Abstract
Objectives
Seroprevalence studies can provide a measure of SARS-CoV-2 cumulative incidence, but a better understanding of spike and nucleocapsid (anti-N) antibody dynamics following infection is needed to assess the longevity of detectability.
Methods
Adults aged ≥18 years, from households enrolled in the Virus Watch prospective community cohort study in England and Wales, provided monthly capillary blood samples, which were tested for spike antibody and anti-N. Participants self-reported vaccination dates and past medical history. Previous polymerase chain reaction (PCR) swabs were obtained through Second Generation Surveillance System linkage data. The primary outcome variables were seropositivity and total anti-N and spike antibody levels after PCR-confirmed infection.
Results
A total of 13,802 eligible individuals provided 58,770 capillary blood samples. A total of 537 of these had a previous positive PCR-confirmed SARS-CoV-2 infection within 0-269 days of antibody sample date, among them 432 (80.45%) having a positive anti-N result. Median anti-N levels peaked between days 90 and 119 after PCR results and then began to decline. There is evidence of anti-N waning from 120 days onwards, with earlier waning for females and younger age categories.
Conclusion
Our findings suggest that anti-N has around 80% sensitivity for identifying previous COVID-19 infection, and the duration of detectability is affected by sex and age.
Keywords: Anti-N, Anti-S, Serosurveillance, COVID-19, Corona virus
Introduction
Antibodies produced following natural infection with SARS-CoV-2, the virus which causes COVID-19, are known to provide some protection against reinfection for at least 6 months in the early stages of the pandemic (Hansen et al., 2021). The proportion of infected individuals who are N-antibody (anti-N) and S-antibody (anti-S) positive and the stability of the antibody response over time are not well established (Siggins et al., 2021). In the United Kingdom, surveillance has been largely through symptomatic testing with reverse transcriptase polymerase chain reaction (RT-PCR) assays or asymptomatic testing through lateral flow device tests. In large-scale population surveys for the Office for National Statistics COVID Infection Study and the REal-time Assessment of Community Transmission (REACT) study, and UK Health Security Agency (formerly Public Health England) blood donation surveys, monitoring through seroprevalence is being carried out (Public Health, 2021; Coronavirus (COVID-19) 2022; Riley et al., 2021). Meta-analyses of the proportion of infections that are asymptomatic show that approximately one-third of cases do not develop symptoms at any point during acute infection (Beale et al., 2020).
Seroprevalence studies can provide a measure of cumulative incidence that accounts for asymptomatic infections, but more information on antibody waning is needed to aid the interpretation of these studies. The widespread use of COVID-19 vaccines in England and Wales (e.g., Pfizer, AstraZeneca, and Moderna) that only stimulate anti-S means that distinguishing antibodies derived from natural infection from those derived through vaccination requires measurement of both anti-N and anti-S (Favresse et al., 2021). A challenge with seroprevalence studies is that it is still unclear how long anti-S and anti-N remain in circulation after infection. The duration of which antibodies are detectable can also inform modeling approaches guiding the pandemic response, especially in countries where vaccine rollout is in the early stages. Much of the current evidence on antibody response or duration of detection is focused on specific occupational or institutional subgroups, with a reversion of anti-N at 242 days (e.g., healthcare, university, nursing homes) (Krutikov et al., 2022; Shrotri et al., 2021; Vusirikala et al., 2021). Assessing longer-term antibody responses across the population is critical to evaluate immune protection at the population level. Furthermore, anti-S are produced in response to both vaccination or previous infection and are therefore not an accurate measure of previous infection in countries that have rolled out widespread COVID-19 vaccination programs. Anti-N, which is only produced in response to SARS-CoV-2 infection or vaccines not available in the United Kingdom, may be a reliable option for serosurveillance, but little is known about the timeline of seroconversion nor duration of detectable levels of antibodies (Bochnia-Bueno et al., 2022).
To improve our understanding of the longevity of anti-N and anti-S responses over time, we evaluated both antibody detection and titers to establish factors that contribute to seropositivity and waning after infection, defined by a previous positive PCR. Specifically, we aimed to investigate:
-
1.
The proportion of individuals who are anti-N positive, within 269 days of PCR-confirmed infection, and associated demographic and clinical characteristics.
-
2.
Anti-N and anti-S detection and titers from 0-540 days since infection.
-
3.
Comparison of anti-N and anti-S detection and titers based on infection, vaccination, or both.
Methods
Study design and setting
The Virus Watch study is a household community cohort of acute respiratory infections in England and Wales that started recruitment in June 2020 (Hayward et al., 2021). As of August 31, 2021, 50,773 participants were recruited using various methods, including posting on social media, SMS messages, and letters from their general practice. Participants provided information on age, sex, ethnicity, household information (e.g., number of household members, postcode), and medical history (e.g., underlying medical conditions, medication history). Participants were followed up weekly by email with a link to a survey that captured information about vaccination status and SARS-CoV-2 infection. Between February and May 2021, invitations to participate in monthly antibody testing were sent to enrolled eligible households. Consenting participants provided capillary blood samples on a monthly basis up until August 31,2021.
Samples
Capillary blood samples (400-600 µl) were self-collected by participants using an at-home kit manufactured by the company, Thriva. Completed kits were returned by participants using prepaid envelopes and priority postage boxes to United Kingdom Accreditation Service-accredited laboratories for serological testing using Roche's Elecsys Anti-SARS-CoV-2 electrochemiluminescence assays targeting total immunoglobulin to the nucleocapsid (N) protein, or to the receptor binding domain in the S1 subunit of the spike protein (S) (Roche Diagnostics, Basel, Switzerland (Public Health, 2020). Results for anti-N were reported as numeric values in the form of a cut-off index (COI). The detection limit is determined by the ratio of the luminescence of the sample relative to the predefined negative threshold. This threshold was determined by calibrating against known negative samples (Egger et al., 2020). At the manufacturer-recommended seropositivity thresholds (≥1.0 COI for N and ≥0.8 units per milliliter [U/ml] for S), the N assay has a sensitivity of 97.2-99.5% and specificity of 99.8%, whereas the S assay has a sensitivity of 97.9-98.8% and a specificity of 100%, with a high agreement between the assays for samples from previously infected individuals (Ainsworth et al., 2020; Public Health England, n.d.; Riester et al., 2021). All samples processed between February 24, 2021 (first samples) to August 31, 2021 were included in the analysis of anti-N and, depending on vaccination status, anti-S results. For anti-S levels, samples processed before July 1 (with the exception of a 2-day pilot) had an upper limit of 250 U/ml. After this date, if sample results exceeded the upper limit of the analytical measuring interval, they underwent dilution with a diluent universal of 1: 10 up to 1: 100, providing an upper limit of 25,000 U/ml (Roche Diagnostics GmbH, 2020)
Covariates
Age, sex, ethnicity, underlying health conditions, body mass index, and whether participants were taking immunosuppressants were self-reported during study registration. Age was grouped into the following categories: 18-34, 35-49, 50-64, 65-79, and ≥80 years. Ethnicity data were grouped into Black, White, South Asian, Other Asian, Mixed categories, and Other/missing. Sex was limited to male and female categories. Participants were asked to report specific health conditions, which were grouped as part of the analysis (further details in Appendix Table 1 ).
Table 1.
Proportion of those with positive anti-N result, grouped by age, sex and vaccination status
| Anti-N Positive (n / N) | Percentage Positive (%) | 95% CI | |
|---|---|---|---|
| Age category | |||
| 18-34 | 58 / 79 | 73∙42% | (63∙68, 83∙16) |
| 35-49 | 121 / 143 | 84∙62% | (78∙7, 90∙53) |
| 50-64 | 151 / 193 | 78∙24% | (72∙42, 84∙06) |
| 65-79 | 100 / 120 | 83∙33% | (76∙66, 90) |
| 80+ | 2 / 2 | 100% | (100, 100) |
| Sex | |||
| Female | 253 / 324 | 78∙09% | (73∙58, 82∙59) |
| Male | 177 / 211 | 83∙89% | (78∙93, 88∙85) |
| Missing data | 2 / 2 | 100% | (100, 100) |
| Vaccinated | |||
| No | 101 / 116 | 87∙07% | (80∙96, 93∙18) |
| Yes | 308 / 396 | 77∙78% | (73∙68, 81∙87) |
| Missing data | 23 / 25 | 92% | (81∙37, 102∙63) |
The primary source of data was the Virus Watch dataset linked to the Second Generation Surveillance System (SGSS), which contains SARS-CoV-2 PCR test results. The linkage period for SGSS encompassed data from April 2020 until August 2021 due to data availability. The earliest positive test result was taken as the date for PCR antibody time analysis. We assumed that participants who did not have a positive PCR result in the SGSS never had virologically confirmed SARS-CoV-2 infection and were therefore excluded from the analysis.
Vaccination status was collected through the weekly Virus Watch questionnaire. This question was added to the weekly surveys on January 11, 2021, where individuals reported the dates of their vaccination. Vaccination type was recorded in data collection and used to establish the national licensure date for each type of vaccine (e.g., Pfizer December 2, 2020, AstraZeneca December 30, 2020, and Moderna January 8, 2021). Results were excluded from analysis if the reported vaccination date preceded the national licensure date or if a second dose was reported but not a first dose. This was specifically relevant for comparison of anti-S and anti-N responses after infection among those who had not been vaccinated or provided a sample prevaccination. When analyzing the response to vaccination in those with no evidence of previous infection, only samples with negative anti-N were included.
Primary outcome(s)
The main outcome variable was the detection of anti-N, expressed as a binary variable (≥1.0 COI), and anti-N level, expressed as the semiquantitative COI and subsequently log-transformed to base 10.
Secondary outcomes
The secondary outcome was a comparison of seropositivity to anti-S and anti-N and levels in relation to previous infection and vaccination.
Participants
Within the Virus Watch cohort, eligible households were defined as having at least one adult aged 18 years and older, a valid England or Wales postcode, and a complete postal address registered (Hayward et al., 2021). Individuals that were 18 years and older within eligible households could consent to participate through the provision of valid electronic consent. Individuals were included in this analysis if they had provided at least one finger-prick blood sample. Evidence of a previous infection was defined as seropositivity to the nucleocapsid protein (COI ≥1.0) among vaccinated individuals or seropositivity to either spike (≥0.8 U/ml) or nucleocapsid protein among unvaccinated individuals. Samples with void anti-N or void anti-S results were excluded from the analyses.
Analysis
Analyses were carried out using two approaches: at the individual level and at the sample level. At individual level, samples obtained between 0-269 days after positive SARS-CoV-2 PCR were aggregated, selecting the sample with the first positive anti-N result, if there was more than one sample per participant. Individual-level data was used to compare age, sex, ethnicity, body mass index, or underlying medical conditions and if taking immunosuppressants. Proportions of individuals positive for anti-N or anti-S were calculated with 95% confidence intervals. To calculate the odds of seroconversion after infection, and explore which explanatory variables influence this, univariable and multivariable logistic regression models were created using individual-level data from those with a previous positive PCR result. The multivariable model was adjusted for age and sex at birth, when they were not the explanatory variable being investigated. The explanatory variables included age categories, sex at birth, conditions associated with severe COVID-19 infection, obesity, and whether they were taking any immunosuppressants or not (see Supplementary files; Table 1).
For antibody levels, as they are not normally distributed, median values for each group were calculated with interquartile ranges (IQRs). Due to the available assay platform and dilution capabilities, the dynamic range of the anti-S assay was limited to 0.4-250 U/ml for samples processed before between February and June 2021. Only samples that underwent further dilution from July 2021 onwards were included in anti-S levels analysis. As dilution capabilities did not affect the threshold for a positive result, all anti-S samples were included when calculating the proportion of positivity. For the analysis of antibody positivity and levels by days after PCR-confirmed infection (e.g., 0-29, 30-59, 60-89, 90-119, 120-269, and >270 days after a positive PCR), all samples were included. The proportion of anti-N positive samples, with 95% confidence intervals, and median antibody levels were calculated for each time-period category. Anti-N positivity and levels over time were stratified by age groups (18-49 and >50) and sex. Comparison of anti-N and anti-S positivity levels were made for unvaccinated individuals or those who provided a sample before vaccination.
Comparison of anti-N and anti-S positive levels were made between those with a previous infection only (no vaccination) and those who had both vaccination and previous infection. Further comparison was made on anti-S positivity over time for those with a previous infection only (no vaccination) and previous vaccination only (no infection), using the date of the PCR test or vaccination date as day 0. Antibody levels were not normally distributed; therefore, nonparametric tests were used to compare groups: the Mann-Whitney U (Wilcoxon rank sum) test for two groups and the Kruskal-Wallis test for more than two groups, with Benjamini and Hochberg correction. For the comparison of binary variables, the chi-square test was used. All analyses were carried out with R studio (R 4.0.5.) using packages: ‘tidyverse’, ‘ggplot2’, ‘flextable’, and ‘rstatix’.
Results
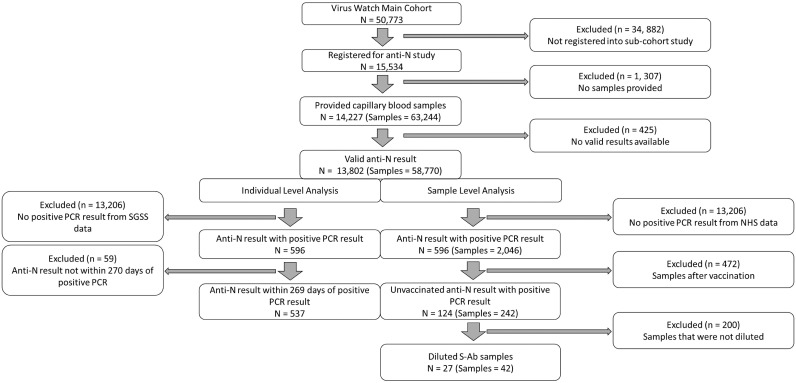
A total of 15,534 participants consented and were eligible to participate in the study, of whom 13,802 participants provided 58,770 valid anti-N samples (Figure 1 ). The median age of participants was 63 (IQR 46, 80) years, and of 13,765 with a recorded sex at baseline survey, 57.07% (n = 7,856) were female. A total of 5,331 samples from 1,479 individuals were anti-N positive.
Figure 1.
Study inclusion flow diagram.
Of the 13,802 individuals with valid antibody results, 596 had a previous positive PCR result from the SGSS linkage data. Of those who did not have a previous positive PCR result in the SGSS linkage data, 8.2% (999/13,206) were positive for anti-N. A total of 537 of those individuals had samples within 269 days (59 excluded) of their PCR result and were therefore included in the individual-level analysis. A total of 80.44% of individuals (432/537) were seropositive for anti-N after PCR-confirmed infection (see Table 1 for groups by age, sex, and vaccination status and Supplementary file Table 2 and 3 for breakdown by medical conditions). A univariable logistic regression model showed no evidence for a difference in being seropositive by age, sex, or health condition. A multivariable logistic regression found higher odds of being seropositive if aged between 35 and 49 than 18 and 34 years (adjusted OR 1.98; p = 0.04), but no association was found for sex, obesity, conditions related to severe COVID-19 infection, or if taking any immunosuppressant (Supplementary file, Table 4).
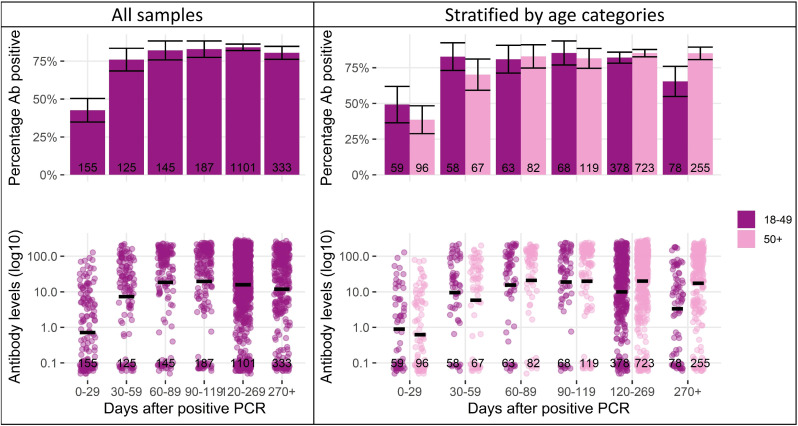
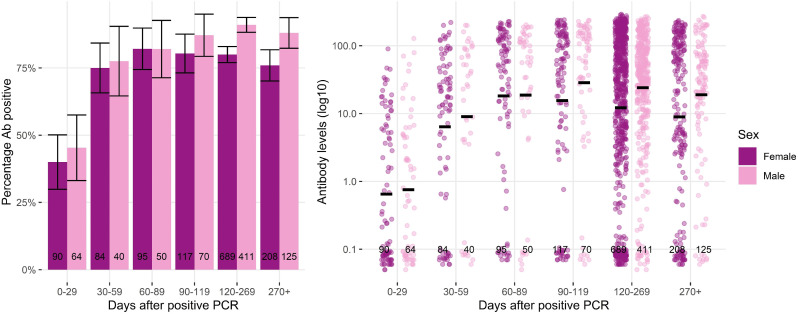
We examined anti-N positivity and titers over time after infection, with PCR test date as day 0 (Figure 2 ). The minimum and maximum days after PCR were 1 day and 520 days (median 168.5 days, IQR 103.63-233.38), respectively. The proportion of samples with detectable levels of anti-N increased from 42.6% at days 0-29 after PCR (66/155, 95% CI 35-50) to 76% at 30-59 days (95/125, 95% CI 69-83), and then remained relatively stable between 80-85% at 60-89, 90-119, 120-269, and >270 days. Median anti-N titers reached a peak at around 60 COI between days 30 and 120, before beginning to decline. Patterns were similar in both sexes and different age groups, although with some evidence of a faster decline in younger adults (aged 18-49 years) and in women (Figure 2 and 3 ). Men had a higher proportion of samples with detectable levels of anti-N on days 120-269 (91%, 95% CI 88-94) and 270+ (88%, 95% 82-94) than women (80%, 95% CI 77-83 and 76%, 95% CI 70-82, respectively). Men also had higher median anti-N levels than in days 120-269 (39.9, IQR 13.32-98.95) than women (30.6, IQR 3.01-116, P = 0.01). Those aged >50 years had a higher proportion of samples, with detectable levels of anti-N on days >270 (85.1%, 95% CI 81-89) than those aged 18-49 years (65.4%, 95% CI 55-76). Median anti-N levels were higher on days 120-269 and >270 for samples from those aged >50 years (48.6 COI, IQR 8.21-128, and 46.5 COI, IQR 4.9-113.75, respectively) than those aged 18-49 years (18.8 COI, IQR 3.08-65.98, P <0.001, and 6.5 COI, IQR 0.18-31.79, P <0.001, respectively).
Figure 2.
Anti-N percentage positivity and levels, stratified by days post positive PCR test and age. Notes: 95% confidence intervals represented by vertical cross bars and anti-N levels underwent Log10 transformation with bars representing the logarithmic mean. PCR = polymerase chain reaction.
Figure 3.
Anti-N percentage positivity and levels by sex and days after positive PCR. Notes: 95% confidence intervals represented by vertical cross bars and anti-N levels underwent Log10 transformation with bars representing the logarithmic mean. PCR = polymerase chain reaction.
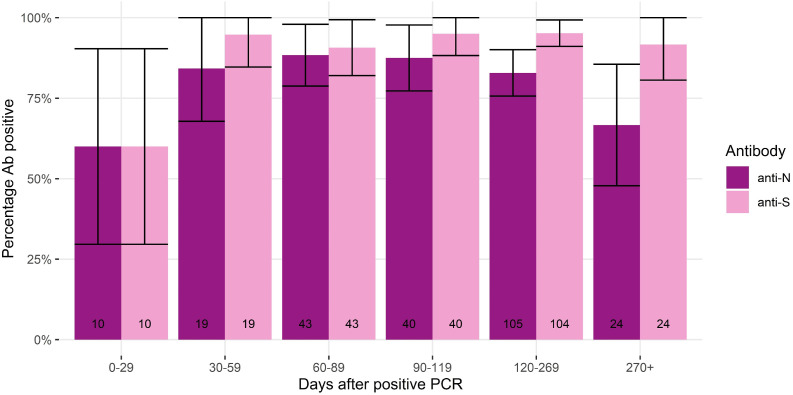
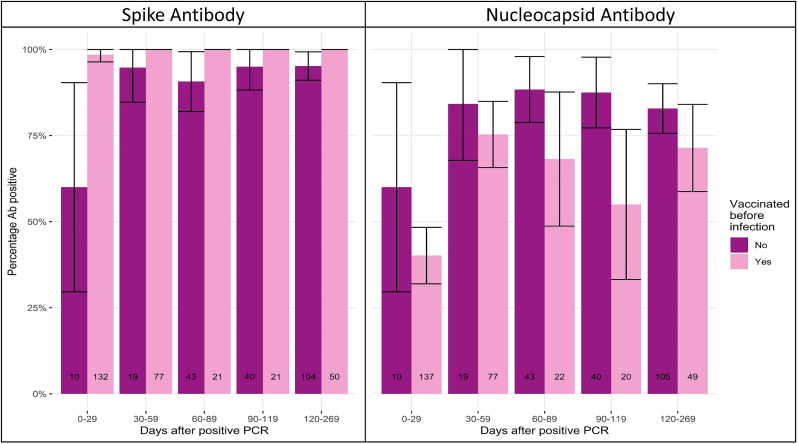
Of the 2,046 samples, 244 belonged to 124 individuals with only a previous infection and no previous vaccination or with only a previous infection and were vaccinated after providing a capillary blood sample. At 0-29 days, 60% (95% CI 30-90%) of samples had both detectable anti-S and anti-N, but anti-S detectability had a sharper increase than anti-N and remained above 90% during the follow-up period (Figure 4 ).
Figure 4.
Percentage antibody positivity anti-N and anti-S among pre-vaccinated samples or samples taken from unvaccinated individuals, stratified by days post virological confirmed infection. Notes: 95% confidence intervals represented by vertical cross bars.
Anti-N positivity was generally higher for samples from those with no previous vaccination (N = 244) than samples from those with vaccination before infection (N = 305), with the most notable difference on days 90-119; 87.8% (95% CI 78-98) versus 55% (95% CI 33-77, P = 0.011). Anti-N titers from samples from unvaccinated individuals had higher median levels than in samples from those who had a previous vaccination (39.6 COI vs 6.53 COI, P <0.0001). The proportion of anti-S positivity was generally higher for samples taken from individuals with a previous vaccination (N = 301) than those without a previous vaccination (N = 244), with the most notable difference on days 0-29; 60% (95% CI 30-90) versus 98.5% (95% CI 96-100), P <0.0001. The proportion of anti-S positivity was higher in samples from individuals with only a previous vaccination (N = 46,647) than those with infection only (N = 244), from 0-269 days. There were no results for >270 days after vaccination and therefore were not included in this section of the analysis. The median age in the vaccination only group was higher than the infection only group (65 vs 43 years old, P <0.0001) (Figure 5 ).
Figure 5.
Proportion of antibody positivity for anti-S and anti-N by days post virological confirmed infection, stratified by vaccination prior to infection. Notes: 95% confidence intervals represented by vertical cross bars.
Discussion
We present the findings of a large community cohort study, demonstrating anti-S and anti-N trends in participants with antibody results from 1-540 days since PCR-confirmed infection. Our study found that approximately 4 of 5 individuals were seropositive for anti-N at any point between 0 and 269 days after a PCR-confirmed SARS-CoV-2 infection. Logistic regression models, both univariable and multivariable, only showed higher odds of a positive ant-N result for those aged 35-49 years than those aged 18-34 years. The peak proportion in positivity for anti-N and median antibody levels was 120-269 days and 90-119, respectively, after infection. The proportion of seropositivity and median antibody levels were significantly higher among men than women from 120 days onwards, which was a similar pattern observed in those aged ≥50 years compared with those aged 18-49 years. Samples of those who were vaccinated before PCR-confirmed infection had a lower median anti-N and higher anti-S 30-269 days after infection. The proportion of anti-S positive samples were higher for vaccination only than infection only over time.
Previous studies have found that anti-N positivity after PCR-confirmed infection ranges from 84.7% at 28 days to 68.2% at 293 days, with proportions being highest among those with severe symptom profiles (Alfego et al., 2021; Thiruvengadam et al., 2021). Antibody positivity may be influenced by the viral load during infection, with higher viral loads causing higher levels of antigen exposure and more severe symptoms (Ali et al., 2021). As our data are from a community cohort study, it can be assumed that the symptom profiles may be less severe among our participants than in hospital-based longitudinal studies, therefore leading to lower antibody levels.
Increasing age and sex in some studies have been associated with higher anti-S and anti-N immunoglobulin G responses (Alfego et al., 2021; Choudhary et al., 2021). The only factor independently associated with seropositivity in the logistic regression model was age, with those aged 35-49 years more likely to be seropositive than those aged 18-34 years. Yet, when assessing anti-N response over time, there was a difference between age categories 18-49 and >50 on days 120-269 and >270, with the former having an earlier peak and earlier antibody waning. Older individuals have a higher frequency of comorbidities, putting them at higher risk of severe disease, which may explain longer duration of anti-N positivity (Zheng et al., 2020). This being said, there were no higher or lower odds of being seropositive if individuals have medical conditions associated with a higher risk of COVID-19-related mortality or taking immunosuppressant therapy. This is likely due to the small number of participants in these groups in our analyses (see Supplementary file Table 5). Although limited to 269 days after infection because of disparity in length of follow-up between individuals, the time interval between PCR and anti-N results was not accounted for in our models. Seropositivity has been shown to be affected by days since PCR, which may also explain why some differences were not found in our regression model (Alfego et al., 2021).
The proportion of seropositive anti-N samples was above 80% from 30 days onwards, but only 42.6% of samples were positive between days 0 and 29. Although this is lower than other studies, where seroconversion rates have been 84.7% at 10-28 days, respectively, these studies followed individuals who had been hospitalized due to COVID-19 (Thiruvengadam et al., 2021). When hospitalized individuals were stratified by symptom severity, the cumulative percentage of anti-N positive asymptomatic individuals was 60% at 22-28 days (Thiruvengadam et al., 2021). Other studies on hospitalized individuals have also demonstrated that symptom severity is associated with an earlier peak in anti-N titers as well as determining longer duration of anti-N positivity (Feng et al., 2021; Van Elslande et al., 2021). A difference in anti-N over time was seen when the data were disaggregated by sex, with earlier peak and waning in females. This may also be related to risk of disease severity, with a meta-analysis showing that men are 2.41 times more at risk of developing severe disease than women (MdA et al., 2020). Higher angiotensin-converting enzyme 2 expression, decreased B cell, and natural killer cell-specific transcripts in men are suggested to be the cause of higher viral loads and therefore, more severe symptoms (Bwire, 2020; Lieberman et al., 2020). Although we have not used symptom severity in our analysis, because Virus Watch is a community cohort study, it can be assumed that in comparison, the symptom profiles are likely to be milder on average. This would explain a lower percentage of anti-N positive samples at 0-29 days but provides a more accurate representation of anti-N longevity in the community setting.
After excluding postvaccination samples, there was evidence of anti-N waning in both proportions of detectable anti-N in samples and median anti-N titers, which was not the case for anti-S. Earlier anti-N waning has also been observed in other studies, which monitored immunoglobulin G (Alfego et al., 2021; Choudhary et al., 2021). A higher proportion of unvaccinated samples were anti-N positive than samples from participants who had vaccination before infection. This trend was also reflected in median anti-N and ant-S titers. This finding is important when considering the use of anti-N as an alternative to anti-S for seroprevalence studies in highly vaccinated populations. The anti-S response seen between vaccinated and unvaccinated is in keeping with the immunological mechanism of the vaccines, which induces an anti-S specific response. The proportion of anti-S positive results in the vaccination only cohort was higher at days 120-269 than anti-S response in the infection only group, indicating a more sustained anti-S response from vaccination. Although anti-S levels may differ between these groups, this is not necessarily an indication of risk of future infection. Comparison of infection risk between vaccination and natural immunity has provided conflicting information, with pooled results of randomized control trials showing no difference and observational studies favoring natural immunity (Choudhary et al., 2021). Our results may be the outcome of confounding, however, because the median age of the vaccine only group was higher than infection only.
The strengths of this study include a large sample size that spans various age groups and captures multiple underlying health conditions (see Supplementary file Table 6). We present serial antibody measurements using a highly sensitive, widely used validated commercial assay that provides quantitative readings (Public Health, 2020). There are limitations, however, because a large proportion of the data is self-reported and therefore susceptible to reporting bias because participants may only volunteer information they feel is relevant or necessary. Self-reporting is also susceptible to data entry errors. This led to some samples being excluded from any analyses based on erroneous reporting of vaccination status. Furthermore, a large proportion of the data in this analysis (e.g., medical conditions, medications) is only collected at registration, so it did not account for changes in participant's health or medications. The dates of PCR-confirmed infection, however, were SGSS data, allowing accurate calculation of time between infection and blood test. Those who did not have a positive PCR-confirmed infection according to the linkage data were excluded from the analysis, therefore further reducing the sample size. The earliest positive PCR result was used in the analysis, and subsequent PCR results were excluded. Therefore, we are unable to report on subsequent asymptomatic reinfections/re-exposure, which may boost antibody levels (Ali et al., 2021; Dispinseri et al., 2021).
Conclusion
As only 4 in 5 participants with a previous PCR-confirmed infection were anti-N positive at any time point up to 269 days after infection, seroprevalence studies on anti-N alone may underestimate the true cumulative incidence of infection. We have demonstrated a decline in anti-N levels from 120 days onwards, providing a better understanding of the limitations of seroprevalence studies. The duration of anti-N positivity is affected by age and sex; therefore, serosurveillance may require a shorter time window of testing after infection.
CRediT authorship contribution statement
Annalan M D Navaratnam: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Madhumita Shrotri: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Vincent Nguyen: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Isobel Braithwaite: Investigation, Methodology, Writing – review & editing. Sarah Beale: Data curation, Investigation, Methodology, Writing – review & editing. Thomas E Byrne: Investigation, Methodology, Writing – review & editing. Wing Lam Erica Fong: Data curation, Investigation, Methodology, Writing – review & editing. Ellen Fragaszy: Investigation, Methodology, Writing – review & editing. Cyril Geismar: Data curation, Investigation, Methodology, Writing – review & editing. Susan Hoskins: Investigation, Methodology, Writing – review & editing. Jana Kovar: Investigation, Methodology, Writing – review & editing. Parth Patel: Investigation, Methodology, Writing – review & editing. Alexei Yavlinsky: Investigation, Methodology, Writing – review & editing. Anna Aryee: Investigation, Methodology, Writing – review & editing. Alison Rodger: Investigation, Methodology, Writing – review & editing. Andrew C Hayward: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. Robert W Aldridge: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Susan Michie: . Pia Hardelid: . Linda Wijlaars: . Eleni Nastouli: . Moira Spyer: . Ben Killingley: . Ingemar Cox: . Vasileios Lampos: . Rachel A McKendry: . Tao Cheng: . Yunzhe Liu: . Jo Gibbs: . Richard Gilson: . Anne M Johnson: .
Acknowledgments
Conflicts of interest
ACH serves on the United Kingdom New and Emerging Respiratory Virus Threats Advisory Group. AMJ was a Governor of Wellcome Trust from 2011-18 and is Chair of the Committee for Strategic Coordination for Health of the Public Research.
Funding Source
The research costs for the study have been supported by the MRC Grant Ref: MC_PC 19070 awarded to UCL on March 30, 2020 and MRC Grant Ref: MR/V028375/1 awarded on August 17, 2020. The study also received $15,000 of Facebook advertising credit to support a pilot social media recruitment campaign on August 18, 2020. The study also received funding from the United Kingdom Government Department of Health and Social Care's Vaccine Evaluation Program to provide monthly Thriva antibody tests to adult participants. This study was supported by the Wellcome Trust through a Wellcome Clinical Research Career Development Fellowship to RA [206602].
Ethical Approval Statement
This study has been approved by the Hampstead NHS Health Research Authority Ethics Committee (ethics approval number – 20/HRA/2320).
Data availability
The authors aim to share aggregate data from this project on our website and through a “Findings so far” section on our website: https://ucl-virus-watch.net/. The authors will also be sharing individual record-level data on a research data sharing service, such as the Office of National Statistics Secure Research Service. In sharing the data, the authors will work within the principles set out in the UKRI Guidance on best practice in the management of research data. Access to use of the data while research is being conducted will be managed by the chief investigators (ACH and RWA) in accordance with the principles set out in the UKRI guidance on best practice in the management of research data. The authors will put the analysis code on publicly available repositories to enable their reuse.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.07.053.
Contributor Information
Virus Watch Collaborative:
Susan Michie, Pia Hardelid, Linda Wijlaars, Eleni Nastouli, Moira Spyer, Ben Killingley, Ingemar Cox, Vasileios Lampos, Rachel A McKendry, Tao Cheng, Yunzhe Liu, Jo Gibbs, Richard Gilson, and Anne M Johnson
Appendix. Supplementary materials
References
- Ainsworth M, Andersson M, Auckland K, Baillie JK, Barnes E, Beer S, et al. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. 2020;20:1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfego D, Sullivan A, Poirier B, Williams J, Adcock D, Letovsky S. A population-based analysis of the longevity of SARS-CoV-2 antibody seropositivity in the United States. EClinicalmedicine. 2021;36 doi: 10.1016/j.eclinm.2021.100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AM, Ali KM, Fatah MH, Tawfeeq HM, Rostam HM. SARS-CoV-2 reinfection in patients negative for immunoglobulin G following recovery from COVID-19. New Microbes New Infect. 2021;43 doi: 10.1016/j.nmni.2021.100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale S, Hayward A, Shallcross L, Aldridge RW, Fragaszy E. A rapid review and meta-analysis of the asymptomatic proportion of PCR-confirmed SARS-CoV-2 infections in community settings. Wellcome Open Res. 2020;5 [Google Scholar]
- Bochnia-Bueno L, De Almeida SM, Raboni SM, Adamoski D, Amadeu LLM, Carstensen S, et al. Dynamic of humoral response to SARS-CoV-2 anti-Nucleocapsid and Spike proteins after CoronaVac vaccination. Diagnostic Microbiology and Infectious Disease. 2022;102(3):115597. doi: 10.1016/j.diagmicrobio.2021.115597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwire GM. Coronavirus: why men are more vulnerable to Covid-19 than women? SN Compr Clin Med. 2020;2:1–3. doi: 10.1007/s42399-020-00341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary HR, Parai D, Dash GC, Peter A, Sahoo SK, Pattnaik M, et al. IgG antibody response against nucleocapsid and spike protein post-SARS-CoV-2 infection. Infection. 2021;49:1045–1048. doi: 10.1007/s15010-021-01651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus (COVID-19). UK: Infection Survey, 2022, p. 16. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/25june2021.
- Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 2021;12:2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Bundschuh C, Wiesinger K, Gabriel C, Clodi M, Mueller T, et al. Comparison of the Elecsys® anti-SARS-CoV-2 immunoassay with the EDITM enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta Int J Clin Chem. 2020;509:18–21. doi: 10.1016/j.cca.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favresse J, Cadrobbi J, Eucher C, Elsen M, Laffineur K, Dogné JM, et al. Clinical performance of three fully automated anti-SARS-CoV-2 immunoassays targeting the nucleocapsid or spike proteins. J Med Virol. 2021;93:2262–2269. doi: 10.1002/jmv.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Yin J, Zhang J, Hu Y, Ouyang Y, Qiao S, et al. Longitudinal profiling of antibody response in patients with COVID-19 in a Tertiary Care Hospital in Beijing. China. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.614436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Fragaszy E, Kovar J, Nguyen V, Beale S, Byrne T, et al. Risk factors, symptom reporting, healthcare-seeking behaviour and adherence to public health guidance: protocol for Virus Watch, a prospective community cohort study. BMJ, (Open) 2021;11 doi: 10.1136/bmjopen-2020-048042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutikov M, Palmer T, Tut G, Fuller C, Azmi B, Giddings R, et al. Prevalence and duration of detectable SARS-CoV-2 nucleocapsid antibodies in staff and residents of long-term care facilities over the first year of the pandemic (VIVALDI study): Prospective cohort study in England. The Lancet Healthy Longevity. 2022;3(1):e13–e21. doi: 10.1016/S2666-7568(21)00282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLOS Biol. 2020;18 doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MdA Barek, MdA Aziz, Islam MS. Impact of age, sex, comorbidities and clinical symptoms on the severity of COVID-19 cases: a meta-analysis with 55 studies and 10014 cases. Heliyon. 2020;6:e05684. doi: 10.1016/j.heliyon.2020.e05684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England. Sero-surveillance of COVID-19 (2021). https://www.gov.uk/government/publications/national-covid-19-surveillance-reports/sero-surveillance-of-covid-19, n.d. (accessed 3 December 2021).
- Public Health England. Evaluation of Roche Elecsys anti-SARS-CoV-2 serology assay for the detection of anti-SARS-CoV-2 antibodies (2020). https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/891598/Evaluation_of_Roche_Elecsys_anti_SARS_CoV_2_PHE_200610_v8.1_FINAL.pdf, n.d. (accessed 1 February 2022).
- Riester E, Findeisen P, Hegel JK, Kabesch M, Ambrosch A, Rank CM, et al. Performance evaluation of the Roche Elecsys anti-SARS-CoV-2 S immunoassay. J Virol Methods. 2021;297 doi: 10.1016/j.jviromet.2021.114271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S, Wang H, Eales O, Haw D, Walters CE, Ainslie KEC, et al. REACT-1 round 12 report: resurgence of SARS-CoV-2 infections in England associated with increased frequency of the Delta variant. MedRxiv. 2021 doi: 10.1101/2021.06.17.21259103v1. 1 June, Accessed 1 February 2022. [DOI] [Google Scholar]
- Roche Diagnostics GmbH (2020). Elecsys Anti-SARS-CoV-2 S. https://www.fda.gov/media/144037/download, n.d. (accessed 1 February 2022).
- Shrotri M, Harris RJ, Rodger A, Planche T, Sanderson F, Mahungu T, et al. Persistence of SARS-CoV-2 N-antibody response in healthcare workers, London, UK. Emerg Infect Dis. 2021;27:1155–1158. doi: 10.3201/eid2704.204554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggins MK, Thwaites RS, Openshaw PJM. Durability of immunity to SARS-CoV-2 and other respiratory viruses. Trends Microbiol. 2021;29:648–662. doi: 10.1016/j.tim.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvengadam R, Chattopadhyay S, Mehdi F, Desiraju BK, Chaudhuri S, Singh S, et al. Longitudinal serology of SARS-CoV-2-Infected individuals in India: a prospective cohort study. Am J Trop Med Hyg. 2021;105:66–72. doi: 10.4269/ajtmh.21-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J, Oyaert M, Ailliet S, Van Ranst M, Lorent N, Vande Weygaerde Y, et al. Longitudinal follow-up of IgG anti-nucleocapsid antibodies in SARS-CoV-2 infected patients up to eight months after infection. J Clin Virol. 2021;136 doi: 10.1016/j.jcv.2021.104765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vusirikala A, Whitaker H, Jones S, Tessier E, Borrow R, Linley E, et al. Seroprevalence of SARS-CoV-2 antibodies in university students: cross-sectional study, December 2020, England. J Infect. 2021;83:104–111. doi: 10.1016/j.jinf.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81:e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors aim to share aggregate data from this project on our website and through a “Findings so far” section on our website: https://ucl-virus-watch.net/. The authors will also be sharing individual record-level data on a research data sharing service, such as the Office of National Statistics Secure Research Service. In sharing the data, the authors will work within the principles set out in the UKRI Guidance on best practice in the management of research data. Access to use of the data while research is being conducted will be managed by the chief investigators (ACH and RWA) in accordance with the principles set out in the UKRI guidance on best practice in the management of research data. The authors will put the analysis code on publicly available repositories to enable their reuse.