Abstract
We have cloned a 3.6-kb genomic DNA fragment from Pseudomonas aeruginosa harboring the rpoA, rplQ, katA, and bfrA genes. These loci are predicted to encode, respectively, (i) the α subunit of RNA polymerase; (ii) the L17 ribosomal protein; (iii) the major catalase, KatA; and (iv) one of two iron storage proteins called bacterioferritin A (BfrA; cytochrome b1 or b557). Our goal was to determine the contributions of KatA and BfrA to the resistance of P. aeruginosa to hydrogen peroxide (H2O2). When provided on a multicopy plasmid, the P. aeruginosa katA gene complemented a catalase-deficient strain of Escherichia coli. The katA gene was found to contain two translational start codons encoding a heteromultimer of ∼160 to 170 kDa and having an apparent Km for H2O2 of 44.7 mM. Isogenic katA and bfrA mutants were hypersusceptible to H2O2, while a katA bfrA double mutant demonstrated the greatest sensitivity. The katA and katA bfrA mutants possessed no detectable catalase activity. Interestingly, a bfrA mutant expressed only ∼47% the KatA activity of wild-type organisms, despite possessing wild-type katA transcription and translation. Plasmids harboring bfrA genes encoding BfrA altered at critical amino acids essential for ferroxidase activity could not restore wild-type catalase activity in the bfrA mutant. RNase protection assays revealed that katA and bfrA are on different transcripts, the levels of which are increased by both iron and H2O2. Mass spectrometry analysis of whole cells revealed no significant difference in total cellular iron levels in the bfrA, katA, and katA bfrA mutants relative to wild-type bacteria. Our results suggest that P. aeruginosa BfrA may be required as one source of iron for the heme prosthetic group of KatA and thus for protection against H2O2.
Bacterial aerobic respiration involves a four-electron reduction of molecular oxygen (O2) to water. Depending upon the environmental conditions, aerobic respiration can be extremely dangerous to the cell. Such is the case when aberrant electron flow from the electron transport chain or cellular redox enzymes to O2 leads to the production of reactive oxygen intermediates (ROIs). These include superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (HO·). The unchecked production of each of these species can lead to cell damage, mutations, or death. The production of HO·, the most destructive of the above compounds, is dependent in part upon the presence of a transition metal, such as iron or copper, and either O2− or H2O2. Relief from ROIs is provided by various defense systems, including antioxidant enzymes (superoxide dismutase [SOD]), catalase, and peroxidase), DNA repair enzymes and binding protein (e.g., Dps [DNA binding protein from starved cells] [33]), and free-radical-scavenging agents (6, 24).
Pseudomonas aeruginosa is a gram-negative bacterium that gains its greatest metabolic energy through aerobic respiration. To counter the production of ROIs, the organism possesses two SODs, with either iron (Fe−; encoded by sodB [18, 20]) or manganese (Mn−; encoded by sodA [18, 20]) as cofactor and whose function is to disproportionate O2− to H2O2 and O2 (34). To remove H2O2, P. aeruginosa possesses three catalases, KatA (10, 17), KatB (10), and KatC (40). KatA activity is the major catalase activity detected in all phases of growth (10, 17). In contrast, KatB activity is detectable in bacteria exposed to H2O2 or paraquat, the latter of which generates a constant flux of H2O2 through SOD-catalyzed dismutation of O2− (10). Unlike KatA and KatB, little is known of the biological role of KatC in P. aeruginosa. In fact, the putative katC gene was only recently discovered fortuitously via the Pseudomonas Genome Project (40).
Most bacterial catalases are multimers (typically dimers, tetramers, or hexamers) that require heme b or heme d for catalytic activity. The final step of heme synthesis is catalyzed by ferrochelatase, which condenses Fe2+ into protoporphyrin IX. Little is known of the cellular source of iron required for heme assembly. One protein that could provide iron for such a process is bacterioferritin A (BfrA, also known as cytochrome b1 or b557), the major iron storage protein in P. aeruginosa (38). Actually, there is evidence in P. aeruginosa for two Bfr proteins (BfrA and BfrB), which differ in their N-terminal amino acid sequences (38, 38a). BfrA is a complex of 24 subunits capable of binding 700 iron atoms (38). It also binds 3 to 9 heme groups per 24 subunits in vivo and 24 heme groups in vitro (25). Recently, Kim et al. (27) identified a bfr gene encoding a bacterioferritin in the related organism P. putida; this gene was located downstream of a gene encoding a group III catalase, CatA. However, the attractive hypothesis that one function of P. putida Bfr is to provide iron for the heme prosthetic group of CatA and thus to contribute to resistance to H2O2 was not pursued. A precedent for such a hypothesis stemmed from research with Campylobacter jejuni, for which mutants deficient in ferritin, a protein related to bacterioferritin, were more sensitive to oxidative stress than wild-type organisms (50).
In this study, we have cloned and characterized the genes encoding KatA and BfrA in P. aeruginosa. Our studies suggest a necessity for BfrA in the maintenance of optimal KatA activity. Hence, we propose that BfrA stores iron that is incorporated into heme, a necessary prosthetic group for KatA activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacteria used in this study are listed in Table 1 and were grown in either Luria (L) broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) or M9 minimal medium (6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 3 mg of CaCl2, 0.25 g of MgSO4 · 7H2O, and 2 g of glucose per liter). Suspensions were grown at 37°C with shaking at 300 rpm or on a roller wheel at 70 rpm. Culture volumes were 1/10 the total flask volume to ensure maximum aeration. Media were solidified with 1.5% Bacto Agar. Frozen stocks were stored indefinitely at −80°C in a 1:1 mixture of 25% glycerol and stationary-phase suspension.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 relA1 supE44 hsdR17(rK− mK+) Δ(lacZYA-argF)U169 | Gibco-BRL |
| SM10 | Mobilizer strain | 47 |
| BL21(λDE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| CSH7 | lacY rpsL thi-1 | 31 |
| UM1 | As CSH7 plus katE1 katG14 | 31 |
| P. aeruginosa | ||
| PAO1 | Wild type; prototroph | 22 |
| PAO1 katA | GmrkatA::Gm | This study |
| PAO1 bfrA | Gmr ΔbfrA::Gm | This study |
| PAO1 katA bfrA | Gmr ΔkatA bfrA::Gm | This study |
| FRD2 katB | algT18 katB::Gm | 10 |
| Plasmids | ||
| pBluescript(KS)- or pBluescript (KS)+ | Extended polylinker pUC derivative | Stratagene |
| pKS-TA | Apr; TA PCR cloning vector that uses EcoRV site for cloning purposes | This study |
| pCRII | Apr; TA PCR cloning vector | InVitrogen |
| pCR2.1 | Apr; TA PCR cloning vector | InVitrogen |
| pUCGM | Apr Gmr; pUC19 + 850-bp Gmr cassette | 45 |
| pPZ30 | Apr; broad-host-range lacZ-based promoter probe vector | 43 |
| pUCP19 | Apr; broad-host-range expression vector | 52 |
| pUCP21T | Apr; broad-host-range expression vector | 52 |
| pEX100T | Apr Cbr; mobilizable oriT sacB vector for construction of mutants | 46 |
| pRK2013 | Kmr Ori (ColE1) OriT (Mob+) Tra+ | 14 |
| pET23a | Apr; overexpression vector | Novagen |
| pJFM12 | Apr; pKS− with 3.6-kb EcoRI-EcoRV fragment containing rpsD′-rpoA-rplQ-katA-bfrA | This study |
| pJFM13 | Apr; pEX100T with blunted 3.6-kb EcoRI-EcoRV fragment containing rpsD′-rpoA-rplQ-katA-bfrA′ in the SmaI site of the vector | This study |
| pJFM14 | Apr Gmr; pJFM13 with 850-bp aaC1 cassette within the SmaI site of katA | This study |
| pJFM15 | Apr Gmr; pEX100T with blunted EcoRI fragment from pBFR1020ΔbfrA::Gm and with 850-bp aaC1 cassette within the deleted 520-bp NdeI-SstII fragment, deleting bfrA | This study |
| pJFM16 | Apr Gmr; pJFM13 with 850-bp aaC1 cassette within the deleted 1,311-bp SmaI-NdeI site, deleting katA and bfrA | This study |
| pJFM17 | Apr; pKS− with 1,450-bp katA PCR product | This study |
| pJFM18 | Apr; pET23a with NcoI-EagI katA | This study |
| pRP411 | Apr; pKS+ containing 411-bp SalI-EcoRV fragment of katA-bfrA intergenic region | This study |
| pBFR1020 | pCR2.1 harboring 1,020-bp PCR fragment containing bfrA | This study |
| pBFR1020ΔbfrA::Gm | pBFR1020 with 520-bp NdeI-SstII deletion of bfrA replaced by aaC1 cassette | This study |
| pPZ-katA | Apr; pPZ30 containing the katA promoter on a 756-bp EcoRI-PstI fragment translationally fused to lacZ | This study |
| pPZ-bfrA | Apr; pPZ30 containing the bfrA promoter on a 389-bp EcoRI-PstI fragment translationally fused to lacZ | This study |
| pBFR4 | pUCP19 with wild-type bfrA gene | This study |
| pBFR18 | pUCP19 with bfrA18 encoding Bfr18 (E18K) | This study |
| PBFR25 | pUCP19 with bfrA25 encoding Bfr25 (Y25I) | This study |
Abbreviations used for genetic markers were those described by Holloway et al. (22). Mob+, mobilization site (ColE1); Tra+, conjugative phenotype; oriT, origin of transfer (RK2); Apr, ampicillin resistance; Cmr, chloramphenicol resistance; TA, thymine-adenine; pKS−, pBluescript KS(−); pKS+, pBluescript (KS)+.
Construction of a P. aeruginosa katB genomic library, cloning steps, and sequence analysis.
Genomic DNA (50 μg) from P. aeruginosa FRD2 katB (10) was digested with 10 U each of EcoRI and EcoRV at 37°C for 2 h. DNA fragments were separated on a 10 to 40% sucrose gradient (3). Purified 2- to 5-kb fragments were ligated into EcoRI-EcoRV-digested pBluescript KS(−) and screened for the presence of P. aeruginosa katA with a heterologous catA gene probe from P. putida (27). Plasmid DNA from positive clones was transformed into catalase-deficient Escherichia coli UM1 (31). Bacterial colonies harboring the P. aeruginosa katA gene bubbled vigorously when coated with 8.8 M H2O2. A selected plasmid, pJFM12, that complemented for catalase activity was sequenced on both strands with a PRISM Dye Deoxy Terminator cycle sequencing kit and analyzed on an ABI model 373A DNA sequencer. Oligonucleotides for sequencing and PCR analysis were synthesized at the DNA Core Facilities in the Department of Molecular Genetics, Biochemistry and Microbiology at the University of Cincinnati College of Medicine or in the Department of Microbiology and Immunology at the University of Colorado Health Sciences Center. Sequence analysis was performed with MacVector 6.5 (Eastman Chemical Co., New Haven, Conn.), Gene Runner (Hastings Software, Inc.), or Sequencer 3.0 (GeneCodes, Madison, Wis.). Amino acid alignments were performed with either the BLASTP program provided by the National Center for Biotechnology Information (1) or the Align Plus 3.0 global alignment program (Sci-Ed Software, Durham, N.C.).
Manipulation of recombinant DNA and genetic techniques.
Plasmid DNA was transformed into either E. coli DH5α-MCR (Gibco-BRL, Gaithersburg, Md.) or E. coli SM10 (47). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml) was often added to agar medium to detect the presence of insert DNA. Restriction endonucleases, the Klenow fragment, T4 DNA polymerase, and T4 DNA ligase were used as specified by the vendor (Gibco-BRL). Plasmid DNA was isolated with plasmid mini-isolation kits (Qiagen Corp.). Restriction fragments were recovered from agarose gels with SeaPlaque low-melting-point agarose (FMC BioProducts, Rockland, Maine). PCRs were performed with Taq DNA polymerase (Gibco-BRL) and appropriate primers by use of a Perkin-Elmer Cetus thermal cycler with 30 cycles of denaturation (1 min, 94°C), annealing (1 min, 54°C), and extension (1 min, 72°C). Amplified DNA fragments were gel purified, cloned into pCRII or pCR2.1 (both from InVitrogen) or a pBluescript KS(−)-based PCR vector (this study), and sequenced.
Phylogenetic analyses.
The aligned amino acid sequences were processed by heuristic parsimonial analyses with PAUP version 3.1.1 (48). In order to minimize the possibility that the algorithm would detect local parsimony (potential monophyly of clusterings comprised of more than one species), 200 bootstrap replicates were generated. A 50% majority-rule consensus tree was constructed from parsimony replicates by use of tree bisection-reconnection and nearest-neighbor branch-swapping methods with stepwise addition of the closest sequence.
Overexpression of KatA in E. coli.
To overexpress P. aeruginosa KatA, PCR primers (sense, 5′-CATATGGAAGAGAAGACCCGCCTGAC-3′; antisense, 5′-CGGCGGCGTCCAGCTTCAGGCCGAGGG-3′) were used to amplify a 1,450-bp katA fragment with pJFM12 as a template. This fragment was cloned into a pBluescript KS(−)-based PCR cloning vector, pKS-TA (Table 1), and the katA fragment was excised with NdeI and EagI and ligated into pET23a (Novagen). After transformation into E. coli BL21(λDE3), bacteria were grown aerobically to the mid-logarithmic phase (optical density at 600 nm, 0.6) and treated with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Bacteria were harvested by centrifugation at 10,000 × g for 10 min at 4°C and washed in 0.9% saline, and the pellet was resuspended in 0.1 M NaH2PO4 (pH 8.0) containing 8 M urea. Six-His-tagged KatA was purified under denaturing conditions using the Qiagen Expressionist kit. The purity of recombinant KatA was assessed after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with 10% acrylamide and staining with Coomassie blue.
Construction of P. aeruginosa katA, bfrA, and katA bfrA mutants.
The strategy for insertional inactivation of the katA and bfrA genes was facilitated by use of the gene replacement vector pEX100T (46), which allowed for the selection of double-crossover events with 6% sucrose (44). To construct a katA mutant, a ∼3.6-kb EcoRI-EcoRV fragment from pJFM12 was filled in with the Klenow fragment and ligated into the unique SmaI site within pEX100T, forming pJFM13. This plasmid was cut with SmaI, a unique site within the katA locus, and ligated to an 850-bp aaC1 (encoding gentamicin resistance [Gmr]) cassette excised from pUCGM (45), forming pJFM14. For the construction of the ΔbfrA mutant, a 1,020-bp fragment containing the bfrA region was generated by PCR with primers having the sequences 5′-ACCGGGTGGACGACGACTACT-3′ and 5′-GCCAACTGGCTGGTCAACCTC-3′ and cloned into pCR2.1, yielding pBFR1020. A 520-bp NdeI-SstII fragment of pBFR1020 comprising the entire bfrA coding sequence was replaced with the aaC1 cassette, resulting in pBFR1020ΔbfrA::Gm. The bfrA::Gm fragment was excised with EcoRI, filled in with the Klenow fragment, and cloned into SmaI-cut pEX100T, forming pJFM15. A katA bfrA double mutant was constructed by replacing the SmaI-NdeI katA′-bfrA′ fragment from pJFM13 with the aaC1 cassette, forming pJFM16. After biparental mating of E. coli SM10 harboring pJFM14, pJFM15, or pJFM16 with recipient P. aeruginosa PAO1, plasmid integration into the genome by homologous recombination was assessed by selection on Pseudomonas isolation agar-gentamicin (300 μg/ml) plates. Isolated Gmr colonies were picked and grown in L broth until the mid-log phase, and serial dilutions were plated on Pseudomonas isolation agar-gentamicin plates containing 6% sucrose. Candidate mutants were confirmed by Southern blot and catalase activity gel analyses (for katA and katA bfrA mutants [19]).
Construction of altered BfrA proteins.
Plasmid pBFR18 (E18K; see below) was constructed as follows. The 5′ 0.4-kb portion of bfrA (fragment A) was amplified by PCR with primer 1 (5′-ACCGGGTGGACGACGACTACT-3′) and HindIII-containing primer 2 (HindIII sequence underlined: 5′-AAGCTTGCCGGTCAACAGCGTATTG-3′). The 3′ 0.63-kb portion of bfrA (fragment B) was amplified with primer 3 (5′-AAGCTTGCCGCGCGCGACCAGT-3′) and primer 4 (5′-GCCAACTGGCTGGTCAACCTC-3′). PCR fragments A and B were cloned into pCR2.1 and sequenced for verification and orientation. Fragment A was excised with EcoRI-HindIII and ligated into pUCP19 linearized with EcoRI-HindIII. The resulting plasmid was linearized with HindIII, and fragment B was ligated into the HindIII site, yielding pBFR18. This plasmid contains the HindIII recognition sequence AAG CTT at codons 18 and 19 (the wild-type sequence at these positions is GAG CTG), resulting in a glutamate-to-lysine change at position 18 (E18K). Plasmid pBFR25 (Y25I; see below) was constructed as follows. The 5′ 0.42-kb portion of bfrA (fragment C) was amplified by PCR with primer 1 and SspI-containing primer 5 (SspI sequence underlined: 5′-AATATTTGGTGCCGCGCGGCCAGCTC-3′). The 3′ 0.61-kb portion of bfrA (fragment D) was amplified with primer 6 (5′-AATATTCATCCACTCGCGCATGTAC-3′) and primer 4. PCR fragments C and D were cloned into pCR2.1 and sequenced. Fragment D was excised with SspI-HindIII and ligated into pCR2.1 containing fragment C linearized with SspI-HindIII. The 1.03-kb fused fragments C and D were excised with EcoRI and ligated into pUCP19 linearized with EcoRI, resulting in pBFR25. This plasmid contains the sequence CAA ATA TTC at codons 24, 25, and 26 (the wild-type sequence at these positions is CAG TAC TTC), resulting in a tyrosine-to-isoleucine change at position 25 (Y25I).
Construction of katA-lacZ and bfrA-lacZ fusions.
The katA promoter was isolated by PCR with primers having the sequences 5′-AAGTGGTCGTCACCTGAGC-3′ and 5′-TCTCGAGGAACCACACGTC-3′, cloned into pCR2.1, sequenced, and directionally cloned as a 756-bp EcoRI-PstI fragment into pPZ30 cut with EcoRI and PstI. The resulting pPZ-katA construct represents an in-frame katA-lacZ translational fusion after 33 codons. Similarly, the bfrA promoter was isolated by PCR with primers having the sequences 5′-ACCGGGTGGACGACGACTACT-3′ and 5′-CTGCAGCGTATTGAGGTAATCG-3′, cloned, and subsequently ligated into pPZ30 as a 389-bp EcoRI-PstI fragment. The resulting construct, pPZ-bfrA, contains the first 14 codons of the bfrA gene fused in frame to lacZ.
Purification of P. aeruginosa KatA.
P. aeruginosa FRD2 katB (10), a nonmucoid algT18 mutant of mucoid cystic fibrosis isolate FRD1 (13), was grown in 10 liters of L broth containing 2 mM FeCl3 for 17 h at 37°C, followed by a 2-h aerobic incubation in the presence of 350 μM paraquat and 10 mM H2O2 to stimulate katA transcription. The bacteria were pelleted by centrifugation at 10,000 × g for 15 min, washed in 0.9% saline, and resuspended in 50 mM Tris-HCl (pH 7.4) containing lysozyme (0.02%) and the protease inhibitors phenylmethylsulfonyl fluoride (0.5 mM), leupeptin (0.5 μM), and pepstatin (0.5 μM). The suspension was subjected to three freeze-thaw (−80°C-37°C) cycles to aid in breakage of the cells and further disrupted three times with a French pressure cell at 12,000 lb/in2 and 4°C. Unbroken cells and cell debris were clarified by ultracentrifugation at 100,000 × g for 1 h at 4°C. The clarified extract was brought to 80% saturation with ammonium sulfate and incubated at 4°C for 17 h, and the precipitated protein was clarified by centrifugation at 10,000 × g for 20 min. The precipitate was dissolved in Tris-HCl (pH 7.4) and dialyzed against six 1-liter changes of the same buffer at 4°C. This solution was filtered through a 0.22-μm-pore-size filter (Nalgene) and concentrated with an Amicon YM-100 membrane. The retentate, containing KatA, was passed over a DE-52 column (2 by 18 cm; Whatman International Ltd., Kent, England) and eluted with a 0 to 200 mM NaCl gradient. After concentration of the catalase-positive fractions and dialysis against distilled water and then 50 mM potassium phosphate (pH 7.4), the sample was loaded on a hydroxyapatite column (2 by 13 cm) equilibrated with potassium phosphate. KatA has previously been found not to bind hydroxyapatite (10). KatA-positive fractions were applied to a Phenyl-Sepharose column, and the enzyme was eluted with a decreasing gradient of ammonium sulfate. Purified KatA fractions were pooled, concentrated, and stored on ice at 0°C. The molecular mass of purified native KatA was estimated by gel filtration with Sephacryl S300 equilibrated with 50 mM Tris-HCl–100 mM NaCl (pH 7.4) and with the known molecular mass standards β-amylase (200 kDa), yeast alcohol dehydrogenase (150 kDa), bovine serum albumin (65 kDa), and carbonic anhydrase (29 kDa). The α- and β-subunit sizes of KatA were determined by denaturing (boiled-sample) SDS-PAGE.
Hydrogen peroxide sensitivity assays. (i) Broth sensitivity.
Bacteria were grown aerobically for 17 h at 37°C in L broth. Suspensions were diluted 1:100 in fresh, prewarmed L broth and grown until the bacteria reached the early logarithmic phase (optical density at 600 nm, 0.6). Organisms were diluted 1:10 in 3 ml of prewarmed L broth and incubated with increasing concentrations of H2O2 (Sigma Chemical Co.) for 15 min. The suspensions were serially diluted in 0.9% saline containing 10 μg of bovine liver catalase (Boehringer Mannehim Biochemicals) per ml, and aliquots were plated on L agar. CFU were enumerated after incubation at 37°C for 24 to 48 h.
(ii) Disk sensitivity.
To assess the role of iron in sensitivity to H2O2, bacteria were grown to the stationary phase in aerobic M9 broth with 0.5% glucose as the carbon source and with or without 50 μM FeCl3. Samples of 100 μl were diluted in 3 ml of molten (50°C) M9 top agar containing 0.6% agar and layered on M9 agar plates. Sterile filter paper disks (7 mm) saturated with 10 μl of 8.8 M H2O2 was determined by measuring the diameter of growth inhibition after aerobic incubation of the plates at 37°C for 24 h.
RNase protection assays.
RNase protection assays were performed with the Riboprobe system (Promega). A 411-bp SalI-EcoRV fragment containing the 3′ end of katA and a portion of the 5′ end of bfrA was cloned behind the T7 promoter of pBluescript (KS)+ cut with SalI and EcoRV, resulting in pRP411. The antisense katA-bfrA riboprobe was generated and radiolabeled by in vitro runoff transcription from the T7 promoter of SalI-linearized pRP411. Normalized (20 μg) sample of total RNA extracted from cells during the exponential (6 h) or stationary (12 h) growth phase under low- or high-iron conditions were hybridized to excess katA-bfrA riboprobe. As a control for RNA integrity and loading accuracy, a constitutively expressed housekeeping gene, omlA, was also used as a riboprobe as previously described (49). RNA protected from single-strand-specific RNase was separated on a denaturing 5% polyacrylamide–8 M urea gel that was dried and analyzed by autoradiography.
Mass spectrometry analysis. (i) Sample preparation for MALDI analysis.
SDS-polyacrylamide gels were stained with 200 mM imidazole for 15 min, followed by a 5-min incubation with 50 mM ZnCl2. The ∼56- and 45-kDa KatA bands were excised from the gels as 1-mm2 sections and destained twice with 200 μl of 50 mM citric acid for 20 min, followed by 300 μl of 10 mM NH4HCO3 in 25% (vol/vol) acetonitrile for 30 min. Gel fragments were rinsed with 100 μl of deionized distilled water for 15 min and dried in a Speed Vac. The dried gel fragments were swollen with 6 μl of a 1-mg/ml solution of sequencing-grade trypsin (Promega) dissolved in 50 mM acetic acid. The mixture was brought to pH 8.0 by the addition of 44 μl of 25 mM NH4HCO3 digestion buffer, digestion was allowed to proceed for 24 h at 37°C, and the reaction was stopped by the addition of 75 to 100 μl of 0.1% trifluoroacetic acid (TFA). As controls, two other samples, one with protein-free gel fragments and the other with trypsin and digestion buffer, were processed in the same fashion. All samples were centrifuged at 13,000 × g for 5 min, and only the supernatant was removed. TFA (200 μl; 0.1%) was added to the remaining gel particles, and the resulting solution was incubated for 20 min with intermittent vortexing. The supernatant was removed, and the gel fragments were resuspended in 60% acetonitrile in 0.1% TFA. After additional incubation and vortexing for 30 min, the supernatant was removed. The last step was repeated twice. All extracts were pooled and dried in a Speed Vac. To the dried extract was added 5 μl of 50% acetonitrile in 0.1% TFA, and the mixture was spotted on a stainless steel target. When the sample was dry, 1 μl of saturated matrix solution (4-hydroxy-α-cyanocinnamic acid in 0.1% TFA) was spotted on the sample and allowed to air dry.
(ii) MALDI analysis.
Matrix assisted laser desorption ionization (MALDI) mass spectra were obtained on a MALDI TOFSPEC SE mass spectrometer in reflectron mode. The laser used for ionization was set at 337 nm with a pulse width of 4 ns and ∼180 μJ per pulse. The spectra presented in this work are averages of 20 to 30 laser shots. The ion acceleration voltage was set at 25 kV. The data were processed and stored on a DEC-3000 α-work station.
(iii) Cellular iron content.
Bacteria were grown aerobically in 400 ml of L broth until the stationary phase. After centrifugation at 10,000 × g for 10 min at 4°C, organisms were washed twice in 200 ml of phosphate-buffered saline (PBS) with 1 mM EDTA (pH 7.4) and resuspended in 200 ml of PBS without EDTA. After centrifugation, the pellet was resuspended in 15 ml of PBS, 10 ml of which was used for iron analysis. Total viable cells and cell dry weight were estimated with the remaining 5-ml suspension. For iron analysis, pelleted bacteria were resuspended in 2 ml of Ultrex II nitric acid (J. T. Baker, Phillipsburg, N.J.) and incubated at 80°C for 1 h, and the volume was brought to 20 ml with deionized distilled water. The samples were analyzed for iron content by inductively coupled plasma-optical emission spectroscopy with a model 965 Plasma Atomcomp apparatus (Thermo Jarrell Ash, Franklin, Md.) at the Chemical Analysis Laboratory, University of Georgia, Athens. All buffers and nitric acid solutions were analyzed as described above to correct for background. The data were calculated as both the number of iron atoms per cell and milligrams of iron per milligram of cell dry weight.
Cell extract preparation, nondenaturing gel electrophoresis, and biochemical assays.
Cell extracts were prepared from cultures harvested by centrifugation at 10,000 × g for 10 min at 4°C. Bacteria were washed twice in ice-cold 50 mM potassium phosphate buffer (pH 7.0) and sonicated in an ice-water bath for 10 s with a model W-225 sonicator (Heat-Systems, Inc., Farmington, N.Y.) at setting 5. The sonicate was clarified by centrifugation at 13,000 × g for 10 min at 4°C. Cell extract preparation for native gel electrophoresis was performed as described above, except that 50 mM Tris-HCl (pH 7.8) was used as the diluent. Catalase activity was determined by monitoring the decomposition of 18 mM H2O2 at 240 nm (5, 10, 19). One unit of activity is that which decomposes 1 μmol of H2O2 min−1 mg−1. Determination of the Km value for purified KatA was accomplished at 22°C with 1 to 80 mM H2O2 and 7 × 10−10 M KatA. β-Galactosidase assays were performed on chloroform-SDS-treated bacteria with o-nitrophenyl-β-d-galactopyranoside, and the results are expressed as Miller units (35). Protein concentrations were estimated by the method of Bradford (8) with bovine serum albumin fraction V (Sigma) as the standard.
Nucleotide sequence accession number.
The DNA and amino acid sequences presented in this work have been assigned GenBank accession no. AF047025.
RESULTS
Cloning and characterization of the katA gene of P. aeruginosa: identification of adjacent loci rpoA, rplQ, and bfrA.
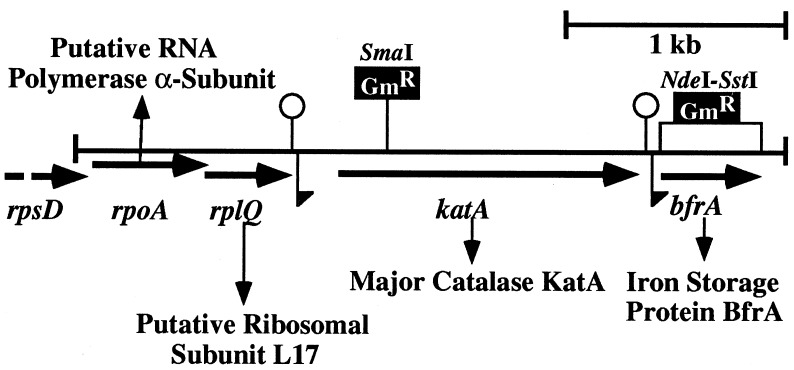
DNA sequence analysis of the 3.6-kb genomic DNA fragment from P. aeruginosa FRD2 that allowed for catalase expression in catalase-deficient E. coli UM1 revealed four open reading frames (ORFs), a map of which is depicted in Fig. 1. The first ORF comprised 999 bp and encoded a predicted protein of 333 amino acids. A BLASTP GenBank search for this protein revealed similarity to the α subunit of the DNA-directed RNA polymerase family (data not shown). The second ORF comprised 390 bp and encoded a putative protein of 129 amino acids which was similar to 50S ribosomal subunit protein L17 of E. coli (GenBank accession no. U18997). The third ORF comprised 1,446 bp and encoded a predicted monomer of 482 amino acids. This protein was 82% identical to the catalase of Proteus mirabilis (GenBank accession no. P42321). The fourth ORF comprised 462 bp and encoded a putative protein of 154 amino acids. This protein was 79% identical to a bacterioferritin of the related organism P. putida (27).
FIG. 1.
Genetic map of the 3.605-kb EcoRI-EcoRV fragment harboring rpoA, rplQ, katA, and bfrA of P. aeruginosa PAO1 and putative gene products. The large loopholes indicate transcriptional terminators. The flags pointing downward indicate the promoter regions upstream of the katA and bfrA genes. The restriction sites from which single katA and bfrA mutants were generated via insertional mutagenesis are given.
Amino acid identity of KatA and BfrA with bacterial catalases and bacterioferritins.
Since this study was focused on the potential relationship between KatA and BfrA in protecting P. aeruginosa against H2O2, we thought it necessary to determine the similarity of KatA and BfrA to other bacterial catalases and bacterioferritins. A BLASTP homology search for KatA and other bacterial catalases revealed the greatest identity with catalases from P. mirabilis (GenBank accession no. P42321; 82% identity over 377 amino acids), Vibrio fischeri (GenBank accession no. AF011784; 79% identity over 371 amino acids), Bordetella pertussis (GenBank accession no. P48062; 77% identity over 370 amino acids), and Bacteroides fragilis (GenBank accession no. U66717; 75% identity over 374 amino acids) (alignment not shown). A search for Bfr proteins revealed the greatest identity with Bfr proteins from P. putida (GenBank accession no. U66717; 79% identity over 154 amino acids), Neisseria gonorrhoeae (GenBank accession no. P72080; 62% identity over 154 amino acids), and Salmonella typhimurium (GenBank accession no. AF058449; 43% identity over 141 amino acids) (Fig. 2).
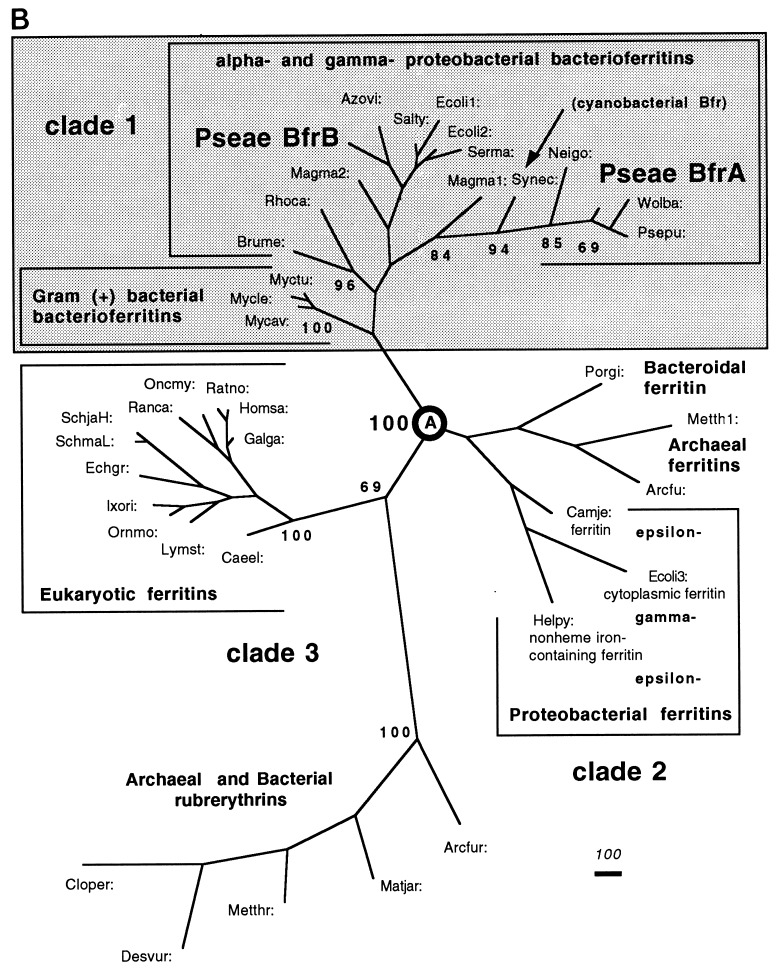
FIG. 2.
Amino acid similarity of BfrA and other Bfr proteins. (A) Proteins were aligned with Align Plus 3.0. Dots indicate identical amino acids, while dashes indicate gaps in the protein sequence relative to P. aeruginosa BfrA. PA, P. aeruginosa BfrA; PP, P. putida Bfr; NG, N. gonorrhoeae Bfr; ST, S. typhimurium Bfr; EC, E. coli Bfr. The amino acids essential for ferroxidase activity in E. coli Bfr are conserved in each Bfr protein and are shown in boldface and marked with an asterisk. (B) Unrooted phylogenetic tree based on the amino acid sequences of 35 (bacterio)ferritins and 5 rubrerythrins and constructed by parsimony methods. The branch lengths (shown in italics) reflect the evolutionary distances calculated as the average number of amino acid changes per 1,000 residues. The numbers in bold in front of the nodes represent the proportion of bootstrap samplings that support the topology shown. Two hundred bootstrap replicates were analyzed. For analysis of the BfrA protein, Bfr sequences were obtained from GenBank (accession no.) for the following organisms: Arcfu, Archaeoglobus fulgidus putative ferritin (AE001047); Arcfur, A. fulgidus rubrerythrin 1 (AE001047); Azovi, Azotobacter vinelandii Bfr (U83692); Brume, Brucella melitensis Bfr (U19760); Caeel, Caenorhabditis elegans ferritin (AF106592); Camje, Campylobacter jejuni ferritin (D64082); Cloper, Clostridium perfringens rubreythrin (X92844); Desvur, Desulfovibrio vulgaris rubrerythrin (U82323); Echgr, Echinococcus granulosus ferritin (Z31712); Ecoli1, Escherichia coli ECOR30 Bfr (AF058450); Ecoli2, E. coli K-12 Bfr (M27176); Ecoli3, E. coli K-12 MG1655 cytoplasmic ferritin (AF000335); Galga, Gallus gallus ferritin heavy (H) chain (Y14698); Helpy, Helicobacter pylori J99 non-heme iron-containing ferritin Pfr (AE00149); Homsa, Homo sapiens apoferritin H chain (X00318); Ixori, Ixodes ricinus ferritin (AF068224); Lymst, Lymnaea stagnalis snail soma ferritin (P42577/X56778); Magma1, Magnetospirillum magnetotacticum Bfr1 (AF001959); Magma2, M. magnetotacticum Bfr2 (AF001959); Metth1, Methanobacterium thermoautotrophicum putative ferritin (AE000804); Metthr, M. thermoautotrophicum rubrerythrin (AE000854); Matjar, Methanococcus jannaschii rubrerythrin (U67520); Mycav, Mycobacterium avium Bfr (X76906); Mycle, Mycobacterium leprae Bfr (P43315); Myctu, Mycobacterium tuberculosis Bfr (Z97193); Neigo, Neisseria gonorrhoeae Bfr (U76633); Oncmy, Oncorhynchus mykiss ferritin-1 H chain (D86625); Ornmo, Ornithodoros moubata ferritin (AF068225); Porgi, Porphyromonas gingivalis ferritin (AB016086); Psepu: Pseudomonas putida Bfr (U66717); Pseae: Pseudomonas aeruginosa Bfr (AF047025); Ranca, Rana catesbeiana ferritin, middle subunit (J02724); Ratno, Rattus norvegicus ferritin H chain (P19132); Rhoca, Rhodobacter capsulatus Bfr (Z54247); Salty, Salmonella typhimurium LT2 Bfr (AF058449); SchjaH, Schistosoma japonicum putative ferritin-1 H chain (AF040385); SchmaL, Schistosoma mansoni ferritin light chain (M64538); Serma, Serratia marcescens Bfr (AF058451); Synec, Synechocystis sp. strain PCC6803 Bfr (D90905); and Wolba, Wolbachia sp. Bfr (21).
The crystal structure (15), iron and heme binding capacities (12), and residues essential for ferroxidase activity of the E. coli Bfr protein are known. P. aeruginosa BfrA possesses the same four glutamate and single tyrosine and histidine residues essential for ferroxidase activity as E. coli Bfr (30). Interestingly, P. aeruginosa BfrA and the other aligned Bfr proteins in Fig. 2A possess the same conserved residues, suggesting that they also possess ferroxidase activity.
Phylogenetic analyses of KatA and BfrA.
Following the alignment of amino acid sequences of KatA and BfrA with similar proteins, unrooted phylogenetic trees were constructed by parsimony methods based on the amino acid sequences of 94 catalases (data not shown) and 35 (bacterio)ferritins (Fig. 2B). The phylogenetic tree generated for catalases was similar to the one constructed recently for 74 eukaryotic and prokaryotic catalases (29). P. aeruginosa KatA is a group III bacterial catalase (29), as is P. putida CatA and Wolbachia sp. Cat, and is most closely related to the major catalase, KatA, of P. mirabilis.
We selected 35 (bacterio)ferritin sequences from bacteria (all available sequences), archaea (3 sequences), and eukarya (12 sequences) for the analysis of the P. aeruginosa BfrA protein. Because of their sequence relatedness (including the glutamate and tyrosine residues that are critical for ferroxidase activity), five archaeal and bacterial rubrerythrin sequences were used as the outgroup. The unrooted tree (Fig. 2B) consists of three clades that are separated at node A by the highest possible confidence. P. aeruginosa BfrA groups in the bacterioferritin-only clade 1, closest to the Bfr proteins from Wolbachia sp. and P. putida. Interestingly, the archaeal and bacterial rubrerythrins group with eukaryal ferritins in clade 2, while clade 3 contains a mixture of archaeal and unusual bacterial ferritins. It is evident that the phylogenetic (bacterio)ferritin tree obtained is not congruent with the tree reflective of the phylogenetic relationships based on 16S rRNA sequences (39).
Complementation of katA in catalase-deficient E. coli.
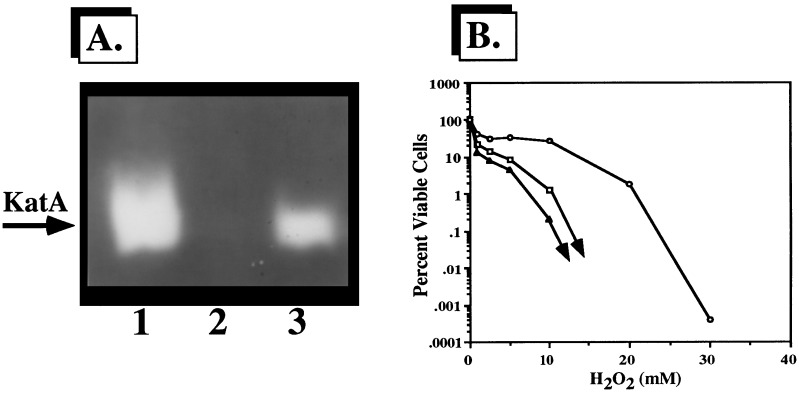
To further assess functional complementation by katA, cell extracts were separated by nondenaturing PAGE and stained for catalase activity. As shown in Fig. 3A, the primary catalase activity of P. aeruginosa, that of KatA, was detectable as a single activity band (lane 1). Provision of plasmid pJFM12 to catalase-deficient E. coli UM1 (Fig. 3A, lane 2) allowed for the expression of KatA activity (lane 3).
FIG. 3.
(A) Complementation of P. aeruginosa katA in E. coli UM1. Cell extracts (20 μg) of aerobically grown, stationary-phase organisms were separated by nondenaturing PAGE and stained for catalase activity (51). Lane 1, P. aeruginosa PAO1; lane 2, E. coli UM1; lane 3, E. coli UM1(pJFM12). (B) Enhanced resistance of E. coli UM1 harboring P. aeruginosa katA to H2O2. Mid-logarithmic-phase bacteria were exposed to various concentrations of H2O2 for 15 min at 37°C (23). The results are typical of three separate experiments. Symbols: □, E. coli CSH7; ▴, E. coli UM1; ○, E. coli UM1(pJFM12).
We next assessed the contribution of P. aeruginosa KatA to the resistance of catalase-deficient E. coli to exogenous H2O2. As shown in Fig. 3B, wild-type E. coli CSH7 was highly resistant to 1 to 10 mM H2O2 but was killed at 20 mM. As predicted, catalase-deficient strain UM1 demonstrated slightly higher sensitivity than wild-type bacteria (31). We did not observe the bimodal killing pattern of wild-type and DNA-repair-deficient strains of E. coli exposed to 1 to 5 mM H2O2 (23). Still, catalase-deficient E. coli UM1 was predictably more sensitive to all tested concentrations of H2O2. The expression of P. aeruginosa KatA in this strain allowed for a higher level of protection against H2O2 than that seen for wild-type E. coli.
Purification and properties of KatA.
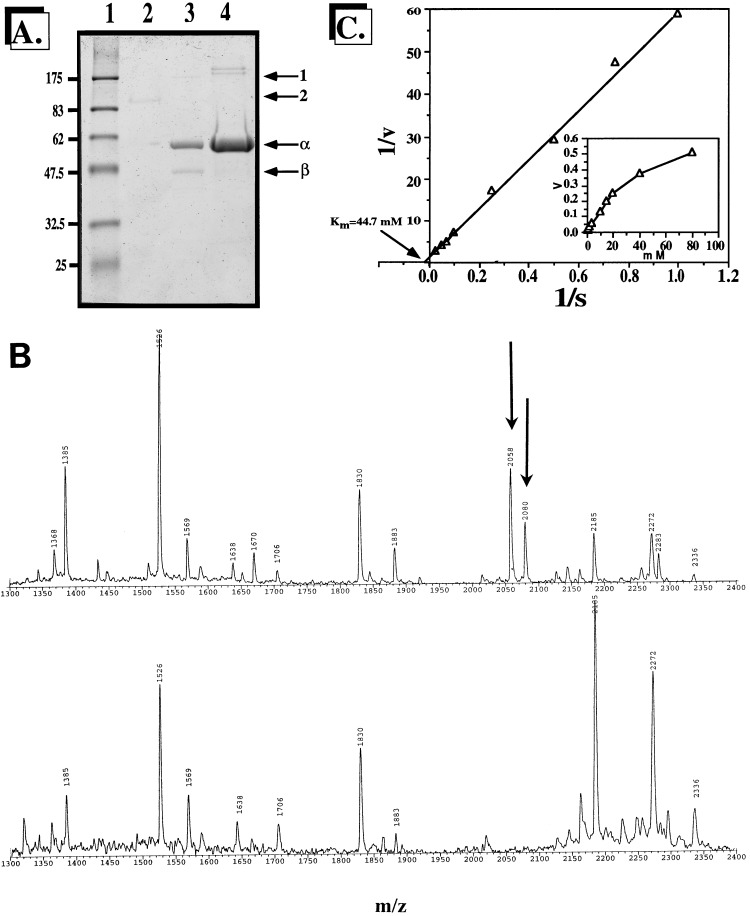
KatA was purified to homogeneity from a katB mutant of P. aeruginosa FRD2 (10) and as a recombinant six-His-tagged protein in E. coli. The molecular mass of native KatA was estimated by gel filtration analysis (data not shown) to be ∼160 to 170 kDa (Fig. 4A, lane 2). Interestingly, this protein retained some catalase activity in the gel prior to staining with Coomassie blue when the gel was coated with H2O2. When denatured, KatA split into its monomeric ∼56-kDa form and 45-kDa form (Fig. 4A, lane 3). Linear scanning densitometry of these bands revealed an approximate 2:1 ratio. Based upon SDS-PAGE (Fig. 4A, lane 2) and gel filtration (data not shown) molecular weight analyses of native KatA, our data suggest that KatA is a heteromultimer. The banding pattern of the six-His-tagged recombinant KatA protein expressed in E. coli was identical to that of native KatA, except that there was less of the smaller subunit (Fig. 4A, lane 4).
FIG. 4.
Purification (A), mass spectrometric analysis (B), and Km measurement (C) of P. aeruginosa KatA. (A) Lane 1, protein molecular mass standards; lane 2, 60 ng of purified unboiled KatA; lane 3, 120 ng of purified boiled KatA; lane 4, 160 ng of purified boiled recombinant six-His-tagged KatA. (B) Mass spectrometric analysis of the 55-kDa subunit (top panel) and smaller subunits (bottom panel) of KatA. The arrows in the top panel indicate peptides that are absent in the bottom panel. (C) Double-reciprocal Lineweaver-Burk plot of KatA activity with various H2O2 concentrations. Experiments were performed as described in Materials and Methods.
To determine whether the origin of the two bands was the native KatA enzyme, we subjected both protein bands to tryp sin digestion and MALDI mass spectrometric analysis. As shown in Fig. 4B, the molecular masses of the major peaks were identical for both the 56-kDa band (top panel) and the 45-kDa band (bottom panel), with the exception of two peptides with molecular masses of 2,079.71 and 2,057.65 Da (arrows). Interestingly, the trypsin cleavage sites for these peptides correspond to amino acids 7 to 27 [(R)LTTAAGAPVVDNQNVQTAGPR(G)] and 58 to 76 [(K)GSAAYHTFTVTHDITPYTRAK(I)], respectively. These data suggest that the smaller band is a second translational product encoded by katA. The first translational start codon (ATG) after nucleotide 228 (76 amino acids) was in frame and located at position 265, with two overlapping ribosome binding sites (253-AAGAAGA-259) preceding it. From these data, we conclude that KatA is a heteromultimer, possibly an α2β-heterotrimer. Gel filtration and SDS-PAGE analyses of CatF from the related species P. syringae suggest that it, too, may be a heteromultimer, although this notion has not yet been proven with mass spectrometry or protein sequencing (28). This is the first demonstration of a heteromultimeric catalase among all three catalase groups (29).
Catalases are typically enzymes with low substrate affinities, with Km values for H2O2 of 2.07 mM for the catalase/peroxidase of Streptomyces cyaneus (36) and 78 mM for the catalase of Bacillus subtilis (32) (the P. aeruginosa KatB Km is 10.6 mM [10]). After double-reciprocal Lineweaver-Burk analysis, we determined an apparent Km of 44.7 mM with H2O2 as the substrate for KatA purified from P. aeruginosa FRD2 katB (10) (Fig. 4C). This rather high apparent Km is similar to that for the structurally similar but phylogenetically different catalase CatF from P. syringae (Km, ∼60 mM) (28).
Phenotypes of katA, bfrA, and katA bfrA mutants.
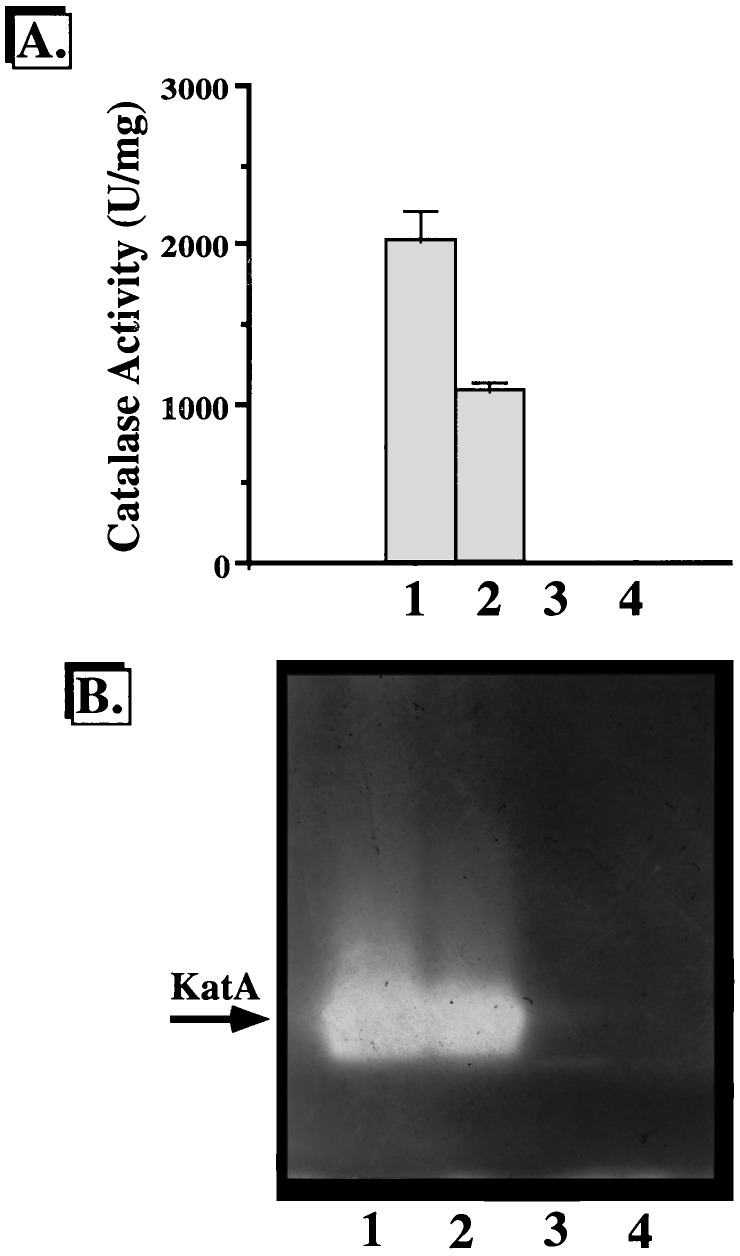
The major catalase activity of P. aeruginosa is that of KatA (10, 17). Since one of our hypotheses was that iron bound to BfrA is necessary for the production of some heme, the prosthetic group of KatA, we predicted that bfrA mutants would have reduced KatA activity. Catalase activity gel staining of cell extracts from the wild type and isogenic bfrA, katA, and katA bfrA mutants revealed that wild-type bacteria produce only KatA (Fig. 5A, lane 1), with a specific activity of 2,018 U/mg (Fig. 5B, lane 1). This finding is consistent with our earlier observation that the second catalase, KatB, is not expressed unless organisms are treated with H2O2 (10). Interestingly, the bfrA mutant produced visibly less KatA than wild-type bacteria in catalase activity gels (Fig. 5A, lane 2); this result correlated with a 47% loss of catalase activity determined spectrophotometrically (Fig. 5B, lane 2). The katA and katA bfrA mutants produced no detectable catalase, as monitored by native gel or spectrophotometric assays (Fig. 5A and B, lanes 3 and 4).
FIG. 5.
Catalase activity (A) and activity staining (B) of P. aeruginosa strains. (A) Catalase activity in cell extracts from stationary-phase organisms was measured as described by Beers and Sizer (5); the values are means ± standard errors for three experiments. 1, P. aeruginosa PAO1; 2, bfrA; 3, katA; 4, katA bfrA. (B) Cell extracts (20 μg) from the above organisms were separated by nondenaturing PAGE in triplicate and stained for catalase activity (51). Lane 1, P. aeruginosa PAO1; lane 2, bfrA; lane 3, katA; lane 4, katA bfrA. The KatA activity band is shown by an arrow.
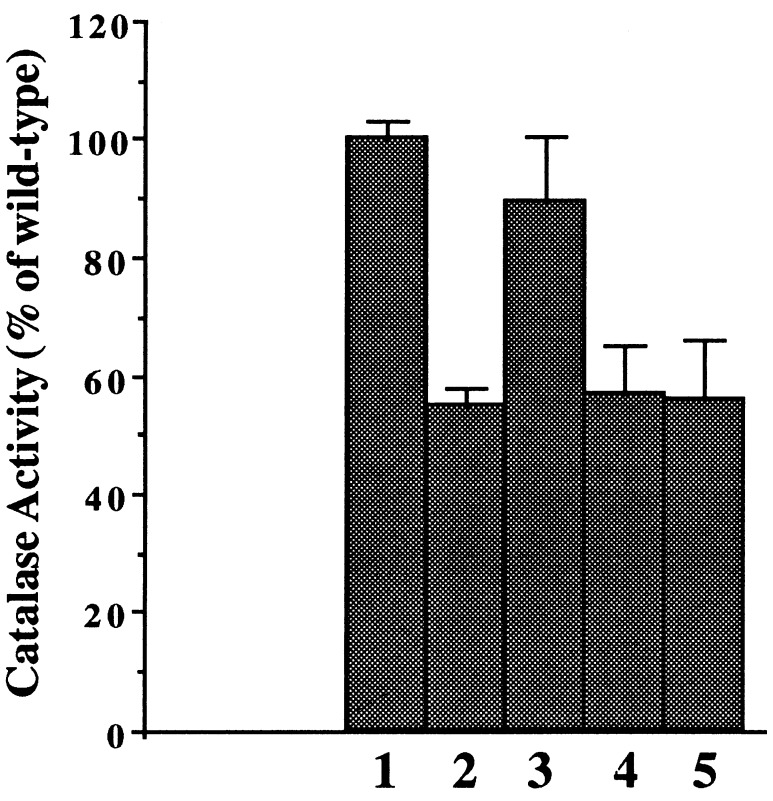
Ferroxidase activity of BfrA is essential for optimal KatA activity.
The Bfr protein of E. coli possesses several amino acids that are critical for the ferroxidase activity that oxidizes Fe2+ to Fe3+ within the BfrA core (30). An alignment of P. aeruginosa BfrA with E. coli Bfr and other bacterioferritins revealed that each protein harbors these residues (Fig. 2A). To test our hypothesis that the ferroxidase activity of BfrA is essential for optimal KatA activity, two bfrA mutant plasmids were constructed. The first, pBFR18, possessed a glutamate-to-lysine change at amino acid 18 (E18K). The second, pBFR25, possessed a tyrosine-to-isoleucine change at amino acid 25 (Y25I). As shown in Fig. 6, wild-type organisms (lane 1) possessed nearly twice the catalase activity of the bfrA mutant (lane 2), consistent with the results shown in Fig. 5B. Provision of pBFR4 harboring the wild-type bfrA gene partially restored catalase activity (Fig. 6, lane 3). In contrast, the catalase activity of the bfrA mutant harboring either pBFR18 (E18K) or pBFR24 (Y25I) remained at bfrA mutant levels (Fig. 6, lanes 4 and 5).
FIG. 6.
Importance of ferroxidase-center amino acids in optimal KatA activity in P. aeruginosa. P. aeruginosa harboring plasmids with wild-type or altered bfrA genes was grown aerobically to the stationary phase in L broth at 37°C. Catalase activity of cell extracts was monitored in triplicate. 1, Wild type plus pUCP19) 2, bfrA plus pUCP19) 3, bfrA plus pBFR4; 4, bfrA plus pBFR18 (E18K); 5, bfrA plus pBFR25 (Y25I).
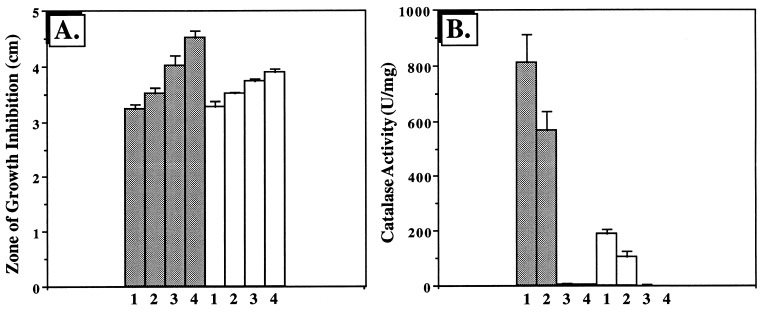
BfrA and KatA are important in optimal resistance to H2O2: role of iron.
Since catalase activity was reduced in the bfrA strain and absent in the katA and katA bfrA strains, we predicted that these mutants would demonstrate enhanced sensitivity to H2O2 and that cellular iron levels might influence H2O2 sensitivity. As shown in Fig. 7A, the bfrA, katA, and especially katA bfrA mutants demonstrated enhanced sensitivity to H2O2 relative to wild-type organisms. The sensitivity of the katA and katA bfrA mutants was, in part, dependent upon cellular iron levels, since these organisms were less sensitive to H2O2 when grown in iron-limiting medium than when grown in iron-rich medium. Catalase activity was also highest when organisms were grown in iron-replete medium (Fig. 7B).
FIG. 7.
H2O2 disk sensitivity (A) and catalase activity (B) of P. aeruginosa strains: influence of iron. (A) P. aeruginosa strains were grown aerobically in M9 minimal medium with or without 50 μM FeCl3 to the stationary phase. Organisms were diluted 30-fold in 3 ml of molten 0.6% M9 top agar and layered on M9 agar plates. Sterile filter paper disks (7 mm) were impregnated with 10 μl of 8.8 M H2O2 and placed in triplicate on the top-agar surface, and the plates were incubated at 37°C for 17 h. Zones of growth inhibition were measured. Shaded columns, M9 medium plus 50 μM FeCl3; open columns, M9 medium alone. The results are expressed as the means ± standard errors for nine different experiments. 1, P. aeruginosa PAO1; 2, bfrA; 3, katA; 4, katA bfrA. (B) Catalase activity (5) was measured in cell extracts from each strain and expressed as the means ± standard errors for three different experiments. Columns are as in panel A.
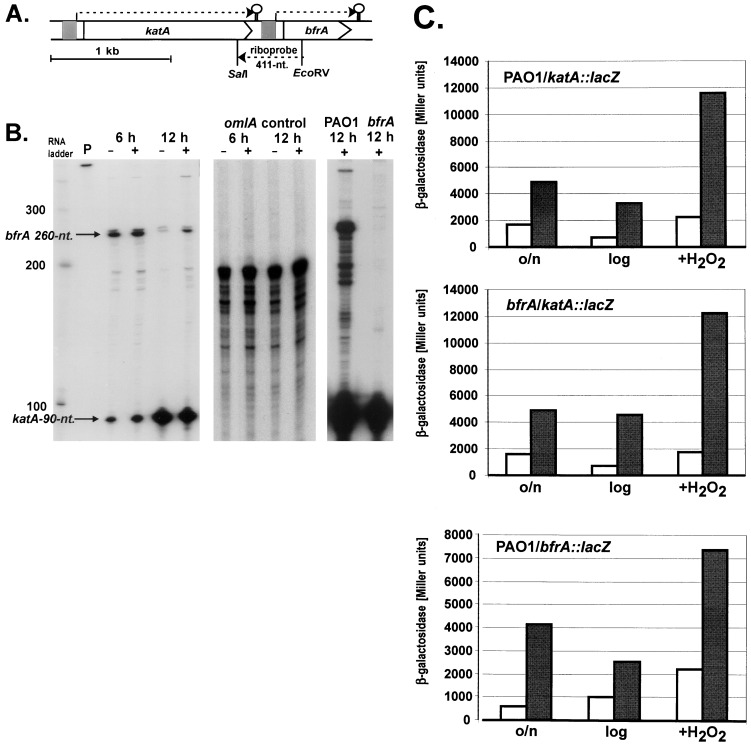
Regulation of katA and bfrA in P. aeruginosa: role of iron and H2O2.
Since the enhanced sensitivity of the bfrA, katA, and katA bfrA mutants to H2O2 was greatest in the presence of iron, we postulated that iron and H2O2 levels might control the transcriptional activation of both katA and bfrA. First, growth-phase- and iron-dependent expression patterns were monitored by RNase protection analysis with a riboprobe that allowed for the simultaneous detection of katA and bfrA transcripts (Fig. 8A). A loading control with a “housekeeping” gene (omlA) was included in Fig. 8B (middle panel). The level of expression of katA was somewhat higher in iron-rich relative to iron-poor medium during the exponential growth phase and was upregulated at least fivefold during the stationary phase (Fig. 8B, left panel). The bfrA gene was expressed at high levels under both low- and high-iron conditions during the exponential growth phase. However, during the stationary phase, the level of bfrA expression was maximal in iron-replete medium but was lower than that expressed in exponential phase. An absence of BfrA had no effect on katA transcription (Fig. 8B, right panel).
FIG. 8.
Regulation of katA and bfrA: effect of growth phase, iron, and H2O2. (A) Genetic organization of katA and bfrA and location of the riboprobe used for the simultaneous detection of the corresponding transcripts. The promoter regions upstream of katA and bfrA are depicted as shaded boxes. Loopholes indicate transcriptional terminators. nt., nucleotides. (B) RNase protection analysis of katA and bfrA. P. aeruginosa was grown aerobically under low (−)- or high (+)-iron conditions. An RNA ladder, undigested probe (P), and detected transcripts for bfrA and katA are indicated (left panel), together with omlA as a constitutive and loading control (middle panel) and the katA transcript in the wild type and the bfrA mutant (right panel). (C) Translational katA-lacZ and bfrA-lacZ activities. All bacteria were grown aerobically in M9 medium. Open columns, M9 low-iron medium (0.2 mM dipyridyl); shaded columns, M9 high-iron medium (50 μg of FeCl3 per ml). The three data pairs in each panel reflect the measured activities during the stationary phase after overnight growth (o/n), during exponential growth (log), and after 1 h of treatment with 1 mM H2O2 every 10 min (+H2O2). The values are the averages for quadruplicate cultures, and error bars (<10%) are omitted for clarity.
A more detailed analysis of katA and bfrA expression with respect to iron concentration and response to H2O2 was performed with translational fusions to the lacZ reporter gene (Fig. 8C). The level of KatA::LacZ expression was enhanced during the stationary phase and was predictably higher in iron-rich versus iron-poor medium. Treatment with exogenous H2O2 caused an ∼3-fold increase in KatA::LacZ activity. The regulation and activity of KatA::LacZ in a ΔbfrA background were virtually identical to those in wild-type bacteria. The level of expression of BfrA::LacZ was highest in the stationary phase and was upregulated at least twofold upon H2O2 treatment. The high level of stationary-phase BfrA expression differed from the pattern observed in the RNase protection analysis. However, it is difficult to compare data from an RNase protection analysis with LacZ reporter data, since the former detects transcripts at a given instant and the latter measures the accumulation of a translated product over the entire time period. Still, taken together, our data suggest that both KatA expression and BfrA expression respond to growth phase, iron, and H2O2.
Iron levels in bfrA, katA, and katA bfrA strains.
Since KatA is the predominant catalase in P. aeruginosa and the bfrA mutant possessed less KatA activity than wild-type organisms, we postulated that iron would be redistributed in the cell, resulting in no net change in total cellular iron levels. To test this hypothesis, iron levels in wild-type and mutant strains were measured by ion-spray mass spectrometry. Not surprisingly, total iron levels varied little in wild-type, katA, bfrA, and katA bfrA strains, ranging from 2.8 × 105 to 4.3 × 105 iron atoms per viable cell and 0.99 × 10−4 to 1.3 × 10−4 mg of iron per mg of cell dry weight. These data are supportive of the view that cellular iron concentrations are regulated at the level of uptake (9).
DISCUSSION
The initial goal of this work was to clone the P. aeruginosa katA gene and determine the contribution of its product to H2O2 detoxification. However, when we discovered the bfrA gene downstream of katA, we wondered whether their close proximity could be extended to a functional relationship of both gene products. Our first attempt at understanding the potential utility of katA and bfrA being so close together on the P. aeruginosa genome was through a phylogenetic analysis of both KatA and BfrA proteins. The phylogenetic analysis of catalases performed in this study will be discussed in more detail elsewhere (31a). However, it is worth mentioning here that P. aeruginosa KatA is a group III bacterial catalase (29) and thus is closely related to the CatA proteins found in P. putida and a Wolbachia sp., a nematode-endosymbiotic member of the Rickettsiales order (21). This fact is important because the genes encoding these catalases are succeeded by bacterioferritin-encoding genes in each of the organisms. Since the Wolbachia sp. contains only one catalase, the proximity of the kat and bfr genes in the three bacteria is intriguing. Although katA and bfrA have their own promoters, which are positively responsive to iron and H2O2, it appears that these genes have not evolved independently due to functional pressures on their expression products. In addition, the constructed ferritin protein tree leads to the conclusion that a bacterioferritin gene was already present in the common ancestor of gram-positive bacteria, cyanobacteria, and proteobacteria, which diverged during further species evolution.
Further compelling evidence for a functional relationship between KatA and BfrA surfaced when we demonstrated that a P. aeruginosa mutant lacking BfrA produces ∼50% of wild-type KatA activity and shows greater H2O2 sensitivity despite possessing wild-type katA transcription and translation. The enhanced log-phase expression of BfrA relative to KatA may serve to sequester labile iron before releasing it for distribution into iron- and/or heme-containing proteins such as KatA. Thus, we believe that BfrA-bound iron is required for optimal KatA activity and, in turn, resistance to H2O2.
P. aeruginosa BfrA is a complex of 24 subunits capable of binding up to 24 heme moieties (25, 38). Because of the hydrophobic nature of heme, it is not likely released from BfrA in vivo but is essential for the optimal release of reduced iron from its core in vitro (37). Because BfrA can also bind ∼700 Fe atoms (11), we postulate that Fe2+ released from BfrA can be incorporated into protoporphyrin IX by ferrochelatase-dependent condensation, forming heme which, in turn, is incorporated into the folding KatA enzyme. Since both iron and H2O2 stimulate katA and bfrA transcription, we postulate that such conditions may cause iron release from the BfrA core. Iron release from the related protein ferritin is mediated, in part, by O2− and not H2O2 (4, 7). However, H2O2 may indirectly produce elevated levels of O2− through its reaction with HO·, a reaction proposed for the resistance of E. coli to mode II killing by 5 to 20 mM H2O2 (23, 24). The mechanism for this event is as follows. First, basal levels of O2− cause the release of Fe2+ from the core of BfrA. In the presence of H2O2, some Fe2+ reacts with O2− in a Fenton reaction to form HO·. In turn, the HO· can react with H2O2 to form even more O2−, thereby reducing additional core BfrA Fe3+ to Fe2+. The release of Fe2+ from Bfr and ferritin in E. coli is also important in the repair of O2−-mediated damage to [4Fe-4S] cluster proteins (26).
Bfr proteins and ferritins possess ferroxidase activity, which oxidizes Fe2+ to Fe3+ to form ferric-oxy-hydroxide-phosphate complexes within their cores (2). Mutagenesis of the conserved and required glutamate, histidine, or tyrosine residues (Fig. 2B) present in P. aeruginosa BfrA was shown to abolish this activity (30). We have demonstrated that P. aeruginosa BfrA with either an E18K or a Y25I substitution does not show wild-type KatA activity (Fig. 5). Ferroxidase activity could benefit the organism by limiting the amount of labile iron available to undergo the Fenton reaction (Fe2+ + H2O2→HO· + Fe3+), thus restricting ensuing damage of biological molecules mediated by H2O2 and HO·. While not yet tested with P. aeruginosa, this hypothesis has been confirmed for murine erythroleukemia cells expressing the ferroxidase-center-containing subunit of the related iron storage protein ferritin; labile iron levels were reduced 2.3-fold in H subunit-overexpressing cells (42). P. aeruginosa also possesses BfrB, and we postulated that it, too, could limit labile iron levels and thus assist in the protection of organisms against oxidative stress. However, a bfrB mutant was not more susceptible to H2O2 and possessed wild-type catalase activity (38a). This result further supports a functional link between BfrA and KatA.
In addition to controlling the level of labile iron within bacteria, BfrA may also indirectly control DNA damage. Recently, it was found that the E. coli Bfr crystal structure contains a four-helix bundle that is nearly identical to the E. coli Dps monomer (DNA binding protein from starved cells (16). Dps has been shown to bind and protect DNA from Fenton reaction-mediated oxidative DNA damage (33). Because of the remarkable structural identity between the E. coli Dps and Bfr proteins, it is conceivable that one mechanism by which Dps and possibly bacterioferritins protect DNA is through their capacity to bind iron. Interestingly, the DpsA protein of a Synechococcus sp. binds DNA, contains heme and catalase activity, and possesses a C-terminal domain that is 55% similar to that of the Azotobacter vinelandii bacterioferritin (41). Although there is no evidence that bacterioferritins bind DNA, they may protect it indirectly via their capacity to sequester reactive iron. Identification of cellular conditions triggering iron and/or heme release from BfrA and determination of whether such conditions also increase KatA activity and oxidative DNA damage are studies currently being carried out. Finally, the trafficking of iron from BfrA to other molecules in bacteria, potentially by iron chaperones, is likely critical for a variety of cellular processes. Future studies will address the mechanism(s) by which iron is released from BfrA and how it is conditionally and preferentially designated for different iron- and/or heme-containing proteins.
ACKNOWLEDGMENTS
We thank P. Loewen (University of Manitoba) for assistance with interpretation of the KatA absorption spectra and for constructive comments regarding KatA structure. We thank A. J. Anderson (Utah State University) for providing a plasmid containing the P. putida catA gene.
This work was supported in part by grants AI-40541 (to D.J.H.) and AI-15940 (to M.L.V.) from the National Institutes of Health (to D.J.H.), Cystic Fibrosis grant HASSET97PO (to D.J.H.), and start-up funds from the Department of Molecular Genetics, Biochemistry and Microbiology at the University of Cincinnati College of Medicine.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andrews S C, Smith J M, Yewdall S J, Guest J R, Harrison P M. Bacterioferritins and ferritins are distantly related in evolution: conservation of ferroxidase-centre residues. FEBS Lett. 1991;293:164–168. doi: 10.1016/0014-5793(91)81177-a. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 5.3.2–5.3.8. [Google Scholar]
- 4.Bando Y, Aki K. Superoxide-mediated release of iron from ferritin by some flavoenzymes. Biochem Biophys Res Commun. 1990;168:389–395. doi: 10.1016/0006-291x(90)92333-u. [DOI] [PubMed] [Google Scholar]
- 5.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 6.Beyer W, Imlay J, Fridovich I. Superoxide dismutases. Prog Nucleic Acid Res. 1991;40:221–253. doi: 10.1016/s0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- 7.Bolann B J, Ulvik R J. Release of iron from ferritin by xanthine oxidase: role of the superoxide radical. Biochem J. 1987;243:55–59. doi: 10.1042/bj2430055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Braun V. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol Chem. 1997;378:779–786. [PubMed] [Google Scholar]
- 10.Brown S M, Howell M L, Vasil M L, Anderson A, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosaencoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheesman M R, Kadir F H A, Al-Basseet J, Al-Massad F, Farrar J, Greenwood C, Thompson A J, Moore G R. E.p.r. and magnetic circular dichroism spectroscopic characterization of bacterioferritin from Pseudomonas aeruginosa and Azotobacter vinelandii. Biochem J. 1992;286:361–367. doi: 10.1042/bj2860361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheesman M R, le Brun N E, Kadir F H, Thomson A J, Moore G R, Andrews S C, Guest J R, Harrison P M, Smith J M, Yewdall S J. Haem and non-haem iron sites in Escherichia colibacterioferritin: spectroscopic and model building studies. Biochem J. 1993;292:47–56. doi: 10.1042/bj2920047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolow F, Kalb A J, Yariv J. Structure of a unique twofold symmetric haem-binding site. Nat Struct Biol. 1994;1:453–460. doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 16.Grant R A, Filman D J, Finkel S E, Kolter R, Hogle J M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 17.Hassett D J, Charniga L, Bean K A, Ohman D E, Cohen M S. Antioxidant defense mechanisms in Pseudomonas aeruginosa: resistance to the redox-active antibiotic pyocyanin and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodBmutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassett D J, Sokol P, Howell M L, Ma J-F, Schweizer H P, Ochsner U, Vasil M L. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosademonstrate defective siderophore-mediated iron uptake and altered aerobic metabolism. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassett D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning of the sodA and sodB genes encoding manganese and iron superoxide dismutase in Pseudomonas aeruginosa: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henkle-Dühren K, Eckelt V H O, Wildenburg G, Blaxter M, Walter R D. Gene structure, activity and localization of a catalase from intracellular bacteria in Onchocerca volvolus. Mol Biochem Parasitol. 1998;96:69–81. doi: 10.1016/s0166-6851(98)00109-1. [DOI] [PubMed] [Google Scholar]
- 22.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imlay J A, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coliby hydrogen peroxide. J Bacteriol. 1986;166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imlay J A, Linn S. Mutagenesis and stress responses induced in Escherichia coliby hydrogen peroxide. J Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadir F H A, Moore G R. Bacterial ferritin contains 24 haem groups. FEBS Lett. 1990;271:141–143. doi: 10.1016/0014-5793(90)80391-u. [DOI] [PubMed] [Google Scholar]
- 26.Keyer K, Imlay J A. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J Biol Chem. 1997;272:27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y C, Miller C D, Anderson A J. Identification of adjacent genes encoding the major catalase and a bacterioferritin from the plant-beneficial bacterium Pseudomonas putida. Gene. 1997;199:219–224. doi: 10.1016/s0378-1119(97)00370-3. [DOI] [PubMed] [Google Scholar]
- 28.Klotz M G, Kim Y C, Katsuwon A J, Anderson A J. Cloning, characterization, and phenotypic expression in Escherichia coli of catF, which encodes the catalytic subunit of catalase isozyme CatF of Pseudomonas syringae. Appl Environ Biotechnol. 1995;43:656–666. doi: 10.1007/BF00164770. [DOI] [PubMed] [Google Scholar]
- 29.Klotz M G, Klassen G R, Loewen P C. Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol Biol Evol. 1997;14:951–958. doi: 10.1093/oxfordjournals.molbev.a025838. [DOI] [PubMed] [Google Scholar]
- 30.Le Brun N E, Andrews S C, Guest J R, Harrison P M, Moore G R, Thomson A J. Identification of the ferroxidase centre of Escherichia colibacterioferritin. Biochem J. 1995;312:385–392. [PMC free article] [PubMed] [Google Scholar]
- 31.Loewen P C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Loewen, P. C., M. G. Klotz, and D. J. Hassett. Catalase—an “old” enzyme that continues to surprise us. Submitted for publication.
- 32.Loewen P C, Switala J. Purification and characterization of a spore-specific catalase-2 from Bacillus subtilis. Biochem Cell Biol. 1988;66:707–714. doi: 10.1139/o88-081. [DOI] [PubMed] [Google Scholar]
- 33.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCord J M, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 35.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. Procedures for working with lac; pp. 72–74. [Google Scholar]
- 36.Mliki A, Zimmermann W. Purification and characterization of an intracellular peroxidase from Streptomyces cyaneus. Appl Environ Microbiol. 1992;58:916–919. doi: 10.1128/aem.58.3.916-919.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore G R, Kadir F H A, Al-Massad F. Haem binding to ferritin and possible mechanisms of physiological iron uptake and release by ferritin. J Inorg Biochem. 1992;47:175–181. doi: 10.1016/0162-0134(92)84063-s. [DOI] [PubMed] [Google Scholar]
- 38.Moore G R, Kadir F H A, Al-Massad F K, Le Brun N E, Thomson A J, Greenwood C, Keen J N, Findlay J B C. Structural heterogeneity of Pseudomonas aeruginosabacterioferritin. Biochem J. 1994;304:493–497. doi: 10.1042/bj3040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Ochsner, U. A., et al. Unpublished data.
- 39.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pathogenesis Corporation and the Cystic Fibrosis Foundation. 15 March 1999, posting date. Pseudomonas Genome Project. [Online.] http://www.pseudomonas.com. [April 1999, last date accessed.]
- 41.Pena M M, Bullerjahn G S. The DpsA protein of Synechococcussp. strain PCC7942 is a DNA-binding hemoprotein: linkage of the Dps and bacterioferritin protein families. J Biol Chem. 1995;270:22478–22482. doi: 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- 42.Picard V, Epsztejn S, Santambrogio P, Cabantchik Z I, Beaumont C. Role of ferritin in the control of the labile iron pool in murine erythroleukemia cells. J Biol Chem. 1998;273:15382–15386. doi: 10.1074/jbc.273.25.15382. [DOI] [PubMed] [Google Scholar]
- 43.Schweizer H P. Improved broad-host-range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene. 1991;103:87–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- 44.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacBmarker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 45.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 46.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 47.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivogenetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 48.Swofford D L. PAUP: phylogenetic analysis using parsimony. Version 3.1.1 for the Macintosh. Washington, D.C: Smithsonian Institute; 1993. [Google Scholar]
- 49.Vasil M L, Ochsner U A, Johnson Z, Colmer J A, Hamood A N. The Fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator PtxR in Pseudomonas aeruginosa. J Bacteriol. 1998;180:6784–6788. doi: 10.1128/jb.180.24.6784-6788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wai S N, Nakayama K, Umene K, Moriya T, Amako K. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol Microbiol. 1996;20:1127–1134. doi: 10.1111/j.1365-2958.1996.tb02633.x. [DOI] [PubMed] [Google Scholar]
- 51.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]
- 52.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]