Abstract
To fight against the devastating coronavirus disease 2019 (COVID-19), identifying robust anti-SARS-CoV-2 therapeutics from all possible directions is necessary. To contribute to this effort, we selected a human metabolites database containing waters and lipid-soluble metabolites to screen against the 3-chymotrypsin-like proteases (3CLpro) protein of SARS-CoV-2. The top 8 hits from virtual screening displayed a docking score varying between ~ − 11 and ~ − 14 kcal/mol. Molecular dynamics simulations complement the virtual screening study in conjunction with the molecular mechanics generalized Born surface area (MM/GBSA) scheme. Our analyses revealed that (HMDB0132640) has the best glide docking score, − 14.06 kcal/mol, and MM-GBSA binding free energy, − 18.08 kcal/mol. The other three lead molecules are also selected along with the top molecule through a critical inspection of their pharmacokinetic properties. HMDB0132640 displayed a better binding affinity than the other three compounds (HMDB0127868, HMDB0134119, and HMDB0125821) due to increased favorable contributions from the intermolecular electrostatic and van der Waals interactions. Further, we have investigated the ligand-induced structural dynamics of the main protease. Overall, we have identified new compounds that can serve as potential leads for developing novel antiviral drugs against SARS-CoV-2 and elucidated molecular mechanisms of their binding to the main protease.
Graphical abstract
Identification of probable hits from human metabolites against SARS-CoV-2 using integrated computational approaches-Missed against MS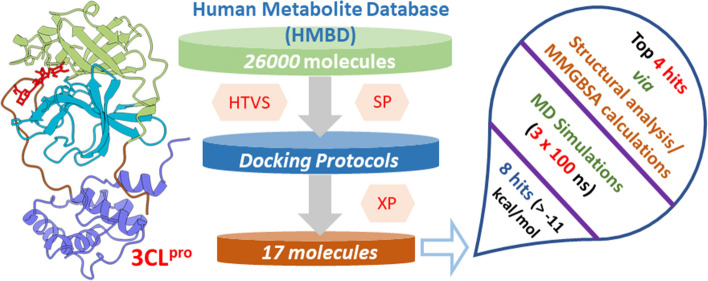
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-022-10513-6.
Keywords: SARS-COV-2, Main protease, Human metabolites, Virtual screening, Molecular dynamics, Free energy
Introduction
In China, a very contagious and severe viral disease was reported at the end of 2019 [1, 2]. This causative agent was later detected as a novel coronavirus (SARS-CoV-2). The disease was subsequently referred to as coronavirus disease 2019 (COVID-19) [3, 4]. This COVID-19 threatened human life and the economy across the globe and affected the lives of millions of people, including more than 6.4 million deaths as of August 5, 2022. It emerged as an epidemic and eventually became a pandemic. As a result, the current COVID-19 pandemic has triggered a medical emergency that is unprecedented in recent global history. There is an urgent need to discover effective therapeutics for treating COVID-19 and find some right immune booster supplements to stay healthy and safe [5].
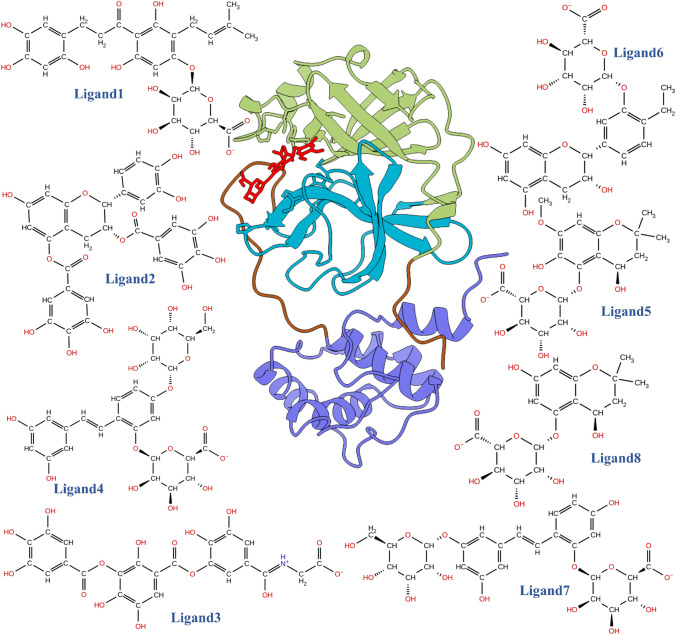
Novel coronaviruses (SARS-CoV-2) are non-segmented, enveloped, positive-sense RNA, single-stranded viruses and are widely distributed in mammals, including humans [6]. Once the cell is infected with SARS-CoV-2, the host cell’s ongoing molecular mechanisms are taken over by the virus to translate its RNA into long protein chains and generate more viral replicas. These viral polyproteins (PP1A) are activated when cut into smaller individual functional components by protease (3-chymotrypsin-like cysteine protease, 3CLpro) [7–9]. Thus, viral proteases (3CLpro) are essential in SARS-CoV-2 virus propagation. Consequently, 3CLpro is regarded as a principal druggable target for SARS-CoV-2. 3CLpro is a heart-shaped protein also known as the main protease (3CLpro). It contains 13 β-strands and nine α-helices and is divided into three domains [10, 11], domain I (residues 8–101), domain II (residues 102–184), a long loop (residues 185–200), and domain III (residues 201–306), shown in Fig. 1. Domain I and II contain primary substrate-binding catalytic dyads, His41 and Cys145, respectively. The C-terminal domain III is mainly involved in the main protease’s dimerization [12]. Several recent computational studies have identified possible small molecules [13–16], plant-derived polyphenols [17, 18], and natural substances [19] to reduce COVID-19 infections by targeting several structural and non-structural proteins of SARS-CoV-2.
Fig. 1.
Ribbon representation of COVID-19 3CLpro complexed with the inhibitor, shown in ball and stick representation. The different part of 3CLpro is shown in different color, i.e., Green: Domain I, Cyan: Domain II, Blue: Domain III, Brown: Inter-domain connecting loop, and Red: Ligand molecules. The top 8 molecules which are screened by the virtual screening workflow are shown in 2D illustration
In recent investigations, metabolomics has been shown to be a promising technique for preventing viral diseases [20, 21]. Metabolomics studies different metabolites in living systems such as bio-fluids, cells, tissues, organisms, etc., which can be small molecules, drug metabolites, protein fragments, intermediates of any processes, etc. Metabolomics is an emerging technology, and very soon, it will be an essential part of precision medicine [22]. The Human Metabolome Database (HMDB) is a freely downloadable human metabolite database and contains various details about different metabolites found in the human body [23]. The HMDB contains 114,214 metabolite entries, including lipid-soluble and water-soluble metabolites [24]. Several medications, such as omeprazole, clopidogrel, cyclophosphamide, and diazepam, are present in the market and may become real pharmaceuticals following metabolism. Thus, metabolites can be an important lead for drug development, for example; (i) 6-Mercaptopurine (HMDB ID-HMDB0015167), azathioprine metabolite, which is an FDA-approved drug used to treat cancer and autoimmune; (ii) Oxazepam (HMDB ID-HMDB0014980), a benzodiazepine derivative which is used to treat anxiety; and (iii) Canrenone (HMDB ID- HMDB0003033), an important active metabolite of spironolactone, which is used as a diuretic agent and treatment of hirsutism [25].
Herein, we have screened the HMDB molecules against SARS-CoV-2 3CLpro to find some potential small molecules that might be useful to combat such deadly diseases. This study may be the first to use in-silico investigations to give an idea of the blocking function of 3CLpro using human metabolites. Finally, we reported promising human metabolites against 3CLpro, which may be further developed as a therapeutic agent against COVID-19 based on their molecular mechanics generalized Born surface area (MM-GBSA) results.
Materials and methods
Preparation of protein
The primary atomic coordinates of 3CLpro (PDB ID- 6LU7, resolution ~ 2.16 Å) [7] were downloaded from the protein data bank and prepared using the protein preparation wizard of the Schrödinger suite [26]. The water molecules and co-factors were removed before minimization, while only the co-crystalized ligand was kept for final energy minimization. The energy minimization was restricted to an RMSD cut-off value of 0.3 Ǻ with the original structure under the OPLS3 force field [27]. The prepared protein was then used for the grid box generation, and a 20 Ǻ box was created, keeping the crystallized ligand in the center. The same gird was used for our entire virtual screening protocol.
Ligand selection, preparation, and virtual screening protocol
We downloaded metabolites from the HMDB database (HMDB 4.0, accessed in May 2021) [24] and converted them to 3D structures using the Schrödinger software. HMDB contains various metabolites such as small molecules, peptides, triglycerides, etc. For curating potential leads from the metabolites, we selected compounds with molecular weights between 70 and 600 Da (~ 26,000 molecules). Since we are interested in studying all the metabolites in this range, we did not filter this database further using Lipinski’s and related filters. Next, we performed the ligand preparation using the LigPrep module of the same suit, which resulted in ~ 36,801 entries. This module helps assign each ligand molecule’s protonation and ionization states. Several structures from each ligand were generated with different ionization states, tautomers, stereochemical information, and ring conformations. Finally, ligands were optimized, yielding low-energy isomers.
The virtual screening was done against 3CLpro with the help of the virtual screening workflow (VSW) of the GLIDE module of Schrödinger [28–30]. The three tiers of virtual screening protocol, namely HTVS (high throughput virtual screening), SP (standard precision), and extra precision (XP), were sequentially employed to obtain potential lead molecules. To reduce the size of the database, we selected the top 30% of the docked complexes obtained from HTVS and used them for screening via Glide-SP. Finally, the top 20% of the SP docking results were considered for the Glide-XP (extra precision) docking. Moreover, the final compound selection was based on a visual inspection of the docking poses and the corresponding XP-docking scores, resulting in 17 unique molecules that were subjected to further analyses and investigations. A similar protocol for the virtual screening study was used in our previous work on COVID-19 3CLpro [15].
Molecular dynamics (MD) simulation
Selected docked structures obtained from the virtual screening were used as an input to the LEaP module of AmberTools19 [31] to generate the input structure for MD simulations. The TIP3P [32] water model was used to solvate the structure in an octahedron water box with a 10 Å buffer region from all directions. An adequate number of ions were also added to neutralize the system. All the complexes were simulated using the Amber ff14SB forcefield [33] for protein and generalized Amber force field (GAFF2) [34] for ligand molecules. A 10 Å cut-off was fixed for calculating the long-range interaction with the help of the particle-mesh Ewald (PME) scheme [35]. The SHAKE [36] algorithm involved the bond length having hydrogen atoms to keep its motion constant. All the systems were simulated at a constant temperature of 300 K, which was maintained using a Langevin thermostat with a collision frequency of 2 ps−1. A detailed description of the simulation protocol was discussed in our previous studies on COVID-19 [11, 37]. All complex structures were subjected to 3 × 100 ns production runs under the NPT ensemble. Trajectories were analyzed using the cpptraj module of AmberTools19, and the last 50 ns trajectories were used for the binding free energy calculation.
Protein–ligand-binding free energy calculation
The molecular mechanics Poisson–Boltzmann (generalized Born) surface area (MM-PB(GB)SA) scheme [38–42] is widely used to determine the binding free energy between protein-inhibitor [14, 43], protein-nucleic acid [44–46] as well as protein-carbohydrate [47, 48] complexes. The total binding free energy comprises internal energy desolvation free energy , and configurational entropy which are related by the following equation [16, 49–53],
| 1 |
To estimate the binding free energy, 2500 frames were selected uniformly from the last 50 ns trajectories, and calculation was done with the help of the MMPBSA.py script available on AmberTools19. The entropic contribution was estimated using the normal mode analysis, and the MM-GBSA pair-wise decomposition scheme also assessed the contribution from each amino acid.
ADMET studies of top ligands
To compute the top lead molecules’ absorption, distribution, metabolism, and excretion properties, the QikProp module of the Schrödinger suite was used. Thirty-five significant pharmaceutical properties were monitored, such as CNS activity, % of human oral absorption, blood–brain barrier prediction, cell permeability, Lipinski rule, etc. The toxicity-related parameters, such as hepatotoxicity, carcinogenicity, mutagenicity, and cytotoxicity, were calculated using the ProTox-II web server [54].
Results and discussions
Virtual screening of metabolites against 3CLpro
To conduct the virtual screening of the human metabolite database, we started with ~ 36,801 entries out of 114,214 HMDB metabolites, which have been further screened using HTVS, Glide-SP, and Glide-XP protocol to get the top 17 lead compounds. These compounds were ranked according to their glide score, and the top 8 ligands were selected by keeping a cut-off value of − 11 kcal/mol. To verify the docking poses, the top 8 ligand molecules were re-docked using Autodock Vina [55] and Glide-XP separately. Superimposition of the docked structures using both methods suggested similar binding poses, as shown in Supporting Information Fig. S1. We also investigated similarities of our principal metabolites by estimating the Tanimoto coefficient.
Further, this estimation was also carried out for five FDA-approved drugs: Remdesivir, Ritonavir, Favipiravir, Indinavir, and Beclabuvir, as shown in Supporting Information, Table S1). Most of the metabolites are chemically distinct except Ligand5 and Ligand8. All other combinations showed a very low similarity like 0.14 (Ligand3 vs. Ligand4) to moderate similarity like 0.58 (Ligand4 vs. Ligand7). On the other hand, FDA-approved drugs showed significantly less similarity, ranging from 0.06 to 0.15. So, this suggests our metabolites are very much exclusive from the commonly used drugs showing a good binding.
The best lead molecule results in a Glide score of − 14 kcal/mol, indicating a promising candidate for drug design. The docking score of the top 8 lead compounds is shown in Table 1. We provided the SMILES of all top 8 ligands in Supporting Information Table S2. One of the best hits in the present study is HMDB0132640, i.e., Ligand1, which is a predicted metabolite of 1-(2,4,6-trihydroxy-3-(3-methyl-2-en-1-yl)phenyl)-3-(2,4,5-trihydroxy phenyl)propan-1-one, a non-cyclic derivative of 2-phenylchromen-4-one flavonoids. It belongs to the class of organic compounds known as flavonoid o-glycosides. It can also be classified as 2′-hydroxy-dihydrochalcones, which is biosynthesized by reducing α,β-unsaturated ketone (chalcone) in the Human gut and is considered to be a flavonoid type of molecule. This type of flavonoid is present in many plants and has shown antiviral properties. The flavonoids have been reported to have a complementary therapeutical role in the treatment of COVID-19 [56]. Ligand1 is one of the predicted metabolites of Isobavachalcone, which is obtained from the seeds of Psoralea corylifolia. Isobavachalcone has proven effective against papain-like protease (PLpro) of SARS-CoV [57]. Isobavachalcone has also been found active against 3C-like protease/main protease (3CLpro/Mpro) of the Middle East respiratory syndrome coronavirus (MERS-CoV) [58]. Thus, HMDB0132640 can be a potential compound for interventional therapy for COVID-19.
Table 1.
Different components of docking scores obtained from the Glide-XP-docking scheme
| Lead molecule | Molecular weight | G-scorea | Glide-lipob | Glide-hbondc | Glide-evdwd |
|---|---|---|---|---|---|
|
Ligand1 (HMDB0132640) |
568.657 | − 14.060 | − 4.160 | 0.000 | − 39.543 |
| Ligand2 (HMDB0030665) | 622.706 | − 12.399 | − 2.859 | − 0.339 | − 61.108 |
| Ligand3 (HMDB0128347) | 573.59 | − 12.151 | − 2.359 | − 0.800 | − 48.830 |
| Ligand4 (HMDB0134117) | 598.64 | − 11.724 | − 2.228 | − 0.769 | − 36.536 |
| Ligand5 (HMDB0125819) | 424.444 | − 11.534 | − 3.702 | − 0.160 | − 47.989 |
| Ligand6 (HMDB0127868) | 492.564 | − 11.134 | − 2.904 | − 0.887 | − 29.007 |
| Ligand7 (HMDB0134119) | 598.64 | − 11.119 | − 3.060 | − 0.430 | − 40.603 |
| Ligand8 (HMDB0125821) | 394.418 | − 11.051 | − 3.369 | − 0.480 | − 44.484 |
aGlide score (kcal/mol)
bLipophilic term derived from hydrophobic grid potential
cHydrogen bonding term in GlideScore
dVan der Waals energy
Another hit from HMDB is HMDB0127868 (Ligand6) which belongs to the 5,7-Dihydroxy flavonoid class. It is a predicted metabolite of 2-(4-ethyl-3-hydroxyphenyl)-3,4-dihydro-2 h-1-benzopyran-3,5,7-triol. It is a polyphenolic compound and can have a potential antiviral property. Its binding energy and docking score are also good (− 10.33 kcal/mol, − 11.134, respectively). HMDB0134119 (Ligand7) is our next hit and is a stilbene glycoside. It is a predicted metabolite of (E)-4-(3,5-dihydroxystyryl)benzene-1,3-diol. Some studies support the potential use of stilbene derivatives in treating SARC-COV infection [19, 59]. Its binding energy and docking score are good (− 9.53 kcal/mol, − 11.119, respectively). Another hit (HMDB0125821/Ligand8) belongs to the 1-benzopyran class of compounds and are polyphenolic compounds. It is a predicted metabolite of 2,2-dimethyl-3,4-dihydro-2 h-1-benzopyran-4,5,7-triol. These benzopyran compounds are found in mushrooms and are the biomarker for consuming these foods. Its binding energy and docking score are good (− 10.28 kcal/mol, − 11.05, respectively).
Structural stability and flexibility of complexes
Overall 3CLpro structure
About eight human metabolites were found to be potential after the virtual screening competition. These compounds were identified as interacting with the binding cavity and catalytic dyads of the SARS-CoV-2 major protease. To further validate the thermodynamics stability and flexibility of the complexes, 100 ns molecular dynamics simulations were carried out, and we monitored each system’s structural and energetic properties during the production simulations.
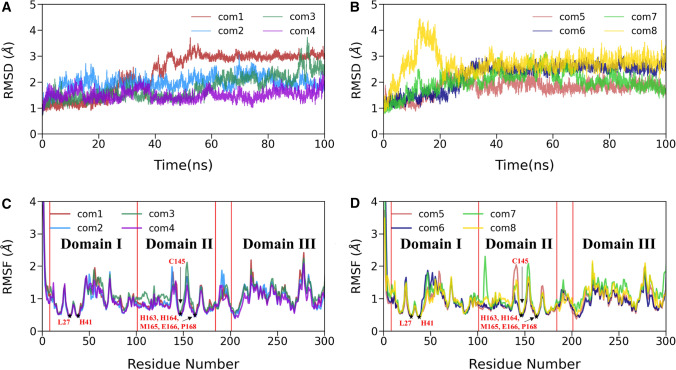
To verify the convergence of simulations, we calculate the time evolution of our system’s receptor backbone atoms root mean square deviation (RMSD) concerning their corresponding initial coordinates. The temporal distribution of RMSD of each system is shown in Fig. 2A, B, and their average values for the last 50 ns are listed in Table 2. We also ran two more replicas for all eight complexes, and the RMSD profile of the other two replica runs is shown in Supporting Information Fig. S2. It is evident from Fig. S2 that the overall RMSD profile was the same in all replica runs, suggesting a converged simulation for all but complex3. In the case of complex3, relatively large fluctuations were observed compared to other complexes. The other seven complex simulations attained stability in the last 50 ns.
Fig. 2.
A, B The time evolution of root means square deviation (RMSD) of backbone atoms of 3CLpro-complex relative to their respective initial coordinates. C, D The root mean square fluctuations (RMSFs) of Cα atoms for all eight 3CLpro-ligand complexes
Table 2.
The average backbone RMSD, the radius of gyration (RoG), and solvent-accessible surface area (SASA) for all eight complexes
| System | RMSD (Å) | RoG (Å) | SASA (nm2) |
|---|---|---|---|
| Complex1 | 3.0 ± 0.1 | 21.9 ± 0.1 | 141.9 ± 2.6 |
| Complex2 | 2.1 ± 0.2 | 22.0 ± 0.1 | 143.9 ± 2.5 |
| Complex3 | 2.1 ± 0.4 | 22.1 ± 0.1 | 144.1 ± 2.7 |
| Complex4 | 1.5 ± 0.2 | 22.0 ± 0.1 | 140.8 ± 2.3 |
| Complex5 | 1.8 ± 0.2 | 21.9 ± 0.1 | 141.6 ± 2.4 |
| Complex6 | 2.6 ± 0.1 | 21.8 ± 0.1 | 137.1 ± 2.3 |
| Complex7 | 2.1 ± 0.3 | 22.0 ± 0.1 | 142.1 ± 3.0 |
| Complex8 | 2.8 ± 0.2 | 21.8 ± 0.1 | 138.2 ± 2.4 |
The data are reported as average ± standard error of the mean (SEM)
Figure 2A, B shows that each system reached a good equilibrium state after 50 ns and maintained it throughout the last 50 ns of production simulations. The average RMSD value was found to vary between (1.5 ± 0.2) Å and (3.0 ± 0.1) Å for all cases. The highest average RMSD value (3.0 ± 0.1) Å was obtained for complex1 and the lowest value (1.5 ± 0.2 Å) for complex4. For the comparison purpose, we also estimated the RMSD of the apo structure from our previous work, which was simulated under the same condition and shown in Supporting Information (Fig. S3A). The average RMSD for the apo form was 2.0 ± 0.01 Å, comparable to the RMSD value of complexes.
We extended our residual fluctuation study, and the root means square fluctuations (RMSFs) of Cα atoms in each protein complex were explored throughout the simulations and displayed in Fig. 2C, D. Figure 2C, D shows that all complexes’ overall atomic fluctuations pattern is the same. Due to the inhibitor binding, we observed lower fluctuations for domain I (residues 8–101) and domain II (residues 102–184). On the other hand, domain III (residues 201–306) showed relatively higher fluctuations than domains I and II. RMSF values rarely crossed 2 Å for most of the Cα atoms, except terminal residues, which is a usual phenomenon. Further, it is evident from Fig. 2C, D that the ligand-binding sites, including Leu27, His41, Cys145, His163, His164, Met165, Glu166, and Pro168, displayed the lowest fluctuation. It can further be observed from Fig. 2C, D that the off-binding site residues like Ser46, Glu47, Leu50, and Pro52 from domain I; Asn151, Ile152, Asp153, Tyr154, and Asp155 from domain II; Met276, Asn277, Gly278, Arg279, Thr280, and Gly302 from domain II showed higher fluctuations compared to other residues. However, our previous study on SARS-CoV-2 3CLpro [60] suggested that the apo 3CLpro protein structure had less atomic fluctuations than the ligand-bound protease, indicating that the apo 3CLpro is less flexible (see Fig. S3B).
Ligand dynamics and binding pocket stability
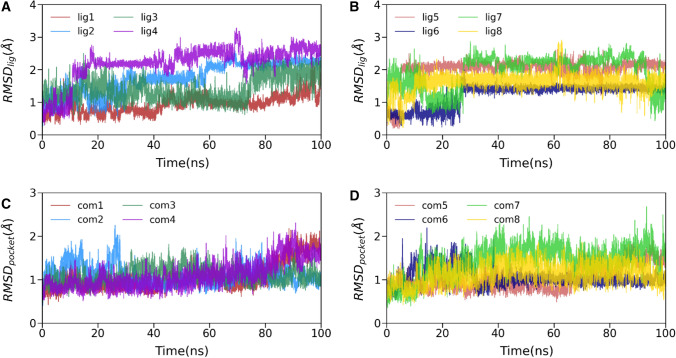
After investigating the overall 3CLpro structure, we also explored the conformation of all the ligands and the respective binding pocket stability in terms of its heavy atoms and backbone atoms RMSD, respectively, as shown in Fig. 3. As shown in Fig. 3A, B, the ligand RMSD values fluctuated within 3 Å, and the average RMSD values ranged from 1.1 to 2.5 Å. Initially, all ligand RMSDs except for ligand3 increased up to 30 ns and stabilized afterward. In the case of ligand3, we observed more fluctuations than other ligands bound to 3CLpro, suggesting that it may make unstable interactions with the binding pocket of 3CLpro. This may weaken its affinity toward 3CLpro. Overall, the flexibility observed from the ligand RMSD profile is evident as these are small molecules. We also estimated the ligand stability by calculating the ligand–protein distance, which is discussed in the subsequent section. However, in the last 30 ns, all ligand RMSDs attain a steady state, which signifies a continuous binding.
Fig. 3.
A, B Time evolution of inhibitors’ heavy atoms root mean square deviations (RMSDs) concerning initial conformations. C, D temporal RMSD variations of backbone atoms around the 5 Å of each ligand (binding pocket)
Similarly, in Fig. 3C, D, we explore the time evolution of backbone atoms deviation at residues of 5 Å around each ligand. As the RMSD plot suggests, the fluctuations of all system’s ligand-binding pockets are lower. The average RMSD values range from 1.0 to 2.0 Å. If we see the fluctuation pattern, all the complexes except (complex1 and complex4) reached stability after 20 ns. The pocket RMSD of complex1 and complex4 revealed the two complexes have almost similar behavior during the entire production simulation and achieved structural stabilities during the initial 80 ns and the final 15 ns. These results suggest that the binding pocket of 3CLpro bound to human metabolites is relatively rigid and compact, which is suitable for better affinity.
Protein compactness and solvent exposure of binding sites
The residual compactness of the protein–ligand structure during molecular dynamics simulation is best described by the radius of gyration (RoG) and the solvent exposure measure in terms of solvent-accessible surface area (SASA). We computed RoG for all the complexes, shown in supporting information, Fig S4A, B, and the last 50 ns trajectories’ average values are listed in Table 2. It is evident from Fig. S4A, B that all the complexes were stable and compact throughout the 100 ns simulations, and the average RoG values range from 21.8 to 22.1 Å for all cases. The initial RoG value for all is high compared to the last 50 ns trajectory. It may indicate that all complexes become more compact due to the binding of human metabolites.
In order to know the solvent exposure of the 3CLpro binding cavity for all the eight simulated systems, we estimated the solvent-accessible surface area (SASA) of protein structures as shown in Fig. S4C, D. The average SASA values of the last 50 ns simulations are listed in Table 2. The initial surface area occupied by each complex is relatively high compared to the final 50 ns. It is evident from Table 2 that SASA values vary between 137.1 nm2 and 144.1 nm2 for all systems. On the other hand, the SASA value for the apo 3CLpro was estimated as 146.5 nm2 [60]. The lower SASA value signifies strong ligand-binding inside the cavity, suggesting the water molecule’s displacement from it. A similar observation was found in earlier studies [61–63].
Human metabolites and 3CLpro domains distance analysis
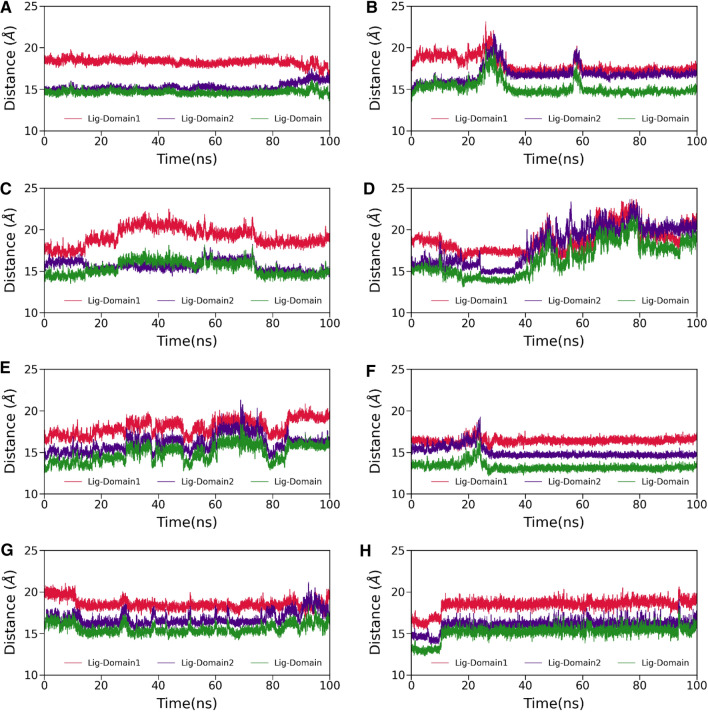
The 3CLpro has divided into three different domains, see Fig. 1. If we talk about the binding of any small molecules on the 3CLpro, then domain I and domain II are mainly responsible for that, and domain III is involved in the dimerization of 3CLpro. Therefore, to explore the structural displacement of the binding pocket and ligand behavior inside the binding cavity, we calculate the center of mass (CoM) distance among human metabolites (ligands) and domains (mainly domain I and domain II) see Fig. 4. As suggested in Fig. 4, except for complex3, complex4, and complex5, the rest have shown no change in their distance plots. It is evident from Fig. 4D that the distance between ligand and domains initially decreases up to 37 ns; afterward, it increases up to 80 ns and finally reaches stable equilibrium and fluctuates around 20 Å for the last 20 ns of simulations. Similarly, we also see some drifting in the distance plot of complex3 and complex5. The binding of human metabolites to 3CLpro is directly affected by these distance deflections.
Fig. 4.
The time evolution of distance between ligand and domain I (in red), ligand and domain II (in violet), ligand and domain I & II (in green). A complex1, B complex2, C complex3, D complex4, E complex5, F complex6, G complex7 and H complex8, respectively
Energetics of human metabolites affinity
To further elucidate the recognition, binding affinity, and specificity of human metabolites against SARS-CoV-2 main protease 3CLpro, we have estimated the total binding affinity and energetic components using molecular mechanics generalized Born surface area (MM-GBSA) approach. A summary of the binding affinity and its components are shown in Supporting Information Fig. S5 and Table 3. As suggested in Fig. S5, the polar solvation and configurational entropic components disfavor the binding of human metabolites while the rest of the components (, ) are favorable toward the protein–ligand complexation. Moreover, Table 3 reveals that the calculated binding affinity of complex1 is the highest (− 18.08 ± 0.87 kcal/mol) followed by complex6 (− 10.33 ± 0.46 kcal/mol), complex8 (− 10.28 ± 0.58 kcal/mol), complex7 (− 9.53 ± 0.85 kcal/mol), complex5 (− 8.75 ± 0.58 kcal/mol) and complex2 (− 5.34 ± 0.68 kcal/mol). On the other hand, the worst binding affinity (due to their high configurational entropic contributions, − T) of complex3 and complex4 suggest that these complexes’ human metabolites are not good binder against 3CLpro. However, the static docking results are promising. It is worth noting that ligand3 displayed an increased configurational entropic contribution as it experienced more fluctuations than other ligands (see Fig. 3A). These results indicate that the complex1’s human metabolite has the strongest binding affinity with 3CLpro.
Table 3.
Energetic components of the binding free energy for 3CLpro of SARS-CoV-2 complexed with human metabolites, estimated using the MM/GBSA (kcal/mol) method
| Systems | ΔEvdW | ΔEelec | ΔGpol | ΔGnp | ΔEMMa | ΔGsolvb | ΔHc | − TΔS | ΔGbindd |
|---|---|---|---|---|---|---|---|---|---|
| Complex1 | − 52.91 (0.10) | − 57.22 (0.35) | 70.40 (0.30) | − 6.79 (0.01) | − 110.13 (0.39) | 63.61 (0.29) | − 46.51 (0.11) | 28.43 (0.86) | − 18.08 (0.87) |
| Complex2 | − 44.26 (0.11) | − 45.62 (0.30) | 62.87 (0.22) | − 6.34 (0.01) | − 89.88 (0.32) | 56.53 (0.21) | − 33.35 (0.14) | 28.01 (0.67) | − 5.34 (0.68) |
| Complex3 | − 32.39 (0.10) | − 79.24 (0.32) | 87.79 (0.29) | − 5.40 (0.01) | − 111.64 (0.35) | 82.79 (0.29) | − 29.25 (0.10) | 30.43 (0.70) | 1.18 (0.71) |
| Complex4 | − 29.93 (0.09) | − 23.86 (0.20) | 38.44 (0.16) | − 4.46 (0.01) | − 53.80 (0.23) | 33.98 (0.15) | − 19.82 (0.11) | 23.99 (0.61) | 4.17 (0.62) |
| Complex5 | − 37.50 (0.11) | − 20.02 (0.30) | 33.29 (0.26) | − 4.59 (0.01) | − 57.52 (0.34) | 28.70 (0.25) | − 28.82 (0.13) | 20.07 (0.57) | − 8.75 (0.58) |
| Complex6 | − 48.53 (0.05) | − 27.72 (0.10) | 48.09 (0.07) | − 5.89 (0.00) | − 76.25 (0.10) | 42.20 (0.07) | − 34.05 (0.05) | 23.67 (0.46) | − 10.33 (0.46) |
| Complex7 | − 44.14 (0.11) | − 32.75 (0.22) | 47.63 (0.16) | − 5.95 (0.01) | − 76.89 (0.28) | 41.68 (0.15) | − 35.21 (0.16) | 25.68 (0.84) | − 9.53 (0.85) |
| Complex8 | − 33.95 (0.06) | − 17.63 (0.17) | 28.26 (0.15) | − 4.18 (0.01) | − 51.58 (0.17) | 24.09 (0.15) | − 27.49 (0.06) | 17.21 (0.58) | − 10.28 (0.58) |
Standard errors of the mean (SEM) are provided in parentheses
aΔEvdW + ΔEelec
bΔGpol+ ΔGnp
cΔEvdW + ΔEelec + ΔGpol+ ΔGnp
dΔEvdW + ΔEelec + ΔGpol+ ΔGnp − TΔS
Han et al. computationally explored the binding affinity of some clinically approved drugs (Chloroquine, Hydroxychloroquine, Remdesivir, Ritonavir, Favipiravir, Beclabuvir, and Indinavir) and recently designed α-ketoamide (13b) inhibitor against SARS-CoV-2 3CLpro [64]. Han et al. suggested that α-ketoamide (13b) is a highly potent inhibitor, and the binding affinity is − 17.75 kcal/mol. Overall, our estimations indicate that human metabolites could be more potent than α-ketoamide (13b), as we get better affinity (− 18.08 kcal/mol) against 3CLpro due to its low value of configuration entropy (28.43 kcal/mol).
Critical residues involved in human metabolites binding with 3CLpro
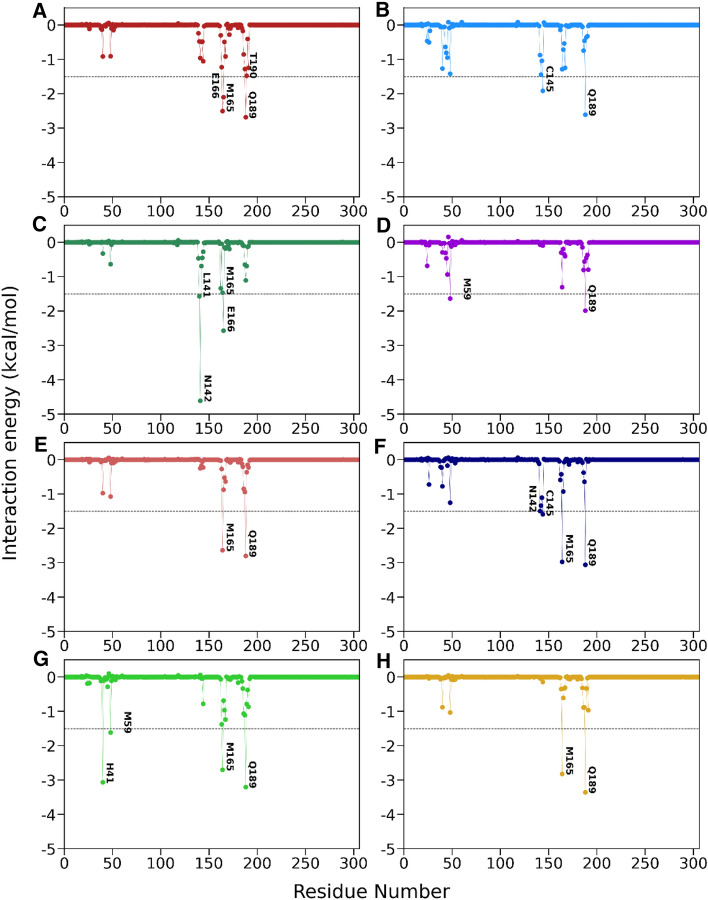
To gain insights into the contribution of each amino acid to the binding, we decomposed the total binding free energy at the residual level using the molecular mechanics generalized Born surface area (MM-GBSA) approach. The protein–ligand interactions spectra are shown in Fig. 5. The binding hotspot residues whose contributions are more than − 1.5 kcal/mol are listed in Supporting Information Table S3. It is clear from Fig. 5 that the favorable binding residues are Met165, His41, Cys145, Glu166, Gln189, Asn142, Leu141, and Met49, and the interaction spectra for all are the same. This agrees with other computational studies [65, 66]. Our RMSF analysis also shows that these binding site residues display low fluctuations due to strong interactions with human metabolites (see Fig. 2C, D). Figure 5 and some recent studies suggest domains I and II are involved favorably in human metabolites like small molecules, and catalytic dyads His41 and Cys145 play an essential role. Moreover, for all cases, the assistance from the backbone and side-chain atoms are also favorable toward the binding, which is a good thing for an inhibitor’s stronger affinity.
Fig. 5.
Per-residue decomposition of the binding free energy for A complex1, B complex2, C complex3, D complex4, E complex5, F complex6, G complex7 and H complex8, respectively. The energy contribution ≥ − 1.5 kcal/mol is shown in each plot
As Table S3 suggested that in the case of complex1, Gln189 (− 2.68 kcal/mol), Met165 (− 2.51 kcal/mol), Glu166 (− 2.10 kcal/mol), and Thr190 (− 1.51 kcal/mol) are the hotspot residues. The van der Waals interactions with these residues have essential contributions to the overall binding affinity of the metabolite. This result implies that the human metabolite in complex1 depicted a tendency to develop hydrophobic interactions with most of these binding site residues. This interaction is thought to reduce binding site residues’ flexibility, as seen in the RMSF analysis (Fig. 2C, D). Similarly, residue Gln189 in complex4 (− 1.99 kcal/mol), complex5 (− 2.80 kcal/mol), complex6 (− 3.06 kcal/mol), complex7 (− 3.20 kcal/mol), and complex8 (− 3.35 kcal/mol) contributed more favorably than the catalytic dyads.
Hydrogen bonds and hydrophobic interactions stability analysis
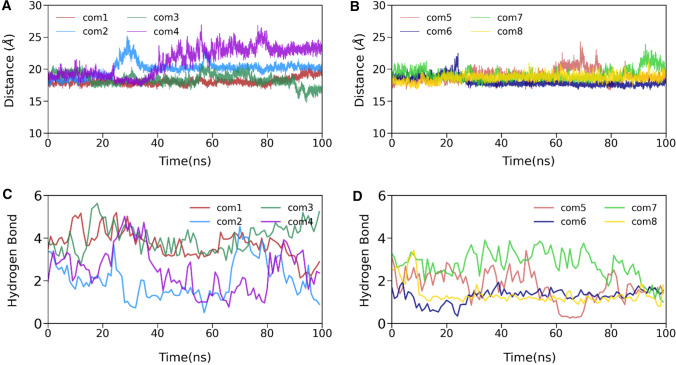
To complement the energetic analysis and understand the complex stability, we determined the time evolution of the center of the mass distance between the 3CLpro and eight human metabolites into their respective complex, shown in Fig. 6A, B. As suggested in Fig. 6A, B, the CoM distance of these complexes is at a stable equilibrium. Although we see some deflection in complex4 up to 80 ns, after that, it fluctuated around 23 Å.
Fig. 6.
The time evolution of the center of the mass distance (A, B) and the number of hydrogen bonds between 3CLpro and human metabolites (C, D). In figure legend, the complex is termed as com in all cases
Next, we estimated the temporal evolution of the number of hydrogen bonds between 3CLpro and metabolites throughout the simulation (see Fig. 6C, D). Detailed profiling of the prominent hydrogen bonds is listed in Supporting Information Table S4. It is evident from Fig. 6C, D that complex3 shows the highest number of hydrogen bonds compared to the rest of the complexes. This high number of hydrogen bonds increases the electrostatic interactions between 3CLpro and human metabolites. However, in complex3, this electrostatic interaction is compensated by polar solvation energy, which is the highest among all complexes. Thus, the number of hydrogen bonds alone is insufficient for obtaining high affinity, as also shown in an earlier study [67]. However, ligand1 has very stable hydrogen between Gln192 and the oxygen (O3) atom (80.39%), which is missing in the case of complex3. The complex2, complex4, complex6, and complex8 have less than two hydrogen bonds in the 100 ns simulation. These results suggest that non-polar solvation and hydrophobic van der Waals interactions are the primary binding force.
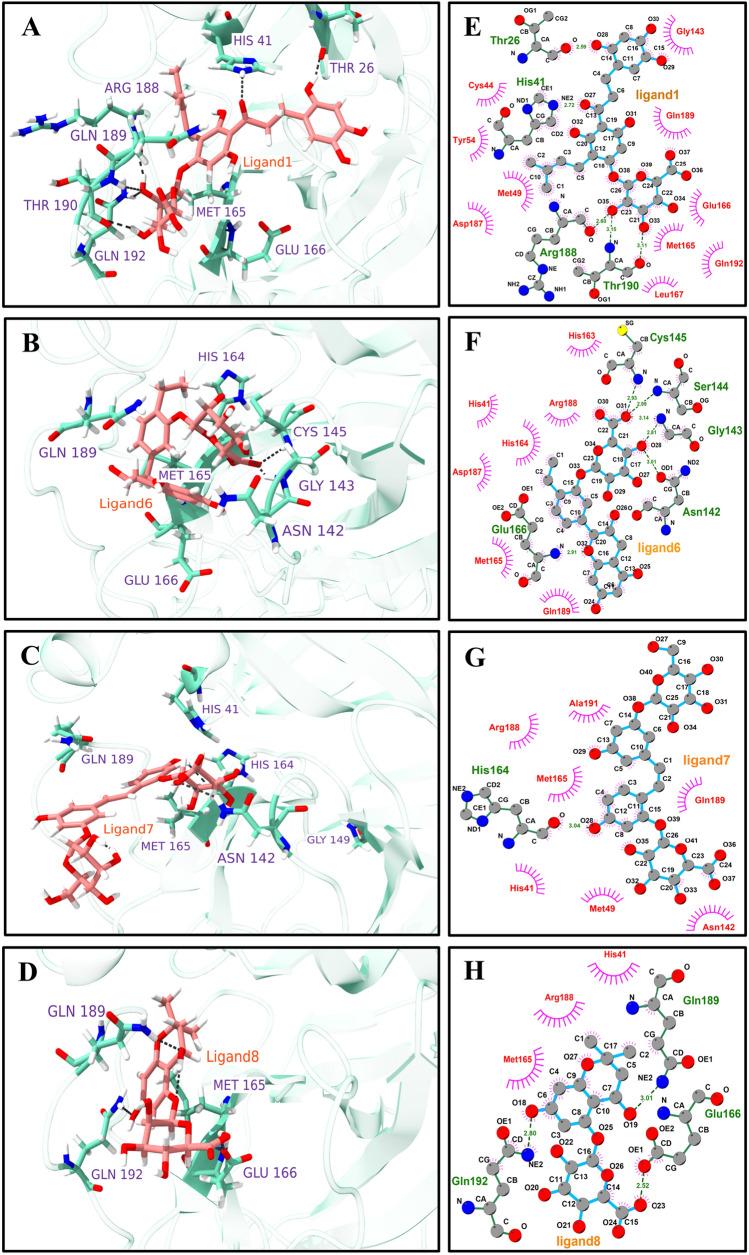
Furthermore, we supplemented the above findings for the top four molecules by exploring various hydrogen bonding and hydrophobic interaction profile between the main protease, 3CLpro, and human metabolites for the final production simulation structure, as shown in Fig. 7. The interaction profiles for complex2, complex3, complex4, and complex5 are shown in Supplementary Information, Fig. S6. The 3D conformation of ligands in the binding site shows the possible orientation of the different ring structures, which leads to the possible set hydrogen bonding pattern (Fig. 7A–D). The detailed interaction profiles were estimated using Ligplot+ (Fig. 7E–H) [68]. For the complex1, Fig. 7E displayed ten hydrophobic interactions with Cys44, Gly143, Tyr54, Gln189, Met49, Asp187, Glu166, Met165, Glm192, and Leu167. These extensive hydrophobic interactions account for the high affinity and stability of human metabolites in complex1. Similarly, residues, including Thr26, His41, Arg188, and Thr190, form strong hydrogen bonding with metabolites. In complex6, seven hydrophobic interactions with residues His163, Arg188, His41, His164, Asp187, Met165, and Gln189 were found, as revealed by Fig. 7F. Figure 7G, H shows that Ala191, Arg188, Met165, Gln189, His41, Met49, and Asn142 are strongly involved in hydrophobic interactions. His164, Gln189, Gln166, and Gln192 participate in the hydrogen bonding interactions for complex7 and complex8. Due to extensive hydrophobic interactions and strong hydrogen bonding, complex1 may be more potent against SARS-CoV-2 3CLpro.
Fig. 7.
The main protease, 3CLpro, and human metabolites interaction profiles (affinity more than − 9 kcal/mol). 3D interaction profile for selected ligands where key residues are shown ball and stick model A Ligand1, B Ligand6, C Ligand7, and D Ligand8. 2D interaction plots are generated by using Ligplot+, E Ligand1, F Ligand6, G Ligand7, and H Ligand8. Pink spike semicircles show the residues involved in the hydrophobic interactions, and hydrogen bonds are shown as green dotted lines
Prediction of pharmacological and toxicological properties
Different physicochemical properties like CNS, Molecular weight, acceptor and donor hydrogen bond cut-off, octanol/water partition coefficient, brain–blood barrier coefficient, and others were calculated and listed in Supporting Information (Table S5). Ligand8 is the least violated Lipinski’s rule among all other top leads. The best lead ligand obtained from the MD simulation studies shows poor druggability as it has the highest number of rule violations. However, recently, it has been found that many drugs do not follow Lipinski’s rule and are presently found on the FDA-approved drug list [69]. Instead of being employed as a therapeutic candidate, these compounds will be used as lead molecules.
Ligand6 and Ligand8 have less than 500 Dalton molecular weights, indicating a promising lead molecule for drug design. Only in the case of human oral absorption, all four ligands were out of the acceptable ranges. Compared with all the parameters, Ligand8 stands out to be the best choice for a lead molecule. Toxicity profiling is one of the critical parameters for successful drug development, which was done in our lead molecules and shown in Supporting Information, Table S6. Ligand1 and Ligand7 were estimated as toxicity group 4, whereas Ligand6 and Ligand8 were estimated as group 5 on a scale of 1 to 5 (higher the number, lower the toxicity). All four lead ligands were evaluated as non-hepatotoxic, non-carcinogen, non-mutagenic, and non-cytotoxic. In comparison to Ligand1, which has the highest binding free energy of all the molecules, Ligand8 exhibits several interesting features.
Conclusion
This study concludes that the natural compounds and their metabolites can play a promising role in managing the SARS-COV-2 infection. The current study is a reference model in which we recommend adjusting the body’s metabolites or eating enough food to develop appropriate metabolites to combat viral infections. We screened the human metabolite database and selected the top 8 lead molecules for further validation. The chosen molecules were further screened using 3 × 100 ns long conventional molecular dynamics simulations and the free energy calculation using the MM/GBSA scheme. Based on our computational study, we identified top four lead compounds to become possible drug candidates. We found that one of the metabolites of isobavachalcone (Ligand1: HMDB0132640) binds very well (− 18.08 kcal/mol) to 3CLpro of SARS-COV-2. Although it does not follow all the druggability rules, it is worth trying a molecule that can bind very well to 3CLpro and may serve as the best lead for this target. On the other hand, Ligand8 is another suitable candidate based on suitable ADMET properties. Therefore, despite some deviations from drug-likeness, four of the eight hits can be taken further as they have ideal interactions with 3CLpro, excellent binding free energy, and good pharmacokinetic properties. Overall, these metabolites have a good chance of being developed as possible COVID-19 protease inhibitors.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was partially supported by the Department of Science and Technology, Govt. of India (grant number DST/NSM/R&D_HPC_Applications/2021/03.18). RR thanks the Indian Institute of Technology Indore for financial assistance. MFS would like to thank the Department of Science and Technology, Govt. of India, for providing a doctoral fellowship under the INSPIRE fellowship scheme (DST/INSPIRE Fellowship/2017/IF170145).
Data availability
The data supporting this study’s findings are available from the corresponding author (PK) upon reasonable request.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Research involving human and animal rights
There is no human or animal experiment in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rajarshi Roy and Md Fulbabu Sk have contributed equally.
References
- 1.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui DS, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madboly WE, Shehata MG, Nashed MSM, Abu-Dief AM. Using safe calculated low power of electrons to cut, analyze and exterminate the outer and inner biological elements of SARS-CoV-2, MERS-CoV-2 and influenza viruses in vitro. J Sci Res Rep. 2022;28(1):1–15. doi: 10.9734/jsrr/2022/v28i130482. [DOI] [Google Scholar]
- 6.Woo PCY, Huang Y, Lau SKP, Yuen K-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Z, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 8.Choudhary MI, Shaikh M, tul-Wahab A, ur-Rahman A. In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS ONE. 2020;15(7):e0235030. doi: 10.1371/journal.pone.0235030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alamri MA, et al. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro. J Biomol Struct Dyn. 2021;39(13):4936–4948. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan RJ, et al. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn. 2021;39(8):2679–2692. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sk MF, Roy R, Jonniya NA, Poddar S, Kar P. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J Biomol Struct Dyn. 2021;39(10):3649–3661. doi: 10.1080/07391102.2020.1768149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Wei Z, Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J Biol Chem. 2004;279(23):24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bzówka M, Mitusińska K, Raczyńska A, Samol A, Tuszyński JA, Góra A. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int J Mol Med. 2020;21(9):3099. doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thurakkal L, Singh S, Roy R, Kar P, Sadhukhan S, Porel M. An in-silico study on selected organosulfur compounds as potential drugs for SARS-CoV-2 infection via binding multiple drug targets. Chem Phys Lett. 2021;763:138193. doi: 10.1016/j.cplett.2020.138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy R, Sk MF, Jonniya NA, Poddar S, Kar P. Finding potent inhibitors against SARS-CoV-2 main protease through virtual screening, ADMET, and molecular dynamics simulation studies. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1897680. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Dief AM, El-Sagher HM, Shehata MR. Fabrication, spectroscopic characterization, calf thymus DNA binding investigation, antioxidant and anticancer activities of some antibiotic azomethine Cu(II), Pd(II), Zn(II) and Cr(III) complexes. Appl Organomet Chem. 2019;33(8):e4943. doi: 10.1002/aoc.4943. [DOI] [Google Scholar]
- 17.Singh S, Sk MF, Sonawane A, Kar P, Sadhukhan S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: an in-silico analysis. J Biomol Struct Dyn. 2021;16:6249–6264. doi: 10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors—an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2021;39(12):4362–4374. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahedi HM, Ahmad S, Abbasi SW. Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn. 2021;39(9):3225–3234. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- 20.Decuypere S, et al. Metabolomics based biomarker discovery for infectious diseases, the case of melioidosis. Int J Infect Dis. 2012;16:e216. doi: 10.1016/j.ijid.2012.05.810. [DOI] [Google Scholar]
- 21.Kumar R, Ghosh M, Kumar S, Prasad M. Single cell metabolomics: a future tool to unmask cellular heterogeneity and virus-host interaction in context of emerging viral diseases. Front Microbiol. 2020;11:1152. doi: 10.3389/fmicb.2020.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1(1):e000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardiner P, et al. Spironolactone metabolism: steady-state serum levels of the sulfur-containing metabolites. J Clin Pharmacol. 1989;29(4):342–347. doi: 10.1002/j.1552-4604.1989.tb03339.x. [DOI] [PubMed] [Google Scholar]
- 26.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 27.Harder E, et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 28.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 29.Friesner RA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 30.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 31.Case DA, et al. AMBER 2018. San Francisco: University of California; 2018. [Google Scholar]
- 32.Price DJ, Brooks CL. A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121(20):10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 33.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 35.Darden T, York D, Pedersen L. Particle mesh Ewald: an N ⋅log( N ) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 36.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22(5):501–508. doi: 10.1002/1096-987X(20010415)22:5<501::AID-JCC1021>3.0.CO;2-V. [DOI] [Google Scholar]
- 37.Sk MF, Jonniya NA, Roy R, Poddar S, Kar P. Computational investigation of structural dynamics of SARS-CoV-2 methyltransferase-stimulatory factor heterodimer nsp16/nsp10 bound to the cofactor SAM. Front Mol Biosci. 2020;7:590165. doi: 10.3389/fmolb.2020.590165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des. 2000;18(1):113–135. doi: 10.1023/A:1008763014207. [DOI] [Google Scholar]
- 39.Kar P, Seel M, Hansmann UHE, Höfinger S. Dispersion terms and analysis of size- and charge dependence in an enhanced Poisson−Boltzmann approach. J Phys Chem B. 2007;111(30):8910–8918. doi: 10.1021/jp072302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C, Greene D, Xiao L, Qi R, Luo R. Recent developments and applications of the MMPBSA method. Front Mol Biosci. 2018;4:87. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang E, et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. 2019;119(16):9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 43.Sk MF, Haridev S, Roy R, Kar P. Investigating potency of TMC-126 against wild-type and mutant variants of HIV-1 protease: a molecular dynamics and free energy study. SAR QSAR Environ Res. 2021;32(11):941–962. doi: 10.1080/1062936X.2021.1999318. [DOI] [PubMed] [Google Scholar]
- 44.Chang S, Zhang D-W, Xu L, Wan H, Hou T-J, Kong R. Exploring the molecular basis of RNA recognition by the dimeric RNA-binding protein via molecular simulation methods. RNA Biol. 2016;13(11):1133–1143. doi: 10.1080/15476286.2016.1223007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y, Sun L, Jia Z, Li L, Alexov E. Predicting protein-DNA binding free energy change upon missense mutations using modified MM/PBSA approach: SAMPDI webserver. Bioinformatics. 2018;34(5):779–786. doi: 10.1093/bioinformatics/btx698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy R, Mishra A, Poddar S, Nayak D, Kar P. Investigating the mechanism of recognition and structural dynamics of nucleoprotein-RNA complex from Peste des petits ruminants virus via Gaussian accelerated molecular dynamics simulations. J Biomol Struct Dyn. 2022;40(5):2302–2315. doi: 10.1080/07391102.2020.1838327. [DOI] [PubMed] [Google Scholar]
- 47.Roy R, Ghosh B, Kar P. Investigating conformational dynamics of Lewis Y oligosaccharides and elucidating blood group dependency of cholera using molecular dynamics. ACS Omega. 2020;5(8):3932–3942. doi: 10.1021/acsomega.9b03398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy R, Jonniya NA, Sk MF, Kar P. Comparative structural dynamics of isoforms of helicobacter pylori adhesin BabA bound to Lewis b hexasaccharide via multiple replica molecular dynamics simulations. Front Mol Biosci. 2022;9:852895. doi: 10.3389/fmolb.2022.852895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kollman PA, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 50.Abu-Dief AM, et al. Synthesis, structural elucidation, DFT calculation, biological studies and DNA interaction of some aryl hydrazone Cr3+, Fe3+, and Cu2+ chelates. Comput Biol Chem. 2022;97:107643. doi: 10.1016/j.compbiolchem.2022.107643. [DOI] [PubMed] [Google Scholar]
- 51.Abu-Dief AM, et al. Synthesis and intensive characterization for novel Zn(II), Pd(II), Cr(III) and VO(II)-Schiff base complexes; DNA-interaction, DFT, drug-likeness and molecular docking studies. J Mol Struct. 2021;1242:130693. doi: 10.1016/j.molstruc.2021.130693. [DOI] [Google Scholar]
- 52.Abu-Dief AM, Abdel-Rahman LH, Abdel-Mawgoud AAH. A robust in vitro anticancer, antioxidant and antimicrobial agents based on new metal-azomethine chelates incorporating Ag(I), Pd (II) and VO (II) cations: probing the aspects of DNA interaction. Appl Organomet Chem. 2020;34(2):e5373. doi: 10.1002/aoc.5373. [DOI] [Google Scholar]
- 53.AbdelRahman LH, Abu-Dief AM, El-Khatib RM, Abdel-Fatah SM, Adam AM, Ibrahim EMM. Sonochemical synthesis, structural inspection and semiconductor behavior of three new nano sized Cu(II), Co(II) and Ni(II) chelates based on tri-dentate NOO imine ligand as precursors for metal oxides. Appl Organomet Chem. 2018;32(3):e4174. doi: 10.1002/aoc.4174. [DOI] [Google Scholar]
- 54.Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46(W1):W257–W263. doi: 10.1093/nar/gky318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solnier J, Fladerer J-P. Flavonoids: a complementary approach to conventional therapy of COVID-19? Phytochem Rev. 2020;20:773–795. doi: 10.1007/s11101-020-09720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim DW, et al. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzym Inhib Med Chem. 2014;29(1):59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 58.Jo S, Kim H, Kim S, Shin DH, Kim M-S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem Biol Drug Des. 2019;94(6):2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y-Q, Li Z-L, Zhao W-J, Wen R-X, Meng Q-W, Zeng Y. Synthesis of stilbene derivatives with inhibition of SARS coronavirus replication. Eur J Med Chem. 2006;41(9):1084–1089. doi: 10.1016/j.ejmech.2006.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sk MF, Roy R, Kar P. Exploring the potency of currently used drugs against HIV-1 protease of subtype D variant by using multiscale simulations. J Biomol Struct Dyn. 2021;39(3):988–1003. doi: 10.1080/07391102.2020.1724196. [DOI] [PubMed] [Google Scholar]
- 61.Nand M, et al. Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci Rep. 2020;10(1):20397. doi: 10.1038/s41598-020-77524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, et al. Discovery of new hydroxyethylamine analogs against 3CL pro protein target of SARS-CoV-2: molecular docking, molecular dynamics simulation, and structure-activity relationship studies. J Chem Inf Model. 2020;60(12):5754–5770. doi: 10.1021/acs.jcim.0c00326. [DOI] [PubMed] [Google Scholar]
- 63.Gupta S, et al. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. 2021;39(12):4334–4345. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han Y, Wang Z, Ren J, Wei Z, Li J. Potential inhibitors for the novel coronavirus (SARS-CoV-2) Briefings Bioinform. 2021;22(2):1225–1231. doi: 10.1093/bib/bbaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiorucci D, Milletti E, Orofino F, Brizzi A, Mugnaini C, Corelli F. Computational drug repurposing for the identification of SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020;39(16):6242–6248. doi: 10.1080/07391102.2020.1796805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pant S, Singh M, Ravichandiran V, Murty USN, Srivastava HK. Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dyn. 2021;39(8):2904–2913. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kar P, Lipowsky R, Knecht V. Importance of polar solvation and configurational entropy for design of antiretroviral drugs targeting HIV-1 protease. J Phys Chem B. 2013;117(19):5793–5805. doi: 10.1021/jp3085292. [DOI] [PubMed] [Google Scholar]
- 68.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 69.Mullard A. Re-assessing the rule of 5, two decades on. Nat Rev Drug Discov. 2018;17(11):777–777. doi: 10.1038/nrd.2018.197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this study’s findings are available from the corresponding author (PK) upon reasonable request.