PURPOSE
ERBB2 copy number (CN), measured using next-generation sequencing, is a predictive biomarker for trastuzumab efficacy in human epidermal growth factor receptor 2 (HER2)–positive advanced esophagogastric and gastric cancer (AGC). We aimed to investigate the association of ERBB2 amplification and gene coalterations with response and resistance to trastuzumab-combined chemotherapy.
METHODS
The SCRUM-Japan GI-SCREEN was a comprehensive genomic profiling project of GI cancer tissues using Oncomine Cancer Research Panel and Oncomine Comprehensive Assay. From 885 patients with AGC who successfully underwent gene profiling, 74 with ERBB2 amplification (CN ≥ 4.0) and who received first-line trastuzumab-combined chemotherapy were selected, and ERBB2 CN and gene coalterations were assessed.
RESULTS
ERBB2 CN did not differ in tumor response to trastuzumab-combined chemotherapy (one-way analysis of variance test, P = .37). Multivariate analysis using the Cox proportional hazard model revealed that ERBB2 CN (continuous log2-converted CN, hazard ratio, 0.76; 95% CI, 0.62 to 0.93; P < .01) and receptor/oncogene amplifications in the HER2 signaling pathway (hazard ratio, 2.5; 95% CI, 1.2 to 5.3; P = .01) were significant predictors for progression-free survival (PFS). ERBB2 variants coexisted in five patients (7%) and were missense mutations. Two patients with low variant allele frequencies (VAFs; 8%, 12%) showed high ERBB2 CN (55, 80) and durable response (≥ 20 months), whereas three patients with high VAFs (66%–90%) showed low ERBB2 CN (8-11) and no response with short PFS (1-10 months).
CONCLUSION
ERBB2 CN and gene coamplification in the HER2 signaling pathway were positive and negative predictors of PFS in trastuzumab-treated HER2-positive AGC patients, respectively. HER2-positive AGC patients with a high VAF of ERBB2 showed poor outcomes and may need HER2 tyrosine kinase inhibitors and trastuzumab deruxtecan.
INTRODUCTION
ERBB2 is an oncogene encoding human epidermal growth factor receptor 2 (HER2), a receptor tyrosine kinase (RTK).1,2 Approximately 15% of advanced esophagogastric and gastric cancer (AGC) harbor ERBB2 amplification that causes overexpression of HER2 and subsequent oncogenesis and tumor progression. Trastuzumab, a humanized immunoglobulin G1 monoclonal antibody against HER2, inhibits ligand-independent activation of overexpressed HER2 RTK.3 Trastuzumab combined with fluoropyrimidine plus platinum showed a survival benefit for HER2-positive AGC and has become the first-line standard of care.4
CONTEXT
Key Objective
There have been a few reports comprehensively investigating ERBB2 copy number (CN) and gene coalterations of the human epidermal growth factor receptor 2 (HER2) signaling pathway in HER2-positive advanced esophagogastric and gastric cancer (AGC). In the SCRUM-Japan GI-SCREEN, a large-scale genomic profiling project of GI cancer tissues using next-generation sequencing, we studied ERBB2 amplification and gene coalterations in association with response and resistance to first-line trastuzumab-combined chemotherapy.
Knowledge Generated
ERBB2 CN and gene coamplification other than ERBB2 in the HER2 signaling pathway were positively and negatively associated with progression-free survival, respectively. A small number of patients with HER2-positive AGC had the ERBB2 sequence variant, and those with high variant allele frequency showed poor prognosis.
Relevance
ERBB2 CN and gene coamplification of the HER2 pathway are useful biomarkers for the prediction of trastuzumab-treatment outcomes, and the ERBB2 covariant should be further studied in the treatment selection for HER2-positive AGC.
Many studies regarding the prediction of trastuzumab efficacy and intrinsic/acquired resistance have demonstrated that amplified ERBB2 copy number (CN) is a predictor of prolonged progression-free survival (PFS) and overall survival (OS) in trastuzumab-combined chemotherapy and that gene coalterations such as oncogenes and tumor-suppressor genes in RTK/RAS/PI3K signaling pathways, cell cycle regulators including cyclin E1 (CCNE1) and cyclin-dependent kinase inhibitor 2A (CDKN2A), TP53, and MYC are resistant to trastuzumab therapy.5-12
Clinically, HER2 positivity is judged on the basis of degree of HER2 protein expression using immunohistochemistry (IHC3+) or in situ hybridization (ISH+) when the expression is equivocal (IHC2+).3 This approach is considered standard of care for the determination of patients with HER2-positive disease amenable to anti–HER2-directed therapy. In recent years, high ERBB2 CN has been reported to be an independent predictor of longer real-world PFS in patients with AGC treated with trastuzumab using next-generation sequencing (NGS) technology, and a significant association of continuous ERBB2 CN with survival was also demonstrated.12-14 Currently, commercial comprehensive genomic profiling can be used to investigate the relevance of gene alterations to the clinical benefits of chemotherapies in clinical practice. Thus, there is a growing need to clarify how information on gene alterations obtained by NGS can be used to better identify biomarker-selected patients uniquely poised to benefit from precision-guided therapies.
This study was a retrospective cohort and analysis that aimed to investigate the ERBB2 CN and gene coalterations in patients with HER2-positive AGC treated with trastuzumab-combined first-line chemotherapy, who were enrolled in a nationwide cancer genome screening project in Japan.
METHODS
Study Design and Patients
SCRUM-Japan GI-SCREEN (Cancer Genome Screening Project for Individualized Medicine in Japan) was a multicenter study of the comprehensive genomic profiling for GI cancer.14 The target gene sequences of tumor tissues were determined using NGS. In the gastric cancer cohort, patients who met the following criteria were eligible for NGS analysis: those with pathologically confirmed esophagogastric and gastric adenocarcinoma, with an Eastern Cooperative Oncology Group performance status (ECOG PS) score of 0-2, whose major organ functions were preserved for systemic chemotherapy, and who provided written informed consent for this study. The ethical, medical, and scientific aspects of this study were reviewed and approved by the institutional review board of each institution, and this study was registered in the University Hospital Medical Information Network Clinical Trials Registry (UMIN000016343). This study was conducted in accordance with the Declaration of Helsinki, as revised in 2000.
In this study, the clinical and genomic data of patients with ERBB2-amplified AGC who received first-line trastuzumab-combined chemotherapy were extracted from those enrolled in the SCRUM-Japan GI-SCREEN. The patients were treated according to the clinical practice guidelines in Japan.15 Trastuzumab was administered at a standard dose and schedule.
Data Collection and Assessments
The primary end point was to determine how ERBB2 amplification and gene coalterations were associated with the response and resistance to trastuzumab-combined chemotherapy. Data on the following clinical characteristics of eligible patients were collected: age, sex, ECOG PS score, presence of primary tumor before chemotherapy, number and location of metastatic organs, presence of ascites, method of tissue collection (tumor biopsy or surgical resection), tumor sites, histologic classification–based Lauren's criteria, HER2 score (IHC3+ or IHC2+ISH+), and concomitant chemotherapy regimen. Response rate, PFS, and OS of trastuzumab-combined first-line chemotherapy were assessed. Tumor response assessment was performed on the basis of the RECIST v1.1, complete response (CR), partial response (PR), stable disease, and progressive disease, if measurable lesions were present. PFS was defined as the time from the initiation of trastuzumab-combined chemotherapy to disease progression and/or death from any cause. OS was defined as the time from the initiation of trastuzumab-combined chemotherapy to death from any cause. Patients who were lost to follow-up were censored on the last response evaluation for PFS and on the last contact for OS.
Target Sequencing
Formalin-fixed and paraffin-embedded tumor tissues were sent to the Life Technologies Clinical Services Laboratory (910 Riverside Parkway, West Sacramento, CA). Tumor DNA and RNA were extracted and analyzed using the Ion Torrent Oncomine Cancer Research Panel (OCP) and Oncomine Comprehensive Assay v3 (OCA; Thermo Fisher Scientific, Waltham, MA). The target regions of 143 cancer-related genes were amplified by OCP analysis and 161 genes were amplified by OCA analysis (Data Supplement). Gene amplification was defined as a CN of ≥ 4.0. Gene mutations were called out if the allele frequency was ≥ 5% and alternate allele observation count of ≥ 10, excluding synonymous mutations. Gene mutations were solely oncogenic mutations determined using the Oncomine Knowledgebase and annotated using Iron Reporter software. The mutation loci were mapped using the MutationMapper of the cBioPortal.16,17 The data files were stored in the SCRUM-Japan Data Center.
Statistical Analyses
The difference between ERBB2 CN and the antitumor effect was evaluated using one-way analysis of variance. The optimal cutoff value of ERBB2 CN was determined by calculating the receiver operating characteristic (ROC) curve on the basis of the PFS rates at 12 and 24 months. Using an ERBB2 CN cutoff value, the background factors in the high and low groups were compared using the Fisher's exact test. Log2-converted ERBB2 CN was also used in every analysis. The patients were classified according to the log2 values from class 2 (CN 22-< 23) to class 7 (CN ≥ 27) for the Kaplan-Meier plots of PFS and OS. For PFS, the prognostic and predictive factors were examined by conducting a multivariate analysis using the Cox proportional hazards model. The log-rank test was used to compare the survival of the patient group. All tests were performed using the JMP version 16.0.0 software program (SAS International, Cary, NC) and EZR version 1.55.18 All P values were calculated using a two-sided test, and a P value of < .05 was considered significant.
RESULTS
Patients
From April 2015 to March 2019, 1,132 patients with AGC were enrolled. Among the 885 patients whose gene sequencing of tumor tissues was completed, 125 (14%) had ERBB2 amplification (Data Supplement). Thirty-four patients whose tissue samples were collected after chemotherapy, 14 who did not receive trastuzumab as first-line chemotherapy, and three with insufficient clinical data were excluded; overall, 74 patients who had ERBB2 amplification and received trastuzumab-combined first-line chemotherapy were finally included in this study. Comprehensive cancer genome profiling was performed using OCP and OCA in 45 and 29 patients, respectively.
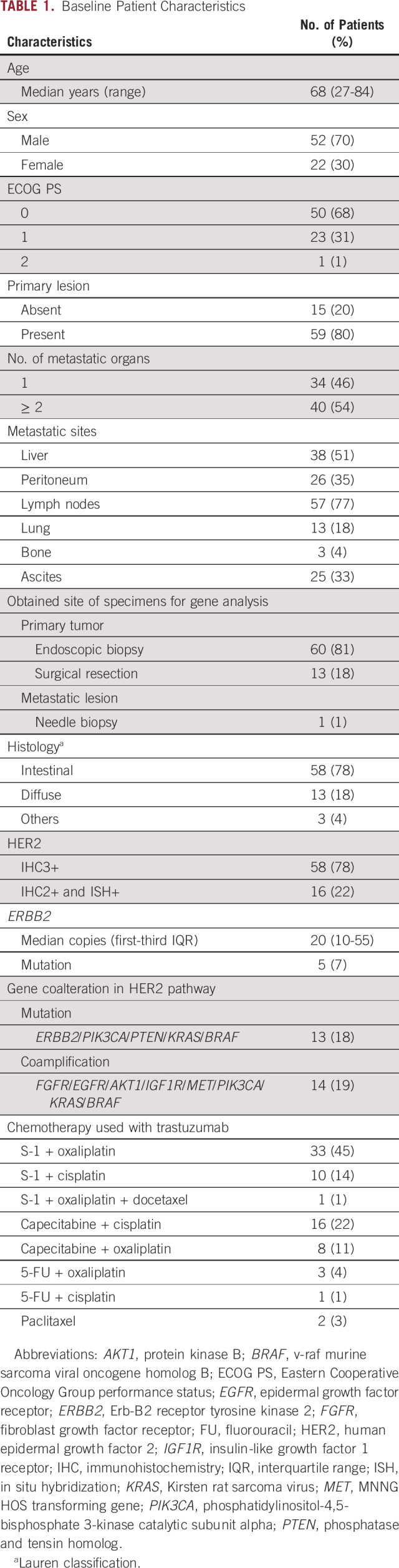
The patient's median age was 68 years (Table 1). Most patients had good general condition, with ECOG PS scores of 0 (68%) and 1 (31%). The tissue samples were mostly obtained from the primary tumor site. The histologic classifications were intestinal type in 58 patients (78%) and diffuse type in 13 patients (18%). HER2 IHC3+ tumors were observed in 58 patients (78%), whereas IHC2+ISH+ were observed in 16 patients (22%). The median ERBB2 CN was 20. Gene comutations (ERBB2, PIK3CA, PTEN, KRAS, and BRAF) and coamplifications (FGFR, EGFR, AKT1, IGF1R, MET, PIK3CA, KRAS, and BRAF) involved in the HER2 pathway were detected in 13 (18%) and 14 (19%) patients, respectively. Other amplifications were observed in CCNE1/CCND1/CDK6 (30%) and MYC (16%). Mutations were commonly observed in the TP53 (72%), PIK3CA/PTEN (8%), ERBB2 (7%), and KRAS/BRAF (3%) genes. All detected gene alterations were common in the OCP and OCA except for BRAF amplification (n = 1), which was not included in the OCP. As for the HER2 scores, none of the patients' characteristics including the proportion of gene coalterations in the HER2 pathway were significantly different between the IHC3+ and IHC2+/ISH+ groups, except for ERBB2 CN (median CN: 29 and 9, respectively; Wilcoxon test, P < .01).
TABLE 1.
Baseline Patient Characteristics
Tumor Response
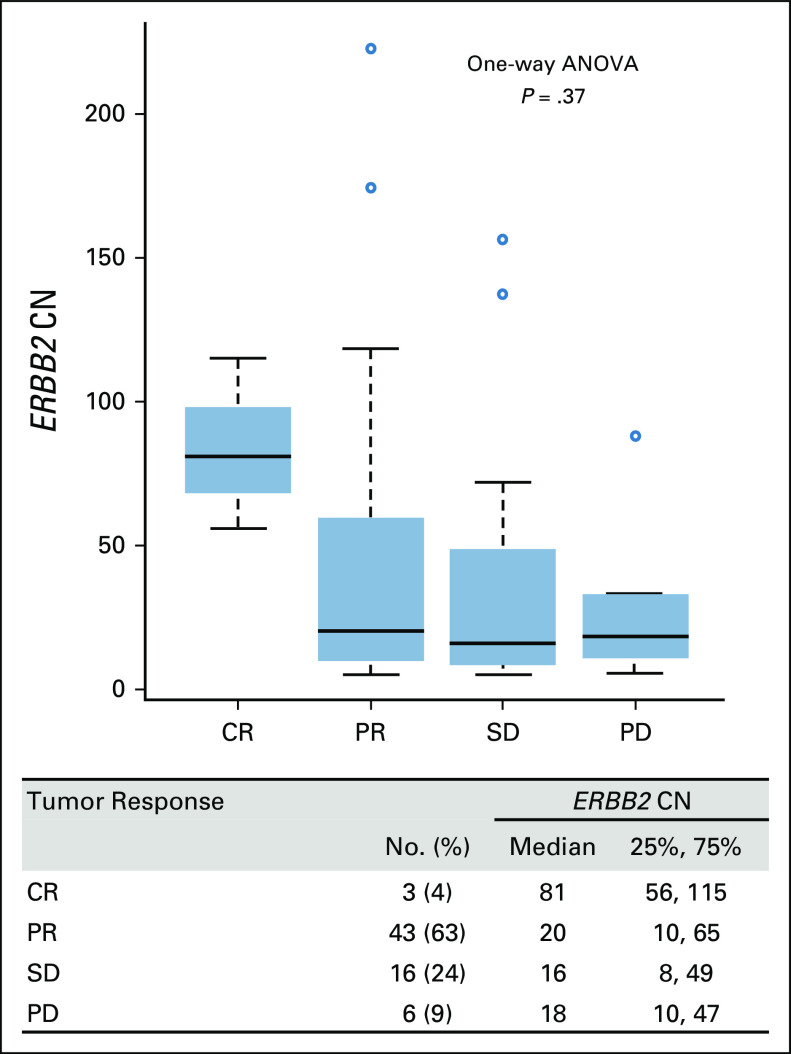
In 68 patients with measurable lesions, three achieved CR (4%), and 43 achieved PR (63%), resulting in a 67% response rate (Fig 1). Neither nonconverted nor log2-converted ERBB2 CN showed a significant difference in tumor response (one-way analysis of variance, P = .37 and P = .46, respectively).
FIG 1.
Tumor responses and ERBB2 gene CN of patients with measurable lesions (n = 68) receiving first-line chemotherapy containing trastuzumab for advanced HER2-positive esophagogastric and gastric cancer. ANOVA, analysis of variance; CN, copy number; CR, complete response; ERBB2, erb-b2 receptor tyrosine kinase 2; HER2, human epidermal growth factor receptor 2; PD, progressive disease; PR, partial response; SD, stable disease.
PFS and OS
The median PFS and OS were 8.8 months (95% CI, 7.4 to 11) and 29 months (95% CI, 20 to 38), respectively. The PFS and OS events occurred in 64 (86%) and 54 (73%) patients, respectively, until the data cutoff date of October 15, 2021.
In Figure 2, all 74 patients are sorted by PFS lengths, and their respective gene coalterations and ERBB2 CN are depicted. Majority of patients with a PFS of > 12 months achieved CR and PR after receiving a trastuzumab-combined chemotherapy. Patients who showed a long PFS of > 24 months had a high ERBB2 CN (mean log2 CN 5.2 v 4.4, Student's t test P = .04) and no gene coamplifications in the HER2 signaling pathway (Fig 2). Mutation of TP53, PTEN/PIK3CA, and ERBB2, and amplification of CCNE1/CCND1 and MYC distributed independently of PFS lengths, showing no association of these alterations with PFS lengths. There was a significant difference in the PFS between patients with and without gene coamplifications other than ERBB2 of the HER2 pathway (P = .038; Data Supplement).
FIG 2.
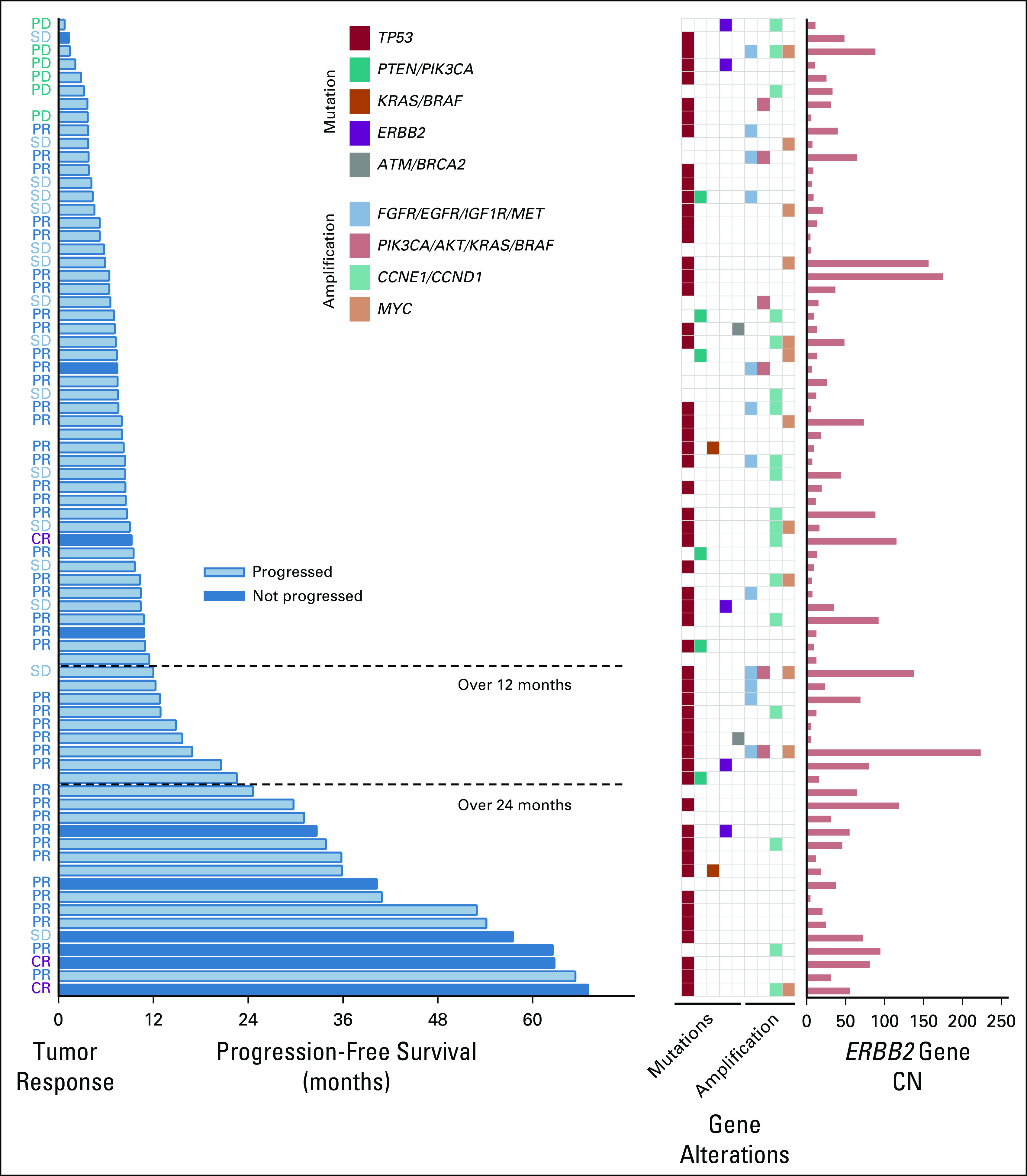
Efficacies and gene alterations in 74 patients treated with first-line chemotherapy containing trastuzumab for advanced HER2-positive esophagogastric and gastric cancer. AKT, protein kinase B; ATM, ataxia-telangiectasia mutated; BRAF, v-raf murine sarcoma viral oncogene homolog B; BRCA2, breast cancer susceptibility gene 2; CCND1, cyclin D1; CCNE1, cyclin E1; CN, copy number; CR, complete response; EGFR, epidermal growth factor receptor; ERBB2, erb-b2 receptor tyrosine kinase 2; FGFR, fibroblast growth factor receptor; HER2, human epidermal growth factor receptor 2; IGF1R, insulin-like growth factor 1 receptor; KRAS, Kirsten rat sarcoma virus; MET, MNNG HOS transforming gene; MYC, myelocytomatosis proto-oncogene; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PD, progressive disease; PR, partial response; PTEN, phosphatase and tensin homolog; SD, stable disease; TP53, tumor protein 53.
The Kaplan-Meier curves were created for each log class of ERBB2 CN. Patients with log classes 2 and 3 appeared to have shorter PFS than those with log class 4 or more except for log class 7 (Fig 3A). Among four patients with log class 7 tumors, two had gene coamplifications of the HER2 signaling pathway. The optimal cutoff values of ERBB2 CN for 12-month PFS and 24-month PFS on the basis of the ROC curve analyses were 15.2 copies (area under the curve: 0.64) and 16.6 copies (area under the curve: 0.68), respectively (Data Supplement). On the basis of these results, the cutoff value was determined to be CN 16 (log2 value = 4.0). Multivariate analyses of HER2 markers associated with PFS were performed using a Cox proportional hazards model. As shown in Figures 4A and 4B, continuous log2-converted ERBB2 CN (hazard ratio [HR], 0.71; 95% CI, 0.57 to 0.88; P < .01) and dichotomized ERBB2 CN at 16 (HR, 0.34; 95% CI, 0.19 to 0.63; P < .01) were significant predictive factors for PFS. In addition, receptor/oncogene amplification in the HER2 signaling pathway was a predictor of poor PFS in both settings (HR, 2.80; 95% CI, 1.38 to 5.69; P < .01, and HR, 2.73; 95% CI, 1.38 to 5.41; P < .01). Similar results were obtained when other clinical characteristics were included as variables (Data Supplement).
FIG 3.
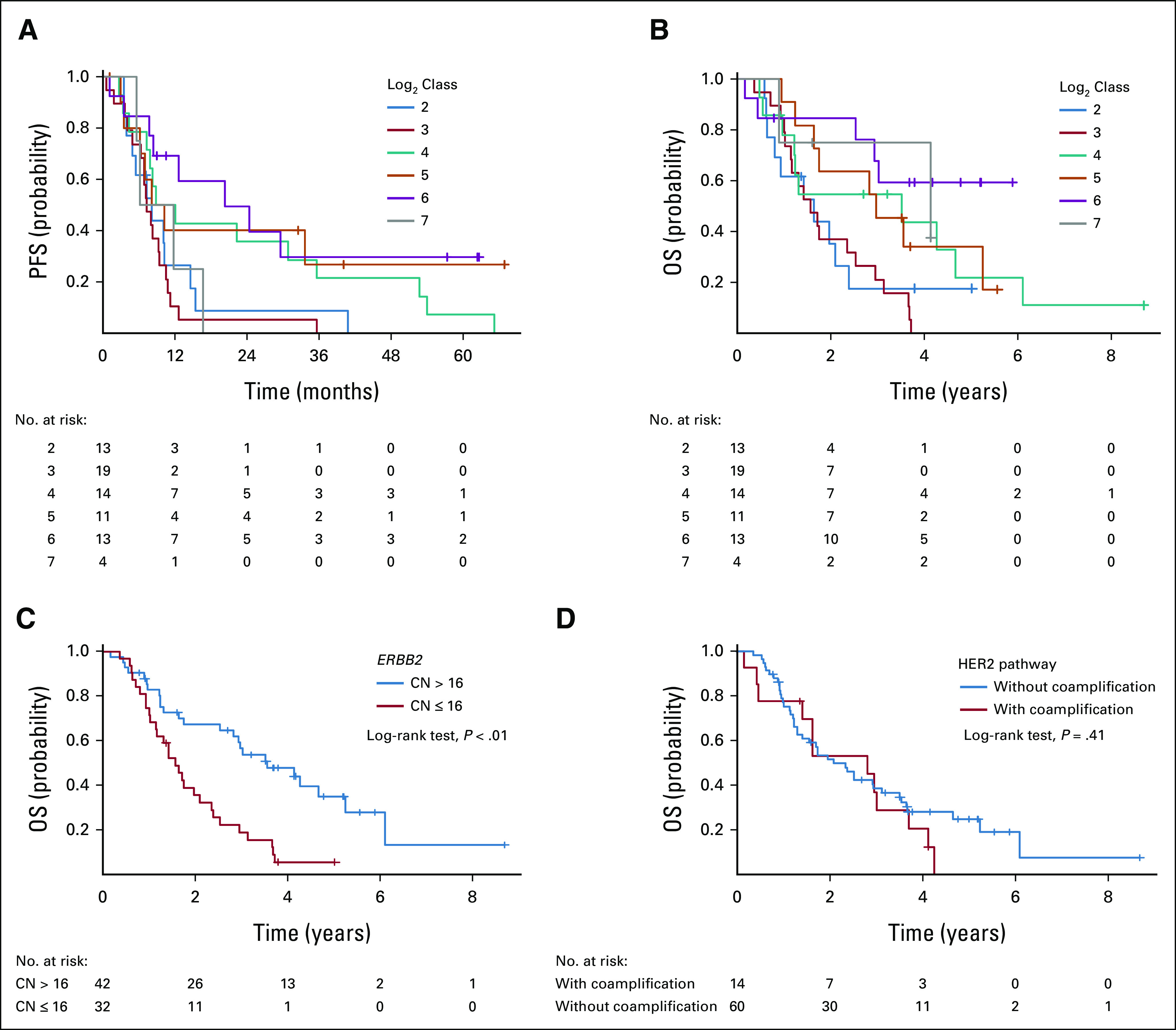
(A) PFS and (B) OS in patients with amplified ERBB2 according to log2-converted ERBB2 CN class. Overall survival according to (C) high/low CN (cutoff 16) and (D) with/without coamplification genes other than ERBB2 in the HER2 signaling pathway. CN, copy number; ERBB2, erb-b2 receptor tyrosine kinase 2; HER2, human epidermal growth factor receptor 2; OS, overall survival; PFS, progression-free survival.
FIG 4.
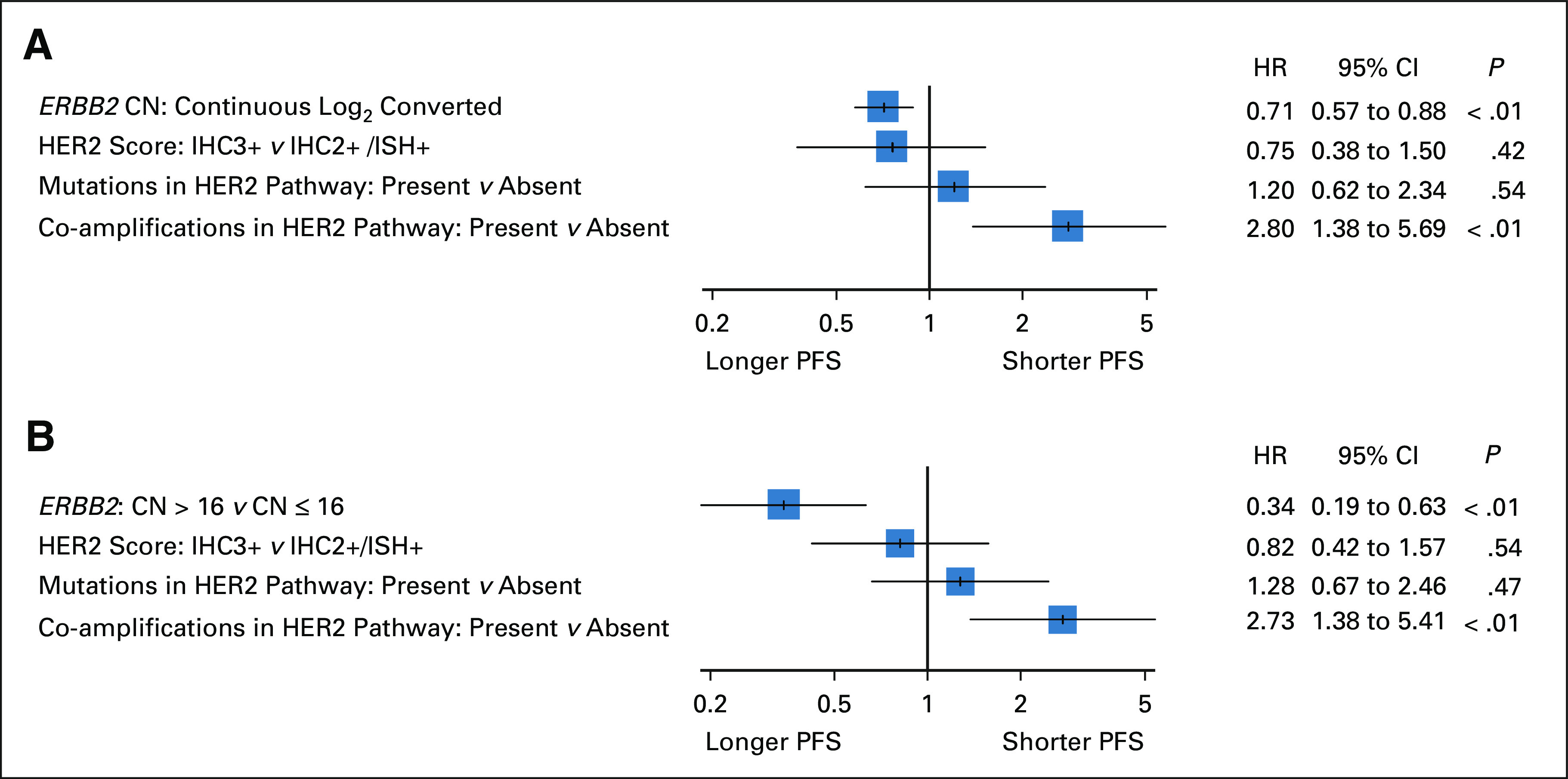
Multivariate analysis of HER2 biomarkers for PFS in 74 patients treated with first-line chemotherapy containing trastuzumab using (A) continuous log2-converted ERBB2 CN and (B) a cutoff value of ERBB2 CN 16. CN, copy number; ERBB2, erb-b2 receptor tyrosine kinase 2; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; IHC, immunohistochemistry; ISH, in situ hybridization; PFS, progression-free survival.
The Kaplan-Meier plots of log class ERBB2 CN for OS resembled those for PFS (Fig 3B). OS was significantly longer in patients with high ERBB2 CN (> 16) than in those with low CN (log-rank test, P < .01), whereas OS was not different between patients with and without amplified HER2 pathway genes (log-rank test, P = .41; Figs 3C and 3D).
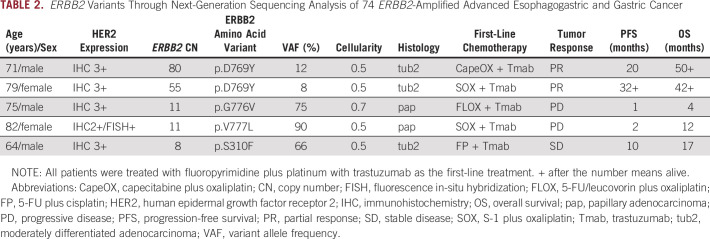
ERBB2 Covariants
Five patients (7%) had ERBB2 covariants (all missense mutations); their details are presented in Table 2. Two patients with low variant allele frequencies (VAFs) showed high ERBB2 CN, PR to trastuzumab-combined treatment, and long PFS and OS, whereas the remaining three patients with high VAFs showed low ERBB2 CN, short PFS and OS, and no response to trastuzumab-combined treatment. Four ERBB2 variants were located in the tyrosine kinase domain, whereas one variant was located in the furin-like cysteine-rich region (Data Supplement). In 42 cases of ERBB2 mutations included in seven publicly available databases of esophagogastric and gastric cancer (1,012 cases; 1,079 samples) from cBioPortal, many missense mutations were found in the tyrosine kinase domain, furin-like cysteine-rich region, and transmembrane/juxtamembrane domain (Data Supplement).12,19-24
TABLE 2.
ERBB2 Variants Through Next-Generation Sequencing Analysis of 74 ERBB2-Amplified Advanced Esophagogastric and Gastric Cancer
DISCUSSION
We demonstrated that ERBB2 CN was a predictor of prolonged PFS, and coamplification of receptor/oncogene that activated the HER2 signaling pathway was an independent predictor of shorter PFS in patients with HER2-positive AGC treated with trastuzumab-combined first-line chemotherapy. In addition, a small subgroup of patients with ERBB2 comutation was found, and those with a high VAF of ERBB2 presented poor outcomes, although it should be noted that the number of patients was limited.
In the ToGA study, which showed prolonged survival in patients treated with trastuzumab-combined chemotherapy, the treatment response rate increased by 12.8% after the addition of trastuzumab (47.3% v 34.5%).4 This finding indicated trastuzumab treatment shrunk tumor. Although this study included a small proportion of patients with HER2 IHC0-1+/fluorescence in-situ hybridization–positive, which was likely a contributor to the more mitigated difference in efficacy, the effect size of trastuzumab would be smaller, compared with the cytotoxic chemotherapy agents combined. This could be the reason why no significant difference in ERBB2 CN was detected in the tumor responses. However, patients with CR tended to have a higher ERBB2 CN (> 50). The remarkable response to trastuzumab may be influenced by the degree of tumor dependency on ERBB2 CN as a driving force for tumor growth. As for the survival data, the median PFS in this study (8.8 months) was similar to that in the ToGA study (6.7 months), whereas the median OS was apparently longer in our study (29 months v 13.8 months). This may be attributed to the different criteria for HER2 positivity and the recent expansion of subsequent treatment options after trastuzumab discontinuation.
ERBB2 CN as a continuous value showed a significant association with PFS in this study, as well as in two recent studies using the nationwide (US-based) deidentified Flartiron Health-Foundation clinic genomic database and Foundation Medicine genomic database for gastroesophageal adenocarcinoma.7,13 The former study demonstrated significant differences in the time to trastuzumab discontinuation and OS between patients with HER2 concordance (HER2+ and ERBB2 amplification+) and those with discordance (HER2+ and ERBB2 amplification–) and between patients with high ERBB2 CN (≥ median value, 25) and those with low ERBB2 CN.7 The latter showed that the association of ERBB2 CN with PFS was not observed in HER2-positive patients who were treated with chemotherapy without trastuzumab.13 Taken together, ERBB2 CN is a robust efficacy predictor of trastuzumab treatment in patients with HER2-positive AGC. Setting a common optimal cutoff value for ERBB2 CN seemed to be difficult, because the trastuzumab effect continuously emerged as the ERBB2 CN increased, and the NGS methods differed across studies. However, some reference values of ERBB2 CN, such as the median, quartile, and ROC-derived optimal values, can be used to predict the efficacy of trastuzumab. In this study, it was 16 copies of ERBB2, and this should be validated in the future. As for poor PFS in patients with log class 7, it might be due to the small number of patients and gene coamplifications on the HER2 signaling pathway.
Gene coalterations in the RTK/RAS/PI3K pathway and cell cycle checkpoints were reported to contribute to the intrinsic resistance to trastuzumab.9-12 Beside them, replacement of clones lacking ERBB2 amplification and newly detected mutation of ERBB2 were implicated in acquired resistance to trastuzumab.12,25 Gene amplification of receptor/oncogene that activated the HER2 pathway was an independent negative predictor of PFS in this study; this finding is in agreement with that of previous studies.9-12 The amplifications fortunately did not affect the OS; from this point of view, ERBB2 CN is a more clinically meaningful biomarker than the RTK, RAS, and PIK3CA amplification. In addition, approximately 40% of patients who had log class 4 or more (ERBB2 CN > 16) showed a PFS of more than 24 months (Fig 3A), and ERBB2 CN was a significant predictor for PFS, whereas IHC/ISH status was not in the multivariate analysis (Fig 4). These results suggested that ERBB2 CN was more useful than the IHC/ISH status in predicting the PFS of patients who were selected by the standard HER2 IHC test combined with ISH.
ERBB2 mutations occur in < 5% of patients with various types of carcinoma and 4.9%-7.7% in those with gastric cancer.26,27 ERBB2 mutation and amplification are mutually exclusive, and the co-occurrence is scarcely reported so far. Of note, co-occurrence was detected in 7% (5/74) of patients with HER2-positive AGC in this study, accounting for 1% of all patients with AGC if the HER2-positive rate was 15%. The mutation loci were distributed in the tyrosine kinase domain and furin-like cysteine-rich region. These were also the case in the cBioPortal database and all known as gain-of-function mutations.12,19-24 This study suggested an inverse correlation between the VAF of comutant ERBB2 and PFS/OS. Trastuzumab is considered ineffective in cells harboring high ERBB2 VAF, and other drugs such as antibody-drug conjugates (trastuzumab deruxtecan)28 and tyrosine kinase inhibitors (lapatinib, afatinib, neratinib, etc)2,26 should be used instead of trastuzumab or in combination with trastuzumab in this case.
This study has some limitations. This was a retrospective study with a small sample size. Validation of the results and comparison of the study group with a control cohort consisting of patients with HER2-positive AGC who did not receive trastuzumab were not performed. Among the examined gene alterations, BRAF amplification was not included in the OCP. HER2-positive disease constitutes a relatively small fraction of AGC; hence, enrollment of patients in clinical studies is difficult. However, similar results using NGS obtained from multiple study groups have been piled up, including those reported in this study, and further collaborations are needed worldwide.
In conclusion, ERBB2 CN and gene coamplification in the HER2 pathway were positive and negative predictors of PFS in trastuzumab-treated HER2-positive AGC patients. The orphan fraction of patients with a coalteration of ERBB2 amplification and mutation was identified, and further investigation is warranted.
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Lilly, Merck Serono, Takeda, Bristol Myers Squibb, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Dainippon Sumitomo Pharma (Inst), Lilly Japan (Inst), Merck Serono (Inst), MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Eisai (Inst)
Takeshi Kajiwara
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Ono Pharmaceutical, Lilly
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Takeda, Chugai Pharma, Sanofi, Yakult Honsha, Merck Serono, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan
Research Funding: Ono Pharmaceutical
Shigenori Kadowaki
Honoraria: Lilly, Bayer, Bristol Myers Squibb, Chugai Pharma, Ono Pharmaceutical, Merck KGaA, Daiichi Sankyo, Eisai, MSD, Taiho Pharmaceutical
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Lilly (Inst), MSD (Inst), Chugai Pharma (Inst), Nobelpharma (Inst), Janssen (Inst)
Keiko Minashi
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Astellas Pharma (Inst)
Satoshi Yuki
Honoraria: Chugai Pharma, Takeda, Lilly Japan, Bayer Yakuhin, Taiho Pharmaceutical, Sanofi, Yakult Honsha, Bristol Myers Squibb Japan, Ono Pharmaceutical, Merck, MSD K.K
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai, Yakult Honsha
Research Funding: Takeda
Hiroki Hara
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Yakult Honsha, Lilly, Ono Pharmaceutical, Takeda, Bristol Myers Squibb, Sanofi, MSD, Daiichi Sankyo, Kyowa Hakko Kirin, Bayer, Asahi Kasei
Consulting or Advisory Role: Ono Pharmaceutical, MSD, Boehringer Ingelheim, Dainippon Sumitomo, Bristol Myers Squibb Japan, Daiichi Sankyo/UCB Japan
Research Funding: AstraZeneca (Inst), Merck Serono (Inst), MSD (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Dainippon Sumitomo Pharma (Inst), Daiichi Sankyo (Inst), BeiGene (Inst), Astellas Pharma (Inst), Bayer (Inst), Amgen (Inst), Chugai Pharma (Inst), Janssen Oncology (Inst)
Takuro Mizukami
Honoraria: Taiho Pharmaceutical, Lilly Japan, Otsuka, Merck, Takeda, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Novartis, Ono Pharmaceutical, Bayer
Research Funding: Taiho Pharmaceutical, Ono Pharmaceutical, Lilly Japan
Toshikazu Moriwaki
Speakers' Bureau: Taiho Pharmaceutical, Chugai Pharma, Yakult Honsha, Takeda, Merck Serono, Sanofi, Lilly Japan, Bayer Yakuhin, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), MSD (Inst), Takeda (Inst), Yakult Honsha (Inst)
Masato Komoda
Honoraria: Eisai, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan, Daiichi Sankyo/UCB Japan, Taiho Pharmaceutical
Seiichiro Mitani
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical
Consulting or Advisory Role: Chugai Pharma
Research Funding: Taiho Pharmaceutical, Lilly
Fumio Nagashima
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Merck Serono, Takeda, Chugai Pharma, Yakult Honsha, Sumitomo Dainippon, Kyowa Hakko Kirin, Janssen
Research Funding: Taiho Pharmaceutical (Inst), Ono Pharmaceutical (Inst), OncoTherapy Science (Inst), Merck Serono (Inst), Zeria Pharmaceutical (Inst), Lilly Japan (Inst), Takeda (Inst), Chugai Pharma (Inst), Yakult Pharmaceutical (Inst), Sumitomo Dainippon (Inst), Daiichi Sankyo (Inst), Shionogi (Inst), Novartis (Inst), J-Pharma (Inst), Bristol Myers Squibb (Inst), Kyowa Hakko Kirin (Inst), Mochida Pharmaceutical Co. Ltd (Inst), Astellas Pharma (Inst), Bayer (Inst), MSD (Inst), Eisai (Inst), NanoCarrier (Inst), Janssen (Inst), Baxalta/Shire (Inst)
Ken Kato
Honoraria: Lilly, BMS, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical, BeiGene, MSD, Oncolys BioPharma, Bayer
Speakers' Bureau: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD
Research Funding: Ono Pharmaceutical (Inst), Shionogi (Inst), MSD Oncology (Inst), Beigene (Inst), Chugai Pharma (Inst), Bayer (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst)
Takanobu Yamada
Honoraria: Taiho Pharmaceutical, Johnson & Johnson/Janssen, Ono Pharmaceutical
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, MSD K.K
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst), Pfizer (Inst), Genomedia (Inst), Sysmex (Inst), Nippon Boehringer Ingelheim (Inst), Chugai Pharma (Inst)
Ichinosuke Hyodo
Honoraria: Taiho Pharmaceutical, Yakult Honsha, Ono Pharmaceutical
Consulting or Advisory Role: Asahi Kasei, Merck, Daiichi Sankyo Co, Ltd, Chugai Pharma, Taiho Pharmaceutical
No other potential conflicts of interest were reported.
SUPPORT
Supported by SCRUM-Japan Funds.
DATA SHARING STATEMENT
All data relevant to the study are included in this article or uploaded in the Data Supplement. All raw data used in the current study are available from the corresponding author on reasonable request.
AUTHOR CONTRIBUTIONS
Conception and design: Kaori Hino, Tomohiro Nishina, Takeshi Kajiwara, Ichinosuke Hyodo
Administrative support: Takeshi Kajiwara, Ken Kato
Provision of study materials or patients: Tomohiro Nishina, Takeshi Kajiwara, Hideaki Bando, Shigenori Kadowaki, Keiko Minashi, Takashi Ohta, Hiroki Hara, Koushiro Ohtsubo, Hiroko Hasegawa, Kentaro Yamazaki
Collection and assembly of data: Kaori Hino, Tomohiro Nishina, Takeshi Kajiwara, Hideaki Bando, Maho Nakamura, Shigenori Kadowaki, Keiko Minashi, Satoshi Yuki, Takashi Ohta, Hiroki Hara, Takuro Mizukami, Toshikazu Moriwaki, Koushiro Ohtsubo, Masato Komoda, Fumio Nagashima, Ken Kato, Takanobu Yamada, Hiroko Hasegawa, Kentaro Yamazaki, Takayuki Yoshino, Ichinosuke Hyodo
Data analysis and interpretation: Kaori Hino, Tomohiro Nishina, Takeshi Kajiwara, Hiroki Hara, Masato Komoda, Seiichiro Mitani, Fumio Nagashima, Kentaro Yamazaki, Ichinosuke Hyodo
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Lilly, Merck Serono, Takeda, Bristol Myers Squibb, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Dainippon Sumitomo Pharma (Inst), Lilly Japan (Inst), Merck Serono (Inst), MSD (Inst), Daiichi Sankyo (Inst), Ono Pharmaceutical (Inst), Eisai (Inst)
Takeshi Kajiwara
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Ono Pharmaceutical, Lilly
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Takeda, Chugai Pharma, Sanofi, Yakult Honsha, Merck Serono, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan
Research Funding: Ono Pharmaceutical
Shigenori Kadowaki
Honoraria: Lilly, Bayer, Bristol Myers Squibb, Chugai Pharma, Ono Pharmaceutical, Merck KGaA, Daiichi Sankyo, Eisai, MSD, Taiho Pharmaceutical
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Lilly (Inst), MSD (Inst), Chugai Pharma (Inst), Nobelpharma (Inst), Janssen (Inst)
Keiko Minashi
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Astellas Pharma (Inst)
Satoshi Yuki
Honoraria: Chugai Pharma, Takeda, Lilly Japan, Bayer Yakuhin, Taiho Pharmaceutical, Sanofi, Yakult Honsha, Bristol Myers Squibb Japan, Ono Pharmaceutical, Merck, MSD K.K
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai, Yakult Honsha
Research Funding: Takeda
Hiroki Hara
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Merck Serono, Yakult Honsha, Lilly, Ono Pharmaceutical, Takeda, Bristol Myers Squibb, Sanofi, MSD, Daiichi Sankyo, Kyowa Hakko Kirin, Bayer, Asahi Kasei
Consulting or Advisory Role: Ono Pharmaceutical, MSD, Boehringer Ingelheim, Dainippon Sumitomo, Bristol Myers Squibb Japan, Daiichi Sankyo/UCB Japan
Research Funding: AstraZeneca (Inst), Merck Serono (Inst), MSD (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Dainippon Sumitomo Pharma (Inst), Daiichi Sankyo (Inst), BeiGene (Inst), Astellas Pharma (Inst), Bayer (Inst), Amgen (Inst), Chugai Pharma (Inst), Janssen Oncology (Inst)
Takuro Mizukami
Honoraria: Taiho Pharmaceutical, Lilly Japan, Otsuka, Merck, Takeda, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Novartis, Ono Pharmaceutical, Bayer
Research Funding: Taiho Pharmaceutical, Ono Pharmaceutical, Lilly Japan
Toshikazu Moriwaki
Speakers' Bureau: Taiho Pharmaceutical, Chugai Pharma, Yakult Honsha, Takeda, Merck Serono, Sanofi, Lilly Japan, Bayer Yakuhin, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), MSD (Inst), Takeda (Inst), Yakult Honsha (Inst)
Masato Komoda
Honoraria: Eisai, Lilly Japan, Ono Pharmaceutical, Bristol Myers Squibb Japan, Daiichi Sankyo/UCB Japan, Taiho Pharmaceutical
Seiichiro Mitani
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical
Consulting or Advisory Role: Chugai Pharma
Research Funding: Taiho Pharmaceutical, Lilly
Fumio Nagashima
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Merck Serono, Takeda, Chugai Pharma, Yakult Honsha, Sumitomo Dainippon, Kyowa Hakko Kirin, Janssen
Research Funding: Taiho Pharmaceutical (Inst), Ono Pharmaceutical (Inst), OncoTherapy Science (Inst), Merck Serono (Inst), Zeria Pharmaceutical (Inst), Lilly Japan (Inst), Takeda (Inst), Chugai Pharma (Inst), Yakult Pharmaceutical (Inst), Sumitomo Dainippon (Inst), Daiichi Sankyo (Inst), Shionogi (Inst), Novartis (Inst), J-Pharma (Inst), Bristol Myers Squibb (Inst), Kyowa Hakko Kirin (Inst), Mochida Pharmaceutical Co. Ltd (Inst), Astellas Pharma (Inst), Bayer (Inst), MSD (Inst), Eisai (Inst), NanoCarrier (Inst), Janssen (Inst), Baxalta/Shire (Inst)
Ken Kato
Honoraria: Lilly, BMS, Ono Pharmaceutical
Consulting or Advisory Role: Ono Pharmaceutical, BeiGene, MSD, Oncolys BioPharma, Bayer
Speakers' Bureau: Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD
Research Funding: Ono Pharmaceutical (Inst), Shionogi (Inst), MSD Oncology (Inst), Beigene (Inst), Chugai Pharma (Inst), Bayer (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst)
Takanobu Yamada
Honoraria: Taiho Pharmaceutical, Johnson & Johnson/Janssen, Ono Pharmaceutical
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, MSD K.K
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst), Pfizer (Inst), Genomedia (Inst), Sysmex (Inst), Nippon Boehringer Ingelheim (Inst), Chugai Pharma (Inst)
Ichinosuke Hyodo
Honoraria: Taiho Pharmaceutical, Yakult Honsha, Ono Pharmaceutical
Consulting or Advisory Role: Asahi Kasei, Merck, Daiichi Sankyo Co, Ltd, Chugai Pharma, Taiho Pharmaceutical
No other potential conflicts of interest were reported.
REFERENCES
- 1. Arteaga CL, Engelman JA. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mitani S, Kawakami H. Emerging targeted therapies for HER2 positive gastric cancer that can overcome trastuzumab resistance. Cancers. 2020;12:400. doi: 10.3390/cancers12020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cutsem EV, Bang Y-J, Feng-Yi F, et al. HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer. 2015;18:476–484. doi: 10.1007/s10120-014-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bang Y-J, Cutsem EV, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 5. Kim J, Fox C, Peng S, et al. Preexisting oncogenic events impact trastuzumab sensitivity in ERBB2-amplified gastroesophageal adenocarcinoma. J Clin Invest. 2014;124:5145–5158. doi: 10.1172/JCI75200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haffner I, Schierle K, Raimúndez E, et al. HER2 expression, test deviations, and their impact on survival in metastatic gastric cancer: Results from the prospective multicenter VARIANZ study. J Clin Oncol. 2021;39:1468–1478. doi: 10.1200/JCO.20.02761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stein SM, Snider J, Ali SM, et al. Real-world association of HER2/ERBB2 concordance with trastuzumab clinical benefit in advanced esophagogastric cancer. Future Oncol. 2021;17:4101–4114. doi: 10.2217/fon-2021-0203. [DOI] [PubMed] [Google Scholar]
- 8. Gomez-Martin C, Plaza JC, Pazo-Cid R, et al. Level of HER2 gene amplification predicts response and overall survival in HER2-positive advanced gastric cancer treated with trastuzumab. J Clin Oncol. 2013;31:4445–4452. doi: 10.1200/JCO.2013.48.9070. [DOI] [PubMed] [Google Scholar]
- 9. Pietrantonio F, Fucà G, Morano F, et al. Biomarkers of primary resistance to trastuzumab in HER2-positive metastatic gastric cancer patients: The AMNESIA case-control study. Clin Cancer Res. 2018;24:1082–1089. doi: 10.1158/1078-0432.CCR-17-2781. [DOI] [PubMed] [Google Scholar]
- 10. Lee JY, Hong M, Kim ST, et al. The impact of concomitant genomic alterations on treatment outcome for trastuzumab therapy in HER2-positive gastric cancer. Sci Rep. 2015;19:9289. doi: 10.1038/srep09289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanchez-Vega F, Hechtman JF, Castel P, et al. EGFR and MET amplifications determine response to HER2 inhibition in ERBB2-amplified esophagogastric cancer. Cancer Discov. 2019;9:199–209. doi: 10.1158/2159-8290.CD-18-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janjigian YY, Sanchez-Vega F, Jonsson P, et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov. 2018;8:49–58. doi: 10.1158/2159-8290.CD-17-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L, Hamdani O, Gjoerup O, et al. ERBB2 copy number as a quantitative biomarker for real-world outcomes to anti–human epidermal growth factor receptor 2 therapy in advanced gastroesophageal adenocarcinoma. JCO Precis Oncol. 2022;6:e2100330. doi: 10.1200/PO.21.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura Y, Fujisawa T, Taniguchi H, et al. SCRUM-Japan GI-SCREEN and MONSTAR-SCREEN: Path to the realization of biomarker-guided precision oncology in advanced solid tumors. Cancer Sci. 2021;112:4425–4432. doi: 10.1111/cas.15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Japanese Gastric Cancer Association Japanese gastric cancer treatment guidelines 2018 (5th edition) Gastric Cancer. 2021;24:1–21. doi: 10.1007/s10120-020-01042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 20. Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 21. Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 22. Chen K, Yang D, Li X, et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112:1107–1112. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoadley KA, Yau C, Hinoue T, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janjigian YY, Maron SB, Chatila WK, et al. First-line pembrolizumab and trastuzumab in HER2-positive oesophageal, gastric, or gastro-oesophageal junction cancer: An open-label, single-arm, phase 2 trial. Lancet Oncol. 2020;21:821–831. doi: 10.1016/S1470-2045(20)30169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Makiyama A, Sukawa Y, Kashiwada T, et al. Randomized, phase II study of trastuzumab beyond progression in patients with HER2-positive advanced gastric or gastroesophageal junction cancer: WJOG7112G (T-ACT study) J Clin Oncol. 2020;38:1919–1927. doi: 10.1200/JCO.19.03077. [DOI] [PubMed] [Google Scholar]
- 26. Hyman DM, Piha-Paul SA, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554:189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park S, Ahn S, Kim DG, et al. High frequency of juxtamembrane domain ERBB2 mutation in gastric cancer. Cancer Genomics Proteomics. 2022;19:105–112. doi: 10.21873/cgp.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shitara K, Bang YJ, Iwasa S, et al. Trastuzumab deruxtecan in previously treated HER2-positive gastric cancer. N Engl J Med. 2020;382:2419–2430. doi: 10.1056/NEJMoa2004413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data relevant to the study are included in this article or uploaded in the Data Supplement. All raw data used in the current study are available from the corresponding author on reasonable request.