Abstract
Membraneless organelles have emerged during the evolution of eukaryotic cells as intracellular domains in which multiple proteins organize into complex structures to perform specialized functions without the need of a lipid bilayer compartment. Here we describe the perinuclear space of eukaryotic cells as a highly organized network of cytoskeletal filaments that facilitates assembly of biomolecular condensates. Using bioinformatic analyses, we show that the perinuclear proteome is enriched in intrinsic disorder with several proteins predicted to undergo liquid-liquid phase separation. We also analyze immunofluorescence and transmission electron microscopy images showing the association between the nucleus and other organelles, such as mitochondria and lysosomes, or the labeling of specific proteins within the perinuclear region of cells. Altogether our data support the existence of a perinuclear dense sub-micron region formed by a well-organized three-dimensional network of structural and signaling proteins, including several proteins containing intrinsically disordered regions with phase behavior. This network of filamentous cytoskeletal proteins extends a few micrometers from the nucleus, contributes to local crowding, and organizes the movement of molecular complexes within the perinuclear space. Our findings take a key step towards understanding how membraneless regions within eukaryotic cells can serve as hubs for biomolecular condensates assembly, in particular the perinuclear space. Finally, evaluation of the disease context of the perinuclear proteins revealed that alterations in their expression can lead to several pathological conditions, and neurological disorders and cancer are among the most frequent.
Keywords: Intrinsically disordered protein/region, Liquid-liquid phase separation, Biomolecular condensates, Membraneless organelles, Perinuclear, Nuclear cloud
1. Introduction
During evolution, different eukaryotic cells have emerged with highly specialized structures and functions. Despite these differences, all eukaryotic cells share common intracellular membranous organelles (MOs), such as the nucleus, endoplasmic reticulum, Golgi apparatus, lysosomes, mitochondria, peroxisomes, and endosomes. MOs are important eukaryotic achievements, as they allow specialized processes, such as energy production and synthesis of macromolecules, to occur concomitantly at different subcellular locations within a single cell. Along with MOs, membraneless organelles (MLOs) have also evolved in eukaryotes to organize the complexity of the intracellular space [1]. Well-characterized examples of MLOs include the nucleolus in the nucleus [2,3], as well as centrosomes, ribosomes, and proteasomes in the cytoplasm [4,5]. Recently, it has become clear that MLOs are liquid compartments assembled through liquid-liquid phase separation (LLPS) [4,6,7]. The unique dynamic properties of liquids (coalesce, deform, show fusion and fission behavior) allow fine spatiotemporal control of diverse cellular reactions (reviewed in Boeynaems et al. [8]).
This prompted us to postulate that the perinuclear space acts as a hub for condensation, favoring multivalent contacts and enhancing phase separation. Several “client” proteins may partition into condensates to perform signaling functions. In addition, membranous organelles located in the perinuclear space such as the endoplasmic reticulum, mitochondria, lysosomes, vesicles and endosomes, can function together with biomolecular condensates intertwined in cytoskeleton networks (reviewed in Koppers et al. [9]). To summarize, when proteins with LLPS ability reach the perinuclear region, they are prone to phase separate in this peculiarly crowded sub-region.
The perinuclear space has different functions, such as regulating the traffic of molecules between the nucleus and the cytosol, providing structural support for the nucleus, controlling nuclear size and position, modulating several intracellular signaling pathways, and allowing a dynamic interaction between the nucleus and other organelles and structures [10]. Therefore, our hypothesis is that to perform these complex functions, this region concentrates intrinsically disordered proteins (IDPs) with phase behavior that is tunable by specific molecules (e.g., protein, nucleic acid (NA), post-translational modifications) in different moments. Collectively, the perinuclear cloud should be a well-organized three-dimensional region favorable to the montage of several biomolecular condensates in eukaryotic cells.
To test this hypothesis, we investigated (i) proteins with a reported perinuclear localization by a systematic bibliometric review; (ii) by in silico analysis, the IDPs that have a perinuclear distribution; and (iii) images from fluorescence and transmission electron microscopy (TEM) showing the contact between the nucleus and other organelles, and the gathering of specific proteins within the juxtanuclear region of cells. Since we hypothesize that the nuclear space has both structural and signaling properties, we discuss the prevalence of predicted intrinsic disorder content and phase separation ability of proteins with known perinuclear distribution.
2. Materials and methods
2.1. Antibodies and probes
Rabbit polyclonal antibodies against desmin (code# D-8281) and LMO7 (code # HPA020923), and mouse monoclonal antibody antisarcomeric alpha-actinin (code# A-7871, clone EA-53) were purchased from Sigma-Aldrich (USA). Rabbit monoclonal anti-Gli-1 (ab134906) antibody was from Abcam (UK). DNA-binding probe 4,6-Diamino-2-phenylindole dihydrochloride (DAPI), Alexa Fluor 488-goat anti-rabbit IgG antibodies and Alexa Fluor 546-goat anti-mouse IgG antibodies were from Molecular Probes (USA).
2.2. Embryonic chick skeletal muscle cell cultures
Primary cultures of myogenic cells were prepared from breast muscles of 11-day-old chick embryos [11]. Embryonated chick eggs were obtained from Granja Tolomei (Rio de Janeiro, Brazil) and handled according to Institutional Animal Care and Use Committee protocols under the number 069/19. Briefly, fragments of pectoral muscle were incubated at 37 °C for 10 min in calcium-magnesium-free solution (CMF, Sigma-Aldrich) containing 0.25% trypsin (Sigma-Aldrich). After removal of the trypsin solution, cells were dispersed by repeated pipetting in culture medium (Minimum Essential Medium with 10% horse serum, 0.5% chick embryo extract, 1% l-glutamine, and 1% penicillin/streptomycin, all from Invitrogen, Brazil). The resulting suspension was filtered, and cells were plated at an initial density of 7.5 × 105 cells/35 mm culture dishes in 2 mL of medium. Cells were cultured on 22-mm Aclar plastic coverslips (Pro-Plastics Inc., USA) previously coated with rat tail collagen. Cells were grown in a humidified 5% CO2 atmosphere at 37 °C. After the first 24 h, cultures were fed with fresh cultured medium.
2.3. COS-7 cells
African green monkey kidney fibroblast-like cell line COS-7 was obtained from ATCC (USA). COS-7 cells were maintained in DMEM supplemented with 10% FBS and a 1% penicillin-streptomycin solution at 37 °C in a humidified 5% CO2-containing atmosphere. COS-7 cells were used in the experiments between passages 3 and 15. Measurements of the size of cytoskeletal filaments and protein aggregates in COS-7 cells were made using Fiji software [12].
2.4. Zebrafish husbandry
Zebrafish (Danio rerio) were maintained in aquaria with recirculating water system at 28 ± 1 °C on a 14:10 light/dark cycle in a vivarium localized at the Institute of Biomedical Sciences, Federal University of Rio de Janeiro (Rio de Janeiro, Brazil). Animals were handled and experimented according to Institutional Animal Care and Use Committee protocols under the number 039/20. Embryos were collected, bleached with 0.05% NaOCl for 3 min and rinsing with water 3 times before chorion removal.
2.5. Immunofluorescence microscopy
Dechorionated zebrafish embryos at 24- or 48-h stages and chick muscle cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Embryos and cells were then permeabilized with 0.5% Triton-X 100 in PBS (PBS/T) three times for 30 min and incubated for 1 h at 37 °C with primary antibodies (all diluted 1:100 in PBS/T). Then, embryos and cells were washed for 30 min with PBS/T and incubated for 1 h at 37 °C with Alexa Fluor-conjugated secondary antibodies (all diluted 1:200 in PBS/T). Nuclei were labeled with 0.1 μg/mL of DAPI in 0.9% NaCl. Embryos and cells were mounted on #1.5 24 × 60-mm glass coverslips (with spacers in the case of embryos) using Prolong Gold (Molecular Probes). Experiments with zebrafish embryos and with chick muscle cells were repeated four times, using fifty zebrafish embryos and five 35 mm chick cell culture dishes each time.
2.6. Fluorescence image acquisition and processing
Cells and embryos were examined with either (i) an Axiovert 100 microscope (Carl Zeiss, Germany) coupled to an Olympus DP71 high-resolution camera, (ii) a Leica TCS SPE laser scanning confocal microscope (Leica, Japan), or (iii) a DSU Spinning Disk confocal scanner mounted on an inverted fluorescent microscope (Olympus, Japan). Control experiments with only secondary antibodies showed only a faint background staining (data not shown). Phase contrast microscopy images of cultured cells were acquired with an Axiovert 100 microscope using a 63 × (NA 1.4) oil-immersion objective lens. Image processing (brightness and contrast adjustments) was performed using Fiji software [12] and figure panels were produced with Adobe Photoshop software (Adobe Systems Inc., USA), where some of the original fluorescence grayscale images were pseudo-colored and superimposed. Some fluorescent images (Gli-1 labeling) were digitally processed by Universal live-cell super-resolution microscopy (SRRF) [13,14].
2.7. Transmission electron microscopy (TEM)
Wild-type C57BL/6J mice of 2- or 4-months old mice were anesthetized with ketamine (50 mg/kg) and xylazine (5 mg/kg) and perfusion-fixed through their ventricles with 2.5% (v/v) glutaraldehyde, 4% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.2). Then the hearts were isolated and kept overnight in a new batch of the same fixative solution at 4 °C. Next day, hearts were washed with the same buffer (3 × 30 min), cut in several 2 × 2 mm fragments, and transferred to glass vials. Experiments with mice hearts were repeated three times.
Eleven-day-old embryonic chick skeletal muscle cells were fixed overnight at room temperature with 2.5% (v/v) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2). Afterwards, the cells were washed three times in the same buffer. Heart samples and cells were postfixed for 40 min in 1% OsO4 in 0.1 M cacodylate buffer containing 5 mM CaCl2 and 0.8% potassium ferricyanide. All samples were dehydrated in acetone and embedded in Epon. Ultrathin sections 60 nm thick were collected on 300-mesh copper grids, stained with 2% uranyl acetate for 30 min and 1% lead citrate for 3 min, and finally the cells observed with a JEOL 1210 (Jeol, Japan) and hearts with a Leo Libra 120 (Zeiss, Oberkochen, Germany) transmission electron microscopes, both operated at 80 kV. Experiments with chick muscle cells were repeated four times.
For cytoskeleton observations of membrane-extracted cells, COS-7 were cultured on nickel grids covered with Formvar film and treated with an extraction solution composed of: phalloidin (50 ng/mL), 10 mM Pipes (pH 6.8), 300 mM sucrose, 3 mM MgCl2, 2 mM EGTA, 1% (v/v) Triton X-100, protease inhibitor cocktail (Roche, USA), and 0.5% glutaraldehyde for 7 min at RT. Then, cells were briefly washed in PBS and fixed in 2.5% (v/v) glutaraldehyde, 4% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.2) for 30 min. Then, cells were washed in PBS (3 × 15 min), dehydrated in ethanol until absolute, critical-point dried (CPD 030, Leica, Japan), and observed in a Leo Libra 120 (Zeiss, Oberkochen, Germany) transmission electron microscope, operated at 80 kV. Experiments with COS-7 cells were repeated three times.
2.8. Bibliometric analysis
Perinuclear proteins were identified by an in-silico bibliometric analysis in PubMed (https://pubmed.ncbi.nlm.nih.gov/). Articles with the descriptor “perinuclear protein” were selected in the entire PubMed collection database (from 1955 to June 2021) and they were then individually examined to check whether each article was describing proteins with a perinuclear localization (Table 1). The disease context of each perinuclear protein was individually examined in PubMed database.
Table 1.
Perinuclear proteins retrieved from PubMed database. Perinuclear proteins were identified by an in-silico bibliometric analysis in the PubMed database (from 1955 to June 2021), and they were then individually examined to check whether each article was describing proteins with a perinuclear localization. Seventy nine perinuclear proteins were found and classified as cytoskeleton, vesicular traffic, and signaling. The disease context of perinuclear proteins was also included in the table.
| Number | Protein | Cellular process | Reference | Disease related | Reference |
|---|---|---|---|---|---|
| 1 | Actin | Cytoskeleton | Khatau et al., 2009 | Congenital myopathies | Clarkson et al., 2004 |
| 2 | Alpha-actinin | Cytoskeleton | [23] | Autoimmune, myopathies, neurodegenerative disorders | Oikonomou et al., 2011 |
| 3 | Alpha-synuclein | Vesicular trafficking | [50] | Parkinson’s disease | Stefanis, 2012 |
| 4 | Alpha-tubulin | Cytoskeleton | Wu and Akhmanova, 2017 | Neurodegenerative disorders | Fourel and Boscheron, 2020 |
| 5 | Annexin A2 | Signaling | Aukrust et al., 2016 | Atherosclerosis, inflammatory disorders | Lim and Hajjar, 2021 |
| 6 | ARP2/3 | Cytoskeleton | Thiam et al., 2016 | Cancer, inflammatory diseases, psoriasis | Kahr et al., 2017; Molinie and Gautreau, 2017 |
| 7 | Beta-catenin | Signaling | Kam and Quaranta, 2009 | Bone diseases, cancer, diabetes, cardiac diseases | Clevers and Nusse, 2012 |
| 8 | Beta-tubulin | Cytoskeleton | Wu and Akhmanova, 2017 | Neurodegenerative disorders | Fourel and Boscheron, 2020 |
| 9 | Brain expressed x-linked 3 (Bex3) | Signaling | Kim et al., 2004 | Cancer, neurodegenerative disorders | Krizman et al., 1999; Navas-Pérez et al., 2020 |
| 10 | Calnexin | Signaling | Lakkaraju et al., 2012 | Cancer, Parkinson’s disease | Kuang et al., 2014; Ryan et al., 2016 |
| 11 | Calpain-3 | Signaling | Andrade Rosa et al., 2020 | Limb-girdle muscular dystrophy type 2A | Richard et al., 1995 |
| 12 | Calreticulin | Signaling | Taguchi et al., 2000 | Cancer, fibrosis, wound healing | Gold et al., 2010 |
| 13 | Centrosome-associated protein | Cytoskeleton | [52] | Cone-rod dystrophy and hearing loss | Kubota et al., 2018 |
| 14 | Coatomer subunit epsilon (COPE) | Vesicular trafficking | [10] | Atherosclerosis, Parvovirus infection | Chen et al., 2021; Matsumura et al., 2017 |
| 15 | CREB binding protein (CBP) | Signaling | [10] | Neurodegenerative disorders | Steffan et al., 2000 |
| 16 | Desmin | Cytoskeleton | [25] | Skeletal and cardiomyopathies | Capetanaki et al., 2015 |
| 17 | Dynactin | Cytoskeleton | Payne et al., 2003 | Neurodegenerative disorders | Puls et al., 2003 |
| 18 | Dynein | Cytoskeleton | Payne et al., 2003 | Neurodegenerative disorders | Hoang et al., 2017 |
| 19 | EHD1 | Vesicular trafficking | Guilherme et al., 2004 | Cardiac diseases | Martins-Marques et al., 2020 |
| 20 | Endophilin B2 | Vesicular trafficking | Vannier et al., 2013 | Influenza A viral infection | Serfass et al., 2017 |
| 21 | epsinR | Vesicular trafficking | Hirst et al., 2003 | Schizophrenia and psychotic disorders | Pimm et al., 2005 |
| 22 | Erk1/2 | Signaling | Asrih et al., 2011 | Cancer, diabetes, inflammatory diseases, obesity | Lawrence et al., 2008 |
| 23 | Filamin | Cytoskeleton | Gay et al., 2011 | Pulmonary diseases, skeletal dysplasia | Bartley et al., 2019; Sasaki et al., 2019 |
| 24 | Formin | Cytoskeleton | Isogai and Innocenti, 2016 | Deafness, neurodegenerative disorders, renal diseases | Labat-de-Hoz and Alonso, 2020 |
| 25 | FOXC1 | cytoskeleton | [10] | Cardiac diseases, congenital glaucoma, leukemia | Khalil et al., 2017; Swaminathan et al., 2016 |
| 26 | Gamma-synuclein | vesicular trafficking | Irina Surgucheva et al., 2006 | Neurodegenerative disorders | Ninkina et al., 2009 |
| 27 | Gamma-tubulin | Cytoskeleton | Moritz et al., 1995 | Brain malformations, cancer, myopathies | Alvarado-Kristensson, 2018; Binarová and Tuszynski, 2019 |
| 28 | Glial fibrillary acidic protein (GFAP) | Cytoskeleton | Buniatian et al., 1996 | Alexander disease, autoimmune astrocytopathy | Brenner et al., 2001; Shan et al., 2018 |
| 29 | Glioma-associated oncogene homolog 1 (Gli1) | Signaling | [22] | Cancer, congenital malformations | Altaba et al., 2007 |
| 30 | GM130 | Signaling | Gangalum et al., 2004 | Lysosomal diseases, neurodegenerative disorders | Liu et al., 2017; Roy et al., 2012 |
| 31 | HSP70 | Signaling | Bodega et al., 2002 | Cancer, lysosomal diseases, neurodegenerative disorders | Kirkegaard et al., 2010; Turturici et al., 2011 |
| 32 | Huntingtin | Signaling | Hoffner et al., 2002 | Huntington’s disease | Tabrizi et al., 2020 |
| 33 | JunB | Signaling | [10] | Inflammatory disorders, myeloproliferative disorders | Passegué et al., 2004; Thomsen et al., 2013 |
| 34 | KASH | Cytoskeleton | Tapley and Starr, 2012 | Hutchinson-Gilford progeria syndrome, laminopathies | Starr, 2011 |
| 35 | Keratin | Cytoskeleton | Lee et al., 2012 | Epidermolysis bullosa and other epithelial disorders | McLean and Moore, 2011 |
| 36 | Kinesin | Cytoskeleton | Cai et al., 2001 | Ciliopathies, neurodegenerative disorders | Asselin et al., 2020 |
| 37 | LIM domain only protein 7 (Lmo7) | Signaling | [19] | Cancer, retinal disorders, skeletal and cardiac myopathies | Nakamura et al., 2011; Semenova et al., 2003 |
| 38 | Microtubule-associated protein (MAP) | Cytoskeleton | Bloom and Vallee, 1983 | Neurodegenerative disorders | D’Andrea et al., 2001; Zhang and Dong, 2012 |
| 39 | Muscle myosin II | Cytoskeleton | Khatau et al., 2009 | Cardiac and skeletal myopathies | Parker and Peckham, 2020 |
| 40 | Myospryn | Cytoskeleton | Kouloumenta et al., 2007 | Muscular dystrophies | Benson et al., 2004 |
| 41 | Nesprin | Cytoskeleton | Lu et al., 2012 | Cancer, hearing loss, myopathies, neurodegenerative disorders | Cartwright and Karakesisoglou, 2014 |
| 42 | Nestin | Cytoskeleton | Lobo et al., 2004 | Cancer, cardiac diseases, neurodegenerative disorders | Bernal and Arranz, 2018; Neradil and Veselska, 2015 |
| 43 | Neurofilament | Cytoskeleton | Lobsiger and Cleveland, 2009 | Neurodegenerative disorders | Khalil et al., 2018 |
| 44 | Non-muscle myosin II | Cytoskeleton | Thomas et al., 2015 | Cancer, inflammatory disorders, neurodegenerative disorders, vascular diseases | Newell-Litwa et al., 2015 |
| 45 | Nuclear pore complex protein | Signaling | Beck and Hurt, 2017 | Cancer, neurodegenerative disorders, viral infection | Sakuma and D’Angelo, 2017 |
| 46 | Nucleoporin (Nup) | Signaling | Ibarra and Hetzer, 2015 | Cancer, cardiac diseases, neurodegenerative disorders, viral infection | Nofrini et al., 2016 |
| 47 | Oxysterol-binding protein 1 | Signaling | Nishimura et al., 2013 | Conjunctivitis, hearing loss, neurodegenerative disorders | Raychaudhuri and Prinz, 2010 |
| 48 | Pericentrin | Cytoskeleton | Doxsey et al., 1994 | Cancer, ciliopathy, dwarfism, neurodegenerative disorders | Delaval and Doxsey, 2010 |
| 49 | Pericentriolar material protein 1 | Cytoskeleton | Dammermann, and Merdes, 2002 | Cancer | Bousquet et al., 2005 |
| 50 | Patronin | Cytoskeleton | Zheng et al., 2020 | Microvillus diseases | Khanal et al., 2016 |
| 51 | p53 | Signaling | [10] | Cancer, neurodegenerative disorders | Vousden and Lane, 2007 |
| 52 | Peripherin | Cytoskeleton | Pedersen et al., 1993 | Retinal degenerative disorders, neurodegenerative disorders | Yuan et al., 2012 |
| 53 | PERF15 | Cytoskeleton | Oko and Morales, 1994 | Sperm abnormalities | Selvara et al., 2010 |
| 54 | PICK1 | Signaling | Staudinger et al., 1995 | Cancer, neurodegenerative disorders, cardiac diseases | Li et al., 2016 |
| 55 | Plectin1 | Cytoskeleton | Staszewska et al., 2015 | Cancer, muscular dystrophies, skin diseases | Bausch et al., 2011; Smith et al., 1996 |
| 56 | Postacrosomal sheath WW domain-binding protein | Cytoskeleton | Wu et al., 2007 | Sperm abnormalities | Nomikos et al., 2014 |
| 57 | Phospholipase D (PLD) | Signaling | Souza et al., 2014 | Autoimmune, cancer, cardiac diseases, infection neurodegenerative disorders | Brown et al., 2017 |
| 58 | Promyelocytic leukemia protein (PML) | Signaling | Daniel et al., 1993 | Cancer, neurodegenerative disorders, viral infection | Bonilla et al., 2002; Rego et al., 2001 |
| 59 | Prion protein (PrP) | Signaling | Nikles et al., 2008 | Neurodegenerative disorders | Watts et al., 2018 |
| 60 | PTEN | Signaling | [10] | Autism, cancer | Post et al., 2020 |
| 61 | Rab11 | Vesicular trafficking | Vossenkämper et al., 2007 | Cancer, diabetes, neurodegenerative disorders | Bhuin and Roy, 2015 |
| 62 | RAB2 | Vesicular trafficking | Oko and Sutovsky, 2009 | Cancer, neurodegenerative disorders | Lőrincz et al., 2017 |
| 63 | Rabring7 | Vesicular trafficking | Mizuno et al., 2003 | Cancer, HIV viral infection, neurodegenerative disorders | Mizuno et al., 2003 |
| 64 | Rac1 | Cytoskeletal | Woroniuk et al., 2018 | Cancer, cardiovascular diseases neurodegenerative disorders | Marei and Malliri, 2017 |
| 65 | Raf1 | Signaling | Prouty et al., 1998 | Cancer, cardiomyopathies, neurodegenerative disorders | Kobayashi et al., 2010 |
| 66 | Ran-binding protein 2 (RanBP2) | Signaling | [10] | Encephalopathies | Levine et al., 2020 |
| 67 | Ras | Signaling | Basu et al., 2017 | Autoimmune diseases, cancer, neuro-cardiofacial-cutaneous syndromes | Schubbert et al., 2007 |
| 68 | Refilin | Cytoskeleton | Gay et al., 2011 | Chondrocyte disorders | Baudier et al., 2018 |
| 69 | Retinoblastoma binding protein 2 (RBP2) | Signaling | [10] | Cancer | Maggi et al., 2016 |
| 70 | Spermatid perinuclear ribonucleic acid-binding protein | Cytoskeleton | Schumacher et al., 1998 | Sperm abnormalities | Schumacher et al., 1998 |
| 71 | Spectraplakin/shot | Cytoskeletal | Sun et al., 2019 | Cancer, neurodegenerative disorders, neuromuscular diseases, skin disorders, viral infection | Zhang et al., 2017 |
| 72 | Proto-oncogene tyrosine-protein kinase (c-Src) | Signaling | Redmond et al., 1992 | Cancer, kidney disorders, lens diseases, neurodegenerative disorders | Irby and Yeatman, 2000; Wang and Zhuang, 2017 |
| 73 | Stat3 | Signaling | Bild et al., 2002 | Immune diseases, infectious diseases, skin disorders | Vogel et al., 2015 |
| 74 | STEF/TIAM2 | Signaling | Woroniuk et al., 2018 | Cancer, immune diseases, neurodegenerative disorders | Maltas et al., 2020 |
| 75 | Stimulator of interferon genes protein (Sting) | Signaling | Barber, 2015 | Cancer, viral infection, inflammatory diseases | Barber, 2015 |
| 76 | Transcriptional coactivator p300 | Signaling | Chen et al., 2007 | Cancer, cardiac diseases, diabetes, HIV infection | Liu et al., 2008 |
| 77 | TAU | Cytoskeleton | Adamec et al., 2002 | Neurodegenerative disorders | Iqbal and Gong, 2016 |
| 78 | VAMP | Vesicular trafficking | Oishi et al., 2005 | Neurodegenerative disorders | Bourassa et al., 2012 |
| 79 | Vimentin | Cytoskeleton | Tolstonog et al., 2002 | Cancer, cataracts, Crohn’s disease, rheumatoid arthritis, HIV infection | Danielsson et al., 2018 |
2.9. Bioinformatic analyses of intrinsic disorder and LLPS propensity
To identify the proteome with perinuclear location, we selected human proteins from the UniProtKB/Swiss-Prot, which is a manually annotated database, using the search engine for subcellular locations (https://www.uniprot.org/locations/198, The UniProt Consortium, release 2021 03). This location is based on Gene Ontology (GO) 0048471 and defined as “the cytoplasmic region just around the nucleus”. The canonical sequences from the 285 identified proteins and proteins gathered from literature (as described in Section 2.8) were retrieved in FASTA format and submitted to analysis by the Predictor of Natural Disordered Region (PONDR)-VSL2 provided by http://www.pondr.com/ [15]. The PONDR-VSL2 was trained by 1327 proteins and showed a predictor accuracy of approximately 81% for both short (below 30 residues) and long (above 30 residues) disordered regions. Thus, it intends to recognize disordered amino acid stretches of any length [15]. We then verified whether the retrieved perinuclear proteins were part of the DisProt database, a reviewed repository of experimentally proven disordered regions [16]. We selected the perinuclear proteins reported by DisProt and the ones with an overall disorder content above one-third of their primary sequences (≥33.3%, named herein as perinuclear IDR-containing proteins) for further analysis. The mean net charge versus mean hydropathy (CH-plot) was obtained by PONDR (http://www.pondr.com), as described in [29]. The LLPS-propensity analysis by the catGRANULE algorithm (http://s.tartaglialab.com/new_submission/catGRANULES) [30] was calculated for the perinuclear IDR-containing proteins. Overall catGRANULE scores above zero indicate LLPS ability [30]. The LLPS prediction potentially mediated by pi-pi contacts was evaluated by the PScore algorithm (http://abragam.med.utoronto.ca/~JFKlab/Software/psp.htm), whereby a PScore ≥4.0 predicts the LLPS propensity [17].
2.10. Bioinformatic analyses of functional aspects, sequence composition and prion-like regions
The UniProt IDs from the human perinuclear IDR-containing subgroups: LLPS (43 proteins) and non-LLPS (74 proteins) were submitted to STRING (available at https://string-db.org/) [51] and enriched keywords were analyzed. The groups were evaluated by nucleic acid (NA) and cytoskeleton related processes by the following UniProt keywords (reviewed): “RNA-binding” (KW-0694), “mRNA transport” (KW-0509), “DNA-binding” (KW-0238), “cytoskeleton” (KW-0206), “cell projection” (KW-0966) and “intermediate filaments” (KW-0403). Sequence composition from the LLPS and non-LLPS were evaluated by the Composition Profiler (available at http://www.cprofiler.org/cgi-bin/profiler.cgi). Results were plot in GraphPad Prism v.6. Prion-like regions were analyzed by PLAAC (available at http://plaac.wi.mit.edu/).
3. Results
3.1. How is the perinuclear space organized?
To evaluate how the perinuclear region of eukaryotic cells is organized, we initially analyzed COS-7 fibroblastic cells after membrane extraction, which reveals the fine ultrastructure of the nuclei and the cytoskeleton system. An intricate three-dimensional network of filaments and protein aggregates was seen occupying the cytoplasmic space and associated with the nuclei (Fig. 1A). This 3D nuclear space extends ~2–5 μm from the outer nuclear surface towards the cytoplasm. High magnification images showed the common cytoskeletal filaments like actin microfilaments (~7 nm) and intermediate filaments (~12 nm) (Fig. 1B). Since we did not use Taxol to preserve the microtubules while extracting the membranes, we assumed that most of the thicker filaments were made of intermediate filaments. In addition, those thicker filaments exhibited few branches, which do not occur with microtubules (Fig. 1B). After lipids were dissolved, round aggregates of proteins were found adhered to cytoskeletal filaments, varying in size from a minimum of 30 nm and maximum of 80 nm in diameter (average = 40 ± 18 nm, N = 8 cells). We cannot determine whether all (or part) of the aggregates were already attached to the filaments before the extraction/fixation. However, we speculate that those complexes floating around without a strong interaction would be washed out. The dense network of cytoskeleton filaments surrounding the nucleus creates the proper environment for biochemical processes, such as the regulation of the traffic of molecules between the nucleus and the cytoplasm, RNA processing, protein synthesis, and modulation of signaling pathways. Interestingly, we noticed in many instances intermediate filaments strongly attached to the outer nuclear membrane (Fig 1B).
Fig. 1.
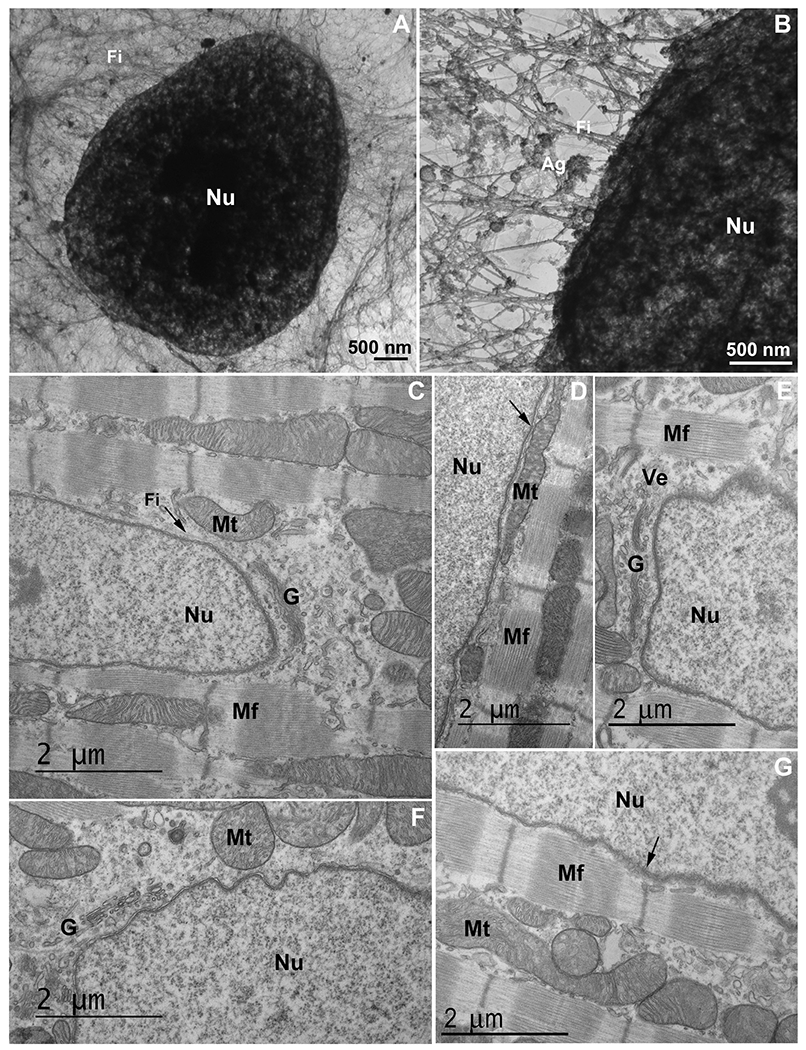
An intricated network of cytoskeletal filaments and several organelles are found in the nuclear space of eukaryotic cells. (A and B) COS-7 cells were extracted and analyzed under transmission electron microscopy. Note the presence of a dense network (in A and B) of cytoskeletal filaments (Fi) linked to the nucleus (Nu). In the higher magnification (B) it is possible to see several protein aggregates (Ag) attached to the cytoskeletal network. Scale bars in A and B = 500 ηm. (C–G) 2-month-old mouse cardiac tissues were processed for transmission electron microscopy and images show mitochondria (Mt), Golgi apparatus (G), vesicles (Ve), myofibers (Mf), and cytoskeletal filaments (Fi) in proximity with the nucleus (Nu). Arrow in (C) shows cytoskeletal filaments in the perinuclear space, arrow in (D) shows mitochondria in close contact with the outer nuclear membrane, and arrow in (G) points to the Z-disk of a myofibril in the vicinity of the nucleus. Scale bars in C-G = 2 μm. N = 3 independent experiments.
Next, we further explored the perinuclear region of cells using ultrathin sections. Mouse cardiomyocytes showed a close interaction between the outer nuclear surface and several organelles, such as mitochondria, Golgi apparatus, vesicles and myofibrils (Fig. 1C–G). Interestingly, we observed layers of myofibrils oriented parallel to the nuclear cloud (Fig. 1D, G), with Z-disks and T-tubules apparently anchored to the nuclear surface through tiny filaments (Fig. 1G). Also, cytoskeletal filaments were seen close to the nucleus, likely to be intermediate filaments (Fig. 1C). Mitochondria, which are mostly intercalated among the myofibrils, were seen in the nuclear cloud (Fig. 1C, D, F, G).
We also analyzed embryonic chick skeletal muscle cells under TEM (Fig. 2A–D). Fig. 3A is a low magnification image where many Golgi cisternae are preferentially localized adjacent to the nucleus. Not only the Golgi, but lysosomes also seem to be attached to the outer nuclear membrane (Fig. 2B). Mitochondria, on the other hand, have a broader distribution, although some are remarkably close to the nucleus (Fig. 2C, D).
Fig. 2.
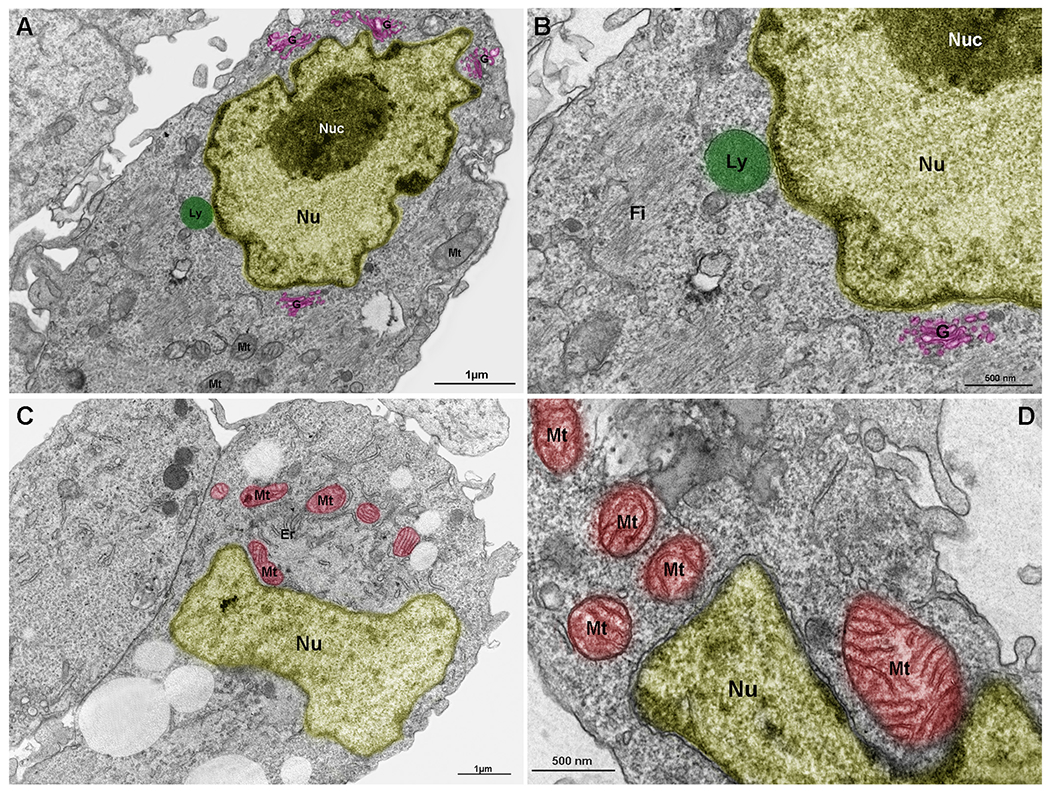
Lysosomes, mitochondria and Golgi are components of the nuclear cloud. Embryonic chick pectoral muscle was processed for transmission electron microscopy and organelles were digitally colored to facilitate the visualization. Images show lysosomes (Ly, in green in A and B), mitochondria (Mt, in red in C and D), endoplasmic reticulum (Er in C), and Golgi (G, in pink in A and B) in proximity with the outer nuclear membrane (Nu). The highly packed nuclear compartment nucleoli (Nuc, in dark yellow) are seen within the nucleus (light yellow) of muscle cells (in A and B). Some lysosomes (in A and B) and mitochondria (in C and D) seem to be adhered to the nuclear surface. Bars in A and C = 1 μm, and bars in B and D = 500 nm. N = 4 independent experiments.
Fig. 3.
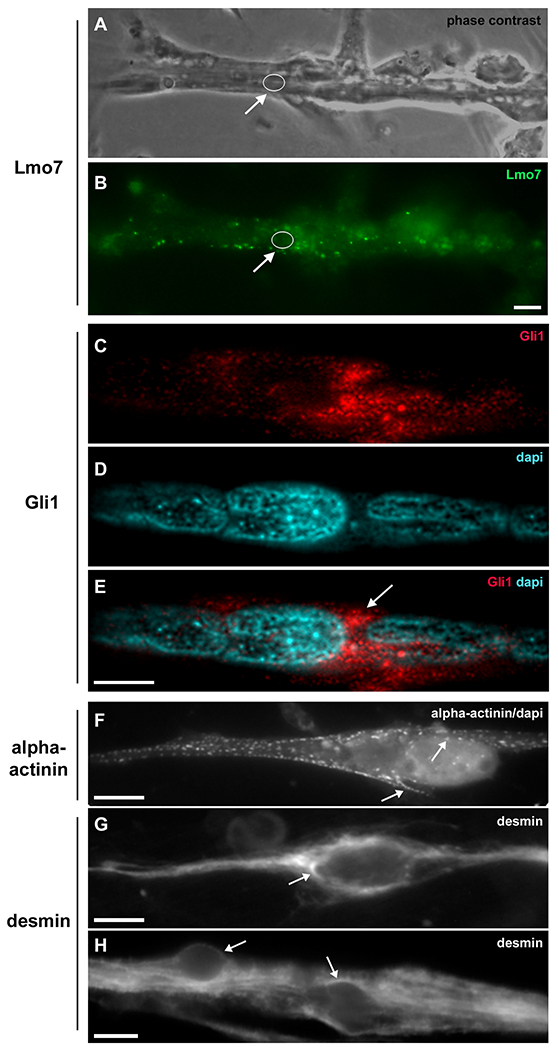
Lmo7, Gli1, alpha-actinin and desmin are concentrated in the nuclear cloud in skeletal muscle fibers. Primary cultures of chick myogenic cells were labeled with antibodies against Lmo7 (B), Gli1 (C–E), alpha-actinin (F) and desmin (G–H), and with the nuclear dye DAPI (D–F). A 72-h chick multinucleated myotube was visualized under phase contrast microscopy (A) and under fluorescence microscopy to show the localization of Lmo7 (green, in B). Arrows in A and B point to Lmo7 distribution near the nuclear surface of a myotube. White open circles (in A and B) mark the region of one nucleus surrounded by Lmo7-positive aggregates. Scale bar in A = 10 μm. Gli-1 (red, in C and E) localizes at the perinuclear region of a 72-h multinucleated myotube (arrow in E). A merged image (with Gli1 and DAPI) is shown in E. Scale bar in E = 5 μm. The intermediate filament desmin and the sarcomeric protein alpha-actinin accumulate at the perinuclear region of mononucleated myoblasts (F and G) and multinucleated myotubes (H). A merged image of sarcomeric alpha-actinin and DAPI is shown in (F). Note the punctate distribution of alpha-actinin (arrows in F) and the continuous distribution of desmin filaments (arrows in G and H) in the juxtanuclear region of cells. Scale bars in F and H = 10 μm, and in G = 5 μm. n = 4 independent experiments.
3.2. Which proteins localize within the perinuclear space?
Since several protein aggregates were observed in close interaction with the external nuclear surface, we decided to evaluate which proteins were reported to be found at the perinuclear region of eukaryotic cells. A bibliometric approach was employed in which the entire PubMed database was scanned for articles reporting proteins with a perinuclear localization. Seventy nine perinuclear proteins were found (Table 1) and classified in three different groups according to their functions: cytoskeleton, vesicular traffic, and signaling. Most of these proteins (60%) were cytoskeletal. All three classes of cytoskeletal filaments were found, namely, microfilaments (MF), microtubules (MT) and intermediate filaments (IF), including the proteins actin (MF), alpha/beta/gamma tubulin (MT), desmin (IF), vimentin (IF), neurofilament (IF), keratin (IF), glial fibrillary acidic protein (IF), nestin (IF) and peripherin (IF). Interestingly, several cytoskeletal-associated proteins, such as alpha-actinin (MF), arp2/3 (MF), filamin (MF), formin (MF), rac1 (MF), plectin (IF), and the motor proteins myosin, dynein and kinesin were among them. The second most represented group (25%) of perinuclear proteins was involved in cell signaling, including Calpain-3, Gli-1, Lmo7, Raf1, Huntingtin, p53, and PrP. Several proteins involved in vesicular trafficking were also identified (15%) among the preferentially nuclear localized proteins, including Rab, alpha- and gamma-synuclein, and VAMP (Table 1).
Fig. 3 provide some examples of the perinuclear localization of reported cytoskeletal and signaling perinuclear proteins, including desmin, alpha-actinin, Lmo7 and Gli-1. Lmo7 is a scaffolding protein that carries Lim and F-box domains, which enables the protein to interact with different partners (in the cytoplasm and within the nucleus) and to act like a signaling hub [18,19]. Lmo7 is concentrated at the perinuclear space of chick skeletal muscle fibers (white arrows in Fig. 3A, B), suggesting that this location might be related to changes in nuclear transport dynamics in muscle cells, as previously described [20].
Gli-1 is an effector protein in the sonic hedgehog signaling pathway in eukaryotes and involved in several processes, including cell fate, proliferation, and differentiation [21]. Gli-1 can move from the perinuclear space to the nucleus and vice-versa, and within the nucleus can regulate gene expression [22]. Fig. 3C–E show Gli-1 perinuclear location in embryonic skeletal muscle fibers.
Alpha-actinin is an actin-associated protein found in multiple actin-containing cellular structures, including cell-matrix adhesion sites, contractile ring in cytokinesis, and perinuclear actin cap [23]. A thin dotted line of sarcomeric alpha-actinin can be found at the perinuclear region of skeletal myoblasts (Fig. 3F), where it is probably acting as an actin crosslinking factor.
Desmin is a muscle-specific IF protein and one of the first cytoskeleton proteins to be expressed in muscle cells [24]. Most IF proteins have a perinuclear distribution in eukaryotic cells and desmin has been shown to be stably associated with the outer nuclear surface in embryonic chick myoblasts (Fig. 3G and H, and [25]), where its function has been associated with providing structural support and shape to the nucleus. Fig. 3G and H show a dense network of desmin IFs around the nuclei of mononucleated myoblasts and multinucleated myotubes, respectively. Interestingly, the web of desmin IFs present in the perinuclear space seems to be denser in myoblasts than in myotubes (Fig. 3G and H).
Next, we analyzed the perinuclear distribution of proteins in eukaryotic cells in vivo, focusing on the distribution of desmin in whole zebrafish embryos. Confocal images of different focal planes (Fig. 4A–K) confirmed the perinuclear localization of desmin in newly formed muscle somites at the most caudal region of 24-h zebrafish embryos (white arrow in Fig. 4D). A 3D reconstruction of desmin IFs distribution at the perinuclear cloud of zebrafish embryos in shown in Supplementary Video 1. Desmin was also found concentrated adjacent to the septa between muscle somites in zebrafish embryos (black arrow in Fig. 4K). These results are in accordance with previous data from our group showing the concentration of desmin around the nucleus in recently born zebrafish somites [26], and support the existence of a perinuclear region in which specific proteins concentrate in both in vitro and in vivo eukaryotic cells.
Fig. 4.
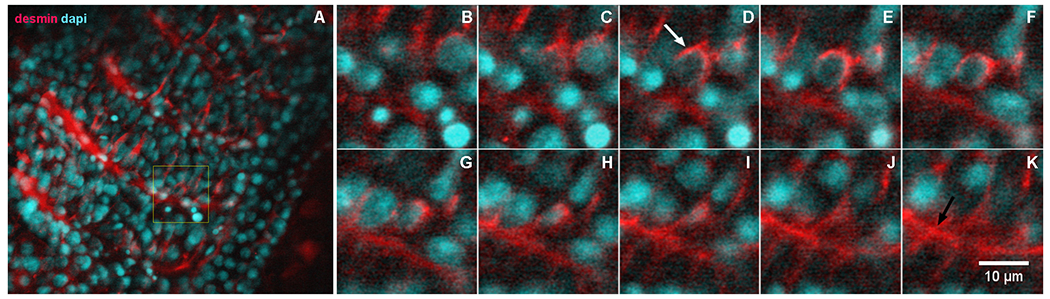
Desmin accumulates at the perinuclear region of early somites in zebrafish embryos. 24-h zebrafish embryos were labeled with an antibody against the muscle-specific intermediate filament protein desmin (red, in A–K) and with the nuclear dye DAPI (cyan, in A–K). Higher magnifications of the area marked in the inset in (A) are shown in images (B–K). Immunofluorescence confocal images of different focal planes (1 μm apart, in B–K) show the perinuclear localization of desmin (white arrow in D) in somite 28 at the most caudal region of a 24-h zebrafish embryo. Desmin is also found at the septa between adjacent muscle somites 28 and 29 in zebrafish embryos (black arrow in K). Scale bar in K = 10 μm. N = 4 independent experiments.
We also evaluated the disease context of all the perinuclear proteins described in Table 1. Our data shows that alterations in the expression of perinuclear proteins can lead to several pathological disorders, including atherosclerosis, autoimmune diseases, cancer, cardiac and skeletal myopathies, diabetes, inflammatory diseases, neurodegenerative disorders, obesity, skin disorders, and viral infection. These results suggest that alterations in their expression and/or intracellular distribution are critical for cell, tissue and organ’s structure and function, and highlight their important role in health and disease. Importantly, among the most prevalent diseases associated with perinuclear proteins were neurodegenerative disorders (50%), cancer (45%), cardiac and skeletal myopathies (27%), immune/inflammatory diseases (17%), viral infection (13%) and skin disorders (10%). These data might be strategic for the development of new therapeutic approaches towards these pathological conditions. Interestingly, it has been described that structural disorder significantly distinguishes proteins up-regulated in neurodegenerative diseases from those linked to cancer [27]. These authors also observed high correlation between structural disorder and age of onset in several neurodegenerative diseases, which strongly supports the role of protein unfolding in neurodegenerative processes [27].
Curiously, many viral proteins and/or RNA have been reported to have a perinuclear distribution during a virus cycle within eukaryotic cells. These data are not included in Table 1, but clearly highlight the important role of the nuclear space for virus-related cellular processes. Hantavirus, rotavirus, hepatitis virus, baculovirus, measles virus, yellow-fever virus, and severe acute respiratory syndrome-associated virus (SARS-CoV) are among the virus proteins and/or virus RNA detected within the nuclear cloud [28]. Importantly, Table 1 shows data related to the perinuclear localization of cellular proteins, but not viral proteins, involved in viral infection. Coatomer subunit epsilon (COPE), endophilin B2, nuclear pore complex protein, nucleoporin (Nup), promyelocytic leukemia protein (PML), Rabring7, spectraplakin/Shot and stimulator of interferon genes protein (Sting) are among the cellular proteins involved in viral infection and detected within the perinuclear space of eukaryotic cells. These data could have valuable impact in new therapeutic strategies targeting viral infections.
3.3. Are IDPs present in the nuclear space?
Since several proteins show a perinuclear localization, we questioned whether they have an enriched intrinsic disorder content, which is an important feature of LLPS “driver” proteins. In addition to data gathered from the literature (Table 1), we retrieved the human mature protein sequences from UniProt described in the subcellular localization of the perinuclear region (Supplementary Table 1, data S1). Only five proteins (STING, Src, oxysterol-binding protein 1, gamma-synuclein and phospholipase D1) from the entire literature group (n = 79) were listed in the perinuclear UniProt dataset (Supplementary Table 1, data S3). Taking together the literature group and UniProt dataset, the perinuclear group contained 359 unambiguously proteins (Supplementary Table 1, data S1 and Table 1).
Using the charge-hydropathy (CH-plot) that enables differentiation of globular proteins versus IDPs based on the peculiar amino acid composition of IDPs [29], we observed 37 proteins from the perinuclear group in the plot space allocated to highly disordered proteins (Fig. 5). The functional annotation of these IDPs is shown in data S4 from Supplementary Table 1. Since this is a binary predictor and the fact that many proteins show a partly disordered nature, a clear differentiation of proteins containing short/long regions of disorder is not possible in the plot.
Fig. 5.
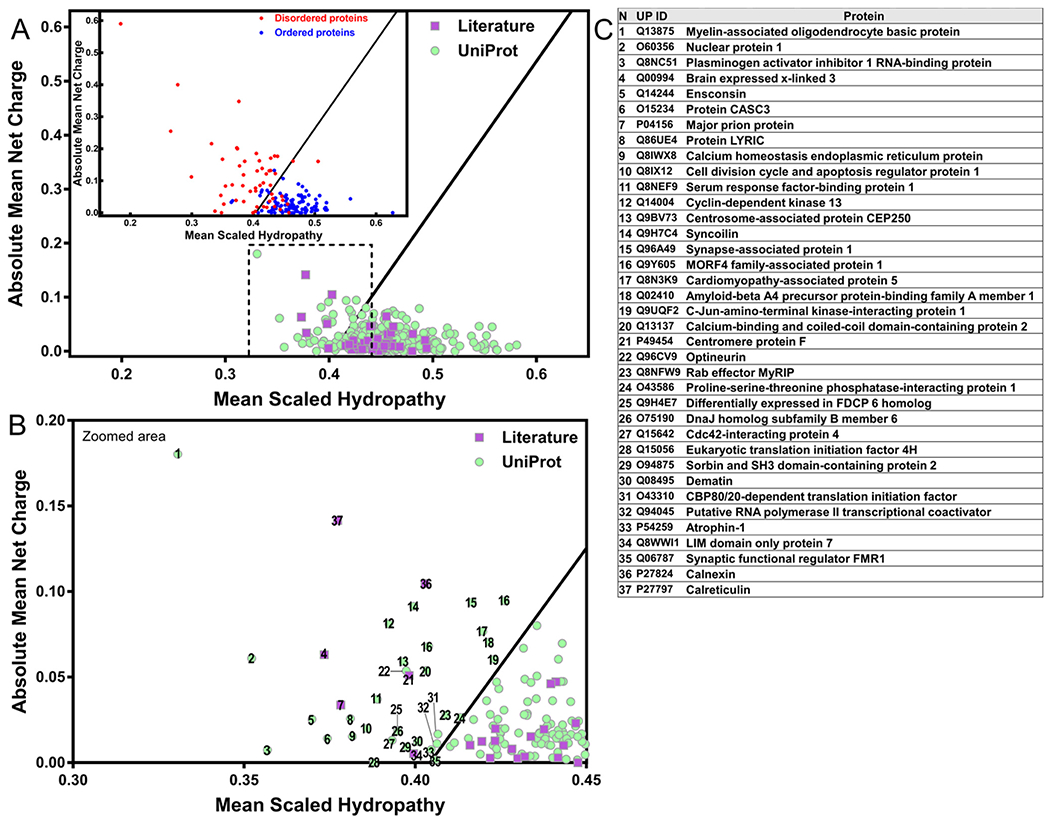
Some proteins of the perinuclear region show the unusual chemical characteristics of IDPs. (A) The charge-hydropathy analysis of the human perinuclear proteins retrieved from the UniProt database (purple squares) and literature (green circles). Inset: globular folded proteins represented as blue circles and natively unfolded proteins as red dots. (B) Zoomed area marked by dashed line from “A”. (C) List of perinuclear proteins with the exquisite charge-hydropathy of IDPs, numbers refer to “B”. Plot described by [29] using 275 globular proteins (blue circles) and 91 IDPs (red circles) that enable differentiation (black line) by their charged and hydrophobic nature at pH 7.0 and obtained by PONDR (available at http://www.pondr.com/).
From the 285 proteins extracted from UniProt, the analysis by the Predictor of Natural Disordered Region (PONDR-VSL2) revealed 60% (171 proteins) with a disorder content above one-third of their primary sequence (>33.3% of overall disorder) (Supplementary Table 1, data S3). The disordered regions with at least 15 residues are listed in Supplementary Table 1, data S1. Moreover, the DisProt database of IDRs/IDPs with experimental biophysical evidence for disorder reported 20 perinuclear proteins that contain one or more regions of disorder (Supplementary Table 1, data S3, highlighted in grey). Regarding the perinuclear dataset from literature, we selected proteins involved in signaling and/or vesicular trafficking for intrinsic disorder analysis, since cytoskeleton components generally have a well-defined 3D fold (Supplementary Table 1, data S2). Among the 41 proteins analyzed, 22 (54%) show a total disorder content above 33.3% or are annotated in the DisProt database. Taken together, we identified 193 perinuclear proteins with enriched intrinsic disorder (≥33.3% of disorder by PONDR-VSL2 or IDPs/IDRs annotated in DisProt), as shown in Table 2.
Table 2.
Perinuclear proteins predicted to be fully or partly disordered show predicted liquid-liquid phase separation propensity and/or experimental evidence. Perinuclear human protein sequences from UniProt and from the literature search with disorder content above 33.33% (PONDR-VLS2) and/or reported on DisProt database were evaluated by the catGRANULE algorithm (catG.) and PScore. Text inside brackets in the fifth column indicate the function associated with the disordered regions reported in DisProt. Bold font: perinuclear proteins selected by both UniProt search and literature. Marked by asterisk: Disorder content predicted by PONDR-VSL2 < 33.33% but present on DisProt database. Blue shade: catGRANULE score ≥ 0.5. Yellow shade: PScore ≥ 4.0. Last column: experimentally proven LLPS as verified in Dataset S1 from [31] or present in original articles cited throughout. HTS, evidence of LLPS based on high-throughput screening. PSP, proteins that contain regions that can mediate LLPS as verified in PhaSePro (https://phasepro.elte.hu/). Proteins organized by decreasing overall PONDR-VSL2 disorder.
| UniProt proteins | |||||||
|---|---|---|---|---|---|---|---|
| N | UniProt ID | Protein | %VSL2 | %DisProt (disorder function) | catG. | PScore | Evidence of LLPS |
| 1 | A1KXE4 | Myelin-associated neurite-outgrowth inhibitor (Mani) | 100.00 | −0.4 | 2.90 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 2 | O15234 | Cancer susceptibility candidate gene 3 protein (MLN 51) | 100.00 | 3.00 (protein binding) | 1.7 | 4.67 | Hardenberg et al., 2020 (dataset S1) *HTS |
| 3 | O76070 | Gamma-synuclein | 100.00 | 100.00 (protein binding; molecular recognition assembler) | 0.3 | <140 aa | |
| 4 | Q15004 | PCNA-associated factor (PAF15) | 100.00 | 100.00 (protein binding) | 0.5 | <140 aa | |
| 5 | P54259 | Atrophin-1 | 97.90 | 1.0 | 5.48 | ||
| 6 | P49454 | Centromere protein F (CENP-F) | 96.98 | 1.1 | 2.82 | ||
| 7 | Q8NC51 | Plasminogen activator inhibitor 1 RNA-binding protein (PAI-RBP1) | 96.32 | 2.8 | 5.39 | ||
| 8 | Q9BV73 | Centrosome-associated protein CEP250 (Cep250) | 95.45 | 0.6 | 0.41 | ||
| 9 | O75190 | DnaJ homolog subfamily B member 6 | 92.94 | 2.2 | 8.81 | ||
| 10 | O60356 | Nuclear protein 1 | 91.46 | 100.00 (regulation of phosphorylation and molecular recognition assembler) | 0.8 | <140 aa | |
| 11 | Q96CV9 | Optineurin | 90.81 | 0.3 | 1.49 | ||
| 12 | Q14244 | Ensconsin | 90.52 | 0.7 | 7.30 | ||
| 13 | Q86UE4 | Protein LYRIC | 90.38 | 1.5 | 3.26 | ||
| 14 | Q9NRR5 | Ubiquilin-4 | 88.85 | 0.5 | 3.70 | Gerson et al., 2021 (PMID: 33431932) | |
| 15 | Q8NEF9 | Serum response factor-binding protein 1 | 87.88 | 1.0 | −0.08 | ||
| 16 | Q07065 | Cytoskeleton-associated protein 4 | 87.71 | 0.8 | 4.26 | ||
| 17 | Q08495 | Dematin | 86.91 | 0.3 | 2.26 | ||
| 18 | Q9H7C4 | Syncoilin | 86.31 | −0.1 | −0.63 | ||
| 19 | Q8N3K9 | Cardiomyopathy-associated protein 5 | 86.21 | 1.1 | 1.52 | ||
| 20 | Q9Y6F6 | Inositol 1,4,5-triphosphate receptor associated 1 | 85.40 | 0.8 | 3.68 | ||
| 21 | Q13875 | Myelin-associated oligodendrocyte basic protein | 84.7 | −0.3 | 2.31 | Aggarwal et al., 2013 (PMID: 23762018) | |
| 22 | Q9NZ56 | Formin-2 | 84.44 | 1.4 | 3.38 | ||
| 23 | Q9H201 | Epsin-3 | 84.34 | 0.6 | 3.64 | ||
| 24 | O15027 | Protein transport protein Sec16A | 84.09 | 1.2 | 5.44 | ||
| 25 | Q14677 | Clathrin interactor 1 | 82.88 | 1.1 | 3.84 | ||
| 26 | Q8TB68 | Proline-rich protein 7 | 82.12 | −1.1 | 2.56 | ||
| 27 | O95996 | Adenomatous polyposis coli protein 2 | 81.07 | 1.0 | 2.64 | ||
| 28 | O94875 | Sorbin and SH3 domain-containing protein 2 | 81.00 | 0.9 | 3.21 | ||
| 29 | Q13625 | Apoptosis-stimulating of p53 protein 2 | 78.90 | 52.10 (protein binding) | 0.9 | 2.17 | |
| 30 | Q8IWX8 | Calcium homeostasis endoplasmic reticulum protein | 78.49 | 0.9 | 5.02 | ||
| 31 | Q9BWF2 | E3 ubiquitin-protein ligase TRAIP | 78.46 | 0.0 | 1.15 | ||
| 32 | Q93045 | Stathmin-2 | 77.65 | −0.6 | 3.92 | ||
| 33 | O75821 | Eukaryotic translation initiation factor 3 subunit G | 77.50 | 0.8 | −0.05 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 34 | O95999 | B-cell lymphoma/leukemia 10 | 76.82 | −0.2 | 1.85 | ||
| 35 | Q8NFW9 | Rab effector MyRIP | 76.48 | 0.4 | 0.89 | ||
| 36 | Q15642 | Cdc42-interacting protein 4 | 76.21 | 0.7 | 3.22 | ||
| 37 | Q8WY41 | Nanos homolog 1 | 75.00 | 0.2 | 3.16 | ||
| 38 | Q9NZU7 | Calcium-binding protein 1 | 74.59 | 1.0 | 2.66 | ||
| 39 | Q969Z4 | Tumor necrosis factor receptor superfamily member 19L | 74.19 | 0.3 | 3.10 | ||
| 40 | Q6ZMQ8 | Serine/threonine-protein kinase LMTK1 | 73.29 | 1.0 | 3.79 | ||
| 41 | Q00613 | Heat shock factor protein 1 | 72.97 | 0.2 | 1.45 | ||
| 42 | Q7Z3Z2 | Protein RD3 | 72.82 | −1.4 | 0.08 | ||
| 43 | Q15056 | Eukaryotic translation initiation factor 4H | 72.18 | 2.3 | 5.75 | Hardenberg et al., 2020 (dataset S1) | |
| 44 | Q96A49 | Synapse-associated protein 1 | 72.16 | 0.2 | −0.87 | ||
| 45 | Q8IX12 | Cell division cycle and apoptosis regulator protein 1 | 70.35 | 0.7 | 2.09 | ||
| 46 | A2IDD5 | Coiled-coil domain-containing protein 78 | 70.09 | 0.3 | 1.47 | ||
| 47 | Q9UGF2 | C-Jun-amino-terminal kinase-interacting protein 1 | 69.90 | 0.8 | 2.75 | ||
| 48 | O60583 | Cyclin-T2 | 69.04 | 0.5 | 1.49 | ||
| 49 | O15055 | Period circadian protein homolog 2 | 68.37 | 0.6 | 3.28 | ||
| 50 | Q08AE8 | Protein spire homolog 1 | 67.72 | 0.2 | 1.17 | ||
| 51 | Q96FS4 | Signal-induced proliferation-associated protein 1 | 67.47 | 0.8 | 1.26 | ||
| 52 | Q8IZ41 | Ras and EF-hand domain-containing protein | 67.30 | 0.8 | 1.09 | ||
| 53 | P05783 | Keratin, type I cytoskeletal 18 | 67.21 | 0.5 | 5.43 | ||
| 54 | Q17RY0 | Cytoplasmic polyadenylation element-binding protein 4 | 66.26 | 1.3 | 4.47 | ||
| 55 | O43310 | CBP80/20-dependent translation initiation factor | 66.22 | 0.6 | 2.56 | ||
| 56 | O60271 | C-Jun-amino-terminal kinase-interacting protein 4 | 65.93 | 1.2 | 2.26 | ||
| 57 | O75112-6 | Isoform 6 of LIM domain-binding protein 3 | 65.72 | 70.30 (protein binding) | −0.1 | 2.53 | |
| 58 | O14770 | Homeobox protein Meis2 | 65.62 | 0.8 | 3.58 | ||
| 59 | Q13137 | Calcium-binding and coiled-coil domain-containing protein 2 | 65.47 | 0.3 | 1.86 | ||
| 60 | O94972 | E3 ubiquitin-protein ligase TRIM37 | 65.46 | 0.5 | 1.65 | ||
| 61 | Q9Y605 | MORF4 family-associated protein 1 | 65.35 | −1.1 | <140 aa | ||
| 62 | Q8IXB3 | Trafficking regulator of GLUT4 1 | 64.97 | −1.0 | −0.85 | ||
| 63 | Q27J81 | Inverted formin-2 | 64.93 | 0.4 | 2.99 | ||
| 64 | O43586 | Proline-serine-threonine phosphatase-interacting protein 1 | 64.66 | 0.2 | 1.2 | ||
| 65 | Q7Z6J0 | E3 ubiquitin-protein ligase SH3RF1 | 64.53 | 0.6 | 1.88 | ||
| 66 | Q86UE8 | Serine/threonine-protein kinase tousled-like 2 | 63.60 | 26.00 (targeting to the nucleus) | 0.9 | 2.22 | |
| 67 | Q5U651 | Ras-interacting protein 1 | 62.93 | 1.1 | 2.00 | ||
| 68 | P20749 | B-cell lymphoma 3 protein | 62.78 | 0.0 | 3.41 | ||
| 69 | Q9HCI5 | Melanoma-associated antigen E1 | 62.38 | 0.6 | 2.70 | ||
| 70 | Q9H4E7 | Differentially expressed in FDCP 6 homolog | 61.97 | 0.2 | 3.32 | ||
| 71 | P31689 | DnaJ homolog subfamily A member 1 | 61.46 | 1.6 | 0.72 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 72 | Q9UPT6 | C-Jun-amino-terminal kinase-interacting protein 3 | 60.85 | 1.2 | 3.45 | ||
| 73 | Q02410 | Amyloid-beta A4 precursor protein binding family A member 1 | 60.81 | 0.8 | 1.43 | ||
| 74 | Q7Z6J2 | Protein TAMALIN | 60.76 | 0.8 | 4.56 | ||
| 75 | Q15569 | Dual specificity testis-specific protein kinase 1 | 60.54 | 0.5 | 2.87 | ||
| 76 | Q99704 | Docking protein 1 | 60.50 | 0.5 | 1.90 | ||
| 77 | Q9BX97 | Plasmalemma vesicle-associated protein | 59.73 | −0.1 | 2.20 | ||
| 78 | P10909 | Clusterin | 59.69 | −0.1 | 1.06 | ||
| 79 | Q8TB72 | Pumilio homolog 2 | 59.57 | 1.2 | 3.33 | ||
| 80 | O75146 | Huntingtin-interacting protein 1-related protein | 59.46 | 0.4 | 0.51 | ||
| 81 | P60880 | Synaptosomal-associated protein 25 (SNAP25) | 59.22 | 0.4 | −1.12 | ||
| 82 | Q15633 | RISC-loading complex subunit TARBP2 | 58.74 | 0.2 | 1.39 | ||
| 83 | Q9H6X4 | Transmembrane protein 134 | 58.46 | 0.2 | 0.32 | ||
| 84 | P50479 | PDZ and LIM domain protein 4 | 58.18 | 0.8 | 2.71 | ||
| 85 | Q9Y4G8 | Rap guanine nucleotide exchange factor 2 | 57.37 | 1.0 | 2.78 | ||
| 86 | Q5SQN1 | Synaptosomal-associated protein 47 | 57.33 | −0.2 | 1.25 | ||
| 87 | Q6P5Z2 | Serine/threonine-protein kinase N3 | 56.92 | 0.3 | 1.61 | ||
| 88 | P27815 | cAMP-specific 3′,5′-cyclic phosphodiesterase 4A | 55.19 | 0.1 | 3.23 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 89 | O96018 | Amyloid-beta A4 precursor protein binding family A member 3 | 55.13 | 0.3 | 1.12 | ||
| 90 | O94827 | Pleckstrin homology domain-containing family G member 5 | 54.77 | 0.6 | 3.42 | ||
| 91 | P35240 | Merlin | 54.62 | 0.0 | 1.07 | ||
| 92 | O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | 53.72 | 0.8 | 3.40 | ||
| 93 | Q5JSH3 | WD repeat-containing protein 44 | 53.45 | 0.9 | 2.13 | ||
| 94 | Q8IX03 | Protein KIBRA | 52.74 | 0.5 | 3.32 | ||
| 95 | P19525 | Interferon-induced, double-stranded RNA-activated protein kinase (eIF-2a protein kinase) | 52.63 | 1.2 | 0.20 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 96 | Q07912 | Activated CDC42 kinase 1 | 52.41 | 4.00 (protein binding) | 0.6 | 3.11 | |
| 97 | Q9UBP0 | Spastin | 50.81 | 0.7 | 1.79 | ||
| 98 | Q99828 | Calcium and integrin-binding protein 1 | 50.79 | −0.6 | 1.76 | Hardenberg et al., 2020 (dataset S1) *PSP | |
| 99 | Q06787 | Fragile X mental retardation protein 1 (FMRP) | 50.63 | 41.00 | 1.5 | 4.70 | Hardenberg et al., 2020 (dataset S1) |
| 100 | P22059 | Oxysterol-binding protein 1 | 50.31 | 1.3 | 2.54 | ||
| 101 | Q8IWQ3 | Serine/threonine-protein kinase BRSK2 | 50.14 | 0.9 | 3.09 | ||
| 102 | A1L4K1 | Fibronectin type III and SPRY domain-containing protein 2 | 50.07 | 0.4 | 1.21 | ||
| 103 | Q9BYT3 | Serine/threonine-protein kinase 33 | 50.00 | 0.4 | 1.08 | ||
| 104 | Q8IWE4 | DCN1-like protein 3 | 49.01 | 0.6 | 2.77 | ||
| 105 | Q8NFU3 | Thiosulfate:glutathione sulfurtransferase | 48.70 | −0.7 | <140 aa | ||
| 106 | Q9H2J4 | Phosducin-like protein 3 | 48.54 | 0.1 | −0.68 | ||
| 107 | Q13459 | Unconventional myosin-IXb | 48.49 | 1.0 | 2.89 | ||
| 108 | O14976 | Cyclin-G-associated kinase | 48.44 | 0.9 | 2.29 | ||
| 109 | Q12840 | Kinesin heavy chain isoform 5A | 48.26 | 0.9 | 3.08 | ||
| 110 | P32456 | Guanylate-binding protein 2 | 47.21 | 0.4 | −0.09 | ||
| 111 | P60321 | Nanos homolog 2 | 47.10 | 0.7 | <140 aa | ||
| 112 | Q9H0R5 | Guanylate-binding protein 3 | 47.06 | 0.3 | 0.00 | ||
| 113 | O43426 | Synaptojanin-1 | 46.73 | 0.9 | 3.36 | ||
| 114 | Q6PIW4 | Fidgetin-like protein 1 | 46.59 | 0.7 | 1.18 | ||
| 115 | Q12982 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 | 46.5 | 0.3 | −0.12 | ||
| 116 | Q14764 | Major vault protein | 45.35 | 0.6 | 2.10 | Hardenberg et al., 2020 (dataset S1) *PSP | |
| 117 | Q9NQI0 | DEAD-Box Helicase 4 (DDX4) | 45.03 | 1.8 | 6.77 | Hardenberg et al., 2020 (dataset S1) | |
| 118 | O75312 | Zinc finger protein ZPR1 | 44.66 | 0.2 | 0.49 | ||
| 119 | Q96S99 | Pleckstrin homology domain-containing family F member 1 | 44.44 | −0.3 | 0.99 | ||
| 120 | Q9HAU5 | Regulator of nonsense transcripts 2 | 44.42 | 16.00 (protein binding) | 1.0 | 2.39 | |
| 121 | Q8IYI6 | Exocyst complex component 8 | 44.00 | 0.4 | 0.52 | ||
| 122 | Q8NI35 | InaD-like protein | 43.70 | 1.5 | 1.36 | ||
| 123 | O95295 | SNARE-associated protein Snapin | 43.38 | −0.1 | <140 aa | ||
| 124 | Q9BUZ4 | TNF receptor-associated factor 4 | 43.19 | 0.1 | 1.14 | ||
| 125 | Q684P5 | Rap1 GTPase-activating protein 2 | 43.01 | 1.2 | 1.95 | ||
| 126 | Q15078 | Cyclin-dependent kinase 5 activator 1 | 42.67 | −0.2 | −0.74 | ||
| 127 | Q92993 | Histone acetyltransferase KAT5 | 42.50 | 0.5 | 0.46 | ||
| 128 | Q96QA5 | Gasdermin-A | 41.80 | 0.3 | 0.24 | ||
| 129 | Q9UBF8 | Phosphatidylinositol 4-kinase beta | 41.54 | 0.5 | 0.36 | ||
| 130 | Q01628 | Interferon-induced transmembrane protein 3 | 41.35 | −1.4 | <140 aa | ||
| 131 | O75140 | GATOR complex protein DEPDC5 | 41.24 | 0.7 | 1.76 | ||
| 132 | Q9Y6R4 | Mitogen-activated protein kinase kinase kinase 4 | 40.8 | 0.8 | 2.56 | ||
| 133 | Q2TAA8 | Translin-associated factor X-interacting protein 1 | 40.43 | 0.4 | 0.32 | ||
| 134 | Q9UI30 | Multifunctional methyltransferase subunit TRM112-like protein | 40.00 | −0.8 | <140 aa | ||
| 135 | A1A4S6 | Rho GTPase-activating protein 10 | 39.95 | 0.6 | 1.83 | ||
| 136 | Q13177 | Serine/threonine-protein kinase PAK 2 | 39.89 | 0.4 | 0.89 | ||
| 137 | O60711 | Leupaxin | 39.64 | −0.2 | 1.09 | ||
| 138 | Q9NV29 | Transmembrane protein 100 | 39.55 | −1.3 | <140 aa | ||
| 139 | Q9NZI8 | insulin-like growth factor 2 mRNA-binding protein 1 | 39.17 | 0.8 | 0.50 | Hardenberg et al., 2020 (dataset S1) | |
| 140 | Q96HP0 | Dedicator of cytokinesis protein 6 | 38.59 | 0.5 | 1.7 | ||
| 141 | O95415 | Brain protein I3 | 38.40 | −0.7 | <140 aa | ||
| 142 | O95248 | Myotubularin-related protein 5 | 37.47 | 0.7 | 2.37 | Hardenberg et al., 2020(dataset S1) *HTS | |
| 143 | O76050 | E3 ubiquitin-protein ligase NEURL1 | 37.46 | 0.2 | 1.69 | ||
| 144 | O76083 | High affinity cGMP-specific 3’,5’-cyclic phosphodiesterase 9A | 37.44 | −0.2 | 1.09 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 145 | Q14289 | Protein-tyrosine kinase 2-beta | 37.36 | 0.3 | 1.54 | ||
| 146 | P48730 | Casein kinase I isoform delta | 37.35 | 0.9 | 2.53 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 147 | O43709 | Probable 18S rRNA (guanine-N (7))-methyltransferase | 37.01 | 0.7 | 0.39 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 148 | Q14524 | Sodium channel protein type 5 subunit alpha | 36.61 | 0.5 | 3.69 | ||
| 149 | Q96PP9 | Guanylate-binding protein 4 | 36.41 | 0.3 | 0.42 | ||
| 150 | Q8TEB7 | E3 ubiquitin-protein ligase RNF128 | 36.21 | 0.5 | 0.72 | ||
| 151 | P07101 | Tyrosine 3-monooxygenase | 35.98 | 0.1 | 0.04 | ||
| 152 | Q9Y6D6 | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 | 35.59 | 0.6 | 1.43 | ||
| 153 | Q9UK39 | Nocturnin | 35.50 | −0.4 | 1.03 | ||
| 154 | Q9UM54 | Unconventional myosin-VI | 35.32 | 1.0 | 1.24 | ||
| 155 | Q9UN36 | Protein NDRG2 | 35.31 | 0.3 | 0.14 | ||
| 156 | P04626 | Receptor tyrosine-protein kinase erbB-2 | 35.14 | 21.40 | 0.6 | 2.79 | |
| 157 | Q9Y2K6 | Ubiquitin carboxyl-terminal hydrolase 20 | 35.01 | 0.6 | 1.08 | ||
| 158 | O14966 | Ras-related protein Rab-7L1 | 34.98 | −0.4 | −0.79 | ||
| 159 | Q14999 | Cullin-7 | 34.98 | 0.6 | 1.72 | ||
| 160 | Q9UDY8 | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | 34.71 | 0.4 | 3.00 | ||
| 161 | Q8TEY7 | Ubiquitin carboxyl-terminal hydrolase 33 | 34.61 | 0.6 | 0.93 | ||
| 162 | Q9UPY3 | Endoribonuclease Dicer (DICER) | 34.44 | 0.7 | 1.58 | ||
| 163 | P26572 | Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase | 34.38 | 0.0 | 0.62 | ||
| 164 | O15162 | Phospholipid scramblase 1 | 33.65 | 0.0 | 4.02 | ||
| 165 | *Q86WV6 | Stimulator of interferon genes protein (STING) | 27.97 | 9.50 (protein binding) | −0.2 | −0.48 | Yu et al., 2021 (PMID: 33833429) |
| 166 | *Q92905 | COP9 signalosome complex subunit 5 | 27.84 | 9.90 (protein binding) | 0.3 | −0.23 | |
| 167 | *P12931 | Proto-oncogene tyrosine-protein kinaske Src | 27.43 | 15.70 (protein binding; lipid binding and regulation of phosphorylation) | 0.6 | 1.06 | |
| 168 | *P20591 | Interferon-induced GTP-binding protein Mx1 | 25.53 | 7.70 | 0.4 | 0.05 | |
| 169 | *P01112 | GTPase HRas | 22.22 | 10.60 | 0.0 | −0.29 | |
| 170 | *P07948 | Tyrosine-protein kinase Lyn | 21.29 | 12.10 (protein binding and molecular recognition effector) | 0.6 | 0.49 | Hardenberg et al., 2020 (dataset S1) *HTS |
| 171 | *Q06609 | DNA repair protein RAD51 homolog 1 | 15.63 | 18.29 (NA binding) | 0.2 | 0.52 | Hardenberg et al., 2020 (dataset S1) |
| Literature proteins | |||||||
| 172 | Q00994 | Brain expressed x-linked 3 (Bex3) | 100 | 0.8 | <140 aa | Do Amaral et al., 2020 (PMID: 32142787) | |
| 173 | P08151 | Glioma-associated oncogene homolog 1 (Gli1) | 96.75 | 1.4 | 5.01 | ||
| 174 | Q08379 | GM130 | 92.22 | 0.6 | 1.21 | ||
| 175 | Q14677 | EpsinR | 82.88 | 1.1 | 3.84 | ||
| 176 | P17275 | JunB | 81.56 | 1.8 | 3.66 | ||
| 177 | Q09472 | Histone acetyltransferase p300 (p300) | 77.55 | 6.90 | 1.1 | 4.67 | |
| 178 | Q92793 | CREB-binding protein (CBP)/p300 | 76.58 | 25.10 (protein binding) | 1.1 | 3.67 | Zhang et al., 2021 (PMID: 34326347) |
| 179 | P04637 | P53 | 68.19 | 48.10 (DOT; protein binding) | 0.1 | 1.99 | Hardenberg et al., 2020 (dataset S1) |
| 180 | P04156 | Prion (PrP) | 67.79 | 40.30 (entropic chain) | 2.1 | 12.9 | Hardenberg et al., 2020 (dataset S1) |
| 181 | P23763 | VAMP | 62.71 | −0.7 | <140 aa | Hardenberg et al., 2020 (dataset S1) *PSP | |
| 182 | P52948 | Nuclear pore complex protein (Nup98) | 61.81 | 8.20 | 1.3 | 5.81 | Hardenberg et al., 2020 (dataset S1) |
| 183 | P27824 | Calnexin | 54.39 | 0.8 | 0.59 | ||
| 184 | P29590 | Promyelocytic leukemia protein (PML) | 53.06 | −0.1 | 2.18 | Hardenberg et al., 2020 (dataset S1) *HTS | |
| 185 | Q8WWI1 | LIM domain only 7 (Lmo7) | 52.53 | 1.0 | 3.66 | ||
| 186 | P27797 | Calreticulin | 52.04 | 95.90 | 1.2 | 1.13 | |
| 187 | P29375 | Retinoblastoma-binding protein 2 (RBP2) | 48.28 | 10.50 | 0.6 | 2.90 | |
| 188 | Q8IVF5 | STEF/TIAM2 | 46.38 | 1.2 | 2.62 | ||
| 189 | P37840 | Alpha-synuclein | 37.14 | 100.00 (molecular recognition effector) | 1.1 | −0.31 | Ray et al., 2020 (PMID: 32514159) |
| 190 | Q9Y4L5 | Rabring7 | 35.53 | 0.2 | 2.29 | ||
| 191 | *P20807 | Calpain-3 | 30.33 | 6.30 (inhibitor) | 0.9 | 0.45 | |
| 192 | *P04049 | Raf1 | 26.70 | 2.90 | 0.3 | 1.81 | Hardenberg et al., 2020 (dataset S1) |
| 193 | *P07355 | Annexin A2 | 16.81 | 9.40 (DOT; protein binding; molecular recognition effector) | 0.4 | −1.05 | Hardenberg et al., 2020 (dataset S1) *HTS |
3.4. Are perinuclear IDR-containing proteins predicted to undergo LLPS?
Given the enrichment of IDPs/IDR-containing proteins localized in the perinuclear region, we asked if they could undergo phase separation using LLPS predictors. The catGRANULE algorithm calculates LLPS ability based on the following features: primary structure composition, structural disorder, and RNA-binding propensity. The algorithm uses as input a database with 120 granule-forming proteins from about 4000 in the yeast proteome [30]. Interestingly, about 83% (161 proteins) of the predicted disordered perinuclear protein dataset showed a minimum threshold for LLPS (score above 0) by the catGRANULE analysis (Table 2, sixth column). Proteins with a more rigorous catGRANULE LLPS score (≥0.5) corresponds to 55% (106 proteins), as shown by a blue shade in Table 2. Vernon et al. [17] reported that low complexity regions of LLPS-forming proteins contain a high number of side chains that establish pi-pi contacts, especially planar such as via the guanidinium from arginine, and aromatic rings (pi-stacking). Thus, the PScore reports phase separation behavior considering the frequency of pi-pi contacts [17]. Analysis by the PScore algorithm detected 19 proteins that might undergo LLPS mostly driven by pi-pi contacts (score ≥ 4.0) (Table 2, highlighted in yellow). To identify published experimental evidence of LLPS in the perinuclear IDR-containing group, we analyzed the datasets from Hardenberg et al. [31], who compiled data from several LLPS-related repositories based on in vitro and/or in vivo evidence. Also, we manually searched for references describing protein liquid behavior as shown in Table 2. From the 31 proteins with LLPS described in published studies, 6 proteins are predicted by both catGRANULE and PScore, while 12 proteins did not show a LLPS score by both algorithms (Fig. 6A). Moreover, 13 proteins with proven LLPS showed an intersection solely with catGRANULE, and one protein is predicted solely by PScore (Fig. 6A). Together with the IDR-containing proteins with evidence of LLPS, we selected the ones predicted by both catGRANULE and PScore for further bioinformatics analysis (total of 43; named herein as perinuclear LLPS-IDR-containing proteins).
Fig. 6.
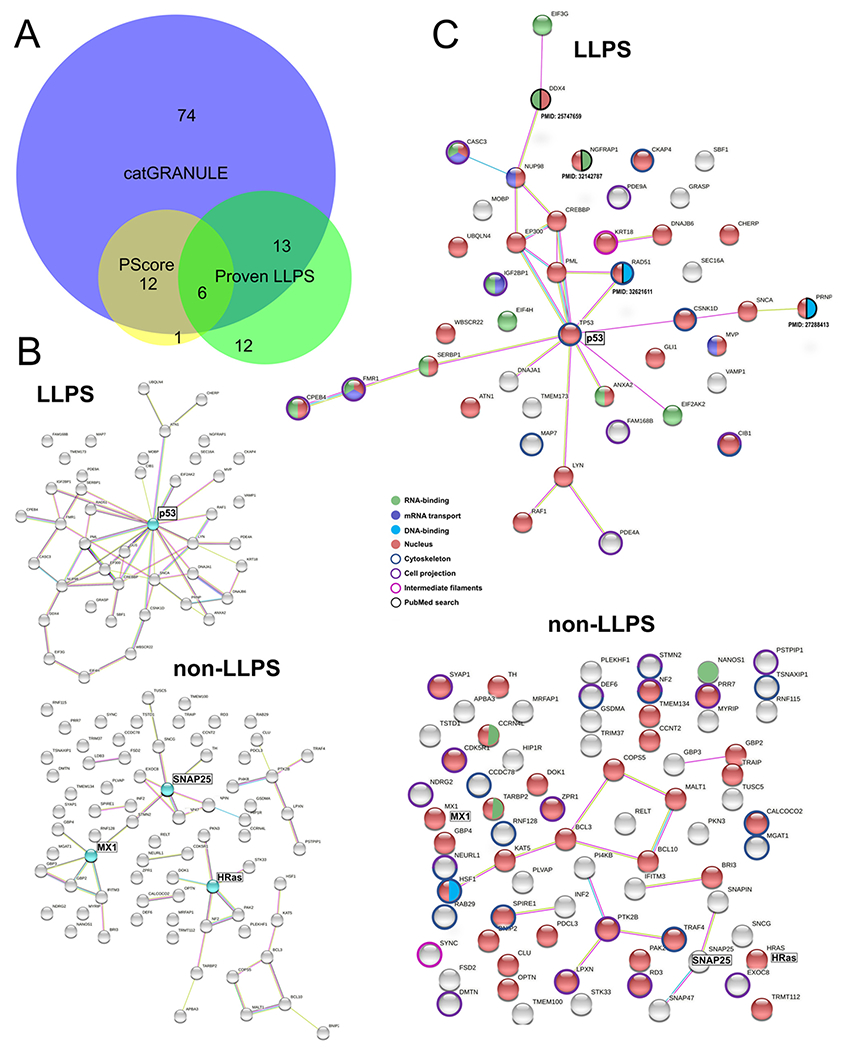
Perinuclear IDR-containing proteins with phase separation ability. (A) Venn diagram (analyzed with BioVenn, available at http://www.biovenn.nl) showing the overlap between predictions by catGRANULE (score ≥ 0.5), PScore (score ≥ 4.0) and perinuclear proteins with peer-reviewed published evidence of phase behavior (in vitro and/or in vivo). (B) STRING network of functional and physical protein-protein interactions. Top: perinuclear LLPS-IDR-containing proteins with 43 members. Bottom: LLPS-IDR-containing proteins with 74 members. Proteins highlighted in cyan are potential hubs using this classification. (C) Combined STRING (only physical interaction shown) and UniProt keywords evaluation of NA-related and cytoskeleton-related processes. Top: LLPS-IDR-containing proteins. Bottom: non-LLPS-IDR-containing proteins. Evidence from literature is assigned with PMID. Legend is shown in the middle.
3.5. Do the perinuclear IDR-containing proteins engage in a network with specific functions?
It is known that liquid condensates can harbor hundreds of ‘client’ proteins, but also many LLPS drivers can conjunctly contribute to condensate assembly through multivalent heterotypic interactions. Based on that, we submitted the LLPS-IDR-containing group and the IDR-containing (not predicted by catGRANULE + PScore; or not present on LLPS literature; total 74 proteins), respectively, to the STRING network database (Fig. 6B). Functional and physical protein interactions are observed in both groups, being p53 in the LLPS-group an important interaction hub whereas MX1, SNAP25 and HRas were the most connected proteins (above 4 edges). Considering only physical interactions, albeit the LLPS-group harbor less members (43), they interact significantly more than the 74 members of the non-LLPS-IDR-containing group (LLPS-group: 26 edges versus IDR-group: 14 edges (Fig. 6C). Additionally, p53 might be a key protein in the perinuclear LLPS group as highlighted by its multiple protein partners. In the non-IDR-group, the proteins assigned in Fig. 6B did not reveal significant physical contacts to other members, and any protein member stand out for several interactions (Fig. 6C).
Interestingly, mRNA transport was the most enriched UniProt keyword for the LLPS-IDR-group (strength 1.34). This was followed by other 15 UniProt keywords, being the most enriched (strength >1.00): biological rhythms (strength 1.20) and host-virus interaction (strength 1.01). Analysis of the IDR-group showed 5 UniProt keywords associated: immunity (strength 0.66); cell projection (strength 0.60); cytoplasm (strength 0.57); coiled-coil (strength 0.46) and phosphoprotein (strength 0.19). However, the top three UniProt keywords strengths are significantly lower (below 0.70) for the IDR-containing group.
3.6. Are perinuclear IDR-containing proteins involved in nucleic acids processes?
Nucleic acids (NA), especially structured regions in RNA/DNA can act as a scaffold for LLPS. In addition, nuclear mRNAs exported to the cytoplasm pass through the perinuclear region where ribonucleoproteins, if present, could be involved in transporting them. LLPS is involved in RNA storage, processing, and transport as well as various processes in the nucleus (transcriptional regulation, DNA repair, organization of chromatin, among others). Thus, we sought to determine if the perinuclear LLPS-IDR-containing are more frequently involved in NA-related metabolism as opposed to the IDR-containing (not predicted by catGRANULE + PScore; or not present on LLPS literature; total 74 proteins). To verify that, we analyzed annotations on UniProt keywords related to NA-processes and merged the results on the STRINGS network. This analysis revealed about 74% (32 proteins) from the LLPS-IDR-containing group related to NA-processes (Fig. 6C, top network). In case of the IDR-containing proteins that would not undergo LLPS, 49% (37 proteins) are related to NA metabolism (Fig. 6C, bottom network). We then used ‘cytoskeleton’, ‘cell projection’ and ‘intermediate filaments’ as UniProt keywords, to investigate the participation on these cytoskeleton-related processes. Proportionally, both groups showed a similar association to cytoskeleton-derived processes (~35% of members from LLPS-IDR group and 39% of members from IDR-group) (Fig. 6C).
3.7. Structural features of perinuclear IDR-containing
The IDR-containing proteins (LLPS and non-LLPS groups) belong to different families and, most of them do not have a three-dimensional structure for the full polypeptide segment. The flexible nature of IDRs together with the phase separation ability can significantly hamper structural determination, especially by nuclear magnetic resonance spectroscopy in solution. In addition, STRING (via searching in SMART tool) did not report any conserved domain among the IDR-containing proteins. To better understand the differences between the perinuclear LLPS-IDR and non-LLPS groups, we analyzed the amino acid composition of the IDR-containing proteins (LLPS and non-LLPS groups) using as background globular proteins from the Protein Data Bank with a sequence identity below 25% (PDBS25). Analysis by the Composition Profiler [32] tool showed that, generally, both groups are enriched in disorder-promoting residues (red bars) and depleted in order-promoting residues (blue bars), as expected (Supplementary Fig. 2). However, the LLPS proteins showed an increased value of proline, glutamine, glycine and histidine compared to the non-LLPS members. In addition, cysteine, glutamate and leucine were increased in the non-LLPS proteins as compared to the LLPS ones. The high content of proline, glutamine and glycine, amino acids, commonly found in prion-like domains, prompted us to investigate the presence of them across the LLPS and non-LLPS groups. While the 74 members of non-LLPS group did not show any predicted prion-forming sequence, the LLPS group showed 8 members (out of the 23) containing potential prion-like domains, as predicted by the PLAAC algorithm [33]. This included CREB-binding protein (UniProt Q92793; PLAAC PRDscore 102.5), HAT p300 (UniProt Q09472; PLAAC PRDscore 102.6), Atrophin-1 (UniProt P54259; PLAAC PRDscore 43.8), PrP (UniProt P04156; PLAAC PRDscore 16.8), CASC3 (UniProt O15234; PLAAC PRDscore 18.3), NUP98 (UniProt P52948; PLAAC PRDscore 13.3), Myelin-associated neurite-outgrowth inhibitor (UniProt A1KXE4; PLAAC PRDscore 16.7) and CPE-binding protein 4 (UniProt Q17RY0; PLAAC PRDscore 14.1). These finding are consonant to the property of low complexity prion-like regions driving LLPS.
4. Discussion
The collection of our findings points to the existence of a highly organized network of cytoskeletal filaments intermingled with several structural and signaling proteins in the perinuclear region of eukaryotic cells. Furthermore, we show that several perinuclear proteins are predicted to be intrinsically disordered. Since a common feature implicated in LLPS is the presence of disordered regions, enabling fluctuation in an ensemble of conformations (reviewed in Uversky [34]), our data point to an important function of IDPs as signaling hubs within the perinuclear cloud. Several perinuclear IDPs might function as scaffolding proteins that regulate “on-off” switches in signaling pathways by their phase separation property.
Disordered regions can form a combination of weak interactions drove by ‘sticker’ residues, that establish intra/intermolecular contacts forming supramolecular structures (reviewed in Boeynaems et al. [8]). These transient interactions can be within monomers (homotypic) or between protein-ligand such as RNA (heterotypic), and the multiple contact points create a 3D network of molecules driving a two-phase regime, a protein/RNA-rich phase (dense) is formed separately from a dispersed (light) phase containing a low concentration of the same molecules (reviewed in Boeynaems et al. [8]). The main property associated to the phase separation ability is multivalency, because of that, not only disordered regions can function as ‘sticky’ patches but molten globule regions (e.g. p53-TAD) [35] and repeated folded domains (e.g. SH3) can also mediate labile crosslinks involved in phase separation (reviewed in Peran and Mittag [36]). However, since IDRs can self-associate at a low concentration, they more often mediate LLPS intracellularly (at a physiological condition). ‘Spacers’ are residues that interleave the sticky patches and whose degree of solvation impacts on the material properties of a phase-separated state and the onset of phase separation [37]. Positive and aromatic side chains are example of stickers that can drive a two-phase regime by cation-pi, and pi-pi contacts, respectively. Spacers are constituted by polar residues, for instance, Gly-rich regions that show an increased flexibility and contribute to liquid condensates [37]. Oppositely, spacers such as serine and proline promote more ordered phase separated states with lower fluidity [37,38]. Structural studies on dissecting the atomic level structure of condensates revealed that determinants of LLPS are complex, and the dissociation between a solely IDP-driven phenomenon is needed since the condensate-forming protein can have a compact 3D structure when phase-separated [39–41]. In many instances, albeit changes in internal dynamics, IDRs can remain disorder in condensates (reviewed by Peran and Mittag, 2019). However, some IDRs enriched in aromatics (not clustered), can transit to reversible β-sheets during LLPS, this folding is transient as opposed to stable steric zippers found in amyloids (reviewed by [36]). In addition, the tumor suppressor protein p53, which arose as an important hub in the perinuclear space (Fig. 6B and C) undergoes LLPS in vitro in a mild denaturing condition (800 mM GndHCl) known to contain a significant population in the molten globule conformation [35]. The pre-molten globule-like structure of retinoid X receptor (hRXRγ) has also been associated to drive LLPS [42].
Interestingly, condensates can recruit specific ‘client’ proteins to catalyze reactions. We hypothesize that the protein aggregates that we observe in the nuclear cloud by electron microscopy of detergent-extracted cells may be protein-rich granules drove by IDR-containing proteins. Together with intrinsic disorder, NA-binding residues and enrichment of pi-stacking side chains are explored by LLPS predictors such as catGRANULE [30] and PScore [17]. The perinuclear IDR-containing proteins (predicted by both algorithms or with proven LLPS) showed an enrichment in mRNA transport function as opposed to the IDPs not predicted to undergo LLPS (Fig. 6). In addition, the perinuclear IDPs from LLPS group were more enriched in specified residues (proline, glutamine, glycine and histidine) whereas the non-LLPS set had glutamic acid and cysteine increased over the PDBS25 background (Supplementary Fig. 2). Furthermore, the LLPS set contains proteins with prion-like domains as opposed to the non-LLPS group (discussed in Section 3.7).
Notably, the signaling proteins studied by immunofluorescence microscopy herein, Gli-1 and Lmo7, show long regions of disorder and high scores for condensation (Table 2). These two proteins are transcription factors (TFs), and likely use its phase separation ability to positively/negatively regulate transcription as reported for other TFs (reviewed in Peng et al. [43]). Indeed, Gli-1 and Lmo7 showed a punctate-pattern reminiscent of liquid-like condensates in the perinuclear region (Fig. 3). Additionally, Gli-1 [21] and Lmo7 [44] are involved in a variety of cancer types, hence, aberrant LLPS of these proteins might contribute to tumor development, configuring an important issue to address in further work.
Seventy nine perinuclear proteins were found through a bibliometric approach using the descriptors “perinuclear protein” in the entire PubMed articles database, while 285 perinuclear proteins were retrieved from UniProtKB/Swiss-Prot, with only five proteins found in both search strategies. Such difference in the number of perinuclear proteins in these two approaches suggest the need for a clear definition and characterization of what defines the limits of the nuclear cloud and which proteins and structures are their constituents. During our survey it became clear that several published articles do not describe perinuclear proteins in their results, even though images of proteins with an undoubted perinuclear localization are present in these papers. For this reason, we foresee that the number and importance of perinuclear proteins (IDPs and non-IDPs) in eukaryotic cells will certainly increase in the next few years.
Our data show that different organelles are positioned within the nuclear cloud. Mitochondria, lysosomes, endoplasmic reticulum, Golgi, and vesicles were found near or in association with the external nuclear surface. Particularly, lysosomes have been reported to be distributed in a rather immobile pool located around the microtubule-organizing center in a cloud, and a highly dynamic pool in the cell periphery [45]. These authors describe that the perinuclear cloud appears to be a site for efficient maturation of endosomes. Lysosomes have recently been associated with cell signaling. Lysosome surfaces serve as a platform to assemble major signaling hubs such as mTORC1, AMPK, Wnt/beta-catenin and the inflammasome [46]. Bagri et al. [47] describe canonical Wnt/beta-catenin activators (BIO and Wnt3a) that induce the perinuclear positioning of lysosomes in cultured muscle cells, suggesting that the Wnt/beta-catenin pathway can modulate the distribution of lysosomes and induce their concentration within the nuclear cloud.
In a broad sense, our findings highlight the structural and signaling role of an intricate 3D network of cytoskeletal filaments and their associated proteins connected to the outer nuclear surface and extending several micrometers into the cytoplasm of eukaryotic cells. These results are in accordance with a previous study showing that the perinuclear cytoskeleton is a region with different mechanical properties than elsewhere in the cytosolic cytoskeleton, with heterogeneously distributed locations exhibiting subdiffusive features [48]. Indeed, we found that roughly 60% of all reported perinuclear proteins are components of the cytoskeleton. Earlier work from our lab has shown that desmin intermediate filaments are stably connected to the outer nuclear surface in skeletal muscle cells in vivo and in vitro [25].
Thus, our findings suggest that the perinuclear cloud is composed of a complex 3D network of cytoskeletal filaments intermingled with multiple structural and signaling proteins, including several IDPs that form biomolecular condensates, and with several organelles bound to the cytoskeleton (Fig. 7 and Table 3). The nuclear cloud has several functions, such as to provide structural support for the nucleus, to control nuclear size and position, to regulate the traffic of molecules between the nucleus and the cytoplasm, to process RNA molecules, to regulate protein synthesis, to modulate several intracellular signaling pathways, and to allow a dynamic interaction between the nucleus and other organelles and cellular structures (Table 3).
Fig. 7.
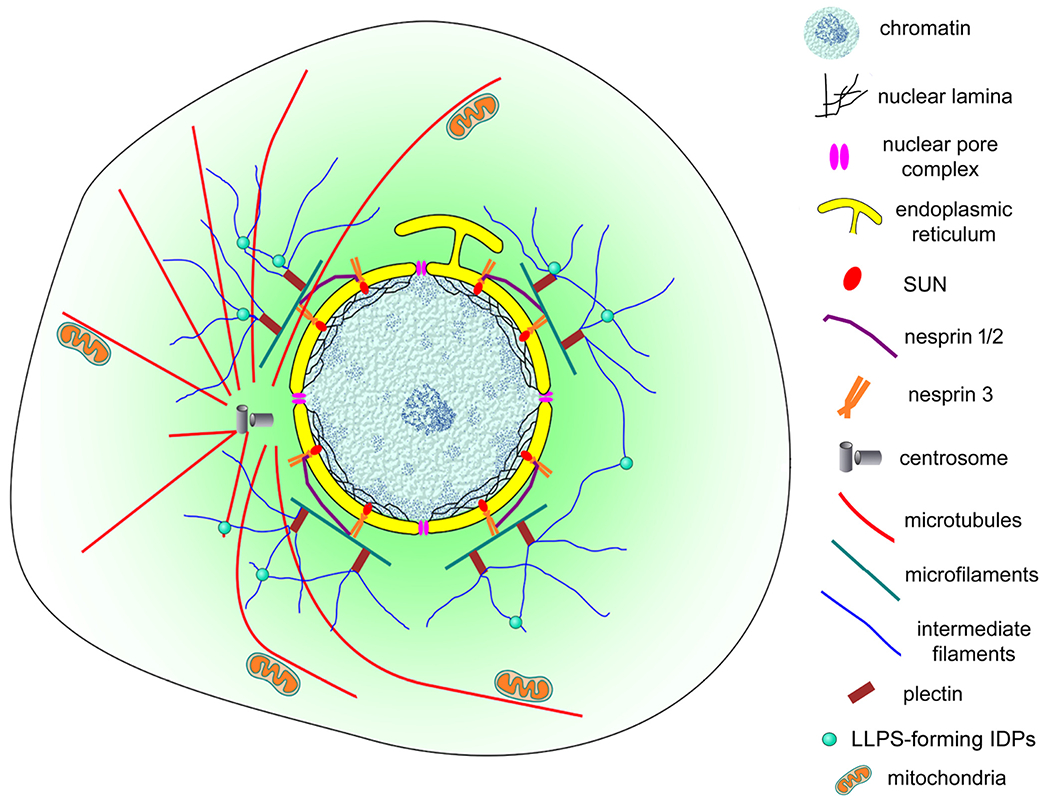
Schematic representation of the nuclear cloud of eukaryotic cells. This model illustrates the complex organization of the perinuclear space (shown in light green) of eukaryotic cells. An intricate 3D network of cytoskeletal filaments is found attached to the nuclear membrane (yellow). Several proteins participate in this organization: cytoskeletal (microfilaments in green, microtubules in red and intermediate filaments in blue) and cytoskeletal associated proteins (plectin, alpha-actinin, and others), nesprins, SUNs, among many others. Dynamic biomolecular condensates drove by intrinsically disordered proteins (IDPs), have a higher propensity to be assembled (small green spheres). Different organelles and cellular structures are also present in the nuclear could: mitochondria (orange), lysosomes, endoplasmic reticulum, Golgi, vesicles, centrosome (grey), and myofibers (in muscle cells).
Table 3.
Main characteristics of the nuclear cloud. The main proteins, structures, organelles and functions of the perinuclear space are described.
| Characteristics and functions of the perinuclear space |
|---|
| Perinuclear cytoskeleton: 3D network of cytoskeletal filaments, including a perinuclear actin cap (MFs) and a high number of actin-associated proteins; MTOC/centrosome, network of MTs and MAPs; web of IFs and plectins (IF-associated proteins) |
| Perinuclear membranous organelles: ER, Golgi, mitochondria, lysosomes, vesicles |
| Perinuclear proteins: structural (cytoskeletal), vesicular trafficking and signaling proteins, including several IDPs |
|
Perinuclear functions: Provide a structural support for the nucleus Spatial organization of the perinuclear space Serve as a hub for the positioning of several organelles Control of nuclear size Control of nuclear position Regulation of the molecular traffic between the nucleus and the cytoplasm Processing of RNA molecules Regulation of protein synthesis and PTMs Regulation of signaling pathways Provide a dynamic interaction between the nucleus and other organelles |
| Disease context: alterations in the expression of perinuclear proteins can lead to several pathological conditions, such as neurodegenerative disorders, cancer, cardiac and skeletal myopathies, immune/inflammatory diseases, and skin disorders |
ER = endoplasmic reticulum, IDPs = intrinsically disordered proteins, IFs = intermediate filaments, MAPs = microtubule-associated proteins, MFs = microfilaments, MTs = microtubules, MTOC = microtubule-organizing center, PTMs = post-translational modification of proteins.
The existence of a membraneless region that concentrates specific structural and signaling proteins at the periphery of the nucleus might be an ancient feature acquired during the evolution of eukaryotic cells. This hypothesis is supported by previous studies showing that the perinuclear concentration of proteins and organelles is present in several eukaryotic unicellular organisms, such as Leishmania [49]. This membrane-free region might have been a key acquisition of a highly organized compartment in ancient eukaryotes providing them with new advantages over competing unicellular organisms.
Supplementary Material
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant 302115/2017-0 for CM, grant 301443/2018-1 for MLC), and by Fundaçço de Apoio à Pesquisa do Estado do Rio de Janeiro (FAPERJ, grant E-26/202.920/2019 for CM and grant E26/210.220/2018 for MLC).
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Claudia Mermelstein reports financial support was provided by National Council for Scientific and Technological Development. Manoel Luis Costa reports financial support was provided by National Council for Scientific and Technological Development.
Footnotes
Special section on “State without borders: Membrane-less organelles and liquid–liquid phase transitions”; Edited by Vladimir N Uversky.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamcr.2021.119161.
Ethics statement
The use of chick embryos and zebrafish embryos was approved by the Ethics Committee for Animal Care and Use in Scientific Research from the Federal University of Rio de Janeiro and received the approval numbers: 069/19 for the chick embryos and 039/20 for the zebrafish embryos.
CRediT authorship contribution statement
CM conceived the work. CM, IAR, LRA, MJA, MLC, SAA and XF contributed to the acquisition of data and their analysis. CM and MJA wrote the first draft of the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Data availability statement
All the datasets generated for this study can be found within the manuscript figures and supplementary material.
References
- [1].Wheeler RJ, Hyman AA, Controlling compartmentalization by non-membrane-bound organelles, Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 373 (2018), 20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Weber SC, Brangwynne CP, Inverse size scaling of the nucleolus by a concentration-dependent phase transition, Curr. Biol 25 (2015) 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stroberg W, Schnell S, On the origin of non-membrane-bound organelles, and their physiological function, J. Theor. Biol 434 (2017. Dec 7) 42–49, 10.1016/j.jtbi.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Woodruff JB, Wueseke O, Viscardi V, et al. Centrosomes: regulated assembly of a supramolecular centrosome scaffold in vitro, Science 348 (2015) 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Hyman AA, Germline P granules are liquid droplets that localize by controlled dissolution/condensation, Science 324 (2009) 1729–1732. [DOI] [PubMed] [Google Scholar]
- [7].Hyman AA, Weber CA, Julicher F, Liquid-liquid phase separation in biology, Annu. Rev. Cell Dev. Biol 30 (2014) 39–58. [DOI] [PubMed] [Google Scholar]
- [8].Boeynaems B, Alberti S, Fawzi NL, Protein phase separation: a new phase in cell biology, Trends Cell Biol. 28 (2018) 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koppers M, Özkan N, Farías GG, Complex interactions between membrane-bound organelles, biomolecular condensates and the eytoskeleton, Front. Cell Dev. Biol 8 (2020), 618733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shaiken TE, Opekun AR, Dissecting the cell to nucleus, perinucleus and cytosol, Sci. Rep 4 (2014. May 12) 4923, 10.1038/srep04923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mermelstein CS, Costa ML, Chagas Filho C, Moura Neto V, Intermediate filament proteins in TPA-treated skeletal muscle cells in culture, J. Muscle Res. Cell Motil 17 (1996) 199–206. [DOI] [PubMed] [Google Scholar]
- [12].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat. Methods 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Culley S, Tosheva KL, Matos Pereira P, Henriques R, SRRF: universal live-cell super-resolution microscopy, Int. J. Biochem. Cell Biol 101 (2018) 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R, Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations, Nat. Commun 7 (2016), 12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peng K, Radivoja P, Vucetic S, Dunker AK, Obradovic Z, Length-dependent prediction of protein intrinsic disorder, BMC Bioinf. 7 (2006) 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hatos A, Hajdu-Soltész B, Monzon AM, et al. DisProt: intrinsic protein disorder annotation in 2020, Nucleic Acids Res. 48 (2020) D269–D276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD, Pi-Pi contacts are an overlooked protein feature relevant to phase separation, eLife 7 (2018), e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Holaska JM, Rais-Bahrami S, Wilson KL, Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes, Hum. Mol. Genet 15 (2006) 3459–3472. [DOI] [PubMed] [Google Scholar]
- [19].Possidonio AC, Soares CP, Fontenele M, Morris ER, Mouly V, Costa ML, Mermelstein C, Knockdown of Lmo7 inhibits chick myogenesis, FEBS Lett. 590 (2016) 317–329. [DOI] [PubMed] [Google Scholar]
- [20].Dedeic Z, Cetera M, Cohen TV, Holaska JM, Emerin inhibits Lmo7 binding to the Pax3 and MyoD promoters and expression of myoblast proliferation genes, J. Cell Sci 124 (2011) 1691–1702. [DOI] [PubMed] [Google Scholar]
- [21].Niewiadomski P, Niedziółka SM, Markiewicz L, Uśpieński T, Baran B, Chojnowska K, Gli proteins: regulation in development and cancer, Cells 8 (2019), 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Teixeira JD, de Andrade Rosa I, Brito J, Maia de Souza YR, de Abreu Manso P, Machado MP, Costa ML, Mermelstein C, Sonic Hedgehog signaling and Gli-1 during embryonic chick myogenesis, Biochem. Biophys. Res. Commun 507 (2018) 496–502. [DOI] [PubMed] [Google Scholar]
- [23].Huelsmann S, Brown NH, Nuclear positioning by actin cables and perinuclear actin. Special and general? Nucleus 5 (2014) 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Costa ML, Escaleira R, Cataldo A, Oliveira F, Mermelstein CS, Desmin: molecular interactions and putative functions of the muscle intermediate filament protein, Braz. J. Med. Biol. Res 37 (2004) 1819–1830. [DOI] [PubMed] [Google Scholar]
- [25].Mermelstein CS, Andrade LR, Portilho DM, Costa ML, Desmin filaments are stably associated with the outer nuclear surface in chick myoblasts, Cell Tissue Res. 323 (2006) 351–357. [DOI] [PubMed] [Google Scholar]
- [26].Costa ML, Escaleira R, Rodrigues VB, Manasfi M, Mermelstein CS, Some distinctive features of zebrafish myogenesis based on unexpected distributions of the muscle cytoskeletal proteins actin, myosin, desmin, alpha-actinin, troponin and titin, Mech. Dev 116 (2002) 95–104. [DOI] [PubMed] [Google Scholar]
- [27].Klus P, Cirillo D, Botta Orfila T, Tartaglia GG, Neurodegeneration and cancer: where the disorder prevails, Sci. Rep 5 (2015), 15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nal B, Chan C, Kien F, Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E, J. Gen. Virol 86 (2005) 1423–1434. [DOI] [PubMed] [Google Scholar]
- [29].Uversky VN, Gillespie JR, Fink AL, Why are natively unfolded proteins unstructured under physiologic conditions? Proteins 41 (2000) 415–427. [DOI] [PubMed] [Google Scholar]
- [30].Bolognesi B, Lorenzo Gotor N, Dhar R, Cirillo D, Baldrighi M, Tartaglia GG, Lehner B, A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression, Cell Rep. 16 (2016) 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M, Widespread occurrence of the droplet state of proteins in the human proteome, PNAS USA 117 (2020) 33254–33262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vacic V, Uversky VN, Dunker AK, Lonardi S, Composition profiler: a tool for discovery and visualization of amino acid composition differences, BMC Bioinf. 8 (2007), 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lancaster AK, Nutter-Upham A, Lindquist S, King OD, PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition, Bioinformatics 30 (2014) 2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Uversky VN, Recent developments in the field of intrinsically disordered proteins: intrinsic disorder-based emergence in cellular biology in light of the physiological and pathological liquid-liquid phase transitions, Annu. Rev. Biophys 50 (2021) 135–156. [DOI] [PubMed] [Google Scholar]
- [35].Petronilho EC, Pedrote MM, Marques MA, Passos YM, Mota MF, Jakobus B, de Sousa GDS, Pereira da Costa F, Felix AL, Ferretti GDS, Almeida FP, Cordeiro Y, Vieira TCRG, de Oliveira GAP, Silva JL, Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands, Chem. Sci 12 (2021) 7334–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Peran I, Mittag T, Molecular structure in biomolecular condensates, Curr. Opin. Struct. Biol 60 (2020) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA, A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins, Cell 174 (2018) 688–699, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alshareedah I, Moosa MM, Banerjee PR. 2021. Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Do Amaral MJ, Araujo TS, Díaz NC, Accornero F, Polycarpo CR, Cordeiro Y, Cabral K, Almeida MS, Phase separation and disorder-to-order transition of human brain expressed X-linked 3 (hBEX3) in the presence of small fragments of tRNA, J. Mol. Biol 432 (2020) 2319–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Emmanouilidis L, Esteban-Hofer L, Damberger FF, de Vries T, Nguyen CK, Ibáñez LF, Allain FHT, NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation, Nat. Chem. Biol 17 (2021) 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kostylev MA, Tuttle MD, Lee S, Klein LE, Takahashi H, Cox TO, Strittmatter SM, Liquid and hydrogel phases of PrPC linked to conformation shifts and triggered by alzheimer’s amyloid-β oligomers, Mol. Cell 72 (2018) 426–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sołtys K, Ożyhar A, Ordered structure-forming properties of the intrinsically disordered AB region of hRXRγ and its ability to promote liquid-liquid phase separation, J. Steroid Biochem. Mol. Biol 198 (2020), 105571. [DOI] [PubMed] [Google Scholar]
- [43].Peng L, Li EM, Xu LY, From start to end: phase separation and transcriptional regulation, Biochim. Biophys. Acta 194641 (2020). [DOI] [PubMed] [Google Scholar]
- [44].Hu Q, Guo C, Li Y, Aronow BJ, Zhang J, LMO7 mediates cell-specific activation of the rho-myocardin-related transcription factor-serum response factor pathway and plays an important role in breast cancer cell migration, Mol. Cell. Biol 31 (2011) 3223–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cabukusta B, Neefjes J, Mechanisms of lysosomal positioning and movement, Traffic 19 (2018) 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Inpanathan S, Botelho RJ. 2019. The lysosome signaling platform: adapting with the times. Front. Cell Dev. Biol 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bagri KM, Rosa IA, Corrêa S, Yamashita A, Brito J, Bloise F, Costa ML, Mermelstein C, Acidic compartment size, positioning, and function during myogenesis and their modulation by the Wnt/beta-catenin pathway, Biomed. Res. Int 20 (2020), 6404230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lozoya OA, Gilchrist CL, Guilak F, Universally conserved relationships between nuclear shape and cytoplasmic mechanical properties in human stem cells, Sci. Rep 6 (2016), 23047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Puechberty J, Blaineau C, Meghamla S, Crobu L, Pagès M, Bastien P, Compared genomics of the strand switch region of Leishmania chromosome 1 reveal a novel genus-specific gene and conserved structural features and sequence motifs, BMC Genomics 8 (2007), 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ray S, Singh N, Kumar R, Patel K, Pandey S, Datta D, Mahato J, Panigrahi R, Navalkar A, Mehra S, Gadhe L, Chatterjee D, Sawner AS, Maiti S, Bhatia S, Gerez JA, Chowdhury A, Kumar A, Padinhateeri R, Riek R, Krishnamoorthy G, Maji SK, α-Synuclein aggregation nucleates through liquid-liquid phase separation, Nat. Chem 12 (2020) 705–716. [DOI] [PubMed] [Google Scholar]
- [51].Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, von Mering C, STRING 8—a global view on proteins and their functional interactions in 630 organisms, Nucleic Acids Res 37 (2009) D412–D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Woodruff JB, Wueseke O, Viscardi V, Mahamid J, Ochoa SD, Bunkenborg J, Widlund PO, Pozniakovsky A, Zanin E, Bahmanyar S, Zinke A, Hong SH, Decker M, Baumeister W, Andersen JS, Oegema K, Hyman AA, Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro, Science 348 (2015) 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated for this study can be found within the manuscript figures and supplementary material.


