Abstract
We have found that the Aspergillus nidulans csmA gene encodes a novel protein which consists of an N-terminal myosin motor-like domain and a C-terminal chitin synthase domain (M. Fujiwara, H. Horiuchi, A. Ohta, and M. Takagi, Biochem. Biophys. Res. Commun. 236:75–78, 1997). To clarify the roles of csmA in fungal morphogenesis, we constructed csmA null mutants. The growth rate of the mutant colonies was almost the same as that of the wild-type strain, but hyphal growth was severely inhibited when a chitin-binding reagent, Calcofluor white or Congo red, was added to the medium. Moreover, morphological abnormalities in tip growth and septum formation were identified microscopically. Proliferation of intracellular new hyphae, called intrahyphal hyphae, which behaved as intrinsic hyphae, was the most striking phenotypic feature among them. These phenotypes were not suppressed when the only chitin synthase domain of csmA was expressed under the control of the alcA promoter, whereas they were suppressed when the intact form of csmA was expressed. Therefore, it was concluded that the product of csmA (CsmA) has important roles in polarized cell wall synthesis and maintenance of cell wall integrity and that the myosin motor-like domain is indispensable for these functions.
Chitin, a β-1,4-linked polysaccharide of N-acetylglucosamine, is a major structural component of fungal cell walls. To maintain the physical strength of cell walls and to form their specific shape, chitin metabolism, including synthesis, degradation, assembly, and cross-linking to other cell wall components, is thought to be very important for many fungi (2, 6, 8, 14). Chitin synthases (EC 2.4.1.16; chs gene products) are integral membrane proteins which catalyze polymerization of N-acetylglucosamine from UDP-N-acetylglucosamine as a substrate. In spite of difficulty in enzyme purification, enzymatic aspects and physiological roles of chitin synthases have been well characterized in the budding yeast Saccharomyces cerevisiae, in which chitin is a minor cell wall component and is localized mainly at the mother-daughter cell junction and septum (6, 8). On the other hand, there is little information about chitin synthases in filamentous fungi because of the existence of multiple isozymes in cells and technical limitations for general manipulations of filamentous fungi in molecular biology.
The primary structures of many fungal chitin synthases are mainly deduced from their gene structures, and they have been divided into five groups—classes I to V—on the basis of their structures in the conserved region (3, 5, 29). We have isolated four chitin synthase genes from an ascomycete, Aspergillus nidulans, and designated them chsA, chsB, chsC, and chsD, corresponding to classes II, III, I, and IV, respectively (21, 22, 34). Specht et al. (29) reported the isolation of the class IV chitin synthase gene, which they designated chsE, and it is almost identical to our chsD. To avoid confusion, Specht’s chsE is referred to as ‘chsE’ here). chsB is essential for normal hyphal growth, while the other genes can be disrupted without any morphological defects, at least in the asexual cycle (4, 21, 29, 34). In S. cerevisiae, class IV member Chs3p is responsible for 90% of chitin synthesis while the others, Chs1p and Chs2p, are involved in only small amounts of chitin synthesis at extreme parts of cells (6, 8). Thus, functions of chitin synthases in individual classes appear to be different between yeast and filamentous fungi. Recently, we cloned csmA from A. nidulans, which encodes a novel protein (1,852 amino acids, 206 kDa) consisting of a C-terminal class V chitin synthase domain and an extra N-terminal myosin motor-like domain (10). This suggests that there is a myosin fused to a metabolic enzyme in filamentous fungi. Specht et al. reported the isolation of a class V enzyme from A. nidulans, and they designated it chsD (29) (Specht’s chsD is referred to as ‘chsD’ here). ‘chsD’ would be a part of the csmA gene or its homologue, judging from their sequence similarity. A gene encoding a class V chitin synthase was cloned from Aspergillus fumigatus (1), and a csm-type gene was also isolated by our group (25).
Myosins are known to be motor proteins which can produce mechanical force to move along actin cables. Five myosin genes (MYO1 to MYO5) of S. cerevisiae have been cloned, and their functions have been analyzed intensely (32). Moreover, it was shown that Myo2p is essential for proper localization of Chs3p (28). Only one myosin gene (myoA) of A. nidulans has been cloned and analyzed (18). myoA belongs to myosin class I and is essential for growth and functions in secretion and endocytosis (18, 33). It has not been reported whether the localization of chitin synthases is disturbed in myoA mutants.
The finding of the novel structure of CsmA has generated new insight into the transport and localization of its enzymatic activity. In filamentous fungi, actin is concentrated in the form of fibers or patches at the hyphal tips, the septa, and the branching sites, where cell wall or septal synthesis is active (13, 14). In yeast, spatial organization of the actin cytoskeleton is apparently linked to the distribution of chitin (8). Thus, it is possible that some chitin synthases are incorporated into vesicles which are transported to the cell surface by myosins along actin cables, whereas CsmA is transported by its N-terminal myosin motor-like domain.
In this paper, we describe the functional analysis of csmA by construction of null mutants in which the myosin motor-like domain, as well as the chitin synthase domain, would be inactivated. csmA and csmA chsD null mutants (the latter was constructed to determine functional overlaps between the domains of classes IV and V) showed remarkable abnormalities in polarized growth, hyphal wall integrity, and conidiophore development. Among them, we found an interesting phenomenon of formation of intracellular hyphae, called intrahyphal hyphae. We constructed the strains in which the entire gene (CS4; alc-csm type) or the chitin synthase-encoding domain of csmA (CH5; alc-chs type) was placed under the control of the A. nidulans alcA promoter. These strains showed phenotypes similar to those of the csmA null mutant described above when they were cultured on medium containing glucose as the sole carbon source. CS4 (alc-csm type) grew normally, whereas CH5 (alc-chs type) showed abnormal morphology when cultured on medium containing threonine as the sole carbon source, where the alcA promoter is active. These results suggest that the N-terminal domain of CsmA functions as a myosin motor to localize CsmA to the active sites.
MATERIALS AND METHODS
Strains, media, and transformation.
The A. nidulans strains used in this study are listed in Table 1. Complete medium, YG medium (0.5% yeast extract, 1% glucose, 0.1% trace elements [26]), minimal medium (MM), and minimal medium containing 100 mM threonine instead of glucose (MMT) for A. nidulans (26, 29) were generally used. YG and MMT plates were YG and MMT containing 1.5% agar, respectively. Unless otherwise specified, MM and MMT were supplemented with arginine at 0.25 mg/ml, biotin at 0.2 mg/ml, pyridoxine at 0.25 mg/ml, and uridine at 2.44 mg/ml. In some experiments (see Fig. 9), uracil at 1.12 mg/ml was also added to MM and MMT plates (referred to MMU and MMUT, respectively). Transformation of A. nidulans were carried out as described previously (17). Transformants were grown in MM with appropriate supplements. To test the effects of reagents on the hyphal growth of A. nidulans strains, Calcofluor white (fluorescent brightener 28; Sigma Chemical Co.) and Congo red (Wako Pure Industries) were used to supplement media at about 47°C. Escherichia coli HB101 (27) was used as a host strain for plasmid amplification and was grown in Luria broth (27). E. coli transformation and plasmid extraction were performed by standard methods (27).
TABLE 1.
A. nidulans strains used in this study
| Strain(s) | Genotype | Source or reference |
|---|---|---|
| ABPU1 | pyrG89 biA1 wA3 argB2 pyroA4 | 22 |
| ABPU/A1 | pyrG89 biA1 wA3 argB2 pyroA4(pSS1) | 11 |
| ABPU/AU | pyrG89 biA1 wA3 argB2 pyroA4(pSS1)(pP1) | 22 |
| D3-2 | pyrG89 biA1 wA3 argB2 pyroA4 chsD::argB | 22 |
| M-1, M-9, M-16 | pyrG89 biA1 wA3 argB2 pyroA4 csmA::argB | This study |
| M2-5–M2-9 | pyrG89 biA1 wA3 argB2 pyroA4 csmA::argB | This study |
| CH5, CH9 | pyrG89 biA1 wA3 argB2 pyroA4 alcA(p)::csmAΔN | This study |
| CS3–CS5 | pyrG89 biA1 wA3 argB2 pyroA4 alcA(p)::csmA | This study |
| DM-3 | pyrG89 biA1 wA3 argB2 pyroA4 chsD::argB csmA::pyrG | This study |
| DM2-1–DM2-4 | pyrG89 biA1 wA3 argB2 pyroA4 chsD::argB csmA::pyrG | This study |
FIG. 9.
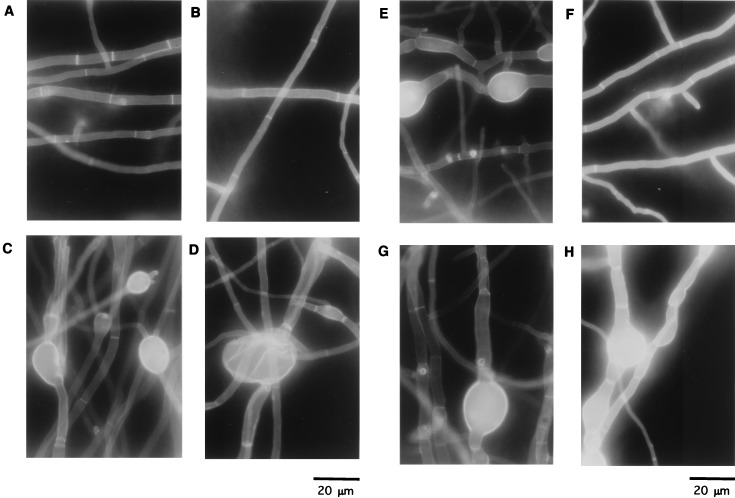
Calcofluor white staining of hyphae of the wild type and strains M2-6, CS4, and CH5. Strains ABPU/A1 (A and B), M2-6 (C and D), CS4 (E and F), and CH5 (G and H) were grown on an MMU plate (A, C, E, and G) or an MMUT plate (B, D, F, and H) for 4 days. Strains were stained with Calcofluor white and observed under a microscope.
Plasmid construction for csmA disruption.
The 1.8-kb BamHI-SphI fragment of pSS1 (21), containing the argB gene, and the 2.0-kb EcoRI-NdeI fragment of pJR15 (23), containing the pyrG gene, were blunted and ligated with BamHI- and SmaI-digested and blunted pMK10 (10) to yield pMΔA15 and pMΔG28, respectively. The 1.6-kb NdeI-XhoI fragment of pJR15 was ligated to NdeI and SalI-digested pMK10 to yield pMΔG1. The 1.0-kb XhoI fragment from pM3X2 (10) was blunted and ligated with SmaI-digested pSS1 to yield pSS-X7. The 2.0-kb StuI fragment from pMK10 was ligated with SphI-digested and blunted pSS-X7 to yield pMΔA16. The 0.4-kb EcoRI-PstI fragment of pAL3 (31; kind gift from N. R. Morris), containing the alcA promoter, was cloned into EcoRI- and PstI-digested pUC119 and designated pF1. The 1.8-kb SphI-XbaI fragment, containing the argB gene, was blunted and ligated with EcoRI-digested and blunted pF1 to yield pALC-arg/X. pM3X2 was digested with BamHI and self-ligated to yield pM3B5′. pM3B5′ was digested with NcoI, blunted, digested with BamHI, and ligated with BamHI- and SmaI-digested pALC-arg/X to yield pM-ALC. pM-ALC was linearized by partial digestion with PstI, blunted, and self-ligated to yield pM-ALCΔP, in which the PstI site on the vector was destroyed. A DNA fragment encoding the chitin synthase domain of CsmA was amplified by PCR using MM1 (5′-CGTTGGATCCGGGCGATATGTTTCAC-3′) and R1-5 (5′-CGATCGTTGCCTTGACCAATGATC-3′) as primers. The amplified fragment was digested with BamHI and HindIII and ligated with BamHI- and HindIII-digested pUC119 to yield pMM1. The 1.2-kb BamHI-HindIII fragment of pMM1 and the 2.3-kb HindIII-XbaI fragment of pMK10 were ligated with BamHI- and XbaI-digested pF1 to yield palc-M2. pSS-X7 was digested with VspI and SphI, blunted, and ligated with EcoRI-digested and blunted palc-M2 to yield pM-ALC-CHS5.
Disruption of the csmA gene and Southern analysis.
All strains were derived from ABPU1 (22), which was auxotrophic for arginine, biotin, pyridoxine, and uridine. A. nidulans argB and pyrG were used as selectable markers to complement arginine and uridine auxotrophy, respectively. ABPU/A1, used as a control strain (11), was obtained from ABPU1 by introducing one copy of the argB gene fragment of pSS1 into the genomic argB locus. Genomic total DNA was extracted as previously described (23). For Southern analysis, DNA labeling and detection were carried out with the enhanced-chemiluminescence system (Amersham Life Sciences, Amersham, United Kingdom). The A. nidulans csmA gene was disrupted in three ways as described below.
(i) Disruption of csmA by pMΔA15.
The 4.2-kb PstI-SphI fragment was used for transformation. By Southern analysis of HindIII- and XhoI-digested total DNA of 18 transformants probed with the 6.7-kb insert of pMK10, integration at the genomic csmA locus was confirmed in three strains named M-1, M-9, and M-16.
(ii) Disruption of csmA by pMΔA16.
The 4.6-kb VspI-SphI fragment was used. By Southern analysis of DraI- and XhoI-digested total DNA of about 100 transformants probed with the 7.0-kb XbaI fragment from pM3X2, integration at the csmA locus was confirmed in five strains named M2-5, M2-6, M2-7, M2-8, and M2-9.
(iii) Disruption of csmA by pMΔG1 or pMΔG28.
csmA of chsD null mutant D3-2 (22) was disrupted by transformation of the 3.7- and 4.4-kb PstI-SphI fragments from pMΔG1 and pMΔG28, respectively. By Southern analysis of XhoI-digested total DNA of about 50 transformants probed with the 6.7-kb insert of pMK10, integration at the csmA locus was confirmed in five strains named DM-3 (disrupted by pMΔG1), DM2-1, D2-2, D2-3, and D2-4 (disrupted by pMΔG28).
Construction of CS3 to 5 (alc-csm type).
Construction of CS3 to 5 was done as follows. PstI-digested pM-ALCΔP was transformed into ABPU1. Strains in which integration of the fragment into the csmA locus occurred were selected by Southern analysis (data not shown) and designated CS3 to CS5.
Construction of CH5 and CH9 (alc-chs type).
Construction of CH5 and CH9 was done as follows. The 6.3-kb NaeI-SpeI fragment of pM-ALC-CHS5 was transformed into ABPU1. Strains in which replacement of the fragment with chromosomal csmA occurred were selected by Southern analysis (data not shown) and designated CH5 and CH9.
Northern analysis.
Total RNA of A. nidulans was isolated with an RNeasy kit (QIAGEN) used in accordance with the manufacturer’s instruction. Northern analysis was done as described previously (10). The 0.4-kb EcoRV fragment of pM-ALC-CHS5 was used as a probe.
Stereomicroscopy and fluorescence microscopy.
Mycelia on plates were observed with a stereomicroscope (SZH10-141; Olympus Co., Tokyo, Japan) equipped with an automatic camera attachment (PM-10AK; Olympus). Mycelia stained with Calcofluor as previously described (35) were observed with a fluorescence microscope (BHS-RFK; Olympus) equipped with an automatic camera (PM-10ADS; Olympus).
Scanning electron microscopy.
Mycelia on plates were fixed for 1 h at 4°C with 2% glutaraldehyde, washed with phosphate-buffered saline (pH 7.4), dehydrated in a graded ethanol series (50, 70, 80, 90, and 99.5%), transferred to isoamyl acetate, and critical-point dried. Samples were coated with gold and observed with a Hitachi scanning electron microscope (HCP-2).
Transmission electron microscopy.
For ultrathin sections, mycelia obtained from plates were fixed for 5 h in 5% buffered glutaraldehyde (pH 7.0) with 0.1 M phosphate buffer and then postfixed for 2 h in buffered 1% osmium tetroxide. The whole fixation procedure was carried out at 4°C. After dehydration with a graded ethanol series, specimens were embedded in epoxy resin. The sections, cut on an ultramicrotome using a glass knife, were stained with uranyl acetate and lead citrate and observed in a JEOL transmission electron microscope (JEM-1010).
RESULTS
Targeting disruption of the csmA gene.
To clarify the function of csmA, we disrupted this gene in several ways (Fig. 1 and 2; see Materials and Methods). Two types of constructs with csmA disrupted were obtained, one which lacked almost the entire coding region of csmA (nullΔ type) and another which lacked the C-terminal 1,220 amino acids, expressing the N-terminal 630 amino acids (N630 type). We analyzed the phenotypes of a nullΔ-type strain (M2-6) and an N630-type strain (M-9). They showed similar phenotypes with respect to growth rate, sensitivities to various reagents, morphology of hyphae, and formation of intrahyphal hyphae (see below). ABPU/A1 (Table 1) was used as a control strain.
FIG. 1.
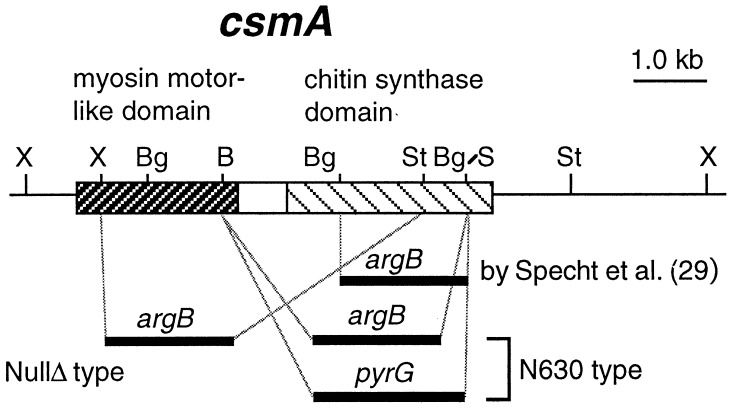
Schematic representation of csmA disruption. The N-terminal myosin motor-like domain and the C-terminal chitin synthase domain are indicated by two hatched boxes. The relative positions of deleted regions of csmA and of ‘chsD’ of Specht et al. (29) are shown with marker genes. Abbreviations for representative endonucleases are as follows: B, BamHI; Bg, BglII; S, SmaI; St, StuI; X, XhoI.
FIG. 2.
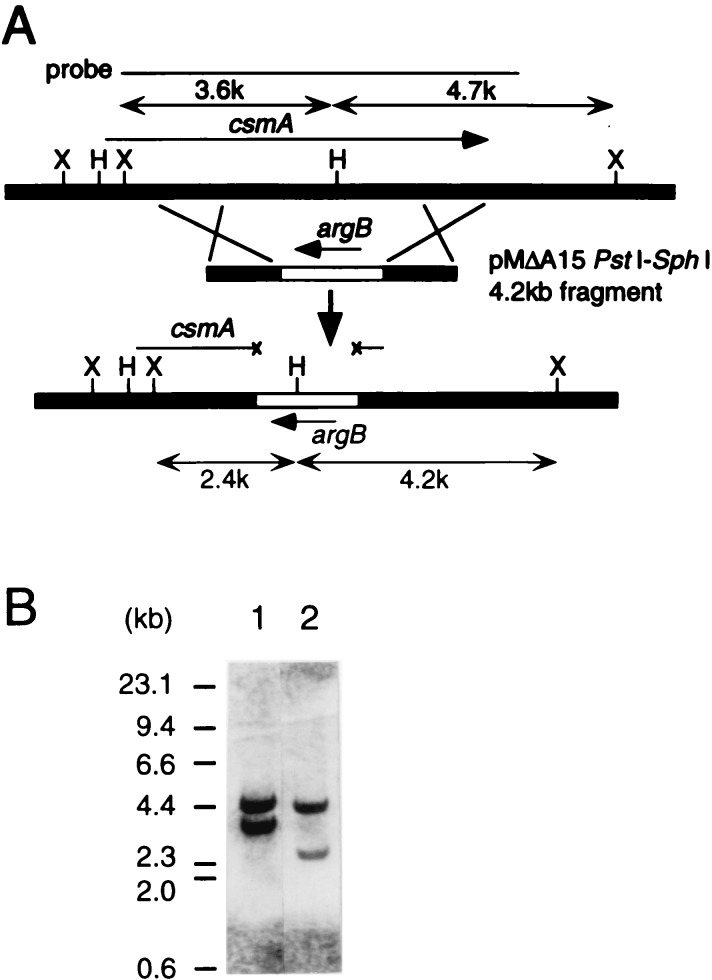
Construction of strain M-9. (A) Disruption of csmA by pMΔA15. The 4.2-kb PstI-SphI DNA fragment from pMΔA15 was used for transformation of A. nidulans ABPU1. The thin line at the top represents the probe used for Southern analysis. Representative restriction sites are abbreviated as follows: B, BamHI; E, EheI; H, HindIII; X, XhoI. (B) Southern analysis of HindIII- and XhoI-digested total DNA of strain M-9 probed with the 6.6-kb EheI-KpnI fragment from pMK10 (10). Band shifts from 3.6 and 4.7 kb (lane 1; ABPU1) to 2.4 and 4.2 kb (lane 2; M-9), respectively, were confirmed.
Growth defects of the csmA null mutant.
The colony growth rate of M-9 (N630 type) on MM plates was about 70 to 90% of that of ABPU/A1. Some hyphae of M-9 repeatedly undulated and extended over and under the agar surface at the edge of the growing colony (Fig. 3B). When mycelia were grown on medium coated with cellophane, this pattern of growth was not seen. Hyphae of M-9 also frequently lysed and excessively secreted pigments into the medium around the colony (Fig. 3D). Moreover, poorly developed aerial hyphae were commonly seen (Fig. 3D). Since these phenotypes indicated certain hyphal abnormalities, we tested the effects of various reagents on the colony growth rate and found that growth of M-9 was severely inhibited on medium containing low concentrations of the chitin-binding dyes Calcofluor white and Congo red (each at 1 to 5 μg/ml). The phenotypes described above were also observed in strain M2-6 (nullΔ type). Taken together, these results indicate that csmA is involved in hyphal wall assembly.
FIG. 3.
Colony edge and surface of the csmA null mutant. Mycelia on MM plates cultured for 3 days at 37°C were observed by stereomicroscopy. Panels: A and C, ABPU/A1 (wild-type strain); B and D, M-9 (ΔcsmA). Hyphal growth pattern, aerial hyphal development, and conidiophore formation are remarkably affected. Bars, 5 mm.
Balloon formation and conidiophore development.
Defects of cell wall assembly in strain M-9 were further analyzed by microscopy (Fig. 4). The hyphal wall should be mainly synthesized at hyphal tips, branching sites, and septa. The thickness of the hyphae of strain M-9 was frequently uneven (Fig. 4B), and depolarized swollen tubes or balloons appeared along hyphae, mainly in old regions (Fig. 4D, E, and H and 5C) and partly at the apical regions of hyphae (Fig. 4C). Frequency of balloon formation was also slightly reduced on plates with osmotic stabilizers. Bundles and fusions among hyphae were also frequently observed (data not shown). M2-6 showed almost the same phenotypes as M-9 in these respects.
FIG. 4.

Calcofluor staining of mycelia identified the pleiotropic morphological defects of the ΔcsmA mutant. Mycelia of M-9 grown on MM plates for 3 days at 37°C were observed. Panels: A, ABPU/A1; B to H, M-9; I, DM-3. Bars, 10 μm.
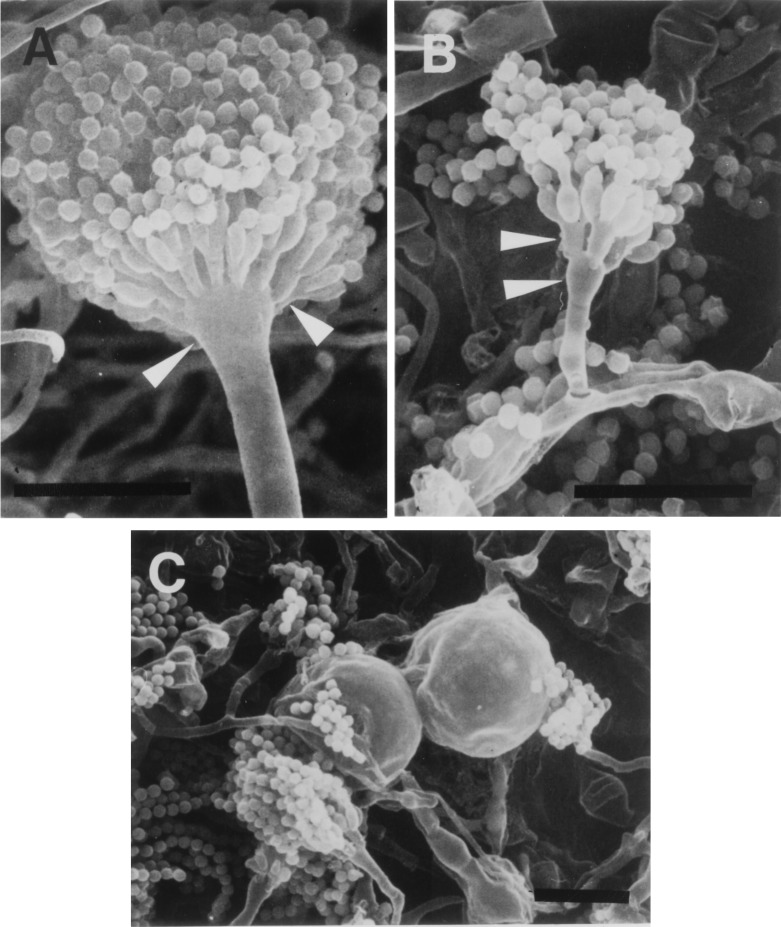
M-9 (N630 type) and M2-6 (nullΔ type) on MM plates formed few conidiophores (less than 0.1% of the population on a plate cultured for 3 days at 37°C). Abnormal morphologies of conidiophores (short stalks and a small population of metulae on vesicles) were observed occasionally in M-9 under this condition (Fig. 5B and C).
FIG. 5.
Scanning electron microscopic images of mycelia and conidiophores of the ΔcsmA mutant. Panels: A, ABPU/A1; B and C, M-9. Conidiophore structure abnormality was found in the short stalk, small vesicle (lower arrowheads), and a small population of metulae (upper arrowheads). Mycelia cultured for 7 days were used. Bars, 20 μm.
The described defects of growth and conidiophore formation of M-9 and M2-6 were remedied, but only slightly, on MM supplemented with osmotic stabilizers, sorbitol (1.2 M), sucrose (1 M), or NaCl (∼1.5 M) and on complete-medium plates.
Intrahyphal hypha and abnormal septum formation.
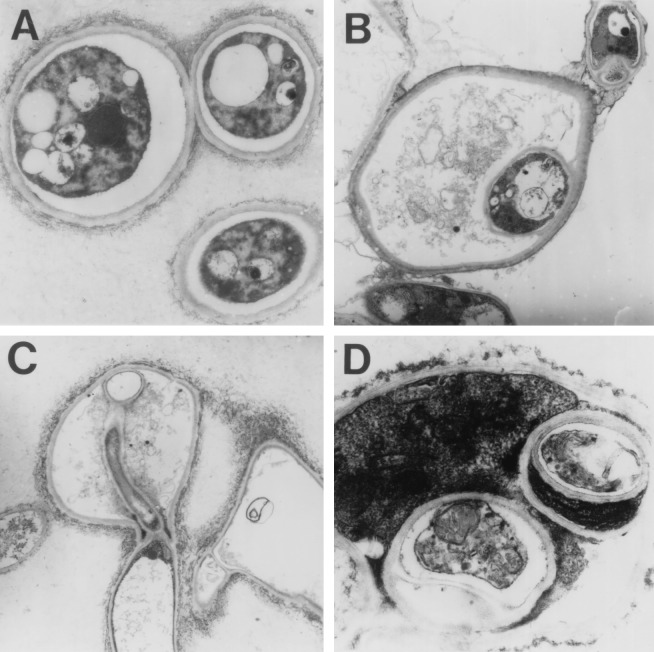
Formation of intrahyphal hyphae was the most striking morphological feature of M-9 and M2-6 (Fig. 4E to I and 6; data not shown). Intrahyphal hyphae, also known as intracellular hyphae and formerly observed in a number of fungi under various conditions (7, 17), were seen in old regions of hyphae. They could extend through other septa, penetrated the parental hyphae (Fig. 4I), and also appeared in balloons (Fig. 4E to H and 6B). Newly produced intrahyphal hyphae were thick and connected with the parental cell wall (Fig. 4F), but the most extended hyphae were thin and separated (Fig. 4G and I). Formation of intrahyphal hyphae may be derived from septa (Fig. 6C). The septa were sometimes structurally abnormal and positioned at irregular intervals (Fig. 4B to G), suggesting a role for CsmA in the control of spatial distribution of septa. Intrahyphal hyphae could grow apically, branch, septate, and also form intraintrahyphal hyphae as a second generation of intrahyphal hyphae (Fig. 6D). Transmission electron microscopic images suggested that in many cases, parental hyphae surrounding intrahyphal hyphae were highly vacuolated and almost dead on the basis of the low electron density (Fig. 6B). Intrahyphal hyphae were also observed, but less frequently, when M2-6 and M-9 were cultured on YG medium (data not shown).
FIG. 6.
Transmission electron microscopic images of intrahyphal hyphae. Panels: A, ABPU/A1; B to D, M-9; C, intrahyphal hyphae, which may have proliferated from septa; D, intraintrahyphal hyphae.
Expression of the chitin synthase domain.
To address the question of whether expression of only the chitin synthase domain could remedy the defects of a csmA null mutant, we constructed strains CH5 and CH9 (alc-chs type), in which the C-terminal part of csmA, encoding the chitin synthase domain, was expressed under the control of the alcA promoter of A. nidulans. Since the expression of the alcA promoter is repressed in the presence of glucose as a carbon source (MM and YG media) and induced in the presence of threonine as a carbon source (MMT), the chitin synthase domain of 992 amino acids was expressed in these strains only on an MMT plate. We also constructed CS strains (CS3 to CS5; alc-csm type) in which the whole protein-coding region of csmA was expressed under the control of the alcA promoter (see Materials and Methods; Fig. 7A and B). Since CH5, CH9, and CS3 to CS5 exhibited the same phenotypes in each construction under the conditions tested, we describe the phenotypes of CH5 and CS4 below. Northern analysis of total RNA from these strains is shown in Fig. 8. Transcripts of appropriate sizes were detected in CH5 (alc-chs type) and CS4 (alc-csm type) only when threonine was used as the sole carbon source (Fig. 8, lanes 1 versus 2 and 3 versus 4). On the other hand, no transcript was observed in M2-6 cultured under both conditions (Fig. 8, lanes 5 and 6). The phenotypes of these strains were quite similar to those of the csmA null mutant (M2-6) on an MMU plate (Fig. 9C, E, and G). The morphology of the hyphae of CS4 was almost the same as that of wild-type strain hyphae on an MMUT plate (Fig. 9F versus E). A few intrahyphal hyphae were observed in the wild-type hyphae on an MMUT plate and were also observed in CS4 (alc-csm type) cells at nearly the same frequency (data not shown). On the other hand, successive balloons and intrahyphal hyphae were frequently seen in CH5 (alc-chs type) on an MMUT plate (Fig. 9H) although their morphology was not identical to that of the null mutant hyphae. The same results were obtained when MM and MMT plates were used instead of MMU and MMUT plates, respectively (data not shown). These results suggest that the myosin motor-like domain is indispensable for the formation of normal-shaped hyphae (a detailed characterization of these strains will be presented elsewhere).
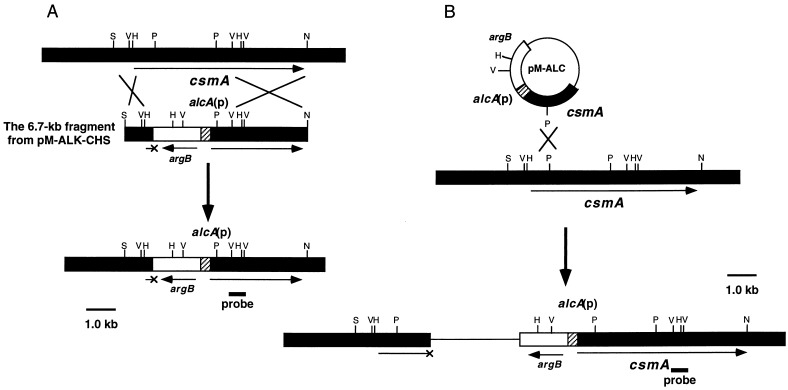
FIG. 7.
Constructions of strains CH5 (alc-chs type) (A) and CS4 (alc-csm type) (B). The 6.7-kb SpeI-NaeI fragment from pM-ALC-CHS5 (A) and PstI-digested pM-ALC (B) were used to transform A. nidulans ABPU1. The alcA promoter is shown as a hatched box. argB is shown as an open box. The small bar indicates the probe used for Northern analysis. Representative restriction site abbreviations: H, HindIII; V, EcoRV; P, PstI; N, NaeI; S, SpeI.
FIG. 8.

Northern analysis of total RNAs from CH5, CS4, and M2-6. CH5 (lanes 1 and 2), CS4 (lanes 3 and 4), and M2-6 (lanes 5 and 6) were grown in MMT medium (lanes 1, 3, and 5) for 5 days or in MM medium (lanes 2, 4, and 6) for 3 days. RNA was isolated as described in Materials and Methods. Approximately 3 μg of RNA was loaded on each lane. The 0.4-kb EcoRV fragment of pM-ALK-CHS was used as a probe. The positions of rRNAs are indicated by the bars on the left.
Double disruption of chsD and csmA.
The chitin synthase domain of CsmA is closely related to the class IV chitin synthases, and their cellular functions may partially overlap. We constructed csmA and nonessential class IV chitin synthase gene chsD double mutants by disruption of csmA in the chsD null mutant D3-2, which has no obvious defects in cell morphology (22) (see Materials and Methods). Disruption of csmA by pMΔG1 (the N-terminal 540 amino acids would be expressed [N540 type]) and pMΔG15 (N630 type) resulted in almost the same phenotype. We mainly analyzed strain DM-3 (N540 type) with ABPU/AU (22) as a control strain. The colony growth rate of strain DM-3 was about the same as that of the wild-type strain. Although inhibition of conidiophore formation by DM-3 was slightly restored compared with that of M-9 and M2-6, DM-3 exhibited the same phenotypes as M-9 and M2-6 with respect to the formation of balloons and intrahyphal hyphae (Fig. 4I).
DISCUSSION
Our finding of a novel structure of the newly isolated chitin synthase gene has suggested a direct link between the cytoskeleton and cell wall chitin synthesis (10). In this study, we constructed A. nidulans strains which lack both the functional myosin motor-like and chitin synthase domains and characterized them with respect to growth and morphology. We also constructed strains CS4 (alc-csm type) and CH5 (alc-chs type) and showed that CS4 formed hyphae with normal morphology whereas CH5 could not do so on an MMT plate.
Strains M-9 (nullΔ type) and M2-6 (N630 type) showed aberrant growth patterns on solid medium and formed balloons in their hyphae. Hyphal growth exhibited hypersensitivity to chitin-binding reagents. These phenotypic changes indicated certain involvement of CsmA in hyphal tip growth. However, abnormalities were predominantly seen in old parts of hyphae, such as balloon formation along hyphae, spatially irregular and structurally aberrant septum formation, small populations of conidiophores, and intrahyphal hypha proliferation. Balloon formation was slightly suppressed by osmotic stabilizers, suggesting that balloons are produced from the swelling of the weakened lateral wall and partially by turgor pressure. Thus, CsmA would be important in the maintenance of hyphal wall integrity and polarized cell wall synthesis, especially under low osmotic pressure conditions. Developmental defects of M-9 and M2-6 on MM and YG plates suggested that CsmA was also involved in asexual development under these conditions. Since inappropriate assembly of the hyphal cell wall may cause uncoordinated expression of the development regulatory genes, these defects may be derived from it. Polyoxin D and nikkomycin Z, competitive inhibitors of chitin synthases (9, 12), cause a marked reduction in conidiophore formation and swellings along hyphae (at concentrations of 1 to 50 μg/ml of medium) of wild-type A. nidulans (data not shown). These phenotypes are very similar to those of M2-6 and M-9. Thus, it is possible that the main target of nikkomycin Z in vivo is the chitin synthase domain of CsmA. Inactivation of other known chitin synthase genes did not show these phenotypes (11, 22, 34), although ChsB activity was sensitive to polyoxin D (30).
As far as we know, this study reports for the first time that disruption of a known gene induces intrahyphal hypha formation. Aberrant septum structures were also seen in M2-6 and M-9. These phenotypes were not observed in CS4 (alc-csm type), but similar phenotypes were detected in CH5 (alc-chs type) on MMUT and MMT plates, so it is suggested that they reflect the failure of spatial control of cell wall synthesis due to loss of the N-terminal function of CsmA. The myosin motor-like domain of CsmA could play an important role in the proper localization of CsmA and other associated factors at sites of cell wall synthesis by association with cytoskeletal structures. Recently, Osherov et al. (24) reported that irregular septa were formed in some myoA mutants. Thus, formation of septa could be regulated by the coordinated functions of MyoA and CsmA. However, formation of intrahyphal hyphae may also be derived from the intracellular condition of old mycelia because the normal septum structure and only a few intrahyphal hyphae were seen in young hyphae. In this case, CsmA might function in chitin metabolism in the septated hyphal compartments of the old regions. Some researchers have reported the presence of intrahyphal hyphae and have discussed their origins (7, 15), including two examples in ascomycetes, A. niger van Tiegh and a “clock” mutant of Neurospora crassa (16, 20), but there is no information at the molecular level on their origins.
There appeared to be little phenotypic difference between csmA and csmA chsD mutants. This suggests that the functional overlap between these products is small. It is in contrast to an apparent functional overlap between chsA and chsD (22). Some functional overlap was observed in this case.
Disruption of the chitin synthase-conserved region of ‘ChsD’ of Specht et al. (Fig. 2) caused severe defects in growth and morphology (29). Colony growth of the ‘chsD’ and ‘chsD chsE’ null mutants on dilute YG medium was severely inhibited. The morphological defects were seen in swellings along hyphae and vesicles. These defects were osmotically remediable. We tested M-9 and M2-6 under the same condition and obtained similar results with regard to growth and conidiophore development. However, the intrahyphal hypha proliferation observed under this condition at low frequency was not reported by Specht et al. (29) (data not shown).
Since CsmA, as well as other chitin synthases, is thought to have transmembrane domains, based on hydropathy analysis, it is possible that the N-terminal domain of CsmA has a role in spatial regulation of the CsmA-containing membrane vesicles, e.g., in exocytotic and/or endocytotic pathways, as well. Moreover, the N-terminal domain of CsmA shows structural differences from that of other members of the myosin superfamily (10), including lack of an IQ motif and of an apparent actin-binding domain in the molecule. Biochemical and genetic characterization of the myosin motor-like domain of CsmA is important.
The CsmA-type and class III chitin synthases have been found only in filamentous fungi. Class III members A. nidulans ChsB, A. fumigatus ChsC and ChsG, and N. crassa Chs-1 have been shown to have critical roles in hyphal growth (4, 19, 34, 35). As shown in this study, CsmA has critical roles in fungal morphogenesis. Recently, we cloned a csmA homologue (csm1) from the rice blast fungus Pyricularia oryzae (25). It is suggested that genes encoding CsmA-type proteins exist in many filamentous fungi which contain chitin as the major cell wall component and have important roles in normal hyphal growth.
ACKNOWLEDGMENTS
We thank Hirofumi Nakamura for assistance with the scanning electron microscope.
This work was performed by using the facilities of the Biotechnology Research Center, The University of Tokyo, and was supported by grants from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Aufauvre-Brown A, Mellado E, Gow N A R, Holden D W. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiaeand important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet Biol. 1997;21:141–152. doi: 10.1006/fgbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 2.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 3.Beth Din A, Specht C A, Robbins P W, Yarden O. chs-4, a class IV chitin synthase gene from Neurospora crassa. Mol Gen Genet. 1996;250:214–222. doi: 10.1007/BF02174181. [DOI] [PubMed] [Google Scholar]
- 4.Borgia P T, Iartchouk N, Riggle P J, Winter K R, Koltin Y, Bulawa C E. The chsB gene of Aspergillus nidulansis necessary for normal hyphal growth and development. Fungal Genet Biol. 1996;20:193–203. doi: 10.1006/fgbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 5.Bowen A R, Chen-Wu J L, Momany M, Young R, Szaniszlo P J, Robbins P W. Classification of fungal chitin synthases. Proc Natl Acad Sci USA. 1992;89:519–523. doi: 10.1073/pnas.89.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulawa C E. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- 7.Buller A H R. Researches on fungi. V. New York, N.Y: Hafner Publishing; 1958. [Google Scholar]
- 8.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debono M, Gordee R S. Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol. 1994;48:471–497. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara M, Horiuchi H, Ohta A, Takagi M. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem Biophys Res Commun. 1997;236:75–78. doi: 10.1006/bbrc.1997.6907. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara, M., T. Motoyama, H. Horiuichi, A. Ohta, and M. Takagi. Unpublished data.
- 12.Georgopapadakou N H, Tkacz J S. The fungal cell wall as a drug target. Trends Microbiol. 1995;3:98–104. doi: 10.1016/s0966-842x(00)88890-3. [DOI] [PubMed] [Google Scholar]
- 13.Harris S D, Morrell J L, Hammer J E. Identification and characterization of Aspergillus nidulansmutants defective in cytokinesis. Genetics. 1993;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath I B. Tip growth in plant and fungal cells. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 15.Lim L L, Fineran B A, Cole A L J. Ultrastructure of intrahyphal hyphae of Glomus fasciculatum (thaxter) Gerdemann and Trappe in roots of white clover (Trifolium repensL.) New Phytol. 1983;95:231–239. [Google Scholar]
- 16.Lowry R J, Sussman A S. Intra-hyphal hyphae in “clock” mutants of Neurospora. Mycologia. 1966;58:541–548. [PubMed] [Google Scholar]
- 17.May G. Fungal technology. In: Kinghorn J R, Turner G, editors. Applied molecular genetics of filamentous fungi. London, England: Chapman & Hall; 1992. pp. 1–27. [Google Scholar]
- 18.McGoldrick C A, Gruver C, May G S. myoA of Aspergillus nidulansencodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellado E, Aufauvre-Brown A, Gow N A R, Holden D W. The Aspergillus fumigatus chsC and chsGgenes encode class III chitin synthases with different functions. Mol Microbiol. 1996;20:667–679. doi: 10.1046/j.1365-2958.1996.5571084.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller C V, Anderson N A. Proliferation of conidiophores and intrahyphal hyphae in Aspergillus niger. Mycologia. 1961;53:433–436. [Google Scholar]
- 21.Motoyama T, Kojima N, Horiuchi H, Ohta A, Takagi M. Isolation of a chitin synthase gene (chsC) of Aspergillus nidulans. Biosci Biotechnol Biochem. 1994;58:2254–2257. doi: 10.1271/bbb.58.2254. [DOI] [PubMed] [Google Scholar]
- 22.Motoyama T, Fujiwara M, Kojima N, Horiuchi H, Ohta A, Takagi M. The Aspergillus nidulans genes chsA and chsDencode chitin synthases which have redundant functions in conidia formation. Mol Gen Genet. 1996;251:442–450. doi: 10.1007/BF02172373. . (Corrected and republished, 253:520–528, 1997). [DOI] [PubMed] [Google Scholar]
- 23.Oakley C E, Weil C F, Kretz P L, Oakley B R. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- 24.Osherov N, Yamashita R A, Chung Y-S, May G S. Structural requirements for in vivo myosin I function in Aspergillus nidulans. J Biol Chem. 1998;273:27017–27025. doi: 10.1074/jbc.273.41.27017. [DOI] [PubMed] [Google Scholar]
- 25.Park I C, Horiuchi H, Hwang C W, Yeh W H, Ohta A, Ryu J C, Takagi M. Isolation of csm1 encoding a class V chitin synthase with a myosin motor-like domain from the rice blast fungus, Pyricularia oryzae. FEMS Microbiol Lett. 1999;170:131–139. doi: 10.1111/j.1574-6968.1999.tb13365.x. [DOI] [PubMed] [Google Scholar]
- 26.Rowlands R T, Turner G. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans. Mol Gen Genet. 1973;126:201–216. doi: 10.1007/BF00267531. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Specht C A, Liu Y L, Robbins P W, Bulawa C E, Iartchouk N, Winter K R, Riggle P J, Rhodes J C, Dodge C L, Culp D W, Borgia P T. The chsD and chsE genes of Aspergillus nidulansand their roles in chitin synthesis. Fungal Genet Biol. 1996;20:153–167. doi: 10.1006/fgbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 30.Tatsuno K, Yamada-Okabe H, Takagi M, Arisawa M, Sudoh M. Properties of yeast expressed Aspergillus nidulanschitin synthase B which is essential for hyphal growth. FEMS Microbiol Lett. 1997;149:279–284. doi: 10.1111/j.1574-6968.1997.tb10341.x. [DOI] [PubMed] [Google Scholar]
- 31.Waring R B, May G S, Morris N R. Characterization of an inducible expression system in Aspergillus nidulans using alcAand tubulin-coding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 32.Winsor B, Schiebel E. An overview of the Saccharomyces cerevisiaemicrotubule and microfilament cytoskeleton. Yeast. 1997;13:399–434. doi: 10.1002/(SICI)1097-0061(199704)13:5<399::AID-YEA126>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita R A, May G S. Constitutive activation of endocytosis by mutation of myoA, the myosin I gene of Aspergillus nidulans. J Biol Chem. 1998;273:14644–14648. doi: 10.1074/jbc.273.23.14644. [DOI] [PubMed] [Google Scholar]
- 34.Yanai K, Kojima N, Takaya N, Horiuchi H, Ohta A, Takagi M. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci Biotechnol Biochem. 1994;58:1828–1835. doi: 10.1271/bbb.58.1828. [DOI] [PubMed] [Google Scholar]
- 35.Yarden O, Yanofsky C. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 1991;5:2420–2430. doi: 10.1101/gad.5.12b.2420. [DOI] [PubMed] [Google Scholar]