PURPOSE
Pancreatic ductal adenocarcinoma (PDAC) is associated with a poor prognosis. Multianalyte signatures, including liquid biopsy and traditional clinical variables, have shown promise for improving prognostication in other solid tumors but have not yet been rigorously assessed for PDAC.
MATERIALS AND METHODS
We performed a prospective cohort study of patients with newly diagnosed locally advanced pancreatic cancer (LAPC) or metastatic PDAC (mPDAC) who were planned to undergo systemic therapy. We collected peripheral blood before systemic therapy and assessed circulating tumor cells (CTCs), cell-free DNA concentration (cfDNA), and circulating tumor KRAS (ctKRAS)–variant allele fraction (VAF). Association of variables with overall survival (OS) was assessed in univariate and multivariate survival analysis, and comparisons were made between models containing liquid biopsy variables combined with traditional clinical prognostic variables versus models containing traditional clinical prognostic variables alone.
RESULTS
One hundred four patients, 40 with LAPC and 64 with mPDAC, were enrolled. CTCs, cfDNA concentration, and ctKRAS VAF were all significantly higher in patients with mPDAC than patients with LAPC. ctKRAS VAF (cube root; 0.05 unit increments; hazard ratio, 1.11; 95% CI, 1.03 to 1.21; P = .01), and CTCs ≥ 1/mL (hazard ratio, 2.22; 95% CI, 1.34 to 3.69; P = .002) were significantly associated with worse OS in multivariate analysis while cfDNA concentration was not. A model selected by backward selection containing traditional clinical variables plus liquid biopsy variables had better discrimination of OS compared with a model containing traditional clinical variables alone (optimism-corrected Harrell's C-statistic 0.725 v 0.681).
CONCLUSION
A multianalyte prognostic signature containing CTCs, ctKRAS, and cfDNA concentration outperformed a model containing traditional clinical variables alone suggesting that CTCs, ctKRAS, and cfDNA provide prognostic information complementary to traditional clinical variables in advanced PDAC.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States with only modest improvements in overall survival (OS) in recent decades.1,2 Clinical characteristics most consistently associated with OS include performance status, disease status (locally advanced pancreatic cancer [LAPC] v metastatic PDAC [mPDAC]), sex, age, liver metastasis, albumin, cancer antigen (CA)19-9, number of metastatic sites, pain, bilirubin, and lactate dehydrogenase,3 although these characteristics are imperfect. There is a great need to identify other noninvasive strategies to more accurately risk stratify patients with PDAC for clinical research, clinical care, and patient counseling. Tissue biomarkers are challenging to obtain as biopsies are often hypocellular, and the majority of patients will not be candidates for surgical resection. Conversely, blood samples are easily obtainable over the course of a patient's therapy.
CONTEXT
Key Objective
Pancreatic adenocarcinoma has poor prognosis. Multianalyte signatures using liquid biopsy variables have shown promise for improving prognostication in patients with other solid tumors but have not been rigorously evaluated to date in patients with pancreatic adenocarcinoma.
Knowledge Generated
Circulating tumor cells and circulating tumor KRAS-variant allele fraction were significantly associated with overall survival in univariable analysis and in multivariable analysis with adjustment for important traditional prognostic variables. Prognostic models containing liquid biopsy variables and traditional clinical variables outperformed models containing traditional clinical variables alone, suggesting that liquid biopsy variables provide complementary information.
Relevance
Multiple liquid biopsy analytes, including circulating tumor KRAS variant allele fraction and circulating tumor cells, may be applied in future clinical research to improve prognostic and predictive models in patients with pancreatic adenocarcinoma.
Cell-free DNA (cfDNA) is shed by cells into the blood while circulating tumor DNA (ctDNA) represents DNA specifically shed by tumor.4 In PDAC, ctDNA is most commonly assessed by the presence or absence of KRAS mutations using polymerase chain reaction or next-generation sequencing as KRAS has been shown to be mutated in > 90% of PDAC cases.5 Rates of circulating tumor KRAS mutation (ctKRAS) detection in the peripheral blood range from 21% to 60% in patients with localized PDAC and 42%-72% in patients with mPDAC.6 Detection of ctKRAS at baseline is consistently associated with worse OS in patients with PDAC of all stages.7-14 Although this suggests that ctKRAS alone may be a sufficient prognostic indicator in PDAC, it would be of great interest to know if the addition of other liquid biopsy analytes, easily obtainable from the same tube of blood, could improve prognostication compared with ctKRAS alone.
Circulating tumor cells (CTCs) are another class of liquid biopsy analytes. In patients with PDAC, rates of CTC detection using various platforms—including antibody-mediated enrichment (ie, CellSearch), methods using size, physical properties, and immunohistochemistry (ie, isolation by size of epithelial tumor cells, or ISET), and microfluidics approaches15-17—have varied from 5% to 89%.18 The presence of CTCs is associated with significantly worse OS in patients with PDAC.18,19 A more recent nonenrichment CTC detection method works by depositing all nucleated cells from a tube of blood onto glass microscope slides and identifies CTCs and other rare cells through immunofluorescence and digital pathology. Although this technology has been clinically qualified in prostate, breast, and lung cancer, this technology has not been evaluated in PDAC.20,21
There is evidence in multiple cancers that multianalyte approaches can provide complementary information compared with that of individual analytes alone. In breast cancer, a multianalyte liquid biopsy signature including CTC mRNA, CTC genomic DNA, extracellular vesicle mRNA, and cfDNA improved prognostication of OS compared with individual analytes, and clustering analysis suggested that liquid biopsy analytes were complementary and offered significant nonoverlapping information.22 In PDAC, a multianalyte panel of tumor-associated extracellular vesicle microRNA and mRNA, cfDNA, ctDNA KRAS mutations, and CA19-9 showed promise for improving diagnosis of PDAC and detecting occult metastases, although these variables were not assessed for prognostic value.24 A study examining baseline ctDNA and exosome DNA in PDAC found that elevation of both markers was a stronger predictor of worse OS compared with their elevation individually.25
Assessing any multianalyte panel in combination with clinical prognostic variables compared with clinical prognostic variables alone is an important step to establish the potential benefit in clinical practice. To our knowledge, the prognostic value of a multianalyte liquid biopsy in patients with advanced PDAC has not been assessed. We hypothesized that a multianalyte prognostic model incorporating liquid biopsy variables with traditional clinical variables would improve discrimination of prognosis in patients with LAPC and mPDAC compared with a model containing traditional clinical variables alone. Additionally, we aimed to evaluate for an independent association of cfDNA, ctKRAS, and CTCs with OS after adjusting for a more robust panel of clinical covariates than has previously been performed.
MATERIALS AND METHODS
Patient Enrollment
Patients with LAPC or mPDAC were screened at presentation to the medical oncology clinic at the University of Pennsylvania after diagnosis. Patients were excluded for having a concurrent cancer or for receiving prior systemic therapy for advanced PDAC. This study was reviewed by the Institutional Review Board at the University of Pennsylvania. Patients provided written informed consent to participate. Systemic treatment decisions were made by clinicians and patients as part of routine clinical care and were unrelated to the present study. Sample size was a convenience sample on the basis of budget for CTC analysis.
CTC Isolation and Identification
Peripheral blood (approximately 10 mL per sample) was collected at the time of patient enrollment and shipped to Epic Sciences (San Diego, CA) for processing. Red blood cells were lysed, and nucleated cells (3 × 106 per slide) were deposited onto 12 microscope slides. Two replicate slides per sample were thawed and stained with antibodies for pan-cytokeratins (CK), CD45, and 4',6-diamidino-2-phenylindole (DAPI). Slides then underwent a rapid fluorescent scanning method and CTCs identified in silico using proprietary digital pathology algorithms.26-28 The resulting images were displayed in a web-based report and confirmed by two trained technicians. Next, CTC enumeration results were compiled and reported as CTCs/mL of blood. The category of interest for the present study was CK+/CD45– cells with an intact nucleus (DAPI+) plus CK+ clusters, counted as one event, defined as two or more adjacent CD45– cells with shared cytoplasmic boundaries, at least one of which was CK+.27
cfDNA Isolation, Quantification, and Detection of ctKRAS Mutation
Isolation and quantification of cfDNA and ctKRAS detection by preamplification droplet digital polymerase chain reaction were performed as previously described (Data Supplement).24 Samples below the assay level of detection (0.01% variant allele fraction [VAF])24 were treated as zero values.
Assessment of Traditional Clinical Variables
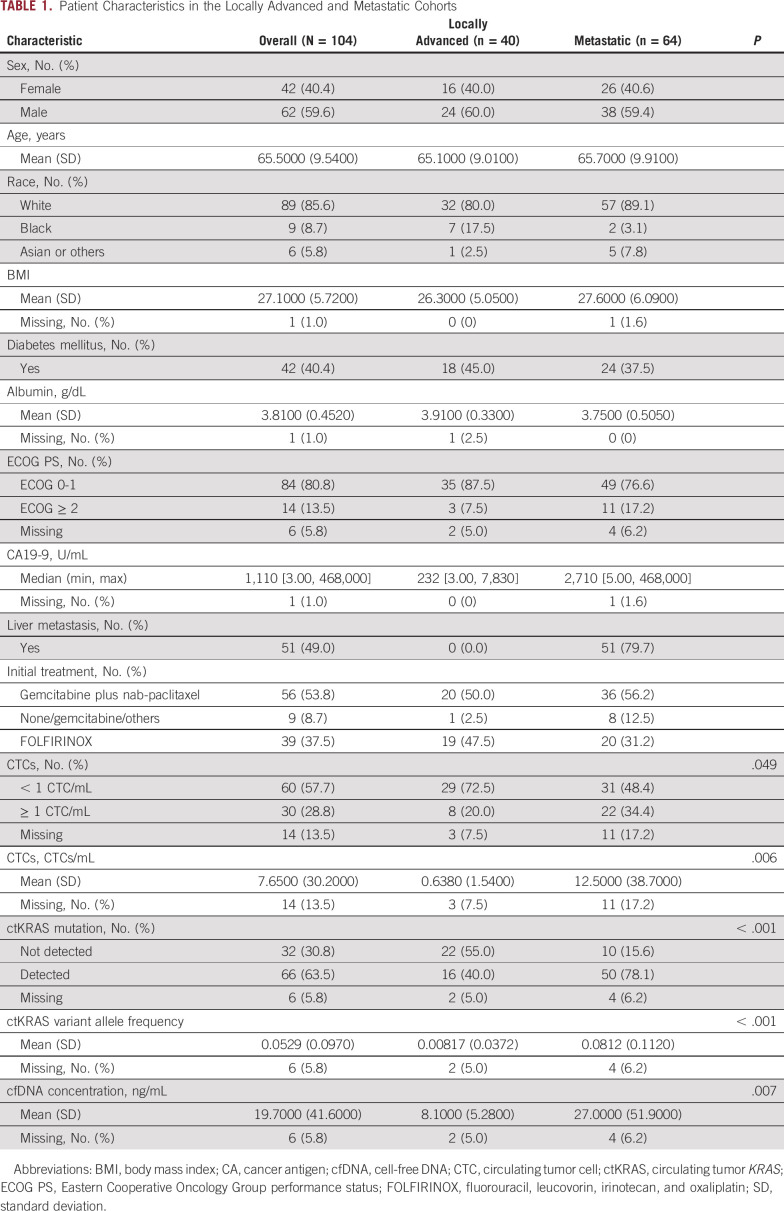
Baseline clinical variables (Table 1) were obtained by chart review. Laboratory variables were considered to be baseline values if collected up to 28 days before enrollment and before first systemic therapy.
TABLE 1.
Patient Characteristics in the Locally Advanced and Metastatic Cohorts
Statistical Methods
Descriptive statistics.
Descriptive statistics were computed as mean and standard deviation for continuous variables and as a proportion of patients for binary or categorical variables. The proportion of missing observations for each variable was reported. To compare values of liquid biopsy variables between LAPC and mPDAC groups, two-tailed t tests or Wilcoxon rank-sum tests were performed (depending on normality of distribution of continuous variables). Pearson's chi-square test or Fisher's exact test was performed to compare categorical variables between LAPC and mPDAC. Fisher's exact test was used to examine the association of CTCs (binary; cut point 1/mL) with ctKRAS (binary; cut point VAF 0.05; rationale described below) across the overall cohort, and linear regression modeling was used to assess the association of CTCs/mL (continuous) with ctKRAS VAF (continuous). Statistical testing for all analyses used an α of .05.
Univariate survival analysis.
Univariate and multivariate survival analysis was performed in R (version 3.6.3) using the rms and survival packages.29-31 OS was defined as the time from study enrollment to death as assessed by clinical records and online obituaries, with censoring at last clinical contact or data cutoff of January 14, 2021. The functional form and transformations of variables used in Cox proportional hazards models are described in the Data Supplement.
Univariate Cox proportional hazards modeling was performed to examine the association of liquid biopsy variables and traditional clinical variables with OS in the overall cohort and within the LAPC and mPDAC cohorts individually. Next, treating ctKRAS VAF and cfDNA as binary variables was explored by examining the spline transformation of each variable in the log relative hazard and selecting a natural cut point. This binary treatment of liquid biopsy variables was used to plot Kaplan-Meier survival curves by levels of the liquid biopsy variable, followed by log-rank testing. Using the binary cutoffs chosen above, an exploratory analysis was performed to examine OS for patients who were dual positive for CTCs and ctKRAS, single positive, or dual negative using the Kaplan-Meier method, followed by log-rank testing.
Multivariate Cox proportional hazards model selection.
Candidate variables for the multivariable prognostic models included variables with a P < .15 in univariate analysis. Our goals were (1) to assess the relationship of liquid biopsy variables with OS after adjusting for clinical covariates and (2) to compare models containing traditional clinical plus liquid biopsy variables to models containing traditional clinical variables alone for model fit and discrimination.
We accomplished these goals with two model selection strategies. First, we included all candidate variables (both liquid biopsy and traditional clinical variables) with a P < .15 in univariate analysis (model A) and compared this with a model that contained only traditional clinical variables that passed the P < .15 threshold in univariate analysis (model B). Second, we performed backward selection by minimizing Akaike's information criterion using the stepAIC function under the MASS package in R.32 We performed this backward selection procedure using all candidate variables with P < .15 in univariate analysis (model C) and ran a second model using this backward selection procedure using traditional clinical variables with P < .15 in univariate analysis alone (model D).
Evaluation of multivariate Cox proportional hazards models.
Evaluation of model assumptions and model fit is described in the Data Supplement. Model discrimination was assessed using Harrel's C-statistic.33 Internal validation with bootstrapping to calculate optimism-corrected Harrel's C-statistics and optimism-corrected calibration curves are further described in the Data Supplement. Sensitivity analysis examining the impact of missing data on the effect estimates in each model is described and presented in the Data Supplement.
RESULTS
Patient Characteristics and Summary Statistics
Forty patients with LAPC and 64 patients with mPDAC were enrolled (104 patients in total; Table 1) between August 14, 2015, and June 22, 2018. Ninety-nine patients (95.2%) died during follow-up. Of patients who were censored, the median follow-up was 40.1 months (range 9.3-44.7 months). The median OS was 11.9 months (95% CI, 9.2 to 15.5) in the overall population, 16.2 months (95% CI, 12.2 to 19.3) in the LAPC cohort, and 8.5 months (95% CI, 7.0 to 12.9 months) in mPDAC. cfDNA, ctKRAS VAF, and CTCs were all significantly higher in patients with mPDAC compared with LAPC (Fig 1).
FIG 1.
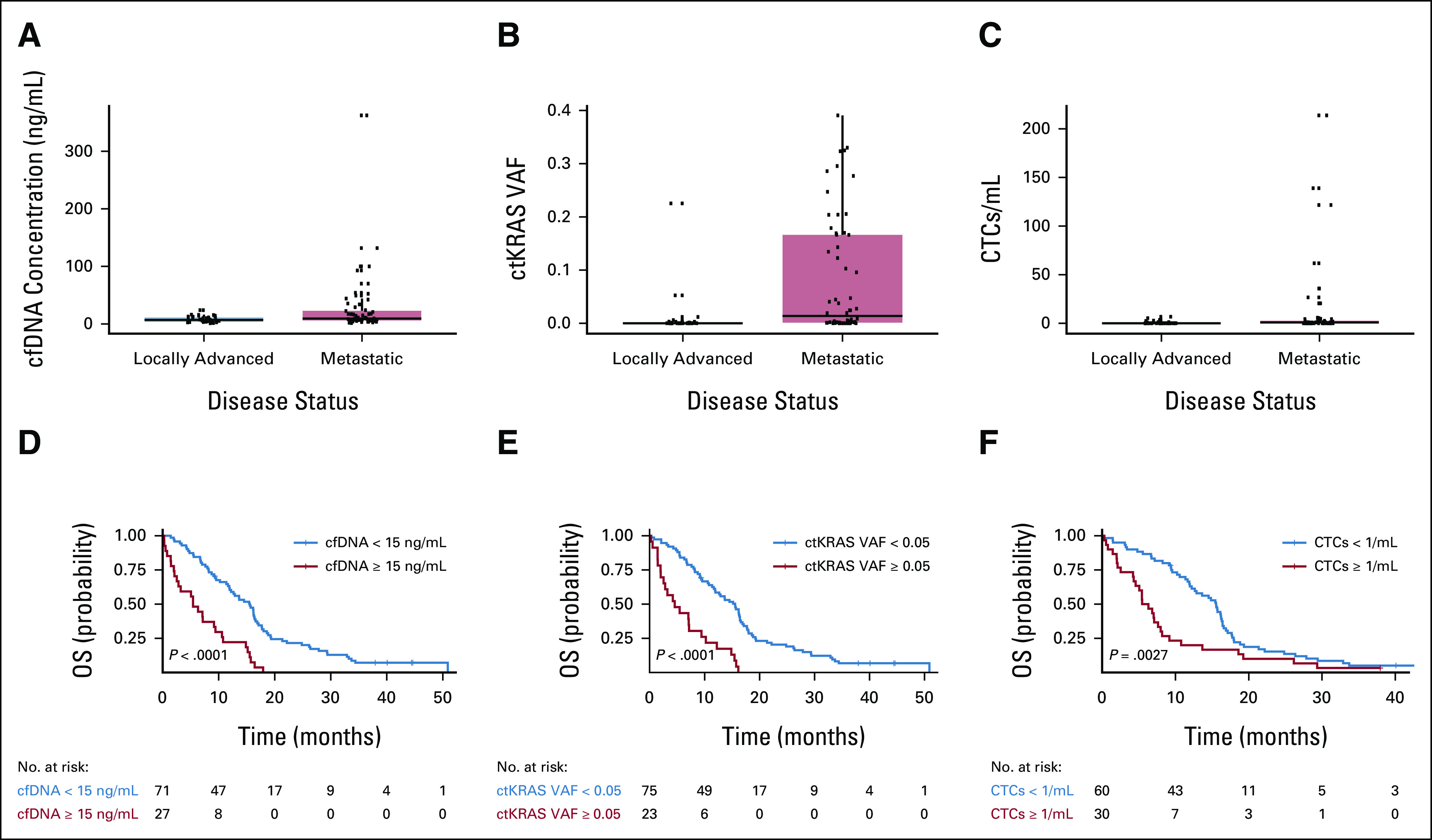
Box plots demonstrating distribution of (A) cfDNA concentration (ng/mL); (B) ctKRAS mutation variant allele frequency (ctKRAS VAF); and (C) CTCs (CTCs/mL of blood) in locally advanced and metastatic cohorts. (A) P values displayed represent the results of a two-tailed, two-sample t-test and (B) and (C) Wilcoxon rank-sum tests as variables were non-normally distributed. Kaplan-Meier survival curves stratified by (D) cfDNA concentration ≥ 15 ng/mL or < 15 ng/mL; (E) ctKRAS VAF ≥ 0.05 or < 0.05; and (F) CTCs ≥ 1/mL or < 1/mL. Binary cutoffs were chosen as discussed in the Statistical Methods section. P values represent the result of a log-rank test comparing the survival curves across reported strata. Numbers of patients at risk at each time point for each group are presented below Kaplan-Meier curves. cfDNA, cell-free DNA; CTC, circulating tumor cell; ctKRAS, circulating tumor KRAS; OS, overall survival; VAF, variant allele fraction.
Univariate Survival Analysis
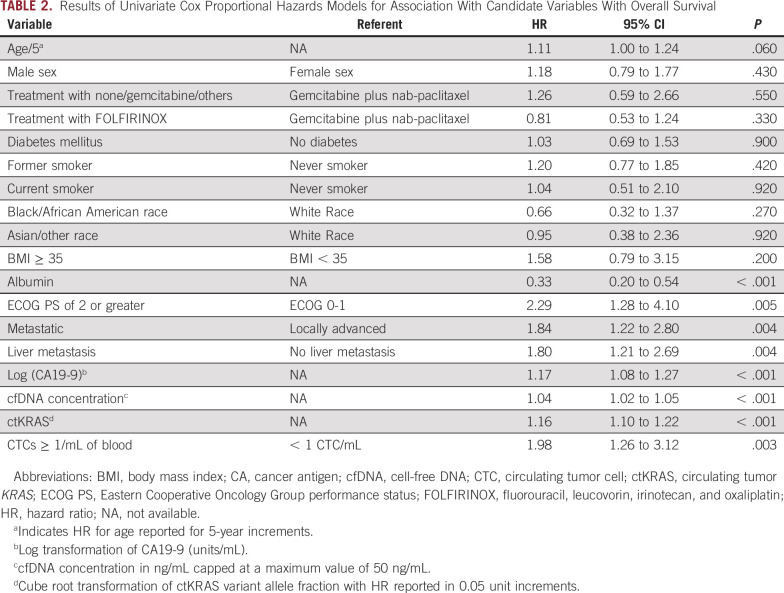
Univariate association of liquid biopsy and traditional clinical variables with OS is presented in Table 2. In addition, we examined the univariate association of liquid biopsy variables with OS within the LAPC and mPDAC subgroups. There was increased hazard of death for each 0.05 unit increase in cube root of ctKRAS VAF that was not statistically significant in the LAPC cohort (hazard ratio [HR], 1.12; 95% CI, 0.99 to 1.27; P = .08) but was statistically significant in the mPDAC cohort (HR, 1.15; 95% CI, 1.08 to 1.22; P < .0001). CTCs ≥ 1/mL was not associated with OS in LAPC (HR, 0.72; 95% CI, 0.31 to 1.69; P = .45) but was significantly associated with increased hazard of death in mPDAC (HR, 5.54; 95% CI, 2.67 to 11.47; P < .0001).Increasing cfDNA concentration was significantly associated with increased hazard of death in both LAPC (HR, 1.08; 95% CI, 1.00 to 1.17; P = .04) and mPDAC cohorts (HR, 1.03; 95% CI, 1.01 to 1.04; P = .0003). When treated as binary variables, CTCs ≥ 1/mL, cfDNA ≥ 15 ng/mL, and ctKRAS VAF ≥ 0.05 were significantly associated with worse OS (Fig 1).
TABLE 2.
Results of Univariate Cox Proportional Hazards Models for Association With Candidate Variables With Overall Survival
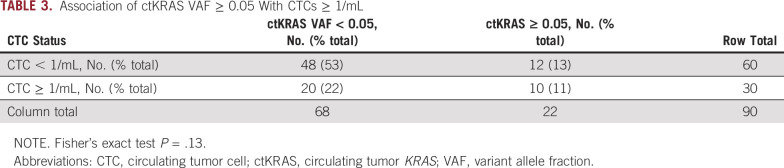
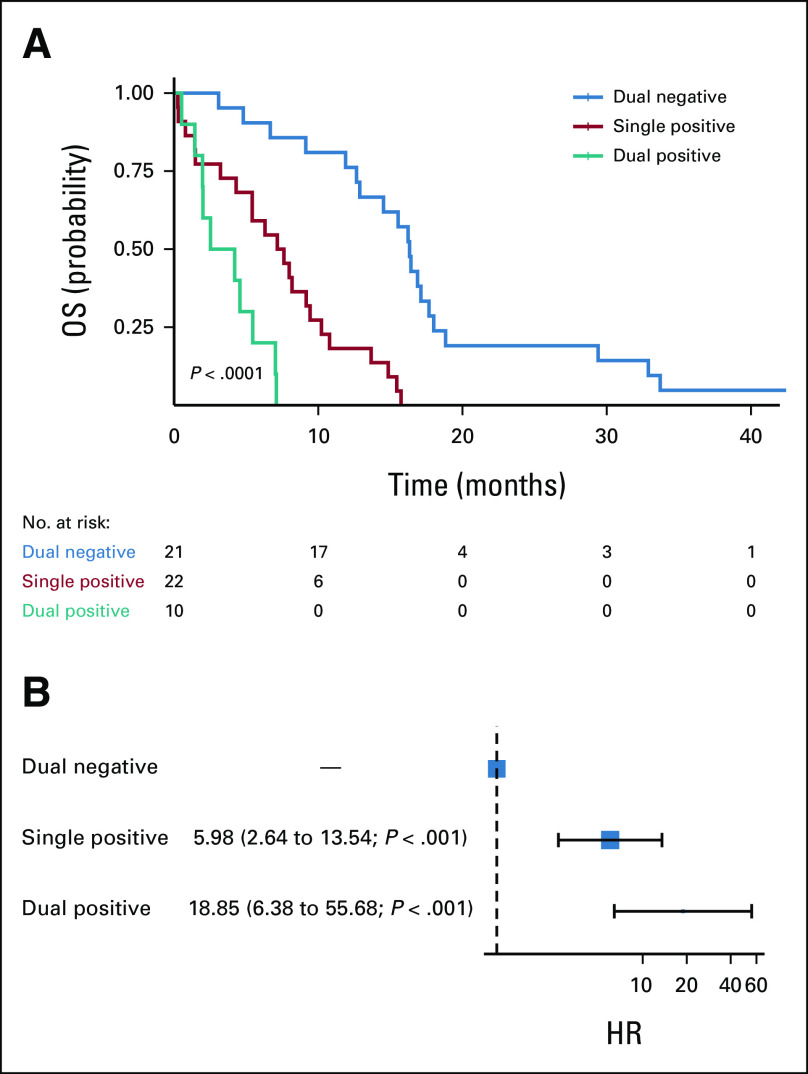
Given that ctKRAS and CTCs are two different measures of tumor shedding, we next examined the frequency with which both analytes were detectable. Interestingly, there was no significant association between CTCs ≥ 1/mL and ctKRAS VAF ≥ 0.05 (P = .13; Table 3) or when treated as continuous variables (P = .47; Data Supplement) suggesting that these analytes may be complementary. Within the LAPC cohort, 27 of 37 patients (73%) with nonmissing data for CTCs and ctKRAS had no detectable CTCs or ctKRAS (dual negative, CTCs < 1/mL and ctKRAS VAF < 0.05), and the remaining 10 of 37 patients (27%) were deemed single positive (CTCs > 1/mL or ctKRAS VAF > 0.05). However, within the mPDAC cohort, 10 of 53 patients (19%) evaluable for both analytes were dual positive, 22 (42%) were single positive, and 21 patients (40%) were dual negative for CTCs and ctKRAS. For the mPDAC cohort, there was a significant difference in OS by log-rank testing and by univariate Cox proportional hazards modeling (Fig 2).
TABLE 3.
Association of ctKRAS VAF ≥ 0.05 With CTCs ≥ 1/mL
FIG 2.
Assessment of OS by CTC and ctKRAS status by (A) the Kaplan-Meier method in patients with mPDAC along with the number of patients at risk and (B) by univariate Cox proportional hazards modeling in patients with mPDAC. Patients were categorized as dual negative for CTCs and ctKRAS (CTCs < 1/mL and ctKRAS VAF < 0.05), single positive (CTCs ≥ 1/mL or ctKRAS VAF ≥ 0.05), or dual positive (CTCs ≥ 1/mL and ctKRAS VAF ≥ 0.05). Twenty-one patients with mPDAC were dual negative, 22 were single positive, and 10 were dual positive. Among patients with mPDAC, dual positivity was associated with worse OS than dual negativity (HR, 18.84; 95% CI, 6.38 to 55.68; P < .001) and single positivity was associated with worse OS than dual negativity (HR, 5.98; 95% CI, 2.64 to 13.54; P < .001). CTC, circulating tumor cell; ctKRAS, circulating tumor KRAS; HR, hazard ratio; mPDAC, metastatic pancreatic ductal adenocarcinoma; OS, overall survival; VAF, variant allele fraction.
Multivariate Cox Proportional Hazards Models
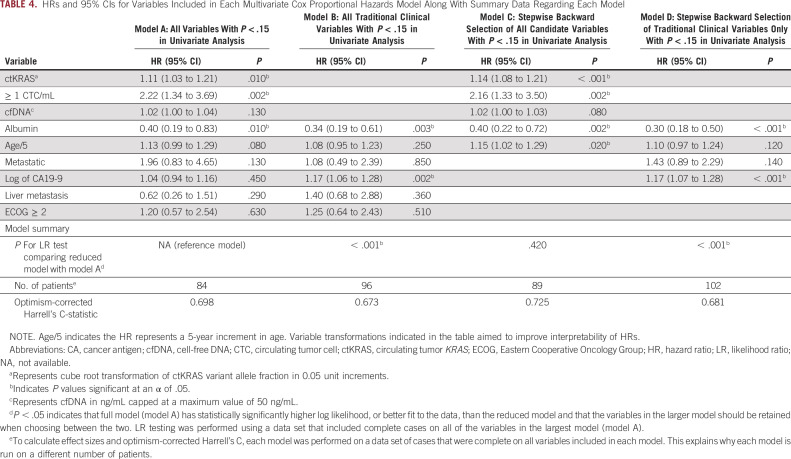
A multivariate model was selected to assess the association of liquid biopsy variables with OS after adjusting for clinical covariates, selected as identified in the Statistical Methods section. CTCs ≥ 1/mL, cube root of ctKRAS VAF, and albumin were independently associated with OS in a model containing all candidate variables that had a P < .15 in univariate analysis (model A; Table 4). cfDNA concentration was not independently associated with OS in this model.
TABLE 4.
HRs and 95% CIs for Variables Included in Each Multivariate Cox Proportional Hazards Model Along With Summary Data Regarding Each Model
Likelihood ratio testing was performed to compare fit of each model with that of model A (Table 4). These comparisons suggest that models containing ctKRAS, CTCs, and cfDNA (models A and C) had statistically significantly improved model fit compared with models which considered traditional clinical variables only (models B and D).
Internal validation using bootstrapping showed optimism-corrected Harrell's C-statistics of 0.698, 0.673, 0.725, and 0.681 for models A-D, respectively. These results suggest that models containing ctKRAS, CTCs, and cfDNA (models A and C) have higher discrimination and are more likely to perform better in external data sets than models containing traditional prognostic variables alone (models B and D). Calibration plots showed reasonable calibration for models A-D (Data Supplement).
DISCUSSION
In a cohort of patients with LAPC or mPDAC, we found that liquid biopsy variables, including ctKRAS VAF and CTCs were significantly associated with OS in univariate analysis. There was no significant association between CTCs and ctKRAS when assessed as continuous or binary variables. In patients with mPDAC, those classified as dual positive for CTCs ≥ 1/mL and ctKRAS VAF ≥ 0.05 had significantly worse OS compared with patients who were single positive and dual negative, respectively. After adjusting for clinical confounders in multivariate analysis, CTCs and ctKRAS VAF remained independently associated with OS. We also found that a prognostic model using liquid biopsy plus traditional clinical variables had significantly improved model fit compared with a model developed using traditional clinical variables alone. In addition, the ability of the models containing liquid biopsy variables plus traditional clinical variables (models A and C) to discriminate between patients with differential survival, as measured by an optimism-corrected Harrell's C-statistic, was higher compared with models using traditional clinical variables only (models B and D). These findings suggest that liquid biopsy variables provide information that is complementary to each other and to that of traditional clinical variables.
One novel aspect of our study is that the association of CTCs detected by a nonenrichment method with OS has not been formally evaluated in PDAC. We found that the presence of CTCs at a concentration of ≥ 1/mL was significantly associated with increased hazard of death in both univariate and multivariate analysis. This result is consistent with that observed in CTCs detected by other methods in patients with advanced PDAC.18,19
We explored whether our liquid biopsy variables might have different associations with OS in patients with LAPC versus mPDAC. Interestingly, for cfDNA and ctKRAS VAF, the effect size and direction of the association with OS was similar in the LAPC and mPDAC cohorts (although the result was not statistically significant for ctKRAS VAF in the LAPC cohort). However, there was no relationship between CTC ≥ 1/mL and OS in the LAPC cohort while there was a very strong relationship between CTC ≥ 1/mL and worse OS in the mPDAC cohort. This result must be considered exploratory as CTC detection rate in LAPC patients was quite low in this study (20%) lending insufficient power to detect an association with survival. This lower detection rate of CTCs in LAPC compared with mPDAC is consistent with previously observed results.34 When examining the combination of CTCs and ctKRAS within the mPDAC cohort, patients with dual positivity for CTCs and ctKRAS had significantly worse OS compared with those with single positivity, who in turn had worse OS compared with those with dual negativity (Fig 2). Overall, these results suggest that although cfDNA concentration and ctKRAS VAF are likely useful prognostic biomarkers in both LAPC and mPDAC, CTC detection and the combination of CTCs and ctKRAS may be more useful when used in patients with more advanced disease.
Limitations include the study of a moderate sample size enrolled at a single institution. Second, although tumor volumetric measurements were outside the scope of our study, previous work has shown an association between ctDNA VAF and tumor volume in patients with mPDAC and that ctDNA VAF and tumor volume were significantly associated with OS.35 Future research may address whether prognostic models including ctDNA VAF, tumor volumetric measurements, or both perform better in patients with PDAC. Third, although methods of ctDNA detection (other than ctKRAS) have also demonstrated prognostic value, a comparison of methods of ctDNA assessment was outside the scope of this study.36 Fourth, although we performed robust internal validation using a bootstrapping approach to estimate the likely performance of the models in an external data set, the models have not undergone external validation and thus are not ready for clinical application. Nevertheless, liquid biopsy variables including ctKRAS, cfDNA, and CTCs have demonstrated promise for improving prognostication beyond traditional clinical variables alone in patients with LAPC and mPDAC.
Future research may include external validation of our prognostic model and assess the utility of ctKRAS, cfDNA, and CTCs in monitoring response to therapy. Additional studies would evaluate the utility of a prognostic model for selecting patients with LAPC who have received neoadjuvant therapy for curative-intent surgery and for stratifying patients with mPDAC onto clinical trials earlier in therapy.
ACKNOWLEDGMENT
We would like to acknowledge, and thank, all of the patients and family members who participated in this study for their contribution to this research.
Jacob E. Till
Stock and Other Ownership Interests: Sorrento Therapeutics, Pfizer (I)
Wei-Ting Hwang
Employment: Janssen (I)
Research Funding: Novartis
Jennifer R. Eads
Employment: Bristol Myers Squibb (I), Janssen Oncology (I)
Stock and Other Ownership Interests: Bristol Myers Squibb/Celgene (I)
Consulting or Advisory Role: Lexicon, Advanced Accelerator Applications, Ipsen
Research Funding: Xencor (Inst), Tarveda Therapeutics (Inst), MedImmune (Inst), Incyte, Oncolys BioPharma (Inst), Seattle Genetics (Inst), Genentech (Inst), Hutchison MediPharma (Inst), AstraZeneca/MedImmune (Inst)
Travel, Accommodations, Expenses: Bristol Myers Squibb (I), Janssen Oncology (I)
Thomas B. Karasic
Honoraria: Exelixis, Incyte, Pfizer, Ipsen, AstraZeneca/MedImmune
Research Funding: Syndax (Inst), Taiho Pharmaceutical (Inst), Celgene (Inst), H3 Biomedicine (Inst), Bristol Myers Squibb (Inst), Lilly (Inst), Sirtex Medical (Inst), Tempest Therapeutics (Inst), Xencor (Inst)
Peter J. O'Dwyer
Consulting or Advisory Role: Genentech
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), Novartis (Inst), Genentech (Inst), Mirati Therapeutics (Inst), Celgene (Inst), GlaxoSmithKline (Inst), BBI Healthcare (Inst), Pharmacyclics (Inst), Five Prime Therapeutics (Inst), Forty Seven (Inst), Amgen (Inst), H3 Biomedicine (Inst), Taiho Pharmaceutical (Inst), Array BioPharma (Inst), Lilly/ImClone (Inst), Syndax (Inst), Minneamrita Therapeutics (Inst)
Expert Testimony: Lilly, Daiichi Sankyo
Amanda Anderson
Employment: Epic Sciences, BioTheranostics
Stock and Other Ownership Interests: Epic Sciences, BioTheranostics
Travel, Accommodations, Expenses: Epic Sciences, BioTheranostics
Megan Slade
Employment: Epic Sciences
Travel, Accommodations, Expenses: Epic Sciences
Michael Lariviere
Honoraria: Curio Science, Alphasights, Gerson Lehrman Group
Research Funding: Merck (Inst)
Kim A. Reiss
Honoraria: MJH Life Sciences
Consulting or Advisory Role: AstraZeneca, Carisma Therapeutics
Research Funding: Lilly (Inst), Clovis Oncology, Bristol Myers Squibb (Inst), Tesaro (Inst), GlaxoSmithKline (Inst)
Mark H. O'Hara
Consulting or Advisory Role: Natera, Geneos, PsiOxus Therapeutics
Research Funding: Bristol Myers Squibb, Celldex, Parker Institute for Cancer Immunotherapy, Lilly, Arcus Biosciences, Natera, PsiOxus Therapeutics, Geneos
Travel, Accommodations, Expenses: AstraZeneca/MedImmune
Erica L. Carpenter
Honoraria: AstraZeneca, Guardant Health
Consulting or Advisory Role: Bristol Myers Squibb
Research Funding: Merck (Inst), Janssen (Inst), Becton Dickinson (Inst), United Health Group, Parker Institute for Cancer Immunotherapy (Inst), AstraZeneca (Inst), ChipDiagnostics (Inst), C2i Genomics (Inst)
Patents, Royalties, Other Intellectual Property: Invention disclosure entitled, “Methods and Compositions for Identifying, Diagnosing and Treating Neuroblastoma,” Invention disclosure entitled, “Methods and Compositions for Identifying, Diagnosing and Treating Neuroblastoma”
Travel, Accommodations, Expenses: AstraZeneca, Foundation Medicine
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Items related to current article were presented at the American Society of Clinical Oncology (ASCO) Annual Meeting, Chicago, IL, June 4-8, 2021.
SUPPORT
Supported by Grant No. 1R01 CA207643-01, by the Penn Center for Pancreatic Research, the Basser Young Leadership Council, the Konner Fund, the Pearl and Philip Basser Award, and An Anonymous Foundation.
W.J.C. and J.E.T. share first authorship to this work. W.-T.H. is the second author on the work. M.H.O. and E.L.C. share senior authorship to this work.
DATA SHARING STATEMENT
The raw data supporting the conclusions of this article will be made available by the authors upon request, without undue reservation.
AUTHOR CONTRIBUTIONS
Conception and design: William J. Chapin, Jacob E. Till, Peter J. O'Dwyer, Kim A. Reiss, Mark H. O'Hara, Erica L. Carpenter
Financial support: Amanda Anderson, Megan Slade, Kim A. Reiss, Erica L. Carpenter
Administrative support: Janae Romeo, Taylor A. Black, Theresa E. Christensen, Colleen Redlinger Tabery, Stephanie S. Yeea
Provision of study materials or patients: Jennifer R. Eads, Thomas B. Karasic, Peter J. O'Dwyer, Charles J. Schneider, Ursina R. Teitelbaum, Kim A. Reiss, Mark H. O'Hara, Amanda Anderson, Megan Slade, Erica L. Carpenter
Collection and assembly of data: William J. Chapin, Jacob E. Till, Janae Romeo, Taylor A. Black, Theresa E. Christensen, Colleen Redlinger Tabery, Amanda Anderson, Megan Slade, Michael LaRiviere, Stephanie S. Yee
Data analysis and interpretation: William J. Chapin, Jacob E. Till, Wei-Ting Hwang, Amanda Anderson, Megan Slade, Kim A. Reiss, Mark H. O'Hara, Erica L. Carpenter
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jacob E. Till
Stock and Other Ownership Interests: Sorrento Therapeutics, Pfizer (I)
Wei-Ting Hwang
Employment: Janssen (I)
Research Funding: Novartis
Jennifer R. Eads
Employment: Bristol Myers Squibb (I), Janssen Oncology (I)
Stock and Other Ownership Interests: Bristol Myers Squibb/Celgene (I)
Consulting or Advisory Role: Lexicon, Advanced Accelerator Applications, Ipsen
Research Funding: Xencor (Inst), Tarveda Therapeutics (Inst), MedImmune (Inst), Incyte, Oncolys BioPharma (Inst), Seattle Genetics (Inst), Genentech (Inst), Hutchison MediPharma (Inst), AstraZeneca/MedImmune (Inst)
Travel, Accommodations, Expenses: Bristol Myers Squibb (I), Janssen Oncology (I)
Thomas B. Karasic
Honoraria: Exelixis, Incyte, Pfizer, Ipsen, AstraZeneca/MedImmune
Research Funding: Syndax (Inst), Taiho Pharmaceutical (Inst), Celgene (Inst), H3 Biomedicine (Inst), Bristol Myers Squibb (Inst), Lilly (Inst), Sirtex Medical (Inst), Tempest Therapeutics (Inst), Xencor (Inst)
Peter J. O'Dwyer
Consulting or Advisory Role: Genentech
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), Novartis (Inst), Genentech (Inst), Mirati Therapeutics (Inst), Celgene (Inst), GlaxoSmithKline (Inst), BBI Healthcare (Inst), Pharmacyclics (Inst), Five Prime Therapeutics (Inst), Forty Seven (Inst), Amgen (Inst), H3 Biomedicine (Inst), Taiho Pharmaceutical (Inst), Array BioPharma (Inst), Lilly/ImClone (Inst), Syndax (Inst), Minneamrita Therapeutics (Inst)
Expert Testimony: Lilly, Daiichi Sankyo
Amanda Anderson
Employment: Epic Sciences, BioTheranostics
Stock and Other Ownership Interests: Epic Sciences, BioTheranostics
Travel, Accommodations, Expenses: Epic Sciences, BioTheranostics
Megan Slade
Employment: Epic Sciences
Travel, Accommodations, Expenses: Epic Sciences
Michael Lariviere
Honoraria: Curio Science, Alphasights, Gerson Lehrman Group
Research Funding: Merck (Inst)
Kim A. Reiss
Honoraria: MJH Life Sciences
Consulting or Advisory Role: AstraZeneca, Carisma Therapeutics
Research Funding: Lilly (Inst), Clovis Oncology, Bristol Myers Squibb (Inst), Tesaro (Inst), GlaxoSmithKline (Inst)
Mark H. O'Hara
Consulting or Advisory Role: Natera, Geneos, PsiOxus Therapeutics
Research Funding: Bristol Myers Squibb, Celldex, Parker Institute for Cancer Immunotherapy, Lilly, Arcus Biosciences, Natera, PsiOxus Therapeutics, Geneos
Travel, Accommodations, Expenses: AstraZeneca/MedImmune
Erica L. Carpenter
Honoraria: AstraZeneca, Guardant Health
Consulting or Advisory Role: Bristol Myers Squibb
Research Funding: Merck (Inst), Janssen (Inst), Becton Dickinson (Inst), United Health Group, Parker Institute for Cancer Immunotherapy (Inst), AstraZeneca (Inst), ChipDiagnostics (Inst), C2i Genomics (Inst)
Patents, Royalties, Other Intellectual Property: Invention disclosure entitled, “Methods and Compositions for Identifying, Diagnosing and Treating Neuroblastoma,” Invention disclosure entitled, “Methods and Compositions for Identifying, Diagnosing and Treating Neuroblastoma”
Travel, Accommodations, Expenses: AstraZeneca, Foundation Medicine
No other potential conflicts of interest were reported.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2. Worni M, Guller U, White RR, et al. Modest improvement in overall survival for patients with metastatic pancreatic cancer: A trend analysis using the surveillance, epidemiology, and end results registry from 1988 to 2008. Pancreas. 2013;42:1157–1163. doi: 10.1097/MPA.0b013e318291fbc5. [DOI] [PubMed] [Google Scholar]
- 3. Ter Veer E, van Rijssen LB, Besselink MG, et al. Consensus statement on mandatory measurements in pancreatic cancer trials (COMM-PACT) for systemic treatment of unresectable disease. Lancet Oncol. 2018;19:e151–e160. doi: 10.1016/S1470-2045(18)30098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh N, Gupta S, Pandey RM, et al. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest. 2015;33:78–85. doi: 10.3109/07357907.2014.1001894. [DOI] [PubMed] [Google Scholar]
- 5. Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Creemers A, Krausz S, Strijker M, et al. Clinical value of ctDNA in upper-GI cancers: A systematic review and meta-analysis. Biochim Biophys Acta Rev Cancer. 2017;1868:394–403. doi: 10.1016/j.bbcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 7. Takai E, Totoki Y, Nakamura H, et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep. 2015;5:18425. doi: 10.1038/srep18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Earl J, Garcia-Nieto S, Martinez-Avila JC, et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 2015;15:797. doi: 10.1186/s12885-015-1779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 10. Pietrasz D, Pécuchet N, Garlan F, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res. 2017;23:116–123. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 11. Lee J-S, Rhee T-M, Pietrasz D, et al. Circulating tumor DNA as a prognostic indicator in resectable pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Sci Rep. 2019;9:16971. doi: 10.1038/s41598-019-53271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L, Zhang Y, Cheng Y, et al. Prognostic value of circulating cell-free DNA in patients with pancreatic cancer: A systemic review and meta-analysis. Gene. 2018;679:328–334. doi: 10.1016/j.gene.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 13. Bachet J-B, Blons H, Hammel P, et al. Circulating tumor DNA is prognostic and potentially predictive of eryaspase efficacy in second-line in patients with advanced pancreatic adenocarcinoma. Clin Cancer Res. 2020;26:5208–5216. doi: 10.1158/1078-0432.CCR-20-0950. [DOI] [PubMed] [Google Scholar]
- 14. Till JE, Abdalla A, Bhagwat N, et al. Baseline level and early on-treatment clearance of circulating mutant KRAS in metastatic pancreatic ductal adenocarcinoma treated with chemotherapy with or without immunotherapy. J Clin Oncol. 2021;39(15 suppl) abstr 4122. [Google Scholar]
- 15. Riethdorf S, Fritsche H, Müller V, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 16. Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells : A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarioglu AF, Aceto N, Kojic N, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stephenson D, Nahm C, Chua T, et al. Circulating and disseminated tumor cells in pancreatic cancer and their role in patient prognosis: A systematic review and meta-analysis. Oncotarget. 2017;8:107223–107236. doi: 10.18632/oncotarget.19928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurihara T, Itoi T, Sofuni A, et al. Detection of circulating tumor cells in patients with pancreatic cancer: A preliminary result. J Hepatobiliary Pancreat Surg. 2008;15:189–195. doi: 10.1007/s00534-007-1250-5. [DOI] [PubMed] [Google Scholar]
- 20. Scher HI, Graf RP, Schreiber NA, et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 2018;4:1179–1186. doi: 10.1001/jamaoncol.2018.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boffa DJ, Graf RP, Salazar MC, et al. Cellular expression of PD-L1 in the peripheral blood of lung cancer patients is associated with worse survival. Cancer Epidemiol Biomark Prev. 2017;26:1139–1145. doi: 10.1158/1055-9965.EPI-17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keup C, Suryaprakash V, Hauch S, et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. Genome Med. 2021;13:85. doi: 10.1186/s13073-021-00902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Radovich M, Jiang G, Hancock BA, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: Preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 2020;6:1410–1415. doi: 10.1001/jamaoncol.2020.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z, LaRiviere MJ, Ko J, et al. A multianalyte panel consisting of extracellular vesicle miRNAs and mRNAs, cfDNA, and CA19-9 shows utility for diagnosis and staging of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2020;26:3248–3258. doi: 10.1158/1078-0432.CCR-19-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bernard V, Kim DU, San Lucas FA, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. 2019;156:108–118.e4. doi: 10.1053/j.gastro.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Werner SL, Graf RP, Landers M, et al. Analytical validation and capabilities of the epic CTC platform: Enrichment-free circulating tumour cell detection and characterization. J Circ Biomark. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scher HI, Armstrong AJ, Schonhoft JD, et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer. 2021;150:83–94. doi: 10.1016/j.ejca.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 30.Harrell FE., Jr. rms: Regression Modeling Strategies. 2020. https://CRAN.R-project.org/package=rms [Google Scholar]
- 31.Therneau TM. A Package for Survival Analysis in R. 2020. https://CRAN.R-project.org/package=survival [Google Scholar]
- 32.Venables WN, Ripley BD. Modern Applied Statistics with S. ed 4. Springer; https://www.stats.ox.ac.uk/pub/MASS4/ [Google Scholar]
- 33. Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34. Ankeny JS, Court CM, Hou S, et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strijker M, Soer EC, de Pastena M, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int J Cancer. 2020;146:1445–1456. doi: 10.1002/ijc.32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uesato Y, Sasahira N, Ozaka M, et al. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PLoS One. 2020;15:e0235623. doi: 10.1371/journal.pone.0235623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors upon request, without undue reservation.