Abstract
Background
Risk stratification or progression in prostate cancer is performed with the support of clinical-pathological data such as the sum of the Gleason score and serum levels PSA. For several decades, methods aimed at the early detection of prostate cancer have included the determination of PSA serum levels. The aim of this systematic review is to provide an overview about recent advances in the discovery of new molecular biomarkers through transcriptomics, genomics and artificial intelligence that are expected to improve clinical management of the prostate cancer patient.
Methods
An exhaustive search was conducted by Pubmed, Google Scholar and Connected Papers using keywords relating to the genetics, genomics and artificial intelligence in prostate cancer, it includes “biomarkers”, “non-coding RNAs”, “lncRNAs”, “microRNAs”, “repetitive sequence”, “prognosis”, “prediction”, “whole-genome sequencing”, “RNA-Seq”, “transcriptome”, “machine learning”, and “deep learning”.
Results
New advances, including the search for changes in novel biomarkers such as mRNAs, microRNAs, lncRNAs, and repetitive sequences, are expected to contribute to an earlier and accurate diagnosis for each patient in the context of precision medicine, thus improving the prognosis and quality of life of patients. We analyze several aspects that are relevant for prostate cancer including its new molecular markers associated with diagnosis, prognosis, and prediction to therapy and how bioinformatic approaches such as machine learning and deep learning can contribute to clinic. Furthermore, we also include current techniques that will allow an earlier diagnosis, such as Spatial Transcriptomics, Exome Sequencing, and Whole-Genome Sequencing.
Conclusion
Transcriptomic and genomic analysis have contributed to generate knowledge in the field of prostate carcinogenesis, new information about coding and non-coding genes as biomarkers has emerged. Synergies created by the implementation of artificial intelligence to analyze and understand sequencing data have allowed the development of clinical strategies that facilitate decision-making and improve personalized management in prostate cancer.
Subject terms: Prognostic markers, Prostate cancer
Introduction
Detection of prostate cancer (PCa) includes the measurement of PSA serum levels and the digital rectal exam (DRE), contributing with the detection of PCa in early stages. The localized disease, when it is confined to the prostate, is treated with radical prostatectomy (RP) [1]. In contrast, localized advanced PCa is treated with surgery, adjuvants, radiotherapy such as external beam radiation or brachytherapy; and hormone therapy such as Luteinizing hormone-releasing hormone (LHRH) agonists and antagonists, abiraterone, and enzalutamide; while metastatic disease is usually treated with hormone therapy such as apalutamide, and chemotherapy [2]. If PCa is diagnosed on time, the treatment can be effective and with minimal morbidity [3]. In order to cover the proportion of indeterminate findings, novel diagnostic biomarkers have been developed such as Prostate Health Index (PHI), 4 K score, SelectMDx, ConfirmMDx and PCA3 [4]. Although several studies have been performed to analyze different molecular biomarkers, such as variant V7 of the androgen receptor [5] or inactivation of the PTEN or c-MYC gene [6], to date, none of them have been approved as a prognostic biomarker for use in clinical settings [7, 8]. Currently, there are a few molecular prognostic biomarkers in clinical use such as OncotypeDX Genomic Prostate Score [9], Prolaris [10], ProMark [11], and Decipher [12], based on cancer-associated gene panels [4]. These molecular tests guide the urologist to establish the appropriate treatment and predict recurrence and progression risk after localized treatment. However, it is important to keep searching for molecular markers that can aid in early diagnosis and prognosis of the patient as well as the establishment of patient response to different treatments, such as new genes, gene fusions, AR variants and non-coding RNAs.
At present, there is an intense debate regarding PSA as a diagnostic, prognostic and screening tool in PCa, and therefore it is especially important to focus on other types of molecular markers that can support clinical outcomes and decision making for therapy [13]. In particular, transcriptome and genomics analysis have contributed to generate new knowledge in the study of PCa and the intracellular signaling pathways that regulate prostate carcinogenesis generating new information about its biology [14]. Otherwise, artificial intelligence and some of its algorithms have been served for clinical application in monitoring, detection, diagnosis, and treatment to generate new clinical predictive models to PCa Management [15]. Alternatively, several studies have combined histology with genomic data, integrating omics information with pathological images in PCa [16, 17] and with implementation of artificial intelligence algorithms such as deep learning and machine learning have served to establish a connection from different branches of omics to get clinical prediction models, thus, creating an integrative perspective that facilitates the discovery of new diagnostic, prognostic and therapeutic molecular biomarkers. Finally, the importance of precision medicine and the fusion between sequencing and artificial intelligence is established with the aim of creating synergies that allow the development of more specific and advanced systems that facilitate obtaining relevant clinical strategies for decision-making and personalized management of PCa patients.
Methods
Aiming to search for new molecular biomarkers involved in the diagnosis, prognosis and prediction, an exhaustive search was conducted by Pubmed, Google Scholar and Connected Papers using keywords relating to the genetics, genomics, transcriptomics and artificial intelligence in PCa, it includes “biomarkers”, “non-coding RNAs”, “lncRNAs”, “miRNAs”, “repetitive sequence”, “risk”, “prognosis”, “prediction”, “therapy”, “exome”, “whole-genome sequencing”, “RNA-Seq”, “transcriptome”, “artificial intelligence”, “machine learning”, and “deep learning”.
Biomarkers and precision medicine in prostate cancer
The National Cancer Institute of the United States of America defines a biomarker as a biological molecule that can be detected in blood, tissue, or bodily fluids that can be measured and whose values allow the identification of a normal or abnormal process, as well as a disease [18]. From the large variety of molecular markers currently in existence, they can be classified according to the clinical context for which they will be used. For example, there are diagnostic, prognostic, and predictive molecular biomarkers [19]. This process has led to the era of precision medicine where the selection of treatment is based on the molecular characteristics of the tumor of each patient [20] (Fig. 1). In the following paragraphs, we will mention and describe some molecular biomarkers that have recently been reported as useful in PCa patient management, including coding and non-coding genes.
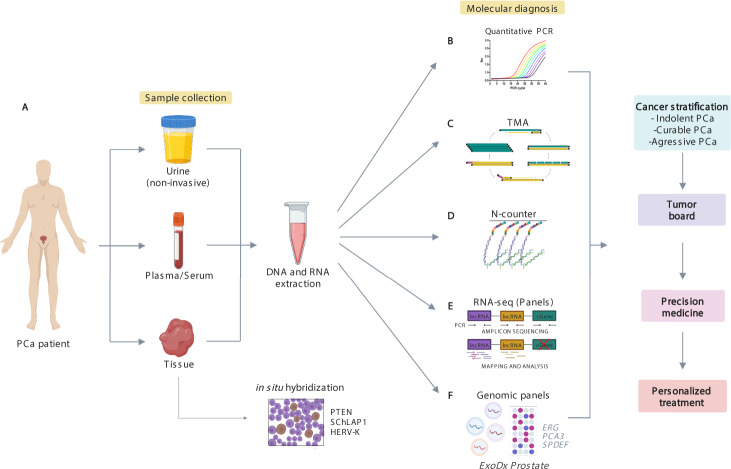
Fig. 1. Landscape of precision medicine and molecular tools in prostate cancer.
A Sample collection. This can be achieved by sampling tissue, blood or even urine (a non-invasive sampling) from the patient and proceeding with a direct detection of the biomarker by in situ hybridization in the tissue sample or a nucleic acid extraction and a molecular assay. B Quantitative PCR (qPCR). This molecular tool can be used to quantify gene expression by determining the amount of a target sequence present in the sample based on fluorescent emission, such as My Prostate Score [119]. C Transcription Mediated Amplification (TMA), PROGENSA is a current test based on a TMA assay. D N-counter. It is a highly multiplexed single-molecule counting system where two probes are used to target the RNA molecule of interest, a capture probe and a reporter probe. Dong et al. used the NanoString nCounter assay to target mRNA transcripts in EVs from PCA cell lines [120]. E RNA Sequencing. The RNA massive sequencing allows analyzing the entire transcriptome and even transcripts yet to be discovered. F There are also genomic panels used for the diagnostic of PCa focused on specific biomarkers, such as the commercial test ExoDx Prostate [121] that detects the expression levels of ERG, PCA3 and SPDEF by qPCR in exosomes from urine samples.
Coding genes as molecular markers of prostate cancer under clinical investigation
Several novel biomarkers for PCa have been proposed, however, their clinical utility remains to be discussed. Nevertheless, coding genes used as biomarkers such as AR, BRCA2, PTEN and the gene fusion TMPRSS2-ERG have predictive value for treatment response and are used in clinical practice [21–23]. Although these molecular markers offer valuable prognostic information for clinical practice, they are only functional in a subset of patients and more clinical trials are needed to validate their utility (Table 1).
Table 1.
PCa-associated biomarkers approached by clinical trials.
| Biomarker | Clinical trial phase | Type of cancer | Patients included | Clinical trial ID |
|---|---|---|---|---|
| TMPRSS2-ERG | Phase II | mCRPC and recurrent PCa | 148 | NCT01576172 |
| Phase II | Recurrent PCa, stage IV PCa | 29 | NCT00330161 | |
| Phase II | Prostatic adenocarcinoma | 148 | NCT01682772 | |
| Phase I | Advanced or metastatic PCa | 113 | NCT00749502 | |
| Phase I | High risk PCa | 65 | NCT02588404 | |
| Phase I | Localized or locally advanced PCa, biochemical recurrent PCa | 84 | NCT03421015 | |
| Phase II | High risk PCa | 208 | NCT02573636 | |
| TP53 | Phase III | mCRPC | 750 | NCT03903835 |
| Phase I/II | Prostatic neoplasia | 36 | NCT00900614 | |
| Phase III | Localized PCa | 7 776 | NCT00001469 | |
| Phase I | Localized or locally advanced PCa, biochemical recurrent PCa | 84 | NCT03421015 | |
| AR | Phase I | Hormone refractory PCa | 140 | NCT00510718 |
| Phase II | PCa | 45 | NCT01990196 | |
| Phase II | Recurrent PCa | 42 | NCT03311555 | |
| Phase I | mCRPC | 58 | NCT01516866 | |
| Phase II | Metastatic PCa, CRPC | 60 | NCT04090528 | |
| Phase I | PCa | 40 | NCT02411786 | |
| Phase II | mCRPC | 8 | NCT02379390 | |
| Phase II | Biochemical recurrent PCa | 90 | NCT01790126 | |
| Phase II | Advanced hormone dependent PCa | 90 | NCT01861236 | |
| BRCA2 | Phase II | High risk PCa | 100 | NCT02154672 |
| Phase III | mCRPC | 408 | NCT03075735 | |
| Phase III | Genetic predisposition to PCa | 1 700 | NCT00261456 | |
| Phase II | mCRPC | 40 | NCT04038502 | |
| Phase II | mCRPC | 70 | NCT03012321 | |
| Phase III | mCRPC | 387 | NCT02987543 | |
| PTEN/P13K/AKT/mTOR | Phase II | High risk PCa | 208 | NCT02573636 |
| Phase III | CRPC | 120 | NCT03580239 | |
| Phase II | PCa previously treated | 108 | NCT01251861 | |
| Phase I | PCa previously treated with enzalutamide | 36 | NCT03310541 | |
| Phase I | Stage III and IV PCa | 62 | NCT01480154 | |
| Phase II | mCRPC | 9 | NCT02091531 | |
| MGMT | NA | NA | NA | NA |
| DNMT1 | Phase I | mCRPC | 19 | NCT05037500 |
| NA | Prostatic adenocarcinoma | 19 | NCT01118741 | |
| Phase III | Prostatic adenocarcinoma | 80 | NCT03535675 | |
| Phase I | Adenocarcinoma of the Prostate, Recurrent PCa, Stage I, IIA, IIB, III and IV PCa | 32 | NCT01912820 | |
| Phase I/II | Prostate Carcinoma | NA | NCT03709550 | |
| JMJD3 | NA | NA | NA | NA |
| KDM4B | NA | NA | NA | NA |
| CDK9 | Phase I | Castrate Resistant Prostate Cancer | 100 | NCT05159518 |
| SF3B2 | NA | NA | NA | NA |
| AR-V7 | Phase III | Castrate Resistant Prostate Cancer | 953 | NCT02438007 |
| AR-V3 | NA | NA | NA | NA |
| HDAC6 | NA | NA | NA | NA |
| PRUNE2 | NA | NA | NA | NA |
| Circulating tumor cells | NA | Prostate Cancer Obesity | 67 | NCT02453139 |
| Phase II | Patients with PSA 4–10 ng/mL | 500 | NCT03488706 | |
| Phase II | Localized PCa | 200 | NCT01961713 | |
| Phase II | mCRPC | 11 | NCT00887640 | |
| Phase II | Advanced PCa | 24 | NCT02552394 | |
| Phase II | mCRPC | 140 | NCT03050866 | |
| Phase I | PCa | 60 | NCT02450435 | |
| cell-free DNA | Phase III | Metastatic PCa | 1038 | NCT00134056 |
| Phase I | PCa | 12 | NCT04081428 | |
| Phase II | Metastatic PCa | 300 | NCT02853097 | |
| Phase II | PCa | 68 | NCT02941029 | |
| Phase II | PCa | 30 | NCT03284684 | |
| Extracellular vesicles | NA | PCa | 108 | NCT04298398 |
| miRNAs | Phase II | mCRPC | 40 | NCT02471469 |
| Phase II | mCRPC | 46 | NCT04188275 | |
| Phase III | High risk PCa | 300 | NCT01220427 | |
| Phase I | PCa | 240 | NCT03911999 | |
| Phase I | PCa | 60 | NCT02366494 | |
| lncRNAS | NA | Prostatic neoplasia | 507 | NCT01024959 |
mCRPC Metastatic castration resistance prostate cancer, CRPC Castration resistance prostate cancer, PCa prostate cancer, PSA Prostate Specific Antigen, NA Not Applicable.
Information obtained from https://clinicaltrials.gov/ (Last accessed: February 25, 2021).
On the other hand, several mechanisms involved in prostate tumorigenesis such as epigenetic changes, alternative splicing, and the presence of gene variants, are possible novel biomarkers based on coding-genes with potential clinical utility [24]. For example, regarding epigenetic regulators, the coding genes MGMT [25], DNMT1 [26], and JMJD3 [27], which are involved in DNA methylation, have been associated with the risk of PCa mortality (HR 0.90; p-value = 3.5 × 102) [25] as well as prostate tumor development (p-values = 0.03 and 0.05, respectively [26, 27]). Similarly, other proteins related to splicing process, such as KDM4B [28], CDK9 [29] and SF3B2 [30] have been recently associated with generation of androgen receptor variant AR-V7 (p-value < 0.05 [28–30]). Furthermore, the splicing variants by themselves are of particular interest for PCa research [31], androgen receptor variants [32] have been described as important factors in PCa development and prognosis, such as variant AR-V3 and its prognostic value (p-value = 0.05) [33]. Likewise, HRAS [34] and PRUNE2 [35] are novel variants related to PCa development with clinical utility yet to be confirmed. Therefore, research focused on finding new coding genes with clinical application as biomarkers, could improve PCa prognosis and treatment.
Another example of coding genes that may have potential clinical utility for PCa correspond to gene mutations involved in hereditary cancer, where it represents the etiology of 5–10% of all neoplasms [36], in which PCa has been associated with family history of cancer [37]. Paradoxically, few high-susceptibility genes consistently related to the hereditary of PCa have been identified, presenting a pattern of dominant autosomal inheritance [38], that has been linked to phenotypic variation and genetic heterogeneity, limiting its association with PCa predisposition [39]. Currently, the analysis of Pathogenic Variants (PV) in predisposition genes associated with defects in homologous recombination and mismatch repair [40] which represents therapeutic targets to PARP1 inhibitors and chemotherapies with platinum compounds, particularly in patients with metastatic and castration-resistant disease [41, 42]. The use of multi-gene panels in germline diagnosis has identified PV in 7% to 12% of PCa patients [43, 44], highlighting BRCA1, BRCA2, ATM, BRIP1, CHEK2, NBN, BARD1, RAD51C, MRE11A and PALB2 (homologous recombination repair); MLH1, MSH2, MSH6 and PMS2 (mismatch repair) as high risk genes, which have clinical guidelines; option for risk reduction surgeries, and personalized treatment, which benefits the PCa patient [45] (Supplementary Table 1). Although hereditary PCa does not imply a generalized molecular diagnosis, it does entail the identification of metastatic disease; early age of onset, and cancer family history, who will have benefit for the therapeutic options and family prevention as a result of the molecular approach [46, 47].
Although all these molecular biomarkers have a potential clinical application, current clinical trials have not been able to determine whether they have sufficient sensitivity and specificity to be considered for clinical purposes, as well as all the genes discussed above are coding genes. Therefore, it is important to focus on the search for new biomarkers, like non-coding genes, which can contribute to the diagnosis and prognosis of PCa patients.
Noncoding genes as molecular markers in prostate cancer
Most of molecular biomarkers in PCa are based in coding genes, but as previous studies have demonstrated; mRNAs tend to have less tissue- and stage-specific expression. In contrast, non-coding RNAs tend to have more tissue-specific and stage-specific expression in disease, which is one of the main reasons noncoding RNAs have been proposed as molecular biomarkers in cancer [48]. In the following paragraphs we describe some of the newest candidates as specific molecular biomarkers in PCa clinical research.
miRNA
One of the most studied small ncRNAs are microRNAs (miRNAs), these are single stranded RNAs of 21–25 nucleotides in length that regulate the post-transcriptional degradation of messenger RNAs and inhibit their translation into proteins. Because of their high stability in body fluids [49] as well as to changes of physical and chemical conditions [50], miRNAs are interesting molecules to be used as biomarkers in cancer. Free miRNAs can be found in several bodily fluids, such as blood, urine, semen, among others [51] and their expression levels are tissue-specific and have been found to be deregulated in cancer [52]. Moreover, they exhibit differential expression between tumor and normal tissues and are useful for tumor classification according to the lineage of origin, differentiation stage, and tumor aggressiveness [53]. It has been reported that circulating miRNAs can be packed in extracellular vesicles (EV) or in association with proteins such as Argonaute2 or lipoproteins in bio-fluids including blood and urine [54–56]. Some miRNAs, such as miR-21, miR-221, miR-1290, and miR-375, have been overexpressed and associated with prognosis in CRPC patients [55, 57]. Yaman and collaborators quantified the levels of miR-21, miR-142, and miR-221 in PCa patients and reported that overexpression of these three miRNAs were associated with an advanced PCa stage [58]. Other groups have identified miRNAs in plasma and serum of patients with locally advanced and metastatic PCa, with BPH and in healthy individuals, showing that differences between each group (i.e., higher levels of miRNAs in patients with locally advanced and metastatic PCa highlight the role of miRNAs as diagnostic biomarkers [59]. Several groups have studied the diagnostic, prognostic, and predictive characteristics of miRNAs circulating in the plasma and serum of PCa patients finding differentially expressed miRNAs according to the Gleason index [60], response to treatment with docetaxel [61], and high blood PSA values [62]. In another study, a panel consisting of four miRNAs was proposed as a biomarker for the diagnosis of PCa [63]. The four miRNAs (miR-4289, miR-326, miR-152-3p and miR-98-5p) were upregulated in plasma of PCa patients compared to healthy controls and was able to differentiate between PCa patients and control individuals with an area under the ROC curve of 0.88, proving their diagnostic accuracy. In the study conducted by Sharova and collaborators [49], a circulating miRNA test consisting of measuring the level of 3 circulating miRNAs (miR-106a, miR-130b and miR-223) was proposed to differentiate between localized PCa and BPH patients. In this test two ratios are calculated: miR-106a/miR-130b and miR-106a/miR-223 ratios, the results showed a better performance (specificity: 0.806, sensitivity: 0.833, accuracy: 0.821) in comparison to PSA (specificity: 0.065, sensitivity: 0.889, accuracy: 0.507), the area under the ROC curve for miRNA test was 0.84 while for PSA was 0.56. This test could be helpful for PCa screening to avoid unnecessary biopsies and assessment of PCa risk. Indeed, the use of miRNAs as biomarkers in PCa has shown promising results for risk assessment, diagnosis, and prognosis. Implementation of miRNA-based tests in combination with gene-based biomarkers could improve the clinical management of PCa patients.
Long non-coding RNAs
As mentioned above, RNA molecules seem to have a critical role in cancer pathways including those within PCa. Long non-coding RNAs are known to be RNA transcripts longer than 200 nucleotides with no protein-coding potential [64], these two major differences distinguish them from mRNA transcripts and any other non-coding RNA. LncRNAs have been implicated in several biological processes such as chromatin-reprogramming, genomic imprinting, transcriptional regulation in cis and trans and post-transcriptional regulation of mRNAs [65–67]. Among some pathological features in which lncRNAs are involved are cell proliferation, tumorigenesis and malignant transformation [68], this is why several studies have proposed lncRNAs as tumor-suppressor genes and oncogenes [69, 70]. Lately, lncRNAs have drawn the attention not only because of their critical role in cancer, but because of their potential as molecular biomarkers due to their tissue-specific and tumor-specific expression [68, 71]. Some lncRNAs, such as PCA3, SChLAP1, and PCAT1 have been proposed as good candidates for biomarkers mainly due to their differential expression in PCa patients [72]. PCA3 is an overexpressed PCa-specific oncogene discovered in 1999 by Bussemakers [73]. PCA3 is already considered a PCa biomarker, and it is measured by the commercial test PROGENSA approved in 2012 by the FDA [74–76] helping to reduce ~40% of unnecessary biopsies providing a great utility in urological diagnosis [77]. PROGENSA PCA3 test has a sensitivity of 62% and a specificity of 75% [78] demonstrating why lncRNAs can be one of the molecular markers with clinical utility. Similarly, SChLAP1 is known for its high expression levels in PCa. This lncRNA antagonizes the SWI/SNF complex promoting aggressiveness and metastasis of the tumor [79]. Its effectiveness as a biomarker has been proved by assays such as RNA in situ hybridization leading to the development of several tests based on the detection of SChLAP1 expression levels and linking them with the patient’s clinical-stage [80]. Therefore, SChLAP1 is considered as a promising biomarker of clinical utility and one of the best genes for prediction of metastasis and biochemical recurrence in PCa patients [79, 81]. Along with these, Luo and collaborators reported that lncRNA-p21 is overexpressed in neuroendocrine PCa (NEPC) and that a treatment based upon enzalutamide increases its expression, and thus, the neuroendocrine differentiation; all of this is caused by the alteration of the Enz/AR/lncRNA-p21/EZH2/STAT3 axis [82]. PCAT1 is another upregulated oncogenic RNA originally identified in PCa by RNA-sequencing analysis [83]. It is related to cell proliferation, apoptosis, migration, and invasion as well as epithelial-mesenchymal transition and cancer progression via the Wnt/β-catenin signaling pathway [84]. Finally, PCAT1 negatively regulates BRCA2 tumor suppressor protein, positively regulates Myc oncoprotein [85] and it might be also acting as a miRNA sponge involved in cell growth [83]. Hence, PCAT1 is considered as a potential biomarker for PCa prognosis and prediction, supporting the statement that lncRNAs represent potential molecular biomarkers in the management of PCa (Table 2). Most of these candidates and a large number of transcriptional units were found due to the breakthrough of the high-throughput massive sequencing technology, specifically, RNA-Seq. Finally, lncRNAs could be used in combination with gene-based biomarkers and gene fusions to increase the sensitivity and specificity of molecular diagnostic tests, which will improve clinical patient management including early detection, diagnosis, prognosis, and prediction of response to treatment [86].
Table 2.
New biomarkers and their clinical potential in prostate cancer.
| Biomarker | Type | Symbol | Validation | Reference |
|---|---|---|---|---|
| miRNAs | Diagnostic | let-7a, miR-145 and miR-155 | Independently validated | [126] |
| miR-21 | Independently validated | [127] | ||
| miR-32-5p | Independently validated | [128] | ||
| miR-141 | Independently validated | [129] | ||
| miR-301a | Research Use Only | [130] | ||
| Prognostic | miR-96-5p, miR-183-5p, miR-145-5p, miR221-5p | Independently validated | [131] | |
| miR-301a | Research Use Only | [130] | ||
| miR-187 | Research Use Only | [132] | ||
| miR-1 | Independently validated | [133] | ||
| miRs-301a, 652, 454, 223 and 139 | Independently validated | [134] | ||
| Therapy response predictive | miR-106-b | Research Use Only | [135] | |
| miR-21 | Independently validated | [136] | ||
| miR-200 | Independently validated | [61] | ||
| miR-890 | Research Use Only | [137] | ||
| miR-34a | Research Use Only | [138] | ||
| lncRNAs | Diagnostic | PCA3 | FDA Approved, 2012 | [139–141] |
| MALAT-1 | Independently validated | [142, 143] | ||
| PCAT14 | Independently validated | [144] | ||
| LOC100287482 | Research Use Only | [145] | ||
| FR0348383 | Independently validated | [146] | ||
| Prognostic | SChLAP1 | Independently validated | [147] | |
| lncRNA-ATB | Independently validated | [148] | ||
| FALEC | Research Use Only | [149] | ||
| TUG1 | Independently validated | [150] | ||
| SNHG9 | Independently validated | [151] | ||
| Therapy response predictive | PCAT1 | Research Use Only | [152] | |
| GAS5 | Research Use Only | [153–155] | ||
| NEAT-1 | Research Use Only | [156, 157] | ||
| DANCR | Research Use Only | [158] | ||
| LOXL1-AS1 | Research Use Only | [159] | ||
| Repetitive sequences | Diagnostic | HERV-K | Research Use Only | [92, 93] |
| MNS16A | Research Use Only | [160] | ||
| Y-STR loci | Research Use Only | [161] | ||
| Prognostic | TG-PCA3 STR | Research Use Only | [162] | |
| CAG repeats | Research Use Only | [163, 164] | ||
| ESR1 TA | Research Use Only | [165] | ||
| MSR1 | Research Use Only | [166] | ||
| microsatellite instability | Research Use Only | [167] | ||
| LINE-1 | Research Use Only | [94] |
Repetitive sequences
Repetitive sequences are large quantities of repeated elements throughout the haploid genome, meaning they are repeated DNA nucleotides found more than twice in the genome that comprises about 55% of the human genome or even more [87]. Their classification can vary from author to author, and it can be based on the origin, function, structure, and genomic distribution of the DNA, but it is mainly based on the latter. The five categories are simple sequence repeats, segmental duplications, tandem repeats and satellite DNA sequences, processed pseudogenes, and transposable elements [88].
Repetitive sequences are also considered as potential molecular biomarkers in diseases like cancer because some of them are overexpressed in different types of tumors cells [89]. Genome sequencing and transcriptome sequencing have improved the discovery and detection of repetitive DNA and RNA elements that cannot be identified by classic biochemical methods [90]. Solovyov and collaborators [91] determined that RNA repetitive sequences are not fully detected when using the poly(A) protocol in RNA-seq procedure, while on the other hand, analyzing the expression of total RNA sequencing can not only identify the repetitive sequences more accurately but delimitate immune phenotypes in cancer and response to immunotherapy [91]. Among the candidates for biomarkers in PCa we can found the HERV-K sequence, which is highly expressed in malignant prostate tissue when comparing it with normal prostate tissue, it is considered as a possible early disease detection biomarker detected in PCa patient blood, and it can even increase PSA test efficiency [92, 93]. Moreover, LINE-1 is a DNA sequence that encodes the RNA-binding protein ORF1p and presents an increased expression in PCa tissues. Its overexpression is associated to cancer tumorigenesis and its hypomethylation to PCa progression [94]. Although the experimental evidence regarding the importance of repeated sequences is not as abundant as other RNA biotypes, these transcripts have the potential to be considered as biomarkers in PCa, nevertheless, more studies are needed to prove its applications as biomarker for diagnosis, prognosis, and treatment management of PCa patients. The contribution of different sequencing methodologies has improved biomarker discovery in the field of non-coding transcripts.
Importance of high-throughput massive sequencing in prostate cancer
DNA and RNA massive parallel sequencing has a large impact on the generation of new knowledge concerning molecular markers in cancer because it explores the whole genome and transcriptome, allowing the detection of global point mutations, insertions, deletions, variations in copy number, translocations, fusion genes, novel-transcript discovery, transcript abundance estimation, differential gene expression and differential splicing of mRNAs [95] (Fig. 2). The application of RNA-Seq provides a quantitative pattern of coding and non-coding genes with transcriptional aberrations within the cell in a disease. This technique is an emerging sequencing technology that has a promising future in disease diagnosis, prognosis, prediction and treatment [96].
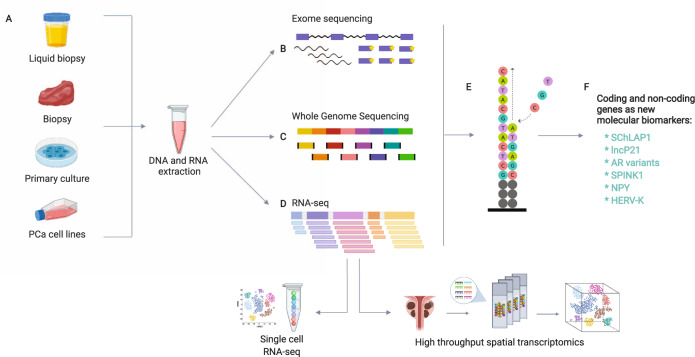
Fig. 2. Basic research towards the discovery of new molecular biomarkers.
There are several sources and molecular approaches for the detection of new biomarkers in PCa. A This can be achieved by using an in vivo model -for which a prostate biopsy should be taken-, a primary culture or a PCa cell line. B Exome sequencing. The DNA samples are first fragmented and then biotinylated oligonucleotide probes -also known as baits- are used to selectively hybridize to target regions in the genome. C Whole-Genome Sequencing. This sequencing technique allows a uniform coverage across the complete genome. D RNA-Seq. RNA samples are synthesized into cDNA once it has been fragmented. Then, adaptors are attached to both ends of each fragment so they can be amplificated by PCR and subsequently sequenced [122]. Within the variants of this technique can be found single-cell RNA-seq, total RNA-seq, targeted RNA-seq, small RNA-seq, spatial transcriptomics, poly-A enrichment, ribosomal RNA depletion, among others. E Illumina next-generation sequencing technology: Individual DNA or cDNA molecules are placed on a flowcell for sequencing by synthesis by using fluorescent labeled nucleotides. PacBio sequencer and Nanopore sequencer can read more than 100 Kb in length of DNA, as well as the disposable sequencer MinION which doesn’t need prior installation [123]. F After a bioinformatic data analysis the results of the sequencing provide new genes as biomarkers candidates in PCa.
Among some studies based on RNA sequencing as a potential tool for finding new PCa biomarkers and drug targets, Berglund and collaborators analyzed the heterogenicity of PCa through a spatial-transcriptomic study in which several expression profiles were identified within a tissue region obtained after RP (Gs 3 + 4, pT3b, PSA = 7.1). These expression profiles allowed the stratification of the tissue regions into cancer components or groups such as cancer, stroma, reactive stroma, normal glands and prostatic intraepithelial neoplasia (PIN). They also found specific genes as potential biomarkers within the results, for example, SPINK1, PGC, and CPP as specific markers of PCa (Gs 3 + 3), NR4A1 as a specific marker of reactive stroma, and NPY as a specific marker of PIN. The fact that these markers are expressed in specific and different locations, demonstrates the level of heterogenicity in prostatic tumors and that studies based on RNA sequencing technologies can open the door to the discovery of novel molecular biomarkers [97].
On the other hand, there is an urgent need to classify patients according to the most appropriate and effective therapy to increase the efficacy of treatment and reduce unnecessary interventions that have no effect on the patients (Fig. 3). An example of this characteristic is a study supporting the use of exomes in precision medicine has been reported by Robinson and collaborators, who demonstrated that actionable mutations detected with the aid of exomes in castration-resistant PCa patients can help determine the best treatment to use and responses of the patients. The results established a mean rate of 4.4 mutations per Mb, in addition to a gain and loss of chromosome regions, with gains in AR and losses in the genes CHD1, PTEN, RB1, and TP53. The relevance of this study is that the molecular changes are actionable in 90% of the samples of CRPC patients, and in particular, patients with mutations in genes such as BRCA2 (12% of cases) and ATM (22% of cases) benefited from treatment with PARP inhibitors (olaparib) [98]. In another study, Armenia and collaborators identified 70 significantly mutated genes that had not been previously associated with PCa, some of them are CUL3 (a ubiquitin ligase that function as a scaffold in the proteasome system), SPEN (a transcription factor involved in repression of gene expression), and KMT2C and KMT2D (epigenetic regulators with histone-lysine N-methyltransferase activity) by analyzing exome sequencing data from 1013 PCa samples [99]. The markers found in this, and other studies could be used as part of gene signatures aimed to stratifying patients with localized and metastatic PCa. Furthermore, recent whole-genome studies have identified mechanisms that generate complex chromosome rearrangements in PCa. Baca and collaborators sequenced the whole genome of 57 prostate tumors and identified several DNA translocations and deletions that arose independently during oncogenesis and progression. They called this phenomenon “chromoplexy” referring to the coordinated and considerable dysregulation of multiple cancer genes supporting a model of punctuated cancer evolution [100]. Therefore, studies based on genomics generate information that could help oncologists to predict the response to treatment, allowing more personalized and effective management of patients with advanced PCa, and considering that not only the coding proportion of the genome has this potential, the non-coding fraction of the genome should also be included. This experimental and clinical approach provides information about the emerging responses that current therapies, such as androgen deprivation, and their effect in PCa patients. Sequencing analysis can also provide the necessary data for a more specific and enriched molecular classification of PCa and could provide delineated subtypes among patients for better management [101].
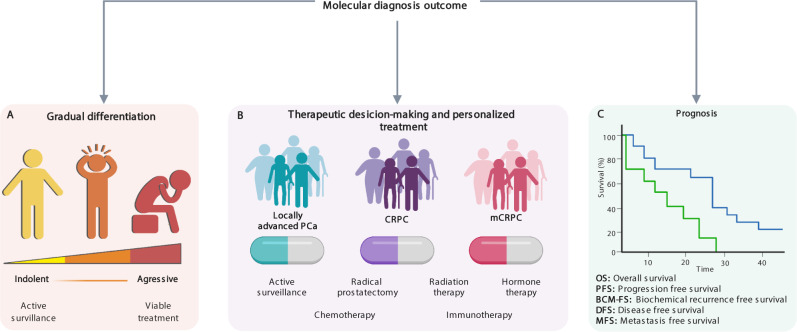
Fig. 3. Molecular diagnosis and precision medicine in cancer patient management.
The advantages of using approaches that target these signatures for disease diagnosis can be many. A PCa-stage discrimination. Each clinical-pathological profile will be stratified using the emerging techniques mentioned earlier, the results obtained could determine whether the patient has an indolent cancer or if it is an aggressive one. B Personalized treatment. The molecular diagnosis can also determine which specific treatment the patient should receive according to their molecular profile and the type of PCa they have, such as active surveillance, hormone therapy, surgery, radiation therapy, chemotherapy or immunotherapy (e.g., PD-1 inhibitors, sipuleucel-T vaccine [124]). C Prognosis. Finishing by getting a prognostic overview of the length of time that the patient will be alive or how well will the patient respond to the treatment he has been given. The prognosis can include overall-survival, progression-free survival, biochemical recurrence-free survival, disease-free survival, cancer-specific survival, and metastasis-free survival.
Artificial intelligence in prostate cancer research
Current methods for the detection of PCa show limitations in its detection [102], representing a need to improve PCa diagnostic and patient stratification with complementary tools. Artificial intelligence (AI) has proven to be an essential implement for clinical diagnoses, and it refers to the ability of a computational process to recognize patterns and make decisions that previously required human intellect to achieve a certain purpose [103].
Machine learning (ML) is a discipline that teaches computers how to build models from the massive data sets that they are assigned with and learn from them. This technologic approach is based on statistic algorithms, most of these algorithms are mathematical models that map the variables (features) of a data sample into a set of outcomes [104, 105]. Then, these algorithms go through a process of training to be able to predict the labels by analyzing the features [15]. The types of learning used in these models are mainly classified as supervised learning and unsupervised learning (Fig. 4A). Supervised learning uses explicit data sets determined by experts, the computer uses the programmed algorithms to minimize the prediction error, which is measured by the difference between the predicted labels and the known labels such as lineal logistic regression and random forest [106]. On the other hand, unsupervised learning relies on samples that are separated into different classes based on the features of the training data such as principal component analysis [106]. It has been suggested that ML could improve some aspects of biomedicine such as disease diagnosis, monitoring, anatomical imaging of organs, tissue biopsies and personalized treatment by using a collection of molecular and phenotypic data [107]. It has also been proved as useful for its application in the human genome project and advances in cancer research and management [106].
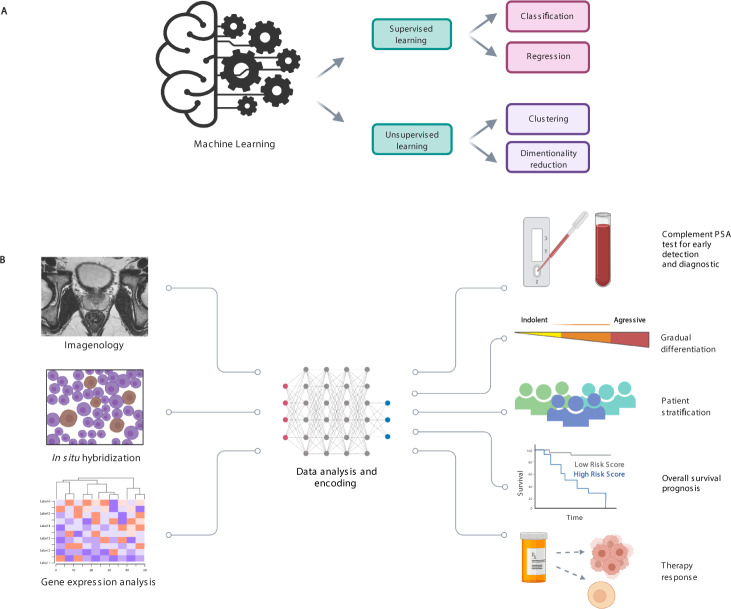
Fig. 4. Artificial intelligence and its application in patient stratification in prostate cancer.
ML is an artificial intelligence approach that can predict a possible outcome in PCa research and improve the patient management. A ML techniques. These algorithms are divided into two main types of learning: supervised learning and unsupervised learning. The former uses pre-determined explicit data, it is the most used in radiology and is based on classification and regression (deep learning, convolutional neural network, random forest, support vector machine, decision tree, logistic regression, among others [125]). The latter uses the features of the training data and doesn’t have a prior division of data in categories, it is based on clustering and dimensional reduction (K-means, hierarchical clustering, among others). B ML applied in PCa management. A recent application of ML is the prediction and analysis of radiomic data. This approach aims to improve the patient stratification and management using imageology, tissue analysis, and molecular data so the clinicians can offer a personalized treatment by differentiating the grade of the disease, stratifying the patients, and determine the therapy response.
The advantageous outcome of ML also applies to PCa research by improving diagnostic and prognostic accuracy, treatment, imaging, surgical interventions, genomics and transcriptomics. It has been reported that machines can be trained to recognize complex patterns in sequencing data together with radiographic images (such as those generated from computed tomography scanning and magnetic resonance) by classifying pixels for segmentation and registration [108]. Its techniques can identify specific genes or sets of genes within expression profiles and specific expression rate that can predict a certain clinical outcome such as progression, biochemical recurrence or metastasis in PCa [109]. There are commercial genomic classifiers available, such as Decipher, that use the random forest algorithm for prediction of PCa metastasis based on the expression analysis of 22 RNA biomarkers of aggressive PCa [15, 110].
Besides, some studies have applied ML algorithms to identify and associate non-coding RNA biomarkers for PCa diagnosis such as lncRNAs [111, 112], and several reports have developed specific algorithms, such as XGBoost by Zhang and collaborators that associate lncRNAs with several cancer types [113], this algorithm is the basis of an improved method called CRlncRC2 which was found to be more sensitive and specific than his previous version CRlncRC. Moreover, miRNAs are another potential biomarker identified through ML algorithms. Bertoli and collaborators used a support vector machine model to detect 29 miRNAs for diagnostic PCa with 97% of accuracy and 7 miRNAs which can be used in prognostic of PCa with about 66% accuracy [114]. Another study group developed a boosted random forest-based algorithm called MEDICASCY to detect cancer drug side effects, indications, efficacy, and mode of action using the chemical structure of the drug. This algorithm showed an 80% precision for detecting drugs that can help inhibit the growth prostatic tumors, as well as ovarian and breast tumors [115].
Likewise, deep learning (DL) is a branch derived from machine learning than can be used to recognize and classify tissue structures in digital information corresponding to a pathology [116]. Tolkach and collaborators developed a trained model based on the technology of deep that recognized tumor tissue from images of 400 histological slides from different patients, as well as a novel algorithm based on three-dimensional reconstruction of PCa architecture that can improve the Gleason grading [117]. Similarly, there are other algorithms that have been applied in clinics, for example, a Support Vector Machine (SVM) model was used for the detection of positive and negative biopsies through dynamic contrast-enhanced and diffusion tensor imaging data [15]. In a 2020 DL study, a deep neural network method was used to identify AR mutations during treatment for PCa. The predictions made by the algorithm can recognize mutants that resist the inhibitor darolutamide and other mutations of pharmacological interest in PCa [118]. Therefore, the development and application of AI using ML in clinical practice could open an infinite landscape of approaches within PCa data analysis (combination of coding and non-coding genes) improving the patient management in a near future.
Conclusions
Evidence based on clinical studies that focuses on finding new biomarkers suggests that there is a wide molecular field that lies unexplored and that could be the key for many clinical challenges nowadays. These markers, such as the coding (AR, BRCA2, PTEN, MLH1, CUL3, SPEN) and non-coding genes (PCA3, SChLAP1, HERV-K and miR-21), the TMPRSS2-ERG gene fusion including their derivatives, and the androgen receptor variant 7 can be found using genomic, transcriptomics and AI approaches. However, these are not the only alterations that can be used for the diagnostic, prognostic and prediction in the management of PCa patients. The clinical evidence mentioned in this review suggests the importance of these types of molecular markers (coding and non-coding genes) and their roles in the decision-making process for establishing the most adequate treatment for patients suffering from this disease. Thus, it is important to establish the types of actionable mutations, or their combinations, in patients with advanced PCa (locally advanced and metastatic), allowing us to determine the type of treatment that will provide a positive response in the PCa patient. In this area, genomic analysis, transcriptome sequencing, and new approaches like spatial transcriptomics, along with the clinical-pathological information, could provide the necessary information. Likewise, the application of ML algorithms will accelerate the identification and discovery of novel molecular biomarkers and it will lead biomedical investigation towards artificial-intelligence-based precision medicine, so it can improve patient management as well as their quality of life, and, in the near future, allow a scientific revolution in medicine for the management of the PCa patient. The new molecular biomarkers mentioned here along with the novel bioinformatic approaches of AI and sequencing techniques will improve biomedical research by complementing PSA test for screening, stratifying patients, and identifying new molecular biomarkers for differentiation of indolent and aggressive disease, prognostic, predictive and surrogate biomarkers with clinical utility (Fig. 4B). Finally, the fusion between sequencing and AI is established with the aim of creating synergies that allow the development of more specific and advanced systems that facilitate obtaining relevant clinical strategies for decision-making and personalized management of PCa patients to combat this global public health problem in men.
Supplementary information
Supplementary Table 1. Main Cancer Susceptibility Genes with Therapeutic Implications for Prostate Cancer
Acknowledgements
We thank to National Cancer Institute of Mexico (INCan) for their support. Rogelio Montiel-Manriquez is a PhD student in the “Programa de Doctorado en Ciencias Biomédicas, UNAM and received a fellowship from CONACYT with Currículum Vitae Único (CVU)- 581151. Figures were created with BioRender.com.
Author contributions
Project development: APAZ, AS, CAC, and LAH. Clinical Information: AS, MAJR, RMAG, MAJD and DPM. Basic research Information: APAZ, RMM, CCH, RGB, FJT, CAC, and LAH. Manuscript writing/editing: APAZ, AS, MAJR, RMAG, RMM, CCH, MAJD, DPM, RGB, FJT, CAC, and LAH.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cristian Arriaga-Canon, Email: carriagac@incan.edu.mx.
Luis A. Herrera, Email: lherrera@inmegen.gob.mx
Supplementary information
The online version contains supplementary material available at 10.1038/s41391-022-00537-2.
References
- 1.Descotes J-L. Diagnosis of prostate cancer. Asian J Urol. 2019;6:129–36. doi: 10.1016/j.ajur.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teo MY, Rathkopf DE, Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–99. doi: 10.1146/annurev-med-051517-011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: an important clinical entity. Nat Rev Urol. 2014;11:317–23. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kretschmer A, Tilki D. Biomarkers in prostate cancer - current clinical utility and future perspectives. Crit Rev Oncol Hematol. 2017;120:180–93. doi: 10.1016/j.critrevonc.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Welti J, Rodrigues DN, Sharp A, Sun S, Lorente D, Riisnaes R, et al. Analytical validation and clinical qualification of a new immunohistochemical assay for androgen receptor splice variant-7 protein expression in metastatic castration-resistant prostate cancer. Eur Urol. 2016;70:599–608. doi: 10.1016/j.eururo.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van der Kwast TH. Prognostic prostate tissue biomarkers of potential clinical use. Virchows Arch. 2014;464:293–300. doi: 10.1007/s00428-014-1540-7. [DOI] [PubMed] [Google Scholar]
- 7.Bazzichetto C, Conciatori F, Pallocca M, Falcone I, Fanciulli M, Cognetti F et al. PTEN as a prognostic/predictive biomarker in cancer: an unfulfilled promise? Cancers. 2019; 11. 10.3390/cancers11040435. [DOI] [PMC free article] [PubMed]
- 8.Lotan TL, Tomlins SA, Bismar TA, Van der Kwast TH, Grignon D, Egevad L, et al. Report from the international society of urological pathology (ISUP) consultation conference on molecular pathology of urogenital cancers. I. Molecular biomarkers in prostate cancer. Am J Surg Pathol. 2020;44:e15–e29.. doi: 10.1097/PAS.0000000000001450. [DOI] [PubMed] [Google Scholar]
- 9.Cullen J, Rosner IL, Brand TC, Zhang N, Tsiatis AC, Moncur J, et al. A Biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol. 2015;68:123–31. doi: 10.1016/j.eururo.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 10.Health Quality Ontario. Prolaris cell cycle progression test for localized prostate cancer: a health technology assessment. Ont Health Technol Assess Ser. 2017;17:1–75. [PMC free article] [PubMed] [Google Scholar]
- 11.Blume-Jensen P, Berman DM, Rimm DL, Shipitsin M, Putzi M, Nifong TP, et al. Development and clinical validation of an in situ biopsy-based multimarker assay for risk stratification in prostate cancer. Clin Cancer Res. 2015;21:2591–2600. doi: 10.1158/1078-0432.CCR-14-2603. [DOI] [PubMed] [Google Scholar]
- 12.Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM, et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–8. doi: 10.1200/JCO.2016.70.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol. 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda S, Elkin SK, Tomson BN, Carter JL, Kurzrock R. Next-generation sequencing of prostate cancer: genomic and pathway alterations, potential actionability patterns, and relative rate of use of clinical-grade testing. Cancer Biol Ther. 2019;20:219–26. doi: 10.1080/15384047.2018.1523849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol. 2019;16:391–403. doi: 10.1038/s41585-019-0193-3. [DOI] [PubMed] [Google Scholar]
- 16.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang E, Zhang M, Shi C, Sun L, Shan L, Zhang H, et al. An overview of advances in multi-omics analysis in prostate cancer. Life Sci. 2020;260:118376. doi: 10.1016/j.lfs.2020.118376. [DOI] [PubMed] [Google Scholar]
- 18.Goossens N, Nakagawa S, Sun X, Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res. 2015; 4. 10.21037/4536. [DOI] [PMC free article] [PubMed]
- 19.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource. Silver Spring (MD): Food and Drug Administration (US); 2016. http://www.ncbi.nlm.nih.gov/books/NBK326791/. Accessed 28 Jun 2021. [PubMed]
- 20.Rubin EH, Allen JD, Nowak JA, Bates SE. Developing precision medicine in a global world. Clin Cancer Res. 2014;20:1419–27. doi: 10.1158/1078-0432.CCR-14-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Disco. 2013;3:1020–9. doi: 10.1158/2159-8290.CD-13-0226. [DOI] [PubMed] [Google Scholar]
- 22.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15:222–34. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macedo-Silva C, Benedetti R, Ciardiello F, Cappabianca S, Jerónimo C, Altucci L. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin Epigenetics. 2021;13:125. doi: 10.1186/s13148-021-01111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.FitzGerald LM, Zhao S, Leonardson A, Geybels MS, Kolb S, Lin DW, et al. Germline variants in IL4, MGMT and AKT1 are associated with prostate cancer-specific mortality: an analysis of 12,082 prostate cancer cases. Prostate Cancer Prostatic Dis. 2018;21:228–37. doi: 10.1038/s41391-017-0029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu A, Hopkins KM, Friedman RA, Bernstock JD, Broustas CG, Lieberman HB. DNMT1 and DNMT3B regulate tumorigenicity of human prostate cancer cells by controlling RAD9 expression through targeted methylation. Carcinogenesis. 2021;42:220–31. doi: 10.1093/carcin/bgaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca F, Bignon Y-J, et al. A new metabolic gene signature in prostate cancer regulated by JMJD3 and EZH2. Oncotarget. 2018;9:23413–25. doi: 10.18632/oncotarget.25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan L, Chen Z, Lu J, Liang Y, Wang M, Roggero CM, et al. Histone lysine demethylase KDM4B regulates the alternative splicing of the androgen receptor in response to androgen deprivation. Nucleic Acids Res. 2019;47:11623–36. doi: 10.1093/nar/gkz1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Q, Poulose N, Girmay S, Helevä A, Doultsinos D, Gondane A, et al. Inhibition of CDK9 activity compromises global splicing in prostate cancer cells. RNA Biol. 2021;18:722–9. doi: 10.1080/15476286.2021.1983287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura N, Nimura K, Saga K, Ishibashi A, Kitamura K, Nagano H, et al. SF3B2-mediated RNA splicing drives human prostate cancer progression. Cancer Res. 2019;79:5204–17. doi: 10.1158/0008-5472.CAN-18-3965. [DOI] [PubMed] [Google Scholar]
- 31.Cao Z-X, Xiao G-A, Zhang W, Ji J, Ye C, Liu D, et al. Comprehensive investigation of alternative splicing and development of a prognostic risk score for prostate cancer based on six-gene signatures. J Cancer. 2019;10:5585–96. doi: 10.7150/jca.31725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanayama M, Lu C, Luo J, Antonarakis ES. AR splicing variants and resistance to AR targeting agents. Cancers. 2021;13:2563. doi: 10.3390/cancers13112563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallio HML, Hieta R, Latonen L, Brofeldt A, Annala M, Kivinummi K, et al. Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases. Br J Cancer. 2018;119:347–56. doi: 10.1038/s41416-018-0172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips J, Pan Y, Tsai B, Xing Y. Pathway-guided analysis identifies Myc-dependent alternative pre-mRNA splicing in aggressive prostate cancers. PNAS. 10. 10.1073/pnas.1915975117. [DOI] [PMC free article] [PubMed]
- 35.Zhao J, Chang L, Gu X, Liu J, Sun B, Wei X. Systematic profiling of alternative splicing signature reveals prognostic predictor for prostate cancer. Cancer Sci. 2020;111:3020–31. doi: 10.1111/cas.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–8. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni Raghallaigh H, Eeles R. Genetic predisposition to prostate cancer: an update. Familial Cancer. 2021. 10.1007/s10689-021-00227-3. [DOI] [PMC free article] [PubMed]
- 38.Pritzlaff M, Tian Y, Reineke P, Stuenkel AJ, Allen K, Gutierrez S, et al. Diagnosing hereditary cancer predisposition in men with prostate cancer. Genet Med. 2020;22:1517–23. doi: 10.1038/s41436-020-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benafif S, Kote-Jarai Z, Eeles RA. PRACTICAL Consortium. A review of prostate cancer genome-wide association studies (GWAS) Cancer Epidemiol Biomark Prev. 2018;27:845–57. doi: 10.1158/1055-9965.EPI-16-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beebe-Dimmer JL, Kapron AL, Fraser AM, Smith KR, Cooney KA. Risk of prostate cancer associated with familial and hereditary cancer syndromes. J Clin Oncol. 2020;38:1807–13. doi: 10.1200/JCO.19.02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez D, Mateo J, Stenzinger A, Rojo F, Shiller M, Wyatt AW, et al. Practical considerations for optimising homologous recombination repair mutation testing in patients with metastatic prostate cancer. J Pathol Clin Res. 2021;7:311–25. doi: 10.1002/cjp2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merseburger AS, Waldron N, Ribal MJ, Heidenreich A, Perner S, Fizazi K et al. Genomic testing in patients with metastatic castration-resistant prostate cancer: a pragmatic guide for clinicians. Eur Urol. 2021. 10.1016/j.eururo.2020.12.039. [DOI] [PubMed]
- 43.AlDubayan SH. Considerations of multigene test findings among men with prostate cancer - knowns and unknowns. Can J Urol. 2019;26:14–16. [PubMed] [Google Scholar]
- 44.Heidegger I, Tsaur I, Borgmann H, Surcel C, Kretschmer A, Mathieu R, et al. Hereditary prostate cancer - primetime for genetic testing? Cancer Treat Rev. 2019;81:101927. doi: 10.1016/j.ctrv.2019.101927. [DOI] [PubMed] [Google Scholar]
- 45.Giri VN, Hegarty SE, Hyatt C, O’Leary E, Garcia J, Knudsen KE, et al. Germline genetic testing for inherited prostate cancer in practice: Implications for genetic testing, precision therapy, and cascade testing. Prostate. 2019;79:333–9. doi: 10.1002/pros.23739. [DOI] [PubMed] [Google Scholar]
- 46.Bono JD, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 47.Scheinberg T, Goodwin A, Ip E, Linton A, Mak B, Smith DP, et al. Evaluation of a mainstream model of genetic testing for men with prostate cancer. JCO Oncol Pr. 2021;17:e204–e216.. doi: 10.1200/OP.20.00399. [DOI] [PubMed] [Google Scholar]
- 48.Grillone K, Riillo C, Scionti F, Rocca R, Tradigo G, Guzzi PH, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter”. J Exp Clin Cancer Res. 2020;39:117. doi: 10.1186/s13046-020-01622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharova E, Grassi A, Marcer A, Ruggero K, Pinto F, Bassi P, et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br J Cancer. 2016;114:1362–6. doi: 10.1038/bjc.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber JA, Baxter DH, Zhang S, Huang DY, How Huang K, Jen, Lee M, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. PNAS. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Endzeliņš E, Melne V, Kalniņa Z, Lietuvietis V, Riekstiņa U, Llorente A, et al. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: a systematic review. Mol Cancer. 2016;15:41. doi: 10.1186/s12943-016-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, De La Rosa-Velázquez IA, González-Barrios R, Contreras-Espinosa L, et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol Ther Nucleic Acids. 2020;20:409–20. doi: 10.1016/j.omtn.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011;32:583–8. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 59.Endzeliņš E, Berger A, Melne V, Bajo-Santos C, Soboļevska K, Ābols A, et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17:730. doi: 10.1186/s12885-017-3737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. PNAS. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin H-M, Castillo L, Mahon KL, Chiam K, Lee BY, Nguyen Q, et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014;110:2462–71. doi: 10.1038/bjc.2014.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W, Meng Y, Liu N, Wen X-F, Yang T. Insights into chemoresistance of prostate cancer. Int J Biol Sci. 2015;11:1160–70. doi: 10.7150/ijbs.11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Australian Prostate Cancer BioResource. Matin F, Jeet V, Moya L, Selth LA, Chambers S, et al. A plasma biomarker panel of four microRNAs for the diagnosis of prostate cancer. Sci Rep. 2018;8:6653. doi: 10.1038/s41598-018-24424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi: 10.1016/j.steroids.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 69.Qiu M-T, Hu J-W, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–20. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 70.Pennisi E. Cell biology. lengthy RNAs earn respect as cellular players. Science. 2014;344:1072. doi: 10.1126/science.344.6188.1072. [DOI] [PubMed] [Google Scholar]
- 71.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Arriaga-Canon C, De La Rosa-Velázquez IA, González-Barrios R, Montiel-Manríquez R, Oliva-Rico D, Jiménez-Trejo F, et al. The use of long non-coding RNAs as prognostic biomarkers and therapeutic targets in prostate cancer. Oncotarget. 2018;9:20872–90. doi: 10.18632/oncotarget.25038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bussemakers MJG, Bokhoven A, van, Verhaegh GW, Smit FP, Karthaus HFM, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–9. [PubMed] [Google Scholar]
- 74.Durand X, Moutereau S, Xylinas E, de la Taille A. ProgensaTM PCA3 test for prostate cancer. Expert Rev Mol Diagn. 2011;11:137–44. doi: 10.1586/erm.10.122. [DOI] [PubMed] [Google Scholar]
- 75.Sartori DA, Chan DW. Biomarkers in prostate cancer: what’s new? Curr Opin Oncol. 2014;26:259–64. doi: 10.1097/CCO.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, Mulders PFA, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–8. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 77.Hessels D, Klein Gunnewiek JMT, van Oort I, Karthaus HFM, van Leenders GJL, van Balken B, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15. doi: 10.1016/S0302-2838(03)00201-X. [DOI] [PubMed] [Google Scholar]
- 78.Xue W-J, Ying X-L, Jiang J-H, Xu Y-H. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: a meta-analysis. J Cancer Res Ther. 2014;10:C218–221. doi: 10.4103/0973-1482.145881. [DOI] [PubMed] [Google Scholar]
- 79.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121–7. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, et al. Nomination and validation of the long noncoding RNA SChLAP1 as a risk factor for metastatic prostate cancer progression: a multi-institutional high-throughput analysis. Lancet Oncol. 2014;15:1469–80. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571. doi: 10.1038/s41467-019-09784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiong T, Li J, Chen F, Zhang F. PCAT-1: a novel oncogenic long non-coding RNA in human cancers. Int J Biol Sci. 2019;15:847–56. doi: 10.7150/ijbs.30970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–60. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Udager AM, Tomlins SA. Molecular biomarkers in the clinical management of prostate cancer. Cold Spring Harb Perspect Med. 2018;8:a030601. doi: 10.1101/cshperspect.a030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian Z, Adhya S. DNA repeat sequences: diversity and versatility of functions. Curr Genet. 2017;63:411–6. doi: 10.1007/s00294-016-0654-7. [DOI] [PubMed] [Google Scholar]
- 88.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011. 10.1126/science.1200801. [DOI] [PMC free article] [PubMed]
- 90.Maloy S, Hughes K. Brenner’s encyclopedia of genetics. San Diego, CA: Academic Press; 2013.
- 91.Solovyov A, Vabret N, Arora KS, Snyder A, Funt SA, Bajorin DF, et al. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Rep. 2018;23:512–21. doi: 10.1016/j.celrep.2018.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rezaei SD, Hayward JA, Norden S, Pedersen J, Mills J, Hearps AC, et al. HERV-K gag RNA and protein levels are elevated in malignant regions of the prostate in males with prostate cancer. Viruses. 2021;13:449. doi: 10.3390/v13030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wallace TA, Downey RF, Seufert CJ, Schetter A, Dorsey TH, Johnson CA, et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis. 2014;35:2074–83. doi: 10.1093/carcin/bgu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nadine Houede, Piazza PV, Pourquier P. LINE-1 as a therapeutic target for castration-resistant prostate cancer. Front Biosci. 2018. 10.2741/4644. [DOI] [PubMed]
- 95.Morganti S, Tarantino P, Ferraro E, D’Amico P, Duso BA, Curigliano G. Next Generation Sequencing (NGS): a revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol. 2019;1168:9–30. doi: 10.1007/978-3-030-24100-1_2. [DOI] [PubMed] [Google Scholar]
- 96.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17:257–71. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berglund E, Maaskola J, Schultz N, Friedrich S, Marklund M, Bergenstråhle J, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9:2419. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50:645–51. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang W, Randhawa R, Jain P, Iczkowski KA, Hu R, Hubbard S, et al. Development and validation of an artificial intelligence-powered platform for prostate cancer grading and quantification. JAMA Netw Open. 2021;4:e2132554. doi: 10.1001/jamanetworkopen.2021.32554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Russell S, Norvig P. Artificial intelligence: a modern approach. Harlow, England: Pearson Education Limited; 2016.
- 104.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bishop CM. Pattern recognition and machine learning. Springer; 2006. [Google Scholar]
- 106.Goecks J, Jalili V, Heiser LM, Gray JW. How machine learning will transform biomedicine. Cell. 2020;181:92–101. doi: 10.1016/j.cell.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shilo S, Rossman H, Segal E. Axes of a revolution: challenges and promises of big data in healthcare. Nat Med. 2020;26:29–38. doi: 10.1038/s41591-019-0727-5. [DOI] [PubMed] [Google Scholar]
- 108.Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, et al. Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal. 2014;18:359–73. doi: 10.1016/j.media.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16:321–32. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–53. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghiam AF, Vesprini D, Liu SK. Long non-coding RNAs: new frontiers for advancing personalized cancer medicine in prostate cancer. Transl Androl Urol. 2017;6:326–30. doi: 10.21037/tau.2017.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Isaev K, Jiang L, Wu S, Lee CA, Watters V, Fort V, et al. Pan-cancer analysis of non-coding transcripts reveals the prognostic onco-lncRNA HOXA10-AS in gliomas. Cell Rep. 2021;37:109873. doi: 10.1016/j.celrep.2021.109873. [DOI] [PubMed] [Google Scholar]
- 113.Zhang X, Li T, Wang J, Li J, Chen L, Liu C. Identification of cancer-related long non-coding RNAs using XGBoost with high accuracy. Front Genet. 2019;10:735. doi: 10.3389/fgene.2019.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bertoli G, Cava C, Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int J Mol Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou H, Cao H, Matyunina L, Shelby M, Cassels L, McDonald JF, et al. MEDICASCY: a machine learning approach for predicting small-molecule drug side effects, indications, efficacy, and modes of action. Mol Pharm. 2020;17:1558–74. doi: 10.1021/acs.molpharmaceut.9b01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tolkach Y, Dohmgörgen T, Toma M, Kristiansen G. High-accuracy prostate cancer pathology using deep learning. Nat Mach Intell. 2020;2:411–8. doi: 10.1038/s42256-020-0200-7. [DOI] [Google Scholar]
- 118.Snow O, Lallous N, Ester M, Cherkasov A. Deep learning modeling of androgen receptor responses to prostate cancer therapies. Int J Mol Sci. 2020;21:E5847. doi: 10.3390/ijms21165847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fujita K, Nonomura N. Urinary biomarkers of prostate cancer. Int J Urol. 2018;25:770–9. doi: 10.1111/iju.13734. [DOI] [PubMed] [Google Scholar]
- 120.Dong L, Huang C-Y, Johnson EJ, Yang L, Zieren RC, Horie K, et al. High-throughput simultaneous mRNA profiling using ncounter technology demonstrates that extracellular vesicles contain different mRNA transcripts than their parental prostate cancer cells. Anal Chem. 2021;93:3717–25. doi: 10.1021/acs.analchem.0c03185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McKiernan J, Donovan MJ, O’Neill V, Bentink S, Noerholm M, Belzer S, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2:882–9. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 122.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–56. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 123.Suzuki Y. Advent of a new sequencing era: long-read and on-site sequencing. J Hum Genet. 2020;65:1–1. doi: 10.1038/s10038-019-0683-4. [DOI] [PubMed] [Google Scholar]
- 124.Handa S, Hans B, Goel S, Bashorun HO, Dovey Z, Tewari A. Immunotherapy in prostate cancer: current state and future perspectives. Ther Adv Urol. 2020;12:1756287220951404. doi: 10.1177/1756287220951404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pineda-Jaramillo JD. A review of Machine Learning (ML) algorithms used for modeling travel mode choice. DYNA. 2019;86:32–41. doi: 10.15446/dyna.v86n211.79743. [DOI] [Google Scholar]
- 126.Kelly BD, Miller N, Sweeney KJ, Durkan GC, Rogers E, Walsh K, et al. A circulating MicroRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015;4:1369–79. doi: 10.3390/jcm4071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou H, Zhu X. MicroRNA-21 and microRNA-30c as diagnostic biomarkers for prostate cancer: a meta-analysis. Cancer Manag Res. 2019;11:2039–50. doi: 10.2147/CMAR.S189026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ambrozkiewicz F, Karczmarski J, Kulecka M, Paziewska A, Cybulska M, Szymanski M, et al. Challenges in cancer biomarker discovery exemplified by the identification of diagnostic MicroRNAs in prostate tissues. Biomed Res Int. 2020;2020:9086829. doi: 10.1155/2020/9086829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li W, Dong Y, Wang KJ, Deng Z, Zhang W, Shen HF. Plasma exosomal miR-125a-5p and miR-141-5p as non-invasive biomarkers for prostate cancer. Neoplasma. 2020;67:1314–8. doi: 10.4149/neo_2020_191130N1234. [DOI] [PubMed] [Google Scholar]
- 130.Kolluru V, Chandrasekaran B, Tyagi A, Dervishi A, Ankem M, Yan X, et al. miR-301a expression: diagnostic and prognostic marker for prostate cancer. Urologic Oncol Semin Original Investig. 2018;36:503.e9–503.e15. doi: 10.1016/j.urolonc.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 131.Larne O, Martens-Uzunova E, Hagman Z, Edsjö A, Lippolis G, den Berg MSV, et al. miQ-a novel microRNA based diagnostic and prognostic tool for prostate cancer. Int J Cancer. 2013;132:2867–75. doi: 10.1002/ijc.27973. [DOI] [PubMed] [Google Scholar]
- 132.Nayak B, Khan N, Garg H, Rustagi Y, Singh P, Seth A, et al. Role of miRNA-182 and miRNA-187 as potential biomarkers in prostate cancer and its correlation with the staging of prostate cancer. Int Braz J Urol. 2020;46:614–23. doi: 10.1590/s1677-5538.ibju.2019.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wei W, Leng J, Shao H. Wang W. MiR-1, a potential predictive biomarker for recurrence in prostate cancer after radical prostatectomy. Am J Med Sci. 2017;353:315–9. doi: 10.1016/j.amjms.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 134.Nam RK, Amemiya Y, Benatar T, Wallis CJD, Stojcic-Bendavid J, Bacopulos S, et al. Identification and validation of a five microRNA signature predictive of prostate cancer recurrence and metastasis: a cohort study. J Cancer. 2015;6:1160–71. doi: 10.7150/jca.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song C-J, Chen H, Chen L-Z, Ru G-M, Guo J-J, Ding Q-N. The potential of microRNAs as human prostate cancer biomarkers: a meta-analysis of related studies. J Cell Biochem. 2018;119:2763–86. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang H-L, Yang L-F, Zhu Y, Yao X-D, Zhang S-L, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–31. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 137.Hatano K, Kumar B, Zhang Y, Coulter JB, Hedayati M, Mears B, et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015;43:4075–86. doi: 10.1093/nar/gkv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–12. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 139.Groskopf J, Aubin SMJ, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–95. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 140.Lemos AEG, Ferreira LB, Batoreu NM, de Freitas PP, Bonamino MH, Gimba ERP. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumour Biol. 2016;37:11339–48. doi: 10.1007/s13277-016-5012-3. [DOI] [PubMed] [Google Scholar]
- 141.Kok JB, de, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–8. [PubMed] [Google Scholar]
- 142.Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091–102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045–55. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yan Y, Liu J, Xu Z, Ye M, Li J. lncRNA PCAT14 is a diagnostic marker for prostate cancer and is associated with immune cell infiltration. Dis Markers. 2021;2021:9494619. doi: 10.1155/2021/9494619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee B, Mazar J, Aftab MN, Qi F, Shelley J, Li J-L, et al. Long noncoding RNAs as putative biomarkers for prostate cancer detection. J Mol Diagn. 2014;16:615–26. doi: 10.1016/j.jmoldx.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang W, Ren S-C, Shi X-L, Liu Y-W, Zhu Y-S, Jing T-L, et al. A novel urinary long non-coding RNA transcript improves diagnostic accuracy in patients undergoing prostate biopsy. Prostate. 2015;75:653–61. doi: 10.1002/pros.22949. [DOI] [PubMed] [Google Scholar]
- 147.Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. 2016;70:549–52. doi: 10.1016/j.eururo.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu S, Yi X-M, Tang C-P, Ge J-P, Zhang Z-Y, Zhou W-Q. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol Rep. 2016;36:10–22. doi: 10.3892/or.2016.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao R, Sun F, Bei X, Wang X, Zhu Y, Jiang C, et al. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate. 2017;77:1107–17. doi: 10.1002/pros.23367. [DOI] [PubMed] [Google Scholar]
- 150.Xu T, Liu C-L, Li T, Zhang Y-H, Zhao Y-H. LncRNA TUG1 aggravates the progression of prostate cancer and predicts the poor prognosis. Eur Rev Med Pharm Sci. 2019;23:4698–705. doi: 10.26355/eurrev_201906_18062. [DOI] [PubMed] [Google Scholar]
- 151.Li C, Hu J, Hu X, Zhao C, Mo M, Zu X, et al. LncRNA SNHG9 is a prognostic biomarker and correlated with immune infiltrates in prostate cancer. Transl Androl Urol. 2021;10:215–26. doi: 10.21037/tau-20-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–8. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu Y-H, Deng J-L, Wang G, Zhu Y-S. Long non-coding RNAs in prostate cancer: functional roles and clinical implications. Cancer Lett. 2019;464:37–55. doi: 10.1016/j.canlet.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 154.Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75:693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]
- 155.Shan Y, Huang Y, Lee AM, Mentzer J, Ling A, Huang RS. A long noncoding RNA, GAS5 can be a biomarker for docetaxel response in castration resistant prostate cancer. Front Oncol. 2021;11:1483. doi: 10.3389/fonc.2021.675215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia Y, et al. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2020;65:109422. doi: 10.1016/j.cellsig.2019.109422. [DOI] [PubMed] [Google Scholar]
- 158.Ma Y, Fan B, Ren Z, Liu B, Wang Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. Onco Targets Ther. 2019;12:5485–97. doi: 10.2147/OTT.S197009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bai T, Liu Y, Li B. LncRNA LOXL1-AS1/miR-let-7a-5p/EGFR-related pathway regulates the doxorubicin resistance of prostate cancer DU-145 cells. IUBMB Life. 2019;71:1537–51. doi: 10.1002/iub.2075. [DOI] [PubMed] [Google Scholar]
- 160.Hofer P, Zerelles J, Baierl A, Madersbacher S, Schatzl G, Maj-Hes A, et al. MNS16A tandem repeat minisatellite of human telomerase gene and prostate cancer susceptibility. Mutagenesis. 2013;28:301–6. doi: 10.1093/mutage/get003. [DOI] [PubMed] [Google Scholar]
- 161.Hameed IH, Jebor MA, Kareem MA. Allelic frequencies for the seventeen Y-STR loci observed in Iraqi male patients with prostate cancer. AJB. 2015;14:1252–60. [Google Scholar]
- 162.Lai J, Moya L, An J, Hoffman A, Srinivasan S, Panchadsaram J, et al. A microsatellite repeat in PCA3 long non-coding RNA is associated with prostate cancer risk and aggressiveness. Sci Rep. 2017;7:16862. doi: 10.1038/s41598-017-16700-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nelson KA, Witte JS. Androgen receptor CAG repeats and prostate cancer. Am J Epidemiol. 2002;155:883–90. doi: 10.1093/aje/155.10.883. [DOI] [PubMed] [Google Scholar]
- 164.Weng H, Li S, Huang J-Y, He Z-Q, Meng X-Y, Cao Y, et al. Androgen receptor gene polymorphisms and risk of prostate cancer: a meta-analysis. Sci Rep. 2017;7:40554. doi: 10.1038/srep40554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McIntyre MH, Kantoff PW, Stampfer MJ, Mucci LA, Parslow D, Li H, et al. Prostate cancer risk and ESR1 TA, ESR2 CA repeat polymorphisms. Cancer Epidemiol Biomark Prev. 2007;16:2233–6. doi: 10.1158/1055-9965.EPI-07-0481. [DOI] [PubMed] [Google Scholar]
- 166.Rose AM, Krishan A, Chakarova CF, Moya L, Chambers SK, Hollands M, et al. MSR1 repeats modulate gene expression and affect risk of breast and prostate cancer. Ann Oncol. 2018;29:1292–303. doi: 10.1093/annonc/mdy082. [DOI] [PubMed] [Google Scholar]
- 167.Leach FS. Microsatellite instability and prostate cancer: clinical and pathological implications. Curr Opin Urol. 2002;12:407–11. doi: 10.1097/00042307-200209000-00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Main Cancer Susceptibility Genes with Therapeutic Implications for Prostate Cancer