-2 antigens. Specialized inflammatory monocytes (CD14+CCR2+ and CD14+CD16+CCR2+) were also positive for SARS-CoV-2 antigens in high frequencies (18.73% ± 18.46% and 14.78% ± 15.5%, respectively) (Figure 3C). We also performed immuno-fluorescence of patients’ PBMCs and detected SARS-CoV-2 antigens in CD4+, CD19+, and CD14+ cells, with no discernible fluo-rescent signal seen in PBMCs from healthy donors (Figure 3D).
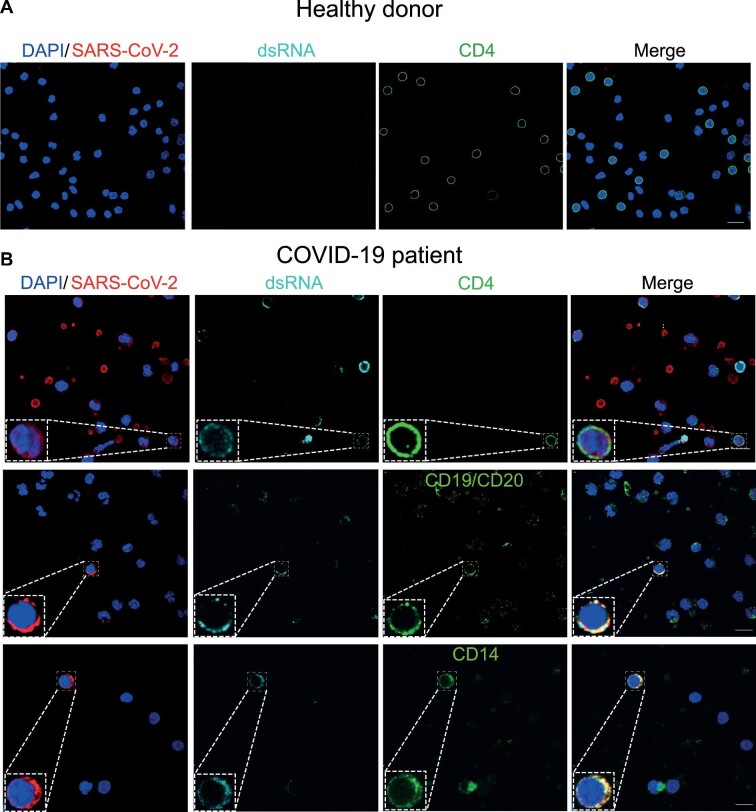
Since rates of SARS-CoV-2-positive cells in PBMCs varied widely (Figure 3C), 15 COVID-19 patients whose clinical information was available were stratified based on the length of time between symptom onset and PBMC collection, and the rates of SARS-CoV-2-positive cells of different immunophenotypes were plotted on a heat map (Figure 3E). The rates of SARS-CoV-2-positive B lymphocytes were high in PBMCs collected at all time points, whereas the rates of SARS-CoV-2-positive monocytes were higher at more extended time points of COVID-19 progression (Figure 3E). Frequencies of SARS-CoV-2-positive cells correlated positively with the length of time of COVID-19 progression, especially for inflammatory CD14+CCR2+ monocytes (r = 0.442, P = 0.044) (Figure 3F). To see whether SARS-CoV-2 was actively replicating in PBMCs from COVID-19 patients, we analyzed the presence of dsRNA in SARS-CoV-2-positive cells of different immunophenotypes by immunofluorescence and confocal microscopy. Staining for dsRNA was found in SARS-CoV-2-positive CD4+ T lymphocytes, B lymphocytes, and monocytes (Figure 4). Similar results were attained by fluorescent in situ hybridization using an N2 oligonucleotide probe specific for the SARS-CoV-2 antigenome, which yielded a strong signal in a pattern typical of viral replication sites in the cytoplasm (Supplementary Figure S4). Altogether, these data confirm that SARS-CoV-2 infects and replicates in circulating lymphomononuclear cells in COVID-19 patients.
Figure 4.
PBMCs from COVID-19 patients present dsRNA, a coronavirus hallmark of replication. PBMCs from healthy donors (A) or COVID-19 patients (B) were isolated and cultivated on coverslips pre-treated with poly-D-lysine. Cells were fixed and stained for SARS-CoV-2 (red), immune phenotypes as CD4, CD19, or CD14 (green), dsRNA (cyan), and nuclei (blue). Immunofluorescence was examined using confocal microscopy. In the bottom left corner of each channel, an inset of the labelling phenotype is shown. Representative images for each immunophenotype are shown, where at least two patients were analyzed. Magnification, 630×. Scale bar, 10 µm.
SARS-CoV-2-positive inflammatory monocytes and lymphocytes are detected postmortem in lung tissues from COVID-19 patients
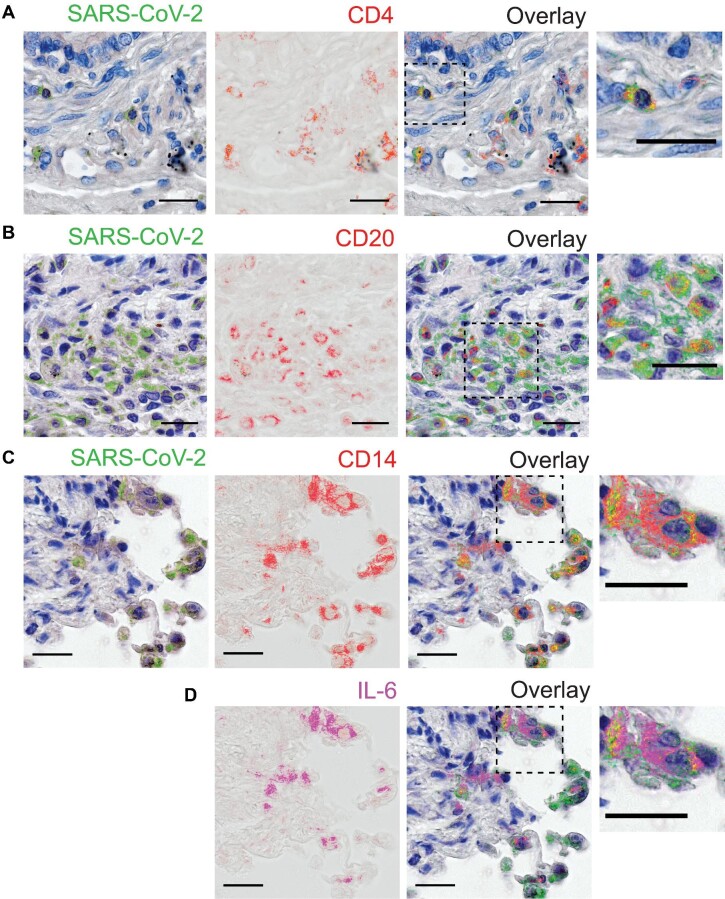
Considering that SARS-CoV-2 is a major respiratory pathogen, we next examined whether SARS-CoV-2-infected cell immunophenotypes found in PBMCs were also present in lung autopsy samples. Postmortem lung specimens from COVID-19 patients revealed abundant staining for SARS-CoV-2, especially throughout the bronchi–vascular axes and alveolar–capillary barriers. No staining was detected in control lung specimens (Supplementary Figure S5). Upon staining for SARS-CoV-2, slides were scanned, then the staining was erased, and slides were re-stained sequentially for the surface antigens CD4, CD20, and CD14. The serial immunolabelling showed positivity for SARS-CoV-2 antigens in CD4+ T lymphocytes (Figure 5A), B lymphocytes (Figure 5B), and monocytes (Figure 5C) in lungs from COVID-19-infected individuals. Based on its role in lung tissue damage in COVID-19 patients, interleukin-6 (IL-6) was also detected by immunostaining, which revealed that several CD14+ monocytes expressing IL-6 were positive for SARS-CoV-2 antigens (Figure 5D), indicating that SARS-CoV-2 also infects inflammatory monocytes in the lungs of COVID-19 patients.
Figure 5.
SARS-CoV-2 is detected in diverse immune cell types in COVID-19 lung autopsies. (A–C) SARS-CoV-2 antigen detection (pseudocolored in green) with sequential staining for CD4 (A), CD20 (B), and CD14 (C) surface markers, pseudocolored in red. (D) Staining for IL-6, pseudocolored in magenta. The overlaid layers from the previous sequential rounds of staining are displayed with superimposed staining indicated in yellow. Dashed line-boxed areas are zoomed in and shown in right panels. Magnification, 400×. Scale bar, 50 µm.
Discussion
SARS-CoV-2 has an extensive tissue tropism accessed following entry from the respiratory tract (Dong et al., 2020). Dissemination of the virus may reflect circulating cells that could serve as ‘Trojan horses’ transporting the virus to secondary target organs and driving extrapulmonary COVID-19-related injury. Consistent with such a model, we recently reported that SARS-CoV-2 infects neutrophils that could carry SARS-CoV-2 into tissues upon recruitment (Veras et al., 2020). SARS-CoV-2 was also detected in monocytes from severe COVID-19 patients with inflammasome activation (Rodrigues et al., 2020). It remained unclear whether SARS-CoV-2 infects other kinds of lymphomononuclear cells in vivo. This study defines the susceptibility of human PBMCs to SARS-CoV-2 infection and validates those findings by direct analysis of cell types from peripheral blood of COVID-19 patients with advanced disease.
We reached our conclusions based on infection studies carried out with the authentic SARS-CoV-2, a SARS-CoV-2 infectious clone (icSARS-CoV-2) engineered to express a replication reporter, and a chimeric VSV-SARS-CoV-2. These three independent strategies have in common the presence of the S glycoprotein on the viral surface, defining it as the key determinant for cell tropism. Our analysis shows that SARS-CoV-2 infection of PBMCs results in a peak of viral replication at 12 hpi, yielding ∼100-fold of the input virus. This modest infection mirrors the kinetics of dsRNA appearance in infected PBMCs and is consistent with previous data showing that SARS-CoV replication in human PBMCs is not sustained for an extended period (Li et al., 2003). This is also in keeping with a study that showed SARS-CoV-2 virus-like particles in CD4+ T cells at as early as 6 and 12 hpi (Banerjee et al., 2020).
Several PBMC immunophenotypes were susceptible to in vitro infection by the three strategies of SARS-CoV-2 in the present study, with monocytes being the most susceptible, followed by B lymphocytes and CD4+ and CD8+ T lymphocytes. The slight variation between results obtained by different strategies may reflect the specificity of viral replication machinery and kinetics. Primary human monocytes have been reported susceptible to MERS-CoV and, more recently, to SARS-CoV-2 (Chu et al., 2016; Rodrigues et al., 2020). In contrast, one study found that among PBMCs, only CD4+ T cells had SARS-CoV-2 virus-like particles (Banerjee et al., 2020). In addition, there have been reports of reduced expression of ACE2 by lymphocytes (Uhlén et al., 2015). We found that blocking ACE2 significantly reduced, but did not abolish, SARS-CoV-2 production by infected PBMCs. This suggests that some ACE2-expressing cell types among PBMCs may contribute disproportionally to the virus progeny production or that, in the absence of ACE2, viral entry is suboptimal in these cells. This suboptimal or alternative entry by SARS-CoV-2 could be through ACE2-independent routes. A recent study showed that a spike mutation confers the ability to infect human airway ACE2-negative cells through heparan sulfates and clathrin-mediated endocytosis (Puray-Chavez et al., 2021). Another alternative entry pathway that has been proposed mentions CD147, which could serve as an alternative virus receptor, as already reported for SARS-CoV (Chen et al., 2005; Wang et al., 2020). CD147, also known as basignin, is expressed in T lymphocyte subsets, although its role in SARS-CoV-2 infection of these cells is unclear (Solstad et al., 2011; Ren et al., 2021).
Intense lymphocytopenia has been considered an indicator of severe clinical outcomes in COVID-19. In the present study, SARS-CoV-2 was detected in PBMCs from COVID-19 patients, more prominently in B lymphocytes and subpopulations of inflammatory monocytes. The predominance of B lymphocytes as targets of SARS-CoV-2 infection in vivo, in contrast to PBMCs infected in vitro, may reflect natural differences in susceptibility of different lymphocyte subsets to SARS-CoV-2 infection due to external cofactors. SARS-CoV entry in B lymphocytes and monocyte-derived cells can occur via a FcγRII-dependent pathway, facilitated by the presence of antibodies (Kam et al., 2007; Jaume et al., 2011). A caveat of the present study was that PBMCs were obtained at different time points during the COVID-19 evolution. The discrepancies in the clinical phases at enrollment could explain the heterogeneity in percentages of SARS-CoV-2-positive cells of different immunophenotypes observed among patients. Accordingly, SARS-CoV-2 RNA was not detected in PBMCs from half of the patients, indicating that SARS-CoV-2 infection in PBMCs may be variable, depending on host factors still unidentified. Another possible scenario would be the presence only in later phases of COVID-19, as suggested by the positive correlation between time from symptom onset and frequency of SARS-CoV-2-positive cells in PBMCs. A possible explanation for an increase in SARS-CoV-2-susceptible cells over time could be an increase in ACE2 expression, triggered by type I interferon (Ziegler et al., 2020). In this context, it is noteworthy that the replication of another coronavirus, SARS-CoV, in PBMCs was not sustained for a long period (Li et al., 2003; Yilla et al., 2005).
An intense T cell depletion in peripheral blood is seen in up to 85% of severe COVID-19 patients (Chen et al., 2020; Huang et al., 2020). Furthermore, T cells from COVID-19 patients show multiple levels of exhaustion markers, and transcriptome analysis of their PBMCs indicated upregulation of genes involved in apoptosis and p53 signaling pathways (Diao et al., 2020; Xiong et al., 2020; Zheng et al., 2020). These data suggest that SARS-CoV-2 infection could induce cell death by apoptosis in PBMCs, which could also happen in inflamed, infected organs of COVID-19 patients. It is important to note that MERS infection also implicates lymphopenia due to direct infection of primary T lymphocytes and induction of apoptosis (Chu et al., 2016). Exposure of human PBMCs to SARS-CoV-2 rapidly induced apoptosis in CD4+ and CD8+ T cell populations, as detected by activation of caspase 3/7, and the translocation of PS, as seen by annexin-V staining. Interestingly, UV inactivation of SARS-CoV-2, as well as a lesser extent of treatment with NH4Cl, prevented apoptosis in these cells. UV irradiation is widely used for viral inactivation by causing pyrimidine nucleotide dimerization while keeping the native structure of proteins (Lo et al., 2021; Loveday et al., 2021). Together, the data indicate that the induction of apoptosis depends on the viral replication process.
Moreover, caspase 3/7 activation was low in monocytes, consistent with monocytes undergoing pyroptosis after caspase 1 activation by SARS-CoV-2 infection (Rodrigues et al., 2020). This rapid induction of apoptosis after SARS-CoV-2 exposure could explain why we did not detect higher numbers of infected CD4+ T cells in COVID-19 patients, since dying cells are quickly removed from circulation. The ability of SARS-CoV-2 to trigger lymphocyte death may explain the observed lymphopenia in COVID-19 patients. The association of lymphopenia with poor prognosis is related to the death of specific T-cell subsets, resulting in loss of immune response regulatory components and driving a cytokine storm that can crosstalk with neutrophil NETosis (Barnes et al., 2020). It is also related to increased IL-6 and Fas–FasL interactions (Diao et al., 2020), resulting in severe lymphoid tissue alterations (Blanco-Melo et al., 2020).
SARS-CoV-2 infection recruits significant inflammatory cell infiltration into the lungs, containing diverse immune cell types that bear close contact with SARS-CoV-2-infected lung cells, such as pneumocytes and alveolar macrophages (Martines et al., 2020). In the present study, we found CD4+ T and B lymphocytes and, importantly, IL-6-expressing inflammatory macrophages positive for SARS-CoV-2 in the lungs from fatal cases of COVID-19.
Inflammatory monocytes play significant roles in the immunopathology of COVID-19 (Mehta et al., 2020; Zhou et al., 2020b), and ICU patients have high levels of circulating CD14+CD16+ inflammatory monocytes, which also correlates with unfavorable outcomes (Zhou et al., 2020b; Zhang et al., 2021). Increased expression of CCR2 and other inflammatory markers in monocytes leads to their infiltration in tissues with high expression of the corresponding CCL2 chemokine (Zhou et al., 2020b). Monocytes with the same profile were found abundantly in BALF from patients with severe COVID-19 (Liao et al., 2020). Our data may suggest that SARS-CoV-2-infected CD14+CCR2+ and CD14+CD16+CCR2+ monocytes could act as ‘Trojan horses’ and traffic viruses to secondary sites of infection, where SARS-CoV-2 causes severe tissue damage. Further studies will be required to clarify whether SARS-CoV-2-positive lymphoid–mononuclear cells in lung tissue become infected in the lung or migrate into the affected tissue from the bloodstream already infected.
Materials and methods
Ethics statement and COVID-19 patients
The study was approved by Conselho Nacional de Ética em Pesquisa (CONEP; Certificados de Apresentação para Apreciação Ética # 30248420.9.0000.5440 and 31797820.8.0000.5440). A total of 29 hospitalized patients were enrolled, all with clinical and radiological features of COVID-19 and confirmed SARS-CoV-2 infection by RT–PCR in respiratory secretions. Clinical features, laboratory results, and therapies are summarized in Supplementary Table S1. For all comparisons, 12 age- and gender-matched healthy controls were enrolled. Written informed consent forms were obtained from both patients and healthy donors.
Viruses and cells
SARS-CoV-2 Brazil/SPBR-02/2020 was kindly provided by Prof. Edison Luiz Durigon (ICB-USP, São Paulo). Virus stocks were made in Vero E6 cells. Virus titration was done by serial 10-fold dilutions inoculated in quadruplicate monolayers of Vero E6 cells and incubated at 37°C in 5% CO2. The presence of cytopathic effect at 4 days post-infection (dpi) similar to that shown in Supplementary Figure S6A was observed, and titers were read and expressed as TCID50 (Reed and Muench, 1938). All experiments with SARS-CoV-2 were done in a biosafety level 3 laboratory (BSL-3) located in the Center for Virus Research from the University of São Paulo in Ribeirão Preto.
Construction and rescue strategies of VSV-eGFP reporter chimeric viruses expressing the S protein from pandemic coronaviruses SARS-CoV, SARS-CoV-2, and MERS-CoV or the hemagglutinin from PIV5 were previously described (Whelan et al., 1995; Case et al., 2020; Kral et al., 2020). Reference sequences to all constructs are described in Supplementary Table S3. Coronavirus chimera stocks were produced in Vero TMPRSS2 cells. All chimeras were titrated by decimal serial dilution, and fluorescent foci were counted using a Typhoon plate reader (Amersham). Experiments were performed at a BSL-2 laboratory at Washington University in St. Louis.
The infectious clone SARS-CoV-2 that expresses the fluorescent protein mNeonGreen (icSARS-CoV-2) was kindly provided by Michael Diamond and is described in Xie et al. (2020). Viral working stocks were grown in Vero hACE2/TMPRSS2 and titrated in Vero E6 by serial 10-fold dilution with a semi-solid overlay. A portion of the stock was innactivated by exposure to an UV-C irradiation lamp (254 nm wavelength) for 10 min inside a biosafety cabinet. Stocks and the inactivation confirmation were done by serial 10-fold dilutions inoculated in Vero E6 monolayers with a semi-solid overlay. At 3 dpi, the read-out was done by fluorescent focus formation and plaque formation assays. Experiments were performed in BSL-3 at Washington University in St. Louis.
Isolation, treatments, and infection of PBMCs
Human PBMCs were isolated from COVID-19 patients or healthy donors by density gradient as previously described (Rios-Santos et al., 2007; Alves-Filho et al., 2009), washed, and resuspended in RPMI 1640 medium. All infections were carried out using 1 × 106 PBMCs at MOI of 1 at room temperature for 1 h under orbital agitation. For progeny quantification experiments, the inoculum was removed by extensive washing with cold phosphate-buffered saline (PBS), replenished with fresh RPMI 1640 medium with 2% fetal bovine serum (RPMI 2% FBS), and incubated at 37°C in 5% CO2. Supernatants from PBMCs were collected at 0, 6, 12, 24, and 48 hpi and subjected to serial 10-fold dilutions to determine virus titers by TCID50. For experiments assessing entry inhibition, 20 mM NH4Cl, 10 μM camostat, anti-ACE2 antibody (0.5 μg/ml, Rhea Biotech, or 20 μg/ml, R&D Systems), and isotype control antibodies IgG anti-digoxin (Vector laboratories) or IgG anti-RSV (Abcam) were used to pre-treat PBMCs for 1 h. If not specified, the treatments were removed by washing twice with Hank's balanced salt solution prior to infection.
Immunofluorescence staining
Coverslips pre-treated with 0.1% poly-lysine were incubated with isolated PBMCs from patients or healthy donors at 37°C for 20 min for cell adherence. After that, coverslips were fixed with 4% paraformaldehyde in PBS for 15 min, washed, and permeabilized with 0.1% saponin in 1% bovine serum album (BSA) solution to access intracellular antigens. Detection of SARS-CoV-2 antigens was done using serum from a convalescent COVID-19 patient (Supplementary Figure S1B). In each experiment using the COVID-19 serum, uninfected PBMCs and SARS-CoV-2-infected Vero cells were used as negative and positive controls, respectively. In addition, a human serum collected in 2016 was used as a negative control. Biotin-conjugated anti-human IgG was used as a secondary antibody, followed by TSA Cyanine 3 System amplification. To determine the types of SARS-CoV-2-infected cells, we used antibodies for CD4, CD8a, CD14, CD19, and CD20. To detect dsRNA complexes indicative of virus replication, we used a mouse monoclonal anti-dsRNA J2 antibody, which binds to dsRNA of 40 bp or longer. Secondary antibodies are described in Supplementary Table S3. The Golgi complex staining and nuclei staining were performed with mouse anti-GM130 antibody and 4′,6-diamidino-2-phenylindole (DAPI) dihydrochloride dye, respectively. Preparations were analyzed in a Zeiss Confocal 780 microscope in a tile 3 × 3 using a single focal plane. After imaging, analysis was performed using Fiji software.
FLICA assay
PBMCs were infected or mock-infected, and at 24 hpi, cells were labelled with FLICA substrate (660-DEVD-FMK; FLICA 660 Caspase-3/7 Assay Kit) as proposed by the manufacturer. Briefly, after incubation, the cells were resuspended in RPMI 2% FBS with FLICA substrate and cultured for 1 h at 37°C. Cells were then washed twice with 1× Apoptosis Buffer (provided in the kit), and proceeded to live/dead and extracellular staining. Finally, cells were fixed with 4% paraformaldehyde and immediately analyzed by flow cytometry. PBMCs were incubated for 4 h at 37°C with 5 µM staurosporine for positive apoptosis control.
Flow cytometry
PBMCs from COVID-19 patients or healthy donors infected in vitro with SARS-CoV-2 were surface-stained with a viability dye and monoclonal antibodies specific for CD3, CD4, CD8a, CD19, CD14, CD16, and CCR2 for 30 min at 4°C. Detection of SARS-CoV-2 intracellular antigens by flow cytometry was done on permeabilized PBMCs with the BD Cytofix/Cytoperm™ kit using the mouse anti-SARS-CoV-2 polyclonal antibody (described in Supplementary Materials and methods) diluted 1:300 in Perm/Wash buffer for 15 min at 4°C. Subsequently, cells were washed with Perm/Wash buffer and incubated with anti-mouse IgG-AlexaFluor488 antibody. In parallel, SARS-CoV-2-infected cells without permeabilization were also stained as controls to verify that virus staining was strictly intracellular. As an additional control, in vitro infected cells were treated with trypsin for 60 min on ice to remove possible surface-bound virus particles (false positives) (Supplementary Figure S7). Surface PS staining and active caspase 3/7 detection were carried out using commercial kits (described in Supplementary Table S3) and following the manufacturer's guidelines. Data were acquired using FACSVerse, FACS Canto (BD Biosciences), or Cytek/Aurora spectral flow cytometers (Cytek Biosciences) for multiparametric cell characterization, virus detection, and caspase activation assays. Subsequent analysis was done using FlowJo v.10 (TreeStar) software. Gating strategies are illustrated in Supplementary Figure S8.
Serial immunohistochemistry
Tissue sections from paraffin-embedded lung fragments obtained postmortem from two COVID-19 fatal cases were tested by immunohistochemistry (IHC) using anti-SARS-CoV-2 mouse polyclonal antibody (described in Supplementary Materials and methods) diluted 1:100 in PBS containing 1% BSA and 0.1% saponin. The primary antibody was detected using a biotinylated secondary antibody. Signal amplification was done by incubation with streptavidin–peroxidase polymer, and signal development was done with peroxidase. Sequential immunoperoxidase labelling and erasing (Glass et al., 2009) was then performed to determine the immunophenotypes of SARS-CoV-2-infected cells in situ. Cell immunophenotyping was performed using antibodies targeting CD4, CD20, CD14, and IL-6. Secondary antibodies were biotinylated. Color development was done as described earlier. Counterstaining was done with Harris hematoxylin, and slides were mounted with coverslips using an aqueous mounting medium. After each staining round, the slides were scanned using a VS120 ScanScope (Olympus) at 400× magnification. Images were then pseudocolored and overlaid in the first hematoxylin-counterstained image of the slide using ImageJ v1.50b (NIH) and Adobe Photoshop CS5 software (Adobe Systems). Lung paraffin-embedded tissue obtained from a fatal case of hantavirus infection from 2016 was used as a negative control for SARS-CoV-2 staining.
Supplementary Material
Acknowledgements
The authors thank Dimensions Sciences, a nonprofit organization that granted a research scholarship to Í.A.C. This study was developed in the framework of Rede Coronaômica MCTI/FINEP, affiliated to RedeVirus/MCTI-Brazil.
Contributor Information
Marjorie C Pontelli, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Ítalo A Castro, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Ronaldo B Martins, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Leonardo La Serra, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Flávio P Veras, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Daniele C Nascimento, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Camila M Silva, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Ricardo S Cardoso, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Roberta Rosales, Department of Cell and Molecular Biology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Rogério Gomes, Regional Blood Center of Ribeirão Preto, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Thais M Lima, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Juliano P Souza, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Brenda C Vitti, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Diego B Caetité, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Mikhael H F de Lima, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Spencer D Stumpf, Department of Biochemistry & Molecular Biology, the University of Texas Medical Branch, Galveston, TX 77555, USA.
Cassandra E Thompson, Department of Biochemistry & Molecular Biology, the University of Texas Medical Branch, Galveston, TX 77555, USA.
Louis-Marie Bloyet, Department of Biochemistry & Molecular Biology, the University of Texas Medical Branch, Galveston, TX 77555, USA.
Juliana E Toller-Kawahisa, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Marcela C Giannini, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Letícia P Bonjorno, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Maria I F Lopes, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Sabrina S Batah, Department of Pathology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Li Siyuan, Department of Pathology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Rodrigo Luppino-Assad, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Sergio C L Almeida, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Fabiola R Oliveira, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Maíra N Benatti, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Lorena L F Pontes, Regional Blood Center of Ribeirão Preto, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Rodrigo C Santana, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Fernando C Vilar, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Maria Auxiliadora-Martins, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Pei-Yong Shi, Department of Biochemistry & Molecular Biology, the University of Texas Medical Branch, Galveston, TX 77555, USA.
Thiago M Cunha, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Rodrigo T Calado, Regional Blood Center of Ribeirão Preto, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
José C Alves-Filho, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Dario S Zamboni, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Department of Cell and Molecular Biology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Alexandre T Fabro, Department of Pathology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Paulo Louzada-Junior, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Rene D R Oliveira, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Divisions of Clinical Immunology, Infectious Diseases and Intensive Care Unit, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Sean P J Whelan, Department of Molecular Microbiology, Washington University in St. Louis, Saint Louis, MO 63110, USA.
Fernando Q Cunha, Center of Research in Inflammatory Diseases, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Eurico Arruda, Virology Research Center, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil; Department of Cell and Molecular Biology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo 14049-900, Brazil.
Funding
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 310100/2017-8, 403201/2020-9, and INCT 465539/2014-9), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; 2013/16349-2 and 2014/02438-6), and the National Institutes of Health (NIH; AI163019). M.C.P and R.G. were funded by CNPq (380849/2020-8).
Conflict of interest: S.P.J.W. and Washington University have filed a patent application on VSV-SARS-CoV-2. S.P.J.W. has received unrelated funding support in sponsored research agreements with Vir Biotechnology and AbbVie. P.-Y.S. has filed a patent on the reverse genetic system of SARS-CoV-2.
References
- Alves-Filho J.C., Freitas A., Souto F.O.et al. (2009). Regulation of chemokine receptor by Toll-like receptor 2 is critical to neutrophil migration and resistance to polymicrobial sepsis. Proc. Natl Acad. Sci. USA 106, 4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.L., Dmytrenko O., Greenberg L.et al. (2021). SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. JACC Basic Transl. Sci. 6, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Nasir J.A., Budylowski P.et al. (2020). Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome Coronavirus 2. Emerg. Infect. Dis. 26, 2054–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B.J., Adrover J.M., Baxter-Stoltzfus A.et al. (2020). Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 217, e20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C.et al. (2020). Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E.et al. (2020). Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X.et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Mi L., Xu J.et al. (2005). Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 191, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.Y.et al. (2016). Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 213, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C.et al. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl Acad. Sci. USA 114, E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y.et al. (2020). Reduction and functional exhaustion of T cells in patients with ccoronavirus disease 2019 (COVID-19). Front. Immunol. 11, 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolhnikoff M., Ferreira Ferranti J., de Almeida Monteiro R.A.et al. (2020). SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet Child Adolesc. Heal. 4, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M., Zhang J., Ma X.et al. (2020). ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 131, 110678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuma A., Tani H., Taniguchi S.et al. (2015). Inability of rat DPP4 to allow MERS-CoV infection revealed by using a VSV pseudotype bearing truncated MERS-CoV spike protein. Arch. Virol. 160, 2293–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass G., Papin J.A., Mandell J.W. (2009). SIMPLE: a sequential immunoperoxidase labeling and erasing method. J. Histochem. Cytochem. 57, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S.et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X.et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y.et al. (2011). Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 85, 10582–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Kien F., Roberts A.et al. (2007). Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcγRII-dependent entry into B cells in vitro. Vaccine 25, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral A.H., Lambdin B.H., Wenger L.D.et al. (2020). Evaluation of an unsanctioned safe consumption site in the United States. N. Engl. J. Med. 383, 589–590. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J.et al. (2020). Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844. [DOI] [PubMed] [Google Scholar]
- Li L., Wo J., Shao J.et al. (2003). SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J. Clin. Virol. 28, 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.W., Matsuura R., Iimura K.et al. (2021). UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci. Rep. 11, 13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday E.K., Hain K.S., Kochetkova I.et al. (2021). Effect of inactivation methods on SARS-CoV-2 virion protein and structure. Viruses 13, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines R.B., Ritter J.M., Matkovic E.et al. (2020). Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg. Infect. Dis. 26, 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M.et al. (2020). COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puray-Chavez M., LaPak K.M., Schrank T.P.et al. (2021). Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 36, 109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S.et al. (2020). Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526, 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. (1938). A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27, 493–497. [Google Scholar]
- Ren X., Wen W., Fan X.et al. (2021). COVID-19 immune features revealed by a large-scale single cell transcriptome atlas. Cell 184, 1895–1913.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Santos F., Alves-Filho J.C., Souto F.O.et al. (2007). Down-regulation of CXCR2 on neutrophils in severe sepsis is mediated by inducible nitric oxide synthase-derived nitric oxide. Am. J. Respir. Crit. Care Med. 175, 490–497. [DOI] [PubMed] [Google Scholar]
- Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y.et al. (2020). Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 218, e20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solstad T., Bains S.J., Landskron J.et al. (2011). CD147 (basigin/emmprin) identifies FoxP3+CD45RO+CTLA4+-activated human regulatory T cells. Blood 118, 5141–5151. [DOI] [PubMed] [Google Scholar]
- Song X., Hu W., Yu H.et al. (2020). Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A, doi:10.1002/cyto.a.24285. [DOI] [PubMed] [Google Scholar]
- Tavazzi G., Pellegrini C., Maurelli M.et al. (2020). Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur. J. Heart Fail. 22, 911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.F., Lo A.W.I. (2004). Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J. Pathol. 203, 740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M.et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vargas-Gandica J., Winter D., Schnippe R.et al. (2020). Ageusia and anosmia, a common sign of COVID-19: a case series from four countries. J. Neurovirol. 26, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.P., Pontelli M.C., Silva C.M.et al. (2020). SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 217, e20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhang Z.et al. (2020). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S.P.J., Ball L.A., Barr J.N.et al. (1995). Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl Acad. Sci. USA 92, 8388–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Feng Z., Diao B.et al. (2021). SARS-CoV-2 induces lymphocytopenia by promoting inflammation and decimates secondary lymphoid organs. Front. Immunol. 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G.et al. (2020). An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L.et al. (2020). Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 9, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Harcourt B.H., Hickman C.J.et al. (2005). SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 107, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T.et al. (2020). TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5, eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Guo R., Lei L.et al. (2021). COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol. 109, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X.et al. (2020). Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 17, 541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. Lou, Wang X.G.et al. (2020a). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X.et al. (2020b). Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci. Rev. 7, 998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K.et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J.et al. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.