Abstract
Background
Altered tumor suppressor genes (TSG-alt) in prostate cancer are associated with worse outcomes. The prognostic value of TSG-alt in metastatic, hormone-sensitive prostate cancer (M1-HSPC) is unknown. We evaluated the effects of TSG-alt on outcomes in M1-HSPC and their prognostic impact by first-line treatment.
Methods
We retrospectively identified patients with M1-HSPC at our institution treated with first-line androgen deprivation therapy plus docetaxel (ADT + D) or abiraterone acetate (ADT + A). TSG-alt was defined as any alteration in one or more TSG. The main outcomes were Kaplan–Meier-estimated progression-free survival (PFS) and overall survival, analyzed with the log-rank test. Clinical characteristics were compared with the χ2 test and Kruskal–Wallis rank sum test. Cox regression was used for univariate and multivariable analyses.
Results
We identified 97 patients with M1-HSPC: 48 (49%) with ADT + A and 49 (51%) with ADT + D. Of 96 patients with data available, 33 (34%) had 1 TSG-alt, 16 (17%) had 2 TSG-alt, and 2 (2%) had 3 TSG-alt. The most common alterations were in TP53 (36%) and PTEN (31%); 6% had RB1 alterations. Median PFS was 13.1 (95% CI, 10.3–26.0) months for patients with normal TSGs (TSG-normal) vs. 7.8 (95% CI, 5.8–10.5) months for TSG-alt (P = 0.005). Median PFS was lower for patients with TSG-alt vs TSG-normal for those with ADT + A (TSG-alt: 8.0 [95% CI, 5.8–13.8] months vs. TSG-normal: 23.2 [95% CI, 13.1–not estimated] months), but not with ADT + D (TSG-alt: 7.8 [95% CI, 5.7–12.9] months vs. TSG-normal: 9.5 [95% CI, 4.8–24.7] months). On multivariable analysis, only TSG-alt predicted worse PFS (hazard ratio, 2.37; 95% CI, 1.42–3.96; P < 0.001).
Conclusions
The presence of TSG-alt outperforms clinical criteria for predicting early progression during first-line treatment of M1-HSPC. ADT + A was less effective in patients with than without TSG-alt. Confirmation of these findings may establish the need for inclusion of molecular stratification in treatment algorithms.
Subject terms: Cancer genetics, Cancer therapy
Introduction
With the advent of comprehensive genomic profiling, many studies have shown the effects of tumor suppressor genes (TSGs) in advanced metastatic prostate cancer [1, 2]. Preclinical and clinical data indicate that cooperative functional losses of TSGs such as TP53, PTEN, and RB1 are associated with worse clinical outcomes in castration-resistant prostate cancer [2–7].
Treatment decisions for newly diagnosed metastatic, hormone-sensitive prostate cancer (M1-HSPC) are usually made on the basis of clinical phenotypes, toxicity, convenience, and cost [8, 9]. Since 2015, 4 agents (docetaxel, abiraterone acetate, enzalutamide, and apalutamide) have shown improved survival for patients with M1-HSPC [10–15]. In the case of docetaxel, the CHAARTED trial (Androgen Ablation Therapy With or Without Chemotherapy in Treating Patients With Metastatic Prostate Cancer) suggested that the benefit is limited to patients with high-volume disease, and all studies showed inferior outcomes in patients with de novo metastatic disease [10, 16, 17]. However, the choice of first-line treatment is still unclear, as is any possible role for introducing molecular characteristics into management decisions.
TSG alterations (TSG-alt) are known to influence the efficacy of cytotoxic chemotherapy and androgen deprivation therapy (ADT). The genomic underpinnings of clinical phenotypes in M1-HSPC are increasingly well defined, and some TSG-alt are known to affect the aggressiveness of prostate cancer. For example, TP53 loss is known to confer a poor response to ADT [1], PTEN loss is a prognostic marker for decreased response to hormonal therapy without affecting docetaxel efficacy [18, 19], and RB1 loss is usually associated with worse outcomes with ADT [6, 20].
In the current study, we aimed to evaluate the prognostic value of TSG-alt of TP53, PTEN, and RB1 in patients with M1-HSPC receiving first-line treatment with abiraterone acetate or docetaxel. Our findings suggest that TSG status could be used in the development of novel treatment algorithms.
Methods
This study was approved by the Mayo Clinic Institutional Review Board. For this retrospective analysis, we searched for the records of patients treated with first-line ADT plus abiraterone acetate (ADT + A) or ADT plus docetaxel (ADT + D) for M1-HSPC between November 1, 2015, and December 23, 2018, at Mayo Clinic in Arizona. We identified consecutive patients with a diagnosis of castration-sensitive prostate cancer who also had baseline genomic testing or tissue available for retrospective analysis. Clinical characteristics, first-line treatment, and pathogenic TSG-alt were recorded.
Our study cohort included patients with either de novo metastatic disease or metastatic relapse after prior local therapy. Only patient outcomes for first-line treatment for the M1-HSPC setting were included. All patients had archival formalin-fixed, paraffin-embedded tumor tissue from a prostate biopsy and/or prostatectomy or other diagnostic biopsies obtained before the initiation of treatment. Sequencing of tumor tissue was performed using Clinical Laboratory Improvement Amendments of 1988–approved commercially available platforms—Tempus (58% of samples), Foundation Medicine (Roche; 37% of samples), and Caris Life Sciences (5% of samples)—for genes including TP53, PTEN, and RB1. TSG-alt was defined as any pathogenic alteration in one or more TSG (TP53, PTEN, or RB1). The definition of TSG-alt was determined according to the testing platform, and allelic status was not examined (Supplementary Table 1). Progression-free survival (PFS) was defined as the time from the start of therapy for M1-HSPC to identification of clinical or radiographic progression defined by the Prostate Cancer Working Group criteria [21]. Radiographic progression, not prostate-specific antigen (PSA) progression, was used to guide therapy, following institutional practice patterns. Overall survival (OS) was defined as the time from start of therapy until death of any cause. Patients who did not have progression of disease or death were considered censored at the date of last follow-up.
PFS and OS were compared between patients with TSG-alt and without gene alterations, TSG-normal. Descriptive statistics were used to compare baseline characteristics between groups according to TSG status and treatment regimen: the χ2 test for categorical data, and the Kruskal–Wallis rank sum test for continuous measures. PFS and OS were estimated with the Kaplan–Meier method, stratified by TSG-alt status and treatment, with censoring at the next therapy or last follow-up. The log-rank test was used to assess differences between groups. Cox regression was used for multivariable analysis, using clinical variables associated with worse prognosis such as high disease volume and metastatic disease at diagnosis. R statistical software version 3.6.2 was used for analysis. P < 0.05 were considered statistically significant.
Results
We identified 97 patients with M1-HSPC who met all inclusion criteria during the study period. Patient baseline characteristics are shown in Table 1. The median age was 70 years (range, 41–89 years). A total of 38 patients (39%) had received prior treatment with either prostatectomy or ADT in combination with localized radiation. The analyzed tissue samples were from radical prostatectomies (n = 11), prostate biopsies (n = 39), transurethral resection of the prostate specimens (n = 9), and biopsy of a local or distant metastatic site (n = 38). Most patients (66%) had de novo metastatic disease at presentation, and 63 (65%) met high-volume disease criteria. The treatment regimen was ADT + D for 49 patients (51%) and ADT + A for 48 patients (49%).
Table 1.
Patient characteristics by treatment groupa.
| Characteristic | All patients (N = 97) | Treatment | P value | |
|---|---|---|---|---|
| Abiraterone acetate (n = 48) | Docetaxel (n = 49) | |||
| Prior treatment | 0.03b | |||
| None | 59 (61) | 23 (48) | 36 (73) | |
| Surgery | 28 (29) | 19 (40) | 9 (18) | |
| RT | 10 (10) | 6 (13) | 4 (8) | |
| Mets at diagnosis | 64 (66) | 27 (56) | 37 (76) | 0.045b |
| TSG alterations | ||||
| TP53 |
35 (36) (n = 96) |
17 (35) |
18 (38) (n = 48) |
0.83b |
| PTEN | 30 (31) | 17 (35) | 13 (27) | 0.34b |
| BRCA | 16 (16) | 11 (23) | 5 (10) | 0.09b |
| APC | 10 (10) | 3 (6) | 7 (14) | 0.19b |
| AR | 11 (11) | 9 (19) | 2 (4) | 0.02b |
| RB1 | 6 (6) | 3 (6) | 3 (6) | 0.98b |
| Total TSG alterations | (n = 96) | (n = 48) | 0.93b | |
| None | 45 (47) | 21 (44) | 24 (50) | |
| 1 | 33 (34) | 18 (38) | 15 (31) | |
| 2 | 16 (17) | 8 (17) | 8 (17) | |
| 3 | 2 (2) | 1 (2) | 1 (2) | |
| MDB | 0.009b | |||
| Low | 34 (35) | 23 (48) | 11 (22) | |
| High | 63 (65) | 25 (52) | 38 (78) | |
| Visceral progression | 29 (30) | 15 (31) | 14 (29) | 0.77b |
| Bone progression |
61 (64) (n = 96) |
29 (62) (n = 47) |
32 (65) | 0.71b |
| Laboratory values | ||||
| Alkaline phosphatase, IU/L |
103.0 (13.0–1380.0) (n = 96) |
99.0 (66.0–260.0) |
108.5 (13.0–1380.0) (n = 48) |
0.050c |
| LDH, U/L |
191.0 (130.0–407.0) (n = 80) |
194.0 (131.0–247.0) (n = 47) |
186.0 (130.0–407.0) (n = 33) |
0.81c |
| Hemoglobin, g/dL |
12.5 (7.4–16.1) (n = 95) |
12.2 (9.0–15.0) (n = 48) |
13.0 (7.4–16.1) (n = 47) |
0.52c |
LDH lactate dehydrogenase, MDB metastatic disease burden, Mets metastases, RT radiotherapy, TSG tumor suppressor gene.
aValues are No. of patients (%) or median (range).
bPearson χ2 test.
cKruskal–Wallis rank sum test.
TSG-alt was present in 51 of 96 patients with data available (53%): TP53 in 36%, PTEN in 31%, and RB1 in 6% (Table 1). Thirty-three patients (34%) had only 1 TSG-alt, whereas 18 (19%) had more than one TSG-alt: TP53 + PTEN in 14 patients, PTEN + RB1 in two patients, and all three in two patients. Clinical characteristics, prior locoregional treatment, disease volume, baseline laboratory values, and metastatic disease at diagnosis were similar between TSG-alt and TSG-normal groups (Supplementary Table 2).
Median follow-up duration was 23.2 months (range, 1.1–65.2 months). Comparison of patients with and without TSG-alt showed that median PFS was lower for patients with TSG-alt (7.8 months [95% CI, 5.8–10.5 months]) than for TSG-normal patients (13.1 months [95% CI, 10.3–26.0 months]; P = 0.005) (Fig. 1). Similarly, median OS differed between the TSG-alt (27.6 months [95% CI, 19.0 months–not estimated]) and TSG-normal group (53.3 months [95% CI, 38.8 months–not estimated]; P = 0.03). Patients without TSG-alt had better PFS than those with any TSG (Fig. 2). There was no difference in median PFS, however, between those having only 1 TSG-alt (7.3 months [95% CI, 5.8–10.8 months]) vs. >1 TSG-alt (8.6 months [95% CI, 4.2–17.6 months]).
Fig. 1. Progression-Free Survival (PFS) by Presence of Tumor Suppressor Gene Alteration (TSG-alt).
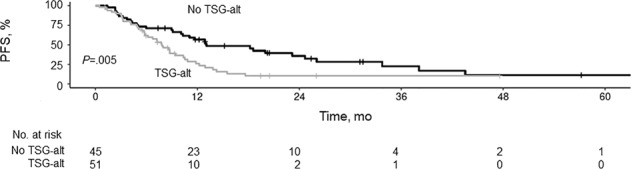
Kaplan–Meier curves showing PFS for patients with and without TSG-alt.
Fig. 2. Progression-Free Survival (PFS) by Number of Tumor Suppressor Gene Alterations (TSG-alt).
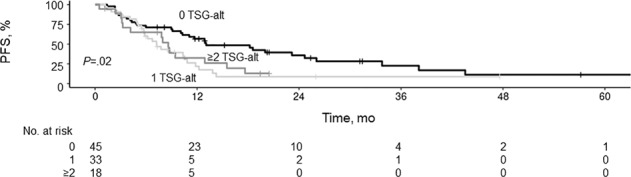
Kaplan–Meier curves showing PFS for patients with 0, 1, or ≥2 TSG-alt.
There was no substantial difference in median PFS in the ADT + A group (11.8 months [95% CI, 9.4–18.2 months]) vs. the ADT + D group (8.6 months [95% CI, 5.7–12.9 months]) for the total cohort. Within treatment groups, however, median PFS was lower for patients with TSG-alt vs. TSG-normal for those with ADT + A (TSG-alt: 8.0 [95% CI, 5.8–13.8] months vs. TSG-normal: 23.2 [95% CI, 13.1–not estimated] months), but not with ADT + D (TSG-alt: 7.8 [95% CI, 5.7–12.9] months vs. TSG-normal: 9.5 [95% CI, 4.8–24.7] months) (Fig. 3A). Similar results were seen for OS, with ADT + A with TSG-normal being superior to the other groups (Fig. 3B).
Fig. 3. Progression-Free Survival (PFS) and Overall Survival (OS) by First-Line Treatment and Tumor Suppressor Gene Alteration (TSG-alt) Status.
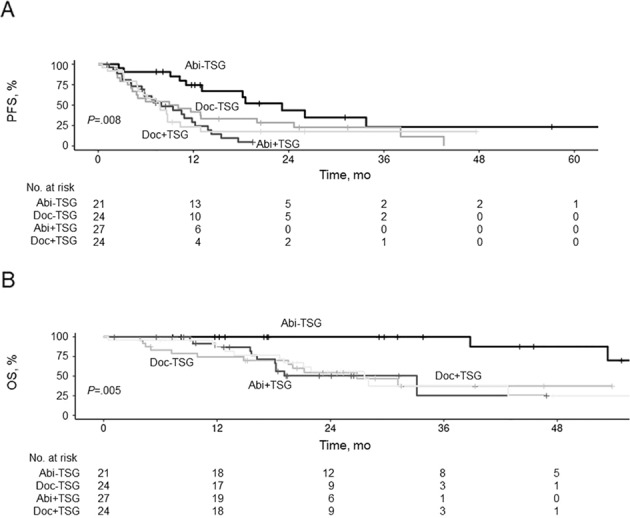
Kaplan–Meier curves showing PFS (A) and OS (B) for patients with (+TSG) and without (−TSG) TSG-alt treated with abiraterone acetate (Abi) or docetaxel (Doc).
On multivariable analysis, TSG-alt and metastatic disease at diagnosis were the only characteristics associated with PFS (Table 2). TSG-alt predicted worse prognosis (hazard ratio, 2.37; 95% CI, 1.42−3.96; P < 0.001), whereas metastatic disease predicted improved PFS (hazard ratio, 0.34; 95% CI, 0.15–0.76; P = 0.009). The association of TSG-alt with OS did not reach statistical significance on multivariable analysis (hazard ratio, 1.94; 95% CI, 0.98–3.86; P = 0.06) (Table 2).
Table 2.
Multivariable cox regression, PFS and OS.
| Characteristic | PFS | OS | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| TSG-alt (vs TSG-normal) | 2.37 (1.42–3.96) | <0.001 | 1.94 (0.98–3.86) | 0.06 |
| Treatment with ADT + D (vs ADT + A) | 1.36 (0.81–2.28) | 0.20 | 1.80 (0.84–3.83) | 0.13 |
| Prior treatment (vs none) | ||||
| Surgery | 1.16 (0.48–2.77) | 0.70 | 0.90 (0.28–2.89) | 0.90 |
| RT | 0.40 (0.13–1.22) | 0.11 | 0.76 (0.15–3.91) | 0.70 |
| High disease volume (vs low) | 1.82 (0.99–3.33) | 0.05 | 0.98 (0.41–2.32) | >0.90 |
| Mets at diagnosis (vs none) | 0.34 (0.15–0.76) | 0.009 | 1.18 (0.41–3.45) | 0.80 |
ADT + A androgen deprivation therapy plus abiraterone acetate, ADT + D androgen deprivation therapy plus docetaxel, HR hazard ratio, Mets metastases, OS overall survival, PFS progression-free survival, RT radiotherapy, TSG-alt tumor suppressor gene alteration.
Discussion
In the current study, we show that the presence of loss-of-function alterations of TP53, PTEN, and RB1 outperforms clinical factors in predicting poor outcomes in M1-HSPC. Using commercially available sequencing assays, we confirmed that the presence of TSG-alt predicts early progression during first-line therapy regardless of treatment selection and has the potential to aid treatment decisions. The presence of TSG-alt also was associated with decreased benefit from abiraterone acetate treatment.
Our findings demonstrated a lower prevalence of TP53, PTEN, and RB1 alterations in M1-HSPC than is seen in metastatic, castration-resistant prostate cancer; a previous report in this cohort showed an alteration frequency of 50% for TP53, 17% for PTEN, and 10% for RB1 [22]. This is consistent with the evolutionary model of treatment pressure in selecting for resistant clones as disease advances. This is also consistent with aggressive phenotypes of androgen receptor–independent anaplastic and neuroendocrine tumors, characterized by loss of TP53, PTEN, and RB1 drivers [23, 24]. Thus, it is not surprising that the early presence of functional TSG losses is prognostic for early progression during first-line treatment with ADT + A or ADT + D.
In our cohort, 18 patients (19%) had evidence of two or more TSG-alt. This rate of TSG coalteration is comparable to the 16% incidence in M1-HSPC, and higher than the 11% incidence in localized hormone-sensitive prostate cancer in the cohorts reported by Hamid et al. [1]. Whether TSG coalteration would confer added prognostic significance or whether loss of a single TSG is enough to confer the full risk of TSG loss will need to be evaluated in larger data sets. Our findings confirm those reported by Stopsack et al. [25] that some genomic alterations were associated with worse prognosis (AR, MYC, and TP53) and some had better outcomes (SPOP and WNT). Our study suggests that M1-HSPC with TSG-normal is best treated with ADT + A rather than ADT + D. In the TSG-alt population, outcomes with ADT-D and ADT-A appear similarly poor, which suggests the need for better treatment options for this group.
Genetic characterization in the first-line treatment setting is feasible because all or nearly all patients should have tissue confirmation of their diagnosis. This was recently demonstrated by Gilson and colleagues [26] in their report on genetic analysis in the STAMPEDE study (Systemic Therapy in Advancing or Metastatic Prostate Cancer: Evaluation of Drug Efficacy). The STAMPEDE trial [26] evaluated the feasibility of performing tumor sequencing to design biomarker-directed trials in metastatic castrate-resistant prostate cancer. In two large cohorts of this subset of patients (185 tumors analyzed retrospectively and 101 prospectively), implementing genomic characterization using residual biopsies was shown to be feasible for directing clinical trial designs in the future. Compared with our cohort, that study showed a similar incidence of genomic alterations for TP53 (33% vs. 36%), PTEN (34% vs. 31%), and RB1 (1% vs. 6%). An advantage of our study is that we were able to compare first-line treatments, instead of comparing randomly assigned therapies in an ongoing clinical trial [26].
Stratification by genetic factors has not yet been included in trials of M1-HSPC. Our current results suggest that TSG status could be a considerable confounding factor for clinical outcomes based on first-line therapy choices. We found a significant difference in PFS and OS between patients with or without TSG-alt who had first-line treatment with abiraterone acetate. In the future, generalized use of broad genomic tumor sequencing in early prostate cancer may be the most accurate method for risk stratification, at least more so than a consensus definition of tumor volume or perhaps even distinguishing de novo vs. relapsed metastatic disease. However, on multivariable analysis, volume of disease and metastatic disease at diagnosis were not particularly associated with PFS and OS.
The main strength of our study is that it includes a real-world patient cohort treated at a tertiary referral center, with sequencing performed using commercially available platforms. This makes our results applicable to most routine oncology practices to identify high-risk patients early in their disease course. In addition, to our knowledge, ours is one of the largest cohorts to date to examine the prognostic significance of TSG status in patients with M1-HSPC [25].
Finally, the different clinical outcomes of our population may result from a slightly higher-risk subset of patients compared with those in historical trials, with regard to known negative prognostic factors such as de novo presentation (61%, vs. 50% in STAMPEDE, 72% in CHAARTED), metastatic disease burden (65%, vs. not reported in STAMPEDE, 64% in CHAARTED), and higher presence of visceral metastases (30%, vs. 6% in STAMPEDE, 15% in CHAARTED) [27]. These differences reflect the variability of highly selected cohorts included in randomized clinical trials in contrast to a heterogeneous real-world patient population.
Limitations
Our study has several limitations. Because this was a retrospective study, significant confounding factors and selection bias cannot be excluded. To limit selection bias, we included all consecutive patients who met inclusion criteria. However, patients without tumor sequencing were excluded, which limits the generalizability of our results. The modest sample size constrains the statistical significance of our findings, and, thus, the data can only be seen as hypothesis generating. Our study reflects some institutional practice patterns. Another limitation is the suboptimal detection and heterogeneity of comprehensive genomic analyses in some of the gene alterations, particularly RB1 and PTEN [28, 29]. We did not compare genomic results with immunohistochemistry. In contrast to the STAMPEDE [26] and CHAARTED [10] studies, which included criteria for PSA progression, we were unable to retroactively apply PSA progression criteria to patients who continued on treatments as directed by their physician.
Conclusions
In summary, we confirmed that pathogenic TSG-alt in TP53, PTEN, and RB1 in patients with M1-HSPC portend a poor prognosis early in the disease course. Loss of function of these TSGs is prognostic for early progression during first-line treatment, regardless of the treatment selection, and identification of TSG-alt outperforms most clinical variables as a predictor of early progression in multivariable analysis. Increased use of tumor sequencing to detect these alterations early in the presentation of prostate cancer may establish the need for inclusion of molecular stratification in prognostic algorithms. Confirmation of these findings holds the potential for improving prognostic tools to better define high-risk patients.
Supplementary information
Author contributions
MGV had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: MGV, HEK, JBE, IBR, AHB. Acquisition, analysis, or interpretation of data: All authors. Drafting of the paper: MGV, HEK, AHB. Critical revision of the paper for important intellectual content: All authors. Statistical analysis: HEK. Administrative, technical, or material support: HEK, JBE, AHB. Supervision: JBE, AHB.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41391-021-00430-4.
References
- 1.Hamid AA, Gray KP, Shaw G, MacConaill LE, Evan C, Bernard B, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76:89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 2.Maughan BL, Guedes LB, Boucher K, Rajoria G, Liu Z, Klimek S, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018;21:260–8. doi: 10.1038/s41391-017-0027-4. [DOI] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50:645–51. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. 2017; 2017. [DOI] [PMC free article] [PubMed]
- 8.VanderWeele DJ, Antonarakis ES, Carducci MA, Dreicer R, Fizazi K, Gillessen S, et al. Metastatic hormone-sensitive prostate cancer: clinical decision making in a rapidly evolving landscape of life-prolonging therapy. J Clin Oncol. 2019;37:2961–7. doi: 10.1200/JCO.19.01595. [DOI] [PubMed] [Google Scholar]
- 9.Gillessen S, Attard G, Beer TM, Beltran H, Bossi A, Bristow R, et al. Management of patients with advanced prostate cancer: the report of the advanced prostate cancer consensus conference APCCC 2017. Eur Urol. 2018;73:178–211. doi: 10.1016/j.eururo.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Kyriakopoulos CE, Chen YH, Carducci MA, Liu G, Jarrard DF, Hahn NM, et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer: long-term survival analysis of the randomized phase III E3805 CHAARTED trial. J Clin Oncol. 2018;36:1080–7. doi: 10.1200/JCO.2017.75.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N. Engl J Med. 2017;377:352–60. doi: 10.1056/NEJMoa1704174. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong AJ, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, Azad A, et al. ARCHES: a randomized, phase iii study of androgen deprivation therapy with enzalutamide or placebo in men with metastatic hormone-sensitive prostate cancer. J Clin Oncol. 2019;37:2974–86. doi: 10.1200/JCO.19.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis ID, Martin AJ, Stockler MR, Begbie S, Chi KN, Chowdhury S, et al. Enzalutamide with standard first-line therapy in metastatic prostate cancer. N. Engl J Med. 2019;381:121–31. doi: 10.1056/NEJMoa1903835. [DOI] [PubMed] [Google Scholar]
- 14.Chi KN, Agarwal N, Bjartell A, Chung BH, Pereira de Santana Gomes AJ, Given R, et al. Apalutamide for metastatic, castration-sensitive prostate cancer. N. Engl J Med. 2019;381:13–24. doi: 10.1056/NEJMoa1903307. [DOI] [PubMed] [Google Scholar]
- 15.James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N. Engl J Med. 2017;377:338–51. doi: 10.1056/NEJMoa1702900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gravis G, Boher JM, Chen YH, Liu G, Fizazi K, Carducci MA, et al. Burden of metastatic castrate naive prostate cancer patients, to identify men more likely to benefit from early docetaxel: further analyses of CHAARTED and GETUG-AFU15 studies. Eur Urol. 2018;73:847–55. doi: 10.1016/j.eururo.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke NW, Ali A, Ingleby FC, Hoyle A, Amos CL, Attard G, et al. Addition of docetaxel to hormonal therapy in low- and high-burden metastatic hormone sensitive prostate cancer: long-term survival results from the STAMPEDE trial. Ann Oncol. 2019;30:1992–2003. doi: 10.1093/annonc/mdz396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rescigno P, Lorente D, Dolling D, Ferraldeschi R, Rodrigues DN, Riisnaes R, et al. Docetaxel treatment in PTEN- and ERG-aberrant metastatic prostate cancers. Eur Urol Oncol. 2018;1:71–7. doi: 10.1016/j.euo.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraldeschi R, Nava Rodrigues D, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol. 2015;67:795–802. doi: 10.1016/j.eururo.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen WS, Aggarwal R, Zhang L, Zhao SG, Thomas GV, Beer TM, et al. Genomic drivers of poor prognosis and enzalutamide resistance in metastatic castration-resistant prostate cancer. Eur Urol. 2019;76:562–71. doi: 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonpavde G, Pond GR, Armstrong AJ, Galsky MD, Leopold L, Wood BA, et al. Radiographic progression by Prostate Cancer Working Group (PCWG)-2 criteria as an intermediate endpoint for drug development in metastatic castration-resistant prostate cancer. BJU Int. 2014;114:E25–31. doi: 10.1111/bju.12589. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sailer V, Eng KW, Zhang T, Bareja R, Pisapia DJ, Sigaras A, et al. Integrative molecular analysis of patients with advanced and metastatic cancer. JCO Precis Oncol. 2019;3. [DOI] [PMC free article] [PubMed]
- 24.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–69. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stopsack KH, Nandakumar S, Wibmer AG, Haywood S, Weg ES, Barnett ES, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res. 2020;26:3230–8. doi: 10.1158/1078-0432.CCR-20-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilson C, Ingleby F, Gilbert DC, Parry MA, Atako NB, Ali A, et al. Genomic profiles of de novo high- and low-volume metastatic prostate cancer: results from a 2-stage feasibility and prevalence study in the STAMPEDE trial. JCO Precis Oncol. 2020;4:882–97. doi: 10.1200/PO.19.00388. [DOI] [PubMed] [Google Scholar]
- 27.Francini E, Gray KP, Xie W, Shaw GK, Valenca L, Bernard B, et al. Time of metastatic disease presentation and volume of disease are prognostic for metastatic hormone sensitive prostate cancer (mHSPC) Prostate. 2018;78:889–95. doi: 10.1002/pros.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lotan TL, Heumann A, Rico SD, Hicks J, Lecksell K, Koop C, et al. PTEN loss detection in prostate cancer: comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget. 2017;8:65566–76. doi: 10.18632/oncotarget.19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nava Rodrigues D, Casiraghi N, Romanel A, Crespo M, Miranda S, Rescigno P, et al. RB1 heterogeneity in advanced metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25:687–97. doi: 10.1158/1078-0432.CCR-18-2068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


