Abstract
Human ABCB6 is an ATP-binding cassette transporter that regulates heme biosynthesis by translocating various porphyrins from the cytoplasm into the mitochondria. Here we report the cryo-electron microscopy (cryo-EM) structures of human ABCB6 with its substrates, coproporphyrin III (CPIII) and hemin, at 3.5 and 3.7 Å resolution, respectively. Metal-free porphyrin CPIII binds to ABCB6 within the central cavity, where its propionic acids form hydrogen bonds with the highly conserved Y550. The resulting structure has an overall fold similar to the inward-facing apo structure, but the two nucleotide-binding domains (NBDs) are slightly closer to each other. In contrast, when ABCB6 binds a metal-centered porphyrin hemin in complex with two glutathione molecules (1 hemin: 2 glutathione), the two NBDs end up much closer together, aligning them to bind and hydrolyze ATP more efficiently. In our structures, a glycine-rich and highly flexible “bulge” loop on TM helix 7 undergoes significant conformational changes associated with substrate binding. Our findings suggest that ABCB6 utilizes at least two distinct mechanisms to fine-tune substrate specificity and transport efficiency.
Keywords: ABCB6, ATP-binding cassette transporter, cryo-electron microscopy, glutathione, porphyrin
INTRODUCTION
Heme (iron protoporphyrin IX) serves as an essential cofactor for signal transduction, oxygen transport, regulation of transcription and translation, drug detoxification, and microRNA processing (Chiabrando et al., 2014; Mense and Zhang, 2006). ATP-binding cassette superfamily B member 6 (ABCB6) translocates the heme precursor coproporphyrinogen III from the cytoplasm into mitochondria, where it is converted to protoporphyrin IX (PPIX), and ferrous iron (Fe2+) is added to the PPIX ring for heme biosynthesis (Krishnamurthy et al., 2006). In addition to its mitochondrial distribution, ABCB6 is also found in the Golgi apparatus (Tsuchida et al., 2008), lysosomes (Kiss et al., 2012) and the plasma membranes (Paterson et al., 2007). ABCB6 is involved in diverse physiological processes such as protection from oxidative stress, tolerance to toxic heavy metals and phenylhydrazine (Chavan et al., 2011; Lynch et al., 2009; Rakvács et al., 2019; Ulrich et al., 2012), acquired multidrug resistance (Heimerl et al., 2007; Park et al., 2006; Yasui et al., 2004), iron homeostasis (Mitsuhashi et al., 2000), and regulation of cellular hemoproteins (Lynch et al., 2009). It is highly expressed in heart, liver, intestine, testis, skeletal muscle, and skin (Chavan et al., 2013; Krishnamurthy et al., 2006; Zhang et al., 2013). Mutations of ABCB6 are associated with various genetic diseases including porphyria and dyschromatosis universalis hereditaria (DUH, a disorder of pigment metabolism) (Zhang et al., 2013), pseudohyperkalemia (chronic renal failure) (Andolfo et al., 2013), coloboma (a developmental defect of the eye) (Wang et al., 2012), and atherosclerosis (Soehnlein, 2014). A high level of ABCB6 is also found in red blood cells where its absence defines the Langereis blood group system (Lan) (Helias et al., 2012). Overexpression of ABCB6 in various tumor cells has been shown to confer drug resistance by reducing the accumulation of chemotherapeutic agents such as paclitaxel, vincristine, 7-ethyl-10-hydroxy-camptothecin (SN-38), and 5-fluorouracil (5-FU), suggesting its physiological and clinical importance (Januchowski et al., 2013; Minami et al., 2014; Park et al., 2006; Varatharajan et al., 2017; Yasui et al., 2004).
In humans, 48 members of the ABC transporter family have been identified, divided into seven subgroups, A to G (Dean et al., 2001; Vasiliou et al., 2009). ABCB6 is a Type IV homodimeric half-transporter of 842 amino acids (Thomas et al., 2020). Each monomer consists of a transmembrane domain (TMD) and a cytoplasmic nucleotide-binding domain (NBD) with an additional N-terminal TMD (TMD0). As expected for an ABC transporter, the two TMDs of the homodimer are responsible for binding various substrates and serve as the translocation pathway across the membrane, while the two NBDs are essential for ATP hydrolysis, which provides the energy for substrate transport (Chavan et al., 2013; Haffke et al., 2010; Polireddy et al., 2012). Although the exact biological role of the TMD0 is controversial, researchers predict that it is involved in protein maturation (Fukuda et al., 2011) or subcellular localization (Kiss et al., 2015).
Recently, cryo-electron microscopy (cryo-EM) structures of human ABCB6 have been reported, with the enzyme in its apo form and in complex with ATP (Song et al., 2021; Wang et al., 2020). However, understanding the mechanisms of substrate recognition and specificity is still challenging due to the lack of substrate-bound structures. Here we present the cryo-EM structures of human ABCB6 in the presence of the porphyrin substrates, coproporphyrin III (CPIII) and hemin, at 3.5 and 3.7 Å resolution, respectively. Our structures reveal that ABCB6 utilizes at least two distinct mechanisms to bind porphyrin substrates depending on the presence or absence of a central metal ion in the porphyrin. Moreover, when taken with the results of functional analyses, our structures demonstrate that a long intervening loop present on TM helix 7 undergoes substrate-specific conformational rearrangements to assist binding and/or transport.
MATERIALS AND METHODS
Cloning, expression, and purification
The core region of human ABCB6 (ABCB6-∆TMD0, residues 206-842) was cloned into the pVL1393 baculovirus transfer vector (BD Biosciences, USA) containing thrombin-cleavable enhanced green fluorescence protein (eGFP) and a decahistidine (10XHis) tag at the C-terminus. Site-directed mutations were generated using overlap polymerase chain reactions, and verified by DNA sequencing. Recombinant virus was produced in Spodoptera frugiperda (Sf9) cells using BestBac 2.0 linearized baculovirus DNA (Expression Systems, USA) and Cellfectin II transfection reagent (Gibco, USA). The proteins were expressed in High Five (Hi5) cells. After harvesting at 72 h post-infection, the cells were broken by sonication in a lysis buffer containing 20 mM HEPES pH 7.0, 300 mM NaCl, 10 mM MgCl2, 10% glycerol, 40 µg/ml DNase I and 1 mM phenylmethylsulfonyl fluoride (PMSF). Proteins were solubilized with 2% (w/v) n-dodecyl-β-D-maltopyranoside (DDM; Anatrace, USA) and 0.2% (w/v) cholesteryl hemisuccinate (CHS; Anatrace) for 2 h. Insoluble cell debris was removed by centrifugation at 300,000 × g for 30 min and the supernatant was loaded onto anti-GFP DARPin-based affinity resin (Hansen et al., 2017). After thorough washing, the C-terminal tag was removed by on-column thrombin cleavage. The eluted proteins were further purified with a Superdex 200 GL 10/300 column (GE Healthcare, USA) in a buffer containing 20 mM HEPES pH 7.0, 200 mM NaCl, 0.056% (w/v) 6-cyclohexyl-1-hexyl-β-D-maltoside (Cymal-6; Anatrace) and 0.0056% (w/v) CHS. The ABCB6-∆TMD0 protein was also reconstituted into peptidisc by the on-beads method as previously described (Angiulli et al., 2020; Carlson et al., 2018; Zeytuni et al., 2020). Briefly, after DDM-solubilization, the supernatant was loaded onto anti-GFP DARPin resin, followed by 10 column volumes of wash buffer containing 20 mM HEPES pH 7.0, 200 mM NaCl and 0.5 mg/ml peptidisc (PEPTIDISC BIOTECH, Canada). The eluate from on-column thrombin cleavage was further purified by Superdex 200 gel filtration chromatography in a buffer containing 20 mM HEPES pH 7.0 and 200 mM NaCl. All purification steps were performed on ice or at 4°C. ABCB6-∆TMD0 and all other mutants exhibited similar expression levels and purification behavior (Supplementary Fig. S1).
ATP hydrolysis assay
ATP hydrolysis was assayed by the molybdate (Chavan et al., 2013; Chifflet et al., 1988; Manolaridis et al., 2018) or NADH-coupled method (Scharschmidt et al., 1979) as previously described, with slight modifications. Briefly, in the molybdate method, the reaction was initiated by addition of 1 mM ATP to 50 µl of reaction buffer (50 mM HEPES pH 7.0, 70 mM KCl, 10 mM MgCl2, 0.056% (w/v) Cymal-6 and 0.0056% (w/v) CHS) in the presence of the purified protein (1.3 µM). For substrate-stimulated ATPase assays, protein samples were incubated with varying concentrations of substrates (Sigma-Aldrich, USA) for 30 min at 37°C. The reaction was quenched with 50 µl of 5% (w/v) sodium dodecyl sulfate (SDS). A 100 µl aliquot of a solution of 5 mM ammonium molybdate, 2 mM zinc acetate and 5.5% (w/v) ascorbic acid was added to each sample and further incubated for 20 min. Pi release was determined by a colorimetric method at 850 nm, with potassium phosphate as a standard.
In the NADH-coupled assay, ATP hydrolysis was measured at 37°C every 10 s for 3 min by monitoring the loss of absorbance at 340 nm (εM = 6,220 cm–1M–1). Briefly, the reaction was initiated by adding 1 µM protein and varying concentrations of substrate to 70 µl of reaction buffer containing 50 mM HEPES pH 8.0, 60 µg/ml pyruvate kinase, 32 µg/ml lactate dehydrogenase, 4 mM phosphoenolpyruvate, 0.3 mM NADH, 2 mM ATP, 10 mM MgCl2, 0.056% (w/v) Cymal-6, and 0.0056% (w/v) CHS. The steady-state kinetic parameters for ATP hydrolysis were determined by fitting the observed initial rates of hydrolysis to the Michaelis–Menten equation using the program GraphPad Prism 7.0.
Proteoliposome preparation
A mixture of porcine brain polar lipid extract dissolved in chloroform and CHS (Avanti Polar Lipids, USA) at a ratio of 4:1 (w/w) was dried under a gentle stream of nitrogen. The lipid thin film was placed under a desiccator overnight to completely remove residual chloroform and then hydrated in reconstitution buffer containing 20 mM HEPES pH 7.5 and 100 mM KCl. After sonication, the lipid mixture was extruded 15 times through a 200 nm polycarbonate membrane filter (Avanti Polar Lipids) to form unilamellar vesicles and then destabilized with 0.056% (w/v) Cymal-6. The purified protein and lipid suspension were mixed in a ratio of 1:25 (w/w). After incubation at 4°C for 2 h, the detergent was removed with Bio-Beads SM-2 resin (Bio-Rad, USA). Proteoliposomes were collected by centrifugation at 200,000 × g for 1 h and resuspended in reconstitution buffer for transport assays. Protein-free liposomes were prepared in parallel.
Transport assay
Transport assay was performed based on the previous study with some modifications (Chavan et al., 2013; Krishnamurthy et al., 2006; Ulrich et al., 2012). Briefly, transport reactions were initiated by addition of proteoliposomes to reaction buffer containing 50 mM HEPES pH 8.0, 70 mM KCl, 10 mM MgCl2 and 2 mM ATP in the presence and absence of 0.2 mM CPIII. After incubation at 37°C for 30 min, the reaction was stopped by cooling the sample to 4°C. Proteoliposomes were collected by centrifugation at 200,000 × g for 30 min, washed twice, and lysed with 10% (w/v) SDS. The fluorescent intensities of CPIII were measured with lasers of 405 nm excitation and 630 nm emission wavelengths, respectively (Synergy H1m; BioTek, USA). The amount of transported CPIII was calculated by making a standard curve with varying concentrations of CPIII prepared under assay conditions.
Microscale thermophoresis (MST) assay
The affinities of CPIII for ABCB6-∆TMD0 and other mutant proteins were measured by MST, based on the changes in initial fluorescence, as described previously (Krossa et al., 2018). Briefly, 5 µM of fluorophore CPIII was added to serial 2-fold dilutions of each protein from 66.6 µM to 2.0 nM. The mixtures were incubated for 20 min at room temperature and loaded onto Monolith NT.115 standard-treated capillaries (NanoTemper Technologies, Germany). Changes of initial fluorescence intensity were monitored with a Monolith NT.115 pico device, and plotted against protein concentration to calculate Kd values, and the results were analyzed with MO.Affinity Analysis software.
Molecular docking
To identify the mode of binding of hemin to ABCB6-∆TMD0, in silico molecular docking was performed, with the GLIDE (Grid-based Ligand Docking with Energetic) module implemented in the Schrödinger Suite (release 2021-3), which uses the Maestro v12.9.123 graphical user interface (Friesner et al., 2004; 2006). As a benchmark test, we used the crystal structure of Pseudomonas aeruginosa hemophore HasA (PDB ID 4O6U, 0.8 Å resolution), which contains a heme ligand (Kumar et al., 2014). Model structures of protein and heme were prepared using the “Protein Preparation Workflow” menu (Preprocess → Optimize H-bond Assignments → Clean Up) with default options. Standard Precision (SP) and Extra Precision (XP) algorithms were used to dock the heme ligand onto the protein grid, with the flexible docking option turned on. The best docking poses were very similar to a co-crystallized ligand (<0.2 Å RMSD [root mean square deviation] for all atoms), suggesting that these docking methods can provide reliable structural models. Adopting the same workflow (Protein Preparation and Ligand Preparation → Receptor Grid Generation → Docking) and options in each procedure, we carried out a molecular docking analysis to dock hemin onto the cryo-EM structure of ABCB6-∆TMD0. A receptor grid with an enclosing box (26 Å × 26 Å × 26 Å) was generated centrally between two glutathione (GSH) molecules, and the hemin was docked into the predicted binding site using SP and XP methods. After visual inspection, the top poses with –5.979, and –5.698 kcal/mol scores, respectively, were selected.
Cryo-EM sample preparation and data acquisition
The purified protein was incubated with 0.2 mM CPIII or 1 mM Hemin:GSH for 30 min at 4°C. Cryo-EM grids were prepared by applying 3 µl of protein at 5 mg/ml onto glow-discharged quantifoil 300-mesh Au R1.2/1.3 holey carbon grids (EM Science, USA). The grids were blotted once for 3-4 s and rapidly cryocooled in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific, USA) with the chamber maintained at 4°C and ~100% humidity.
Cryo-EM data of ABCB6-∆TMD0 in CPIII-bound state was collected using a 300 kV Titan Krios cryo-electron microscope (Thermo Fisher Scientific) with a K3 BioQuantum direct electron detector (Gatan, USA) in super resolution mode. The slit width of the energy filter was set to 20 eV. A total of 5,027 images was recorded by EPU software at a nominal magnification of 130,000×, corresponding to a pixel size of 0.328 Å (physical pixel size 0.656 Å). Each movie was dose-fractionated to 50 frames with a total exposure time of 2 s. The electron dose rate was 15 e–/pix/s (1.0 e–/Å2/frame), leading to a total dose of 50 e–/Å2. The defocus range was from –0.8 to –2.0 µm with 0.2 steps.
For ABCB6-∆TMD0 in hemin:GSH-bound state, the dataset was collected using a 200 kV Talos Arctica cryo-electron microscope (Thermo Fisher Scientific) equipped with a K3 BioQuantum direct electron detector. The slit width of the energy filter was set to 20 eV. A total of 4,066 images was recorded by EPU software at a nominal magnification of 100,000×, corresponding to a pixel size of 0.83 Å. Each movie was dose-fractionated to 50 frames with a total exposure time of 3.52 s. The electron dose rate was 10.36 e–/pix/s(1.0 e–/Å2/frame), leading to a total dose of 50 e-/Å2. The defocus range was set to vary from –0.8 to –2.6 µm with 0.2 steps.
Data processing
The dataset of CPIII-bound structure was processed using cryoSPARC v2.15.0 (Punjani et al., 2017). The recorded super-resolution images were gain-normalized and motion-corrected by dose weighting and binned twice by Fourier cropping. Defocus values were estimated by patch-based contrast transfer function (CTF) estimation. Initially, a small subset of micrographs was used for automated particle picking in template-free manner using blob picker, followed by extraction and 2D classification. Good classes were then used for template-based particle picking from the complete set of micrographs. Ab-initio models were generated from the best 2D classes, which were subjected to two rounds of 3D heterogeneous refinement. The particles from the best 3D classes were further subjected to homogeneous 3D refinement and non-uniform refinement. The final map of ABCB6-∆TMD0 bound to CPIII was refined to 3.5 Å using C1 symmetry based on the gold-standard Fourier shell correlation (FSC) criterion of 0.143 (Henderson et al., 2012). The map was sharpened with automatically calculated B-factor of 85.0 Å2 (Supplementary Figs. S2-S4).
For hemin:GSH-bound structure, beam-induced motion corrections of movie stacks were performed using MotionCor2 to generate dose-weighted micrographs (Zheng et al., 2017). CTF parameters were estimated using CTFFIND4 (Rohou and Grigorieff, 2015). Low-quality micrographs showing low resolution, high defocus and high astigmatism were removed, resulting in a total of 3,965 images for further data processing in RELION v3.1 (Scheres et al., 2012). Approximately 4,000 particles were picked using projections of our 3D reference model and extracted for initial 2D classification. Good classes were used as templates for automated particle picking. After multiple rounds of 2D classification, best groups of particles were subjected to generate an initial 3D model, which was subsequently used as a starting point for 3D classification. One of six 3D classes contained 44% of all the particles (corresponding to 374,957 particles) and gave stronger signals for hemin and GSH. This class was subjected to 3D refinement and post-processing, resulting in a 4.2 Å resolution map. The map was further improved by local CTF refinement and Bayesian polishing in RELION, yielding a map at a resolution of 4.0 Å. The refined particles and map were imported to cryoSPARC (Punjani et al., 2017), where heterogeneous and homogeneous 3D refinement, and non-uniform refinement were performed. The final maps were refined to 3.9 Å using C1 symmetry and 3.7 Å using C2 symmetry. The map was sharpened with automatically calculated B-factors of 178.3 Å2. The local resolution was determined from the two half-maps in cryoSPARC (Supplementary Figs. S5-S7). All details regarding the two datasets are summarized in Supplementary Table S1.
Model building and refinement
The initial model of CPIII-bound ABCB6-∆TMD0 structure was built using the crystal structures of bacterial Atm1 TMD (PDB ID 4MRP) (Lee et al., 2014) and human ABCB6 NBD domains (PDB ID 3NH6) (Haffke et al., 2010) as templates, and was fit into the post-processed maps using UCSF Chimera (Goddard et al., 2007; Pettersen et al., 2004). After a cycle of rigid body fitting and morphing using the phenix.real_space_refine program (Adams et al., 2010), a model of ABCB6-∆TMD0 was built manually according to the density map using COOT (Emsley and Cowtan, 2004), and this was subsequently used as an initial model for the Hemin:GSH dataset. While the cryo-EM density of TMDs was of sufficiently high resolution to reveal side chains, the densities for the NBDs were poorly defined due to their intrinsic heterogeneity. Therefore, the crystal structure of human ABCB6 NBD was docked into the density. Models were further improved by multiple cycles of PHENIX real space refinement with secondary structure restraints, rotamer restraints, Ramachandran restraints, and non-crystallographic symmetry (NCS) restraints (Adams et al., 2010). The refined model was validated using MolProbity (Chen et al., 2010). All figures in the manuscript were produced using PyMOL (https://pymol.org/2/), UCSF Chimera (Pettersen et al., 2004), and Chimera X (http://www.rbvi.ucsf.edu/chimera/).
Data availability
The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-30790 (CPIII-bound state) and EMD-30791 (hemin:GSH-bound state). The atomic coordinates have been deposited in the Protein Data Bank under accession codes 7DNY (CPIII-bound state) and 7DNZ (hemin:GSH-bound state).
RESULTS
Protein preparation for structural and functional studies
For cryo-EM study of human ABCB6, the protein was expressed in Hi5 insect cells and purified in peptidisc (Angiulli et al., 2020; Carlson et al., 2018) or detergent micelles (6-cyclohexyl-1-hexyl-β-D-maltoside, Cymal-6) supplemented with 0.0056% (w/v) CHS. We originally, expressed the full-length protein containing both the TMD0 and core region, but the accumulation of a partially proteolyzed product was detected during purification, generating a heterogeneous mixture of molecules (Supplementary Fig. S8A). SDS-PAGE analysis indicated that the molecular weight of the lower band (~75 kDa) was roughly consistent with that of the truncated mutant containing only the core region (residues 206-842). In a previous study, the TMD0 was shown to be essential for lysosomal targeting, but dispensable for the ATPase and substrate transport activity (Kiss et al., 2015). Therefore, to increase the homogeneity of the protein, we constructed a truncated mutant of ABCB6 in which the entire TMD0 region was eliminated (Supplementary Figs. S8B and S8C). In this article, unless otherwise specified, we refer to this truncated mutant protein as ABCB6-∆TMD0.
Effect of glutathione on porphyrin binding
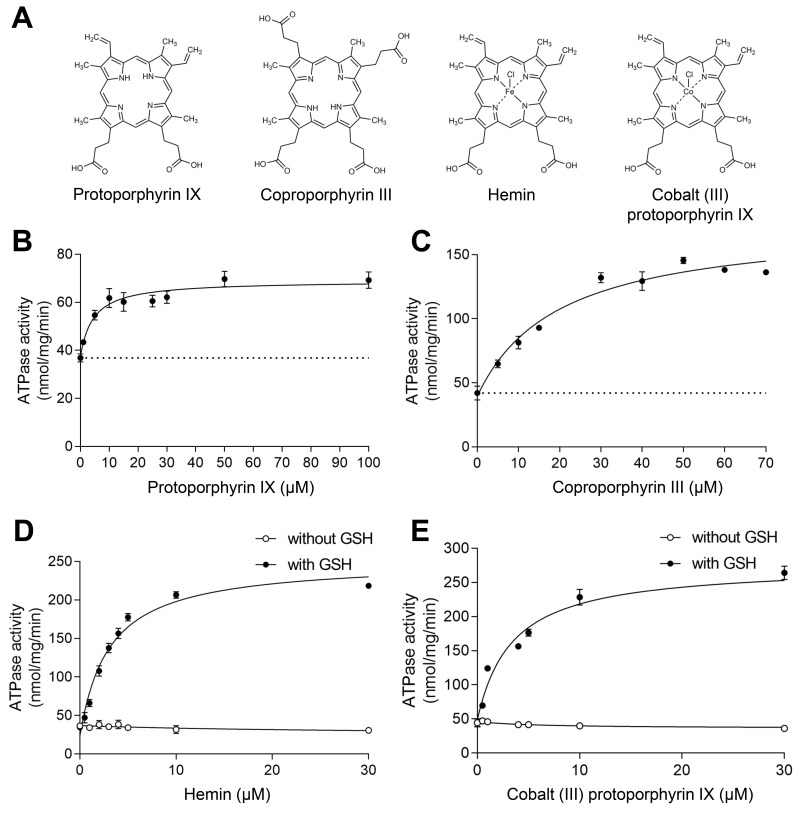
In order to identify the best substrates for structural studies of ABCB6-∆TMD0, in vitro ATP hydrolysis assays were carried out on the detergent-purified proteins using various porphyrins (Fig. 1A). In agreement with the results of previous studies (Song et al., 2021; Wang et al., 2020), our data indicated that at saturating concentrations (100 µM PPIX or 50 µM CPIII), the metal-free porphyrins modestly stimulated the ATPase activity of ABCB6-∆TMD0 approximately 2- to 3-fold (Figs. 1B and 1C). By contrast, metal-centered porphyrins, such as hemin (ferric chloride heme) and cobalt (III) PPIX, markedly stimulated the ATPase activity of ABCB6-∆TMD0 approximately 6-fold only when the cellular antioxidant glutathione (GSH; γ-glu-cys-gly) was included in the reaction buffer (Figs. 1D and 1E).
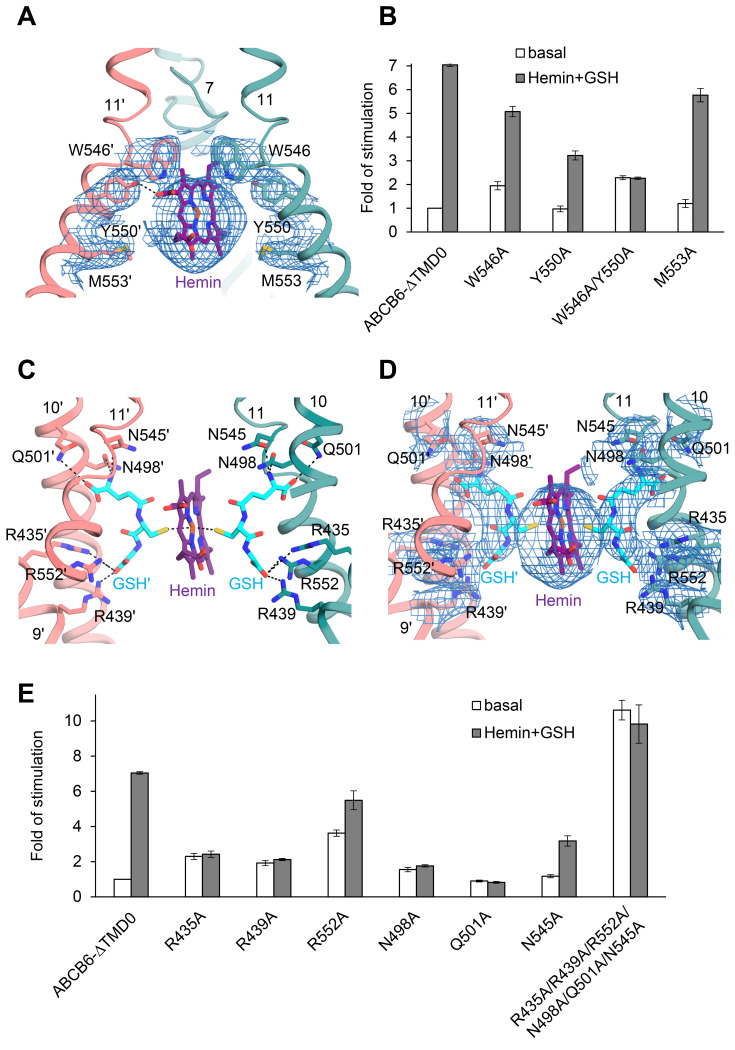
Fig. 1. Functional characterization.
(A) The chemical structures of various porphyrins used in this study. (B) NADH-coupled ATPase activity of the ABCB6-∆TMD0 as a function of PPIX concentration. The Km of PPIX was found to be 4.5 ± 1.2 µM, and the maximal ATPase activity was 69 ± 1.5 nmol/mg/min (or 9.7 min–1). The dashed line indicates the basal ATPase activity in the absence of substrate. Data points represent mean ± SD of triplicate measurements. (C) ATPase activity of the ABCB6-∆TMD0 as a function of CPIII concentration. The Km of CPIII was found to be 19 ± 3.7 µM, and the maximal ATPase activity was 175 ± 7.5 nmol/mg/min (or 25 min–1). (D) ATPase activity of ABCB6-∆TMD0 as a function of hemin concentration with and without 1 mM GSH. The Km for hemin in conjunction with GSH was determined to be 3 ± 0.3 µM and the maximal ATPase activity was 251 ± 7.8 nmol/mg/min (or 36 min–1). (E) ATPase activity of ABCB6-∆TMD0 as a function of cobalt (III) PPIX (Co-PPIX) concentration with and without 1 mM GSH. The Km for Co-PPIX in conjunction with GSH was determined to be 3.4 ± 0.5 µM and the maximal ATPase activity was 276 ± 8.0 nmol/mg/min (or 39 min–1).
Our functional data are entirely consistent with previous work showing that the TMD0 of human ABCB6 can be deleted without effect on the overall folding of the core domain and its ATPase activity (Kiss et al., 2015). The TMD0 domains of ABCB2/3 (TAP1/TAP2) (Koch et al., 2004), ABCB9 (TAPL) (Demirel et al., 2010), and ABCC1 (MRP1) (Bakos et al., 1998) are also known to be dispensable for stability, substrate binding and translocation. A recent study by Wang et al. (2020), however, showed that substrate‐stimulated ATPase activity was completely abolished in the ABCB6-∆TMD0 construct. This discrepancy may have been due to differences in protein purification procedure; Wang et al. (2020) purified ABCB6-∆TMD0 with lauryl maltose neopentyl glycol (LMNG) micelles, whereas we purified it with Cymal-6. This hypothesis is supported by the fact that when we purified the protein in LMNG detergent, it had an elution profile in size-exclusion chromatography similar to that in Cymal-6 (Supplementary Figs. S9A and S9B), but exhibited 1.8-fold higher basal activity that was not significantly stimulated by substrates, as previously observed by Wang et al. (2020) (Supplementary Fig. S9C). Based on these biochemical results, we chose CPIII and hemin as model metal-free and metal-centered porphyrins, respectively, for cryo-EM studies of substrate-bound ABCB6.
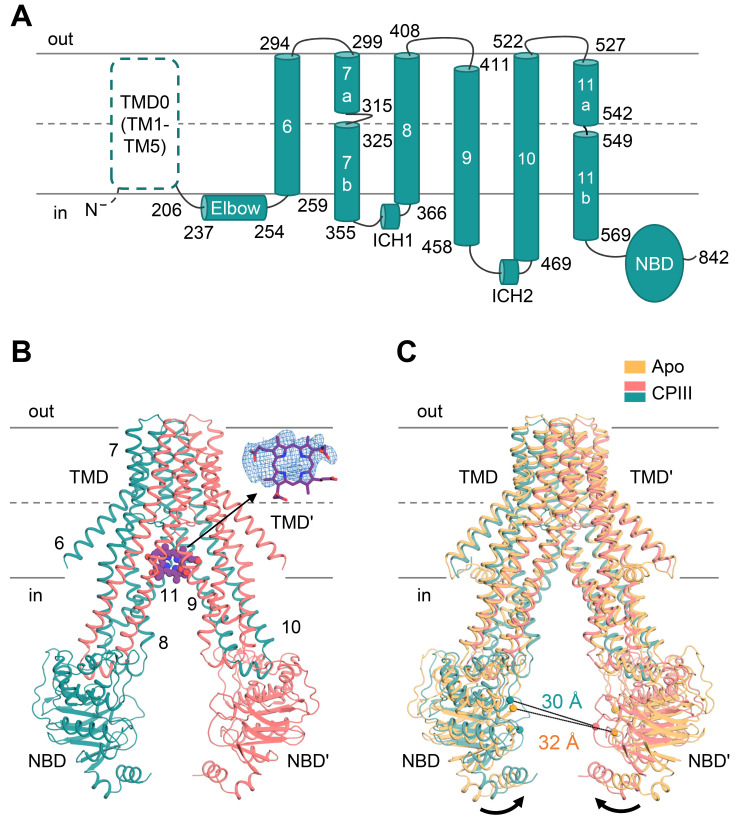
Model building strategy of CPIII bound to ABCB6-∆TMD0
In the presence of CPIII, ABCB6-∆TMD0 has an overall fold similar to the apo form (Figs. 2A and 2B), but the TMDs swing towards each other (~4°), resulting in a somewhat reduced separation between the two NBDs (Fig. 2C) (Song et al., 2021; Wang et al., 2020). The most significant structural changes are concentrated near the central cavity where we could assess the EM density of CPIII at the inner leaflet-cytosol interface (Fig. 2B). Given the large diversity of substrates, one might expect ABCB6 to adopt a flexible and low-selectivity binding strategy for efficient transmembrane substrate transport. In agreement with this argument, the EM density of CPIII was large and globular rather than planar in shape, suggesting the existence of multiple substrate binding modes. Another possible explanation for the ambiguous substrate density is that CPIII is located around the two-fold axis of the ABCB6-∆TMD0 dimer, which would yield a molecularly averaged density map. To define the optimal configuration of CPIII in this ambiguous density, the approach we used was to choose the residues within 6 Å distance of the CPIII map (i.e., W546, Y550, Y386, R552, and M553) and perform alanine scanning mutagenesis. We assumed that if the selected residues were directly involved in CPIII binding, mutations of these residues to alanine would reduce its ability to stimulate ATP hydrolysis and substrate transport due to a lower affinity for CPIII. Based on the results of ATPase-based screening, the most plausible structural model for the bound CPIII molecule was placed into its electron density map, maximizing favorable interactions with the residues found to be important in ATPase activity, while avoiding steric clashes.
Fig. 2. Structure of CPIII-bound ABCB6-∆TMD0.
(A) Schematic representation of the human ABCB6 monomer. The N-terminal TMD0 was not included in the construct, and is drawn with a dashed line. (B) The molecular structure of CPIII-bound ABCB6-∆TMD0. The monomers are shown in pink and teal, respectively. The CPIII is shown as purple spheres. The density corresponding to CPIII (front view) is shown at 4 σ level. The elbow helix was not built due to poor electron density. Single apostrophes are used for the residues (or helices) of one monomer to differentiate them from those of the other monomer. (C) Structure comparison of apo (PDB ID 7D7R) and CPIII-bound ABCB6-∆TMD0. The Cα distance between the conserved G626 of the Walker A motif and the S728 of the signature motif is indicated. Black arrows indicate movements of the NBDs.
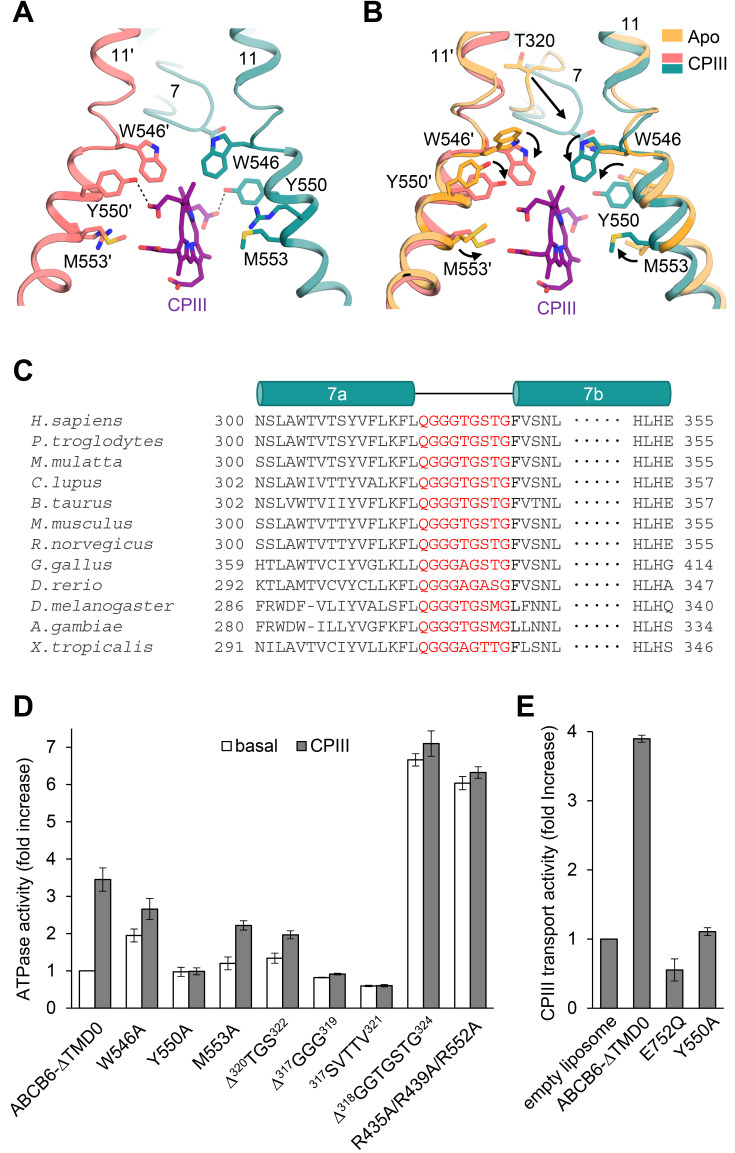
CPIII-bound structure
Our structure reveals that one molecule of CPIII intercalates into the central cavity almost vertical to the membrane, and the propionic acid side chains point towards the cavity surfaces (Fig. 3A). The key residues contributing to CPIII binding are W546, Y550, and M553 on TM 11; of these residues, Y550 is located close to the propionic acids and forms hydrogen bonds with them, while the remaining residues form hydrophobic and van der Waals interactions with the porphyrin ring. Structural comparison demonstrates that upon binding substrate, all the CPIII-coordinating residues orient their side chains towards the bound substrate and secure it in place (Fig. 3B). In particular, a glycine-rich and highly conserved “bulge” loop on TM helix 7 move significantly towards CPIII (Figs. 3B and 3C). The highly conserved T320 on TM 7 bulge loop is not directly involved in the coordination of CPIII, but it alters the orientation of W546 so that the latter avoids colliding with T320 and can interact with CPIII (Fig. 3B).
Fig. 3. Close-up view of the CPIII-bound site.
(A) The view is rotated by 105 degrees along the vertical axis from Fig. 2B. Dashed lines indicate hydrogen bonds between CPIII and Y550. (B) Structure comparison of the substrate binding sites with and without CPIII. Structural changes are marked by black arrows. (C) Sequence alignments of TM 7 of the human ABCB6 homologues. The residues in the TM 7 bulge loop are highlighted in red. (D) ATPase activities of various mutants with 100 µM CPIII. The basal activity of ABCB6-∆TMD0 is set to 100%. Values are mean ± SD of three replicates. (E) CPIII transport activities of the ABCB6-∆TMD0 and other mutants in liposomes. The ATPase-defective mutant E752Q was used as a negative control. Data for protein-free liposomes in the presence of 200 µM CPIII and 2 mM ATP was taken as 100%. Values represent the mean ± SD of triplicate measurements.
Consistent with our structural observations, the Y550A mutation resulted in complete loss of CPIII-stimulated ATP hydrolysis and its transport activity (Figs. 3D and 3E). In the other two mutants, W546A and M553A, there was a slight decrease in the degree of ATPase activation. The structure is consistent with previous biochemical data showing that W546F mutation did not affect ATPase activity for CPIII, while W546V displayed no stimulation (Wang et al., 2020). Two previous studies have described an inhibitory effect of PPIX on W546A activity, and no effect, respectively (Song et al., 2021; Wang et al., 2020). Both CPIII and PPIX are metal-free porphyrins, but there are significant differences between them; CPIII contains four propionic acids, while PPIX contains only two (Fig. 1A). Therefore, we suppose that they may affect the activity of the W546A mutant by different mechanisms. Despite having a large binding cavity, ABCB6-∆TMD0 is not likely to accommodate the two CPIII molecules in a two-layer stacking mode, as observed in the heme-binding protein HmuT (Mattle et al., 2010). The main reason for that is that, unlike heme, which contains two propionic acids (Fig. 1A), CPIII has four, and so could not be stacked without creating a steric collision visible in the electron density map.
The importance of the TM 7 bulge loop and positive surface charges for CPIII binding
The importance of the TM 7 bulge loop for CPIII binding is further supported by deletion analysis. The ∆320TGS322 mutation (deletion of residues 320 to 322) results in ~45% lower maximal ATPase activity relative to ABCB6-∆TMD0 (Fig. 3D). The ∆317GGG319 mutant and the five amino acid substitution (317GGGTG321 → 317SVTTV321) showed no substrate stimulation, possibly owing to a reduction in the loop flexibility needed for CPIII binding. Interestingly, a transporter with a complete deletion of the TM 7 bulge loop (∆318GGTGSTG324) had 6.6-fold higher basal activity than ABCB6-∆TMD0, with no detectable CPIII stimulation. We speculate that deleting the entire TM 7 loop may destabilize the inward-facing conformation, thereby facilitating the transition to the outward-facing conformation. In addition, although the TM 7 loop does not participate directly in substrate binding, its complete deletion resulted in an approximately 1.7-fold increase in dissociation constant (14.1 ± 0.9 µM) for CPIII relative to ABCB6-∆TMD0 (8.2 ± 0.1 µM) (Supplementary Fig. S10).
Previous studies have shown that the entrance to the central cavity is surrounded by a large number of arginines that may play critical roles in recruiting porphyrins bearing negatively charged propionic acids (Song et al., 2021; Wang et al., 2020). Indeed, we observed that a mutant with three arginine substitutions near the bound CPIII (R435A/R439A/R552A) displayed functional properties similar to that of the ∆318GGTGSTG324 mutant, that is, elevated basal ATPase activity, no substrate stimulation (Fig. 3D), and a 2-fold reduction in CPIII affinity (Kd of 17.8 ± 3 µM) (Supplementary Fig. S10). Taken together, our findings suggest that TM 7 loop, along with the positive charges in the vicinity of the substrate binding site, are important for substrate binding, as well as for stability of the inward-facing conformation.
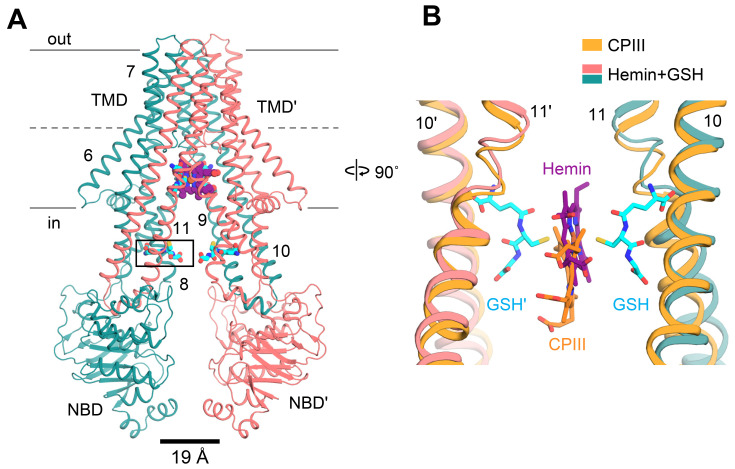
Binding of hemin in concert with GSH primes ABCB6 for high ATP turnover
The molecular structure of ABCB6 in complex with hemin and GSH was determined at 3.9 Å and 3.7 Å resolution, respectively, for C1 and C2 symmetry (Fig. 4A, Supplementary Fig. S11). Binding of hemin and GSH to ABCB6-∆TMD0 induced a global inward movement of the TM helices with local rearrangement of their residues, bringing the two NBDs very close together (Supplementary Fig. S12). As in the case of CPIII, one molecule of hemin was found within the central cavity where its binding site was the same as that of CPIII, but shifted slightly towards the apex (Fig. 4B). In the density map, the TM helices and GSH are well resolved, enabling us to identify their precise locations (Fig. 5A, Supplementary Fig. S11). However, the density map for hemin is not clearly visible at the sites of the propionic acids, making it difficult to determine the orientation of hemin accurately. In principle, three representative binding modes of hemin can be envisaged. However, when we positioned the propionate groups of hemin pointing upwards towards the apex, they exhibited severe steric clashes with W546 (scenario 1, Supplementary Fig. S14A). Thus, taking into account steric factors, the two remaining possibilities are that the propionate groups of hemin point downward to the cytoplasmic entrance of the cavity (scenario 2, Supplementary Fig. S14B) or that they point towards the cavity surface (scenario 3, Fig. 5A).
Fig. 4. Structure of ABCB6- ∆TMD0 in complex with hemin and GSH.
(A) Overall structure of ABCB6-∆TMD0 in complex with hemin and GSH. Hemin and GSH are shown in purple sphere and cyan stick representation, respectively. The Cα distance between the conserved G626 of the Walker A motif and the S728 of the signature motif is shown at bottom. Zoom-in view of the boxed region is presented in Supplementary Fig. S13 (see also Supplementary Discussion). (B) Structure comparison of the substrate binding site of ABCB6-∆TMD0. The CPIII- and hemin:GSH-bound structures are aligned.
Fig. 5. Close-up view of the hemin:GSH binding site.
(A) This view is rotated by 105 degrees along the vertical axis with respect to Fig. 4A. The EM densities of hemin and neighboring residues are shown at 4 σ level. Dashed lines indicate hydrogen bond between hemin and Y550. The GSHs are omitted for simplicity. (B) ATPase activities of various mutants at 10 µM hemin and 1 mM GSH. The basal activity of ABCB6-∆TMD0 is set at 100%. Data points represent mean ± SD of triplicate measurements. (C) Structure of the hemin:GSH binding site in ABCB6-∆TMD0. The view is rotated by 90 degrees along the vertical axis from Fig. 4A. (D) The EM density map of hemin:GSH with neighboring residues is contoured at 4 σ. (E) ATPase activities of the mutants affecting GSH binding. The activity was measured in the presence of 10 µM hemin and 1 mM GSH.
To determine the most likely binding mode, we employed the same mutagenesis-based modelling strategy used for CPIII. Of the changes predicted to affect binding to hemin, the W546A and Y550A substitutions led to 30% and 55% reductions, respectively, in hemin:GSH-stimulated ATPase activity (Fig. 5B). In particular, the Y550A mutant had a 3.3-fold higher Km for hemin (10 µM) and 2-fold lower maximal ATPase activity (136 nmol/mg/min) compared to ABCB6-∆TMD0 (3 µM and 251 nmol/mg/min, respectively) (Supplementary Fig. S15). And the double mutant W546A/Y550A exhibited no hemin:GSH stimulation. Unlike for CPIII, the M553A mutant retained ATPase activity for hemin:GSH (Figs. 3D and 5B). This may be because the upshifted hemin does not form any significant interaction with the M553 residue. In scenario 2, the propionate group is too far away to interact with Y550 (Supplementary Fig. S14B). Hence, our mutagenesis data are in best agreement with scenario 3, in which W546, located above the hemin, is in direct contact with its vinyl and methyl side groups, and Y550 forms a hydrogen bond with the propionate group (Fig. 5A). This analysis is supported by a molecular docking study that pointed to a similar binding mode of hemin to ABCB6-∆TMD0 with docking scores in the range of –5.6 to –5.9 kcal/mol (Supplementary Fig. S16). However, when the same docking analysis was performed with the W546A/Y550A mutant, the top scoring position for hemin (–4.8 kcal/mol) was located outside its density map, creating steric collisions with adjacent residues (Supplementary Fig. S17) (Friesner et al., 2004; 2006).
GSH functions as the primary anchoring site for the ferric iron in hemin
In the structure, a hemin is sandwiched by two molecules of GSH (Figs. 5C and 5D), and the cysteines of the two GSHs interact directly with the ferric ion (Fe3+) of hemin, acting as primary anchoring sites for the metal. This binding scheme is consistent with data showing that when GSH is replaced by ophthalmic acid, in which the cysteine moiety is replaced by 2-aminobutyrate, hemin has no stimulatory effect on ABCB6 activity (Wang et al., 2020). In addition, the GSHs help to stabilize the structure via additional interactions with ABCB6-∆TMD0. The glutamate and glycine of GSH interact with a triad of basic (R435, R439, and R552) and polar (N498, Q501, and N545) residues, respectively, near the hemin (Figs. 5C and 5D). This structure provides important insight into the role of GSH in metal porphyrin binding; (i) the central cysteine residue in the GSH increases substrate selectivity or affinity by forming an axial coordination bond with the metal, (ii) The N-terminal glutamate neutralizes a triad of positive arginines to prevent electrostatic repulsion with the metal ions. (iii) The carboxyl and amide groups of the C-terminal glycine shield the metal ions from a triad of polar residues, so avoiding unwanted electrostatic interactions between the metal and protein. A role for GSH in hemin binding is further supported by site-directed mutagenesis. A series of individual alanine mutations (R435A, R439A, R552A, N498A, Q501A, and N545A) resulted in partial or complete loss of hemin:GSH-stimulated ATPase activity (Fig. 5E). We also observed that combined mutation of all six residues to alanine (R435A/R439A/R552A/N498A/Q501A/N545A) results in marked instability of the mutant, leading to high basal activity and lack of stimulation by hemin:GSH conjugate.
DISCUSSION
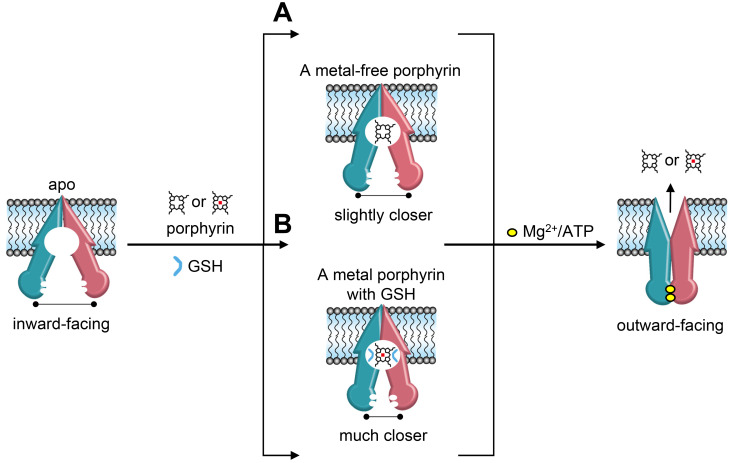
In the present study we establish a structural basis for understanding the different binding mechanisms of ABCB6 depending on the nature of the porphyrin substrates. Our results suggest that the large excess of positive charge at the cavity entrance plays an important role not only in attracting porphyrin molecules, but also in stabilizing the resting state of the ABCB6 transporter. In particular, Y550 is crucial for its interaction with the propionic acid groups of porphyrins. However, loosely-bound metal-free porphyrin is not sufficient to effectively promote the transport cycle (Fig. 6A). In contrast, GSH is a key regulator of the binding of metal porphyrins because it functions as an anchor for the positively charged metal ions (Fig. 6B). Binding of metal porphyrin in conjunction with GSH stabilizes a conformation in which the two NBDs are much closer and positioned to facilitate ATP binding and hydrolysis. During the transport cycle, the TM 7 bulge loops would undergo significant changes in conformation to promote the binding and/or transport of different substrates.
Fig. 6. Proposed transport mechanism of the ABCB6 transporter.
(A) Binding of metal-free porphyrins does not place the NBDs close enough to allow effective ATP hydrolysis. Hence, ABCB6 activity is only modestly stimulated by the presence of metal-free porphyrins. (B) In conjunction with GSH, metal porphyrins prime ABCB6 for effective ATP hydrolysis and substrate translocation. TMD0 is omitted for simplicity.
We also demonstrated the conformational dynamics of ABCB6 in the inward-facing state, highlighting the structural plasticity of the binding cavity. ABCB6 displays a remarkable substrate promiscuity; its substrates range from cadmium (112 g/mol) to hemin (651 g/mol), verteporfin (photosensitizer, 718 g/mol), vincristine (anticancer drug, 824 g/mol), and tomatine (antimicrobial agent, 1,034 g/mol) (Supplementary Fig. S18) (Minami et al., 2014; Polireddy et al., 2012; Rakvács et al., 2019). Therefore, a highly dynamic nature and flexibility in the inward-facing state may be essential to accommodate an extraordinarily broad spectrum of substrates (Lewinson and Bibi, 2001; Loo et al., 2003; Putman et al., 1999). Similarly, P-glycoprotein (Frank et al., 2016; Ward et al., 2013), MsbA (Zou et al., 2009) and BmrA (Mehmood et al., 2012) also show intrinsic conformational dynamics in the inward-facing state, in which they are able to accommodate one or two substrates of various sizes and shapes. Further studies are needed to elucidate the mechanism by which ABCB6 transports many different substrates, and in particular to discover how ABCB6 confers resistance to xenobiotics (Minami et al., 2014; Polireddy et al., 2012) and heavy metals such as cadmium (Cd2+) and arsenic (As3+) (Chavan et al., 2011; Rakvács et al., 2019).
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We are grateful to Dr. Julian Gross for critical reading of the manuscript. We also thank the staff at the following facilities for assistance in grid screening and data collection using cryo-EM: Mrs. Su Jeong Kim in POSTECH, Dr. Hyeongseop Jeong in the Korea Basic Science Institute (KBSI), Dr. Jin-Seok Choi in KAIST Research Analysis Center (KARA). Drs. Ludovic Renault and Wen Yang at The Netherlands Centre for Electron Nanoscopy (NeCEN). This research was supported by grants from the National Research Foundation (NRF) funded by the Ministry of Science, ICT, and Future Planning of Korea (MSIP) (NRF-2017M3A9F6029753, NRF-2019M3E5D6066058, NRF-2019R1A6A1A10073887, and NRF-2021M3A9I4022846), by a grant from the GIST Research Institute (GRI) IIBR funded by the GIST in 2020, and by a postdoctoral fellowship (S.K. [Songwon Kim]) from the NRF funded by MSIP (NRF-2020R1I1A1A01072077).
Footnotes
AUTHOR CONTRIBUTIONS
S.K. (Songwon Kim), S.S.L., J.G.P., S.J., N.J.K., and S.H. expressed and purified proteins. S.K. (Songwon Kim), S.S.L., and J.G.P. performed functional assays. S.K. (Songwon Kim), J.Y.K., and M.S.J. prepared cryo-EM grids, collected and processed cryo-EM data. S.K. (Subin Kim), S.S.L., J.G.P., and S.H.C prepared the figures. J.W.K. and S.K. (Songwon Kim) assisted in data analysis. S.K. (Songwon Kim), J.Y.K., and M.S.J. wrote the manuscript. M.S.J. supervised the project.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo I., Alper S.L., Delaunay J., Auriemma C., Russo R., Asci R., Esposito M.R., Sharma A.K., Shmukler B.E., Brugnara C., et al. Missense mutations in the ABCB6 transporter cause dominant familial pseudohyperkalemia. Am. J. Hematol. 2013;88:66–72. doi: 10.1002/ajh.23357. [DOI] [PubMed] [Google Scholar]
- Angiulli G., Dhupar H.S., Suzuki H., Wason I.S., Duong Van Hoa F., Walz T. New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins. Elife. 2020;9:e53530. doi: 10.7554/eLife.53530.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakos È, Evers R., Szakács G., Tusnády G.E., Welker E., Szabó K., de Haas M., van Deemter L., Borst P., Váradi A., et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J. Biol. Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- Carlson M.L., Young J.W., Zhao Z., Fabre L., Jun D., Li J., Li J., Dhupar H.S., Wason I., Mills A.T., et al. The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife. 2018;7:e34085. doi: 10.7554/eLife.34085.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan H., Khan M.M., Tegos G., Krishnamurthy P. Efficient purification and reconstitution of ATP binding cassette transporter B6 (ABCB6) for functional and structural studies. J. Biol. Chem. 2013;288:22658–22669. doi: 10.1074/jbc.M113.485284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan H., Oruganti M., Krishnamurthy P. The ATP-binding cassette transporter ABCB6 is induced by arsenic and protects against arsenic cytotoxicity. Toxicol. Sci. 2011;120:519–528. doi: 10.1093/toxsci/kfr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., 3rd, Keedy D.A., 3rd, Immormino R.M., 3rd, Kapral G.J., 3rd, Murray L.W., 3rd, Richardson J.S., 3rd, Richardson D.C., 3rd MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiabrando D., Vinchi F., Fiorito V., Mercurio S., Tolosano E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 2014;5:61. doi: 10.3389/fphar.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chifflet S., Torriglia A., Chiesa R., Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal. Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- Dean M., Rzhetsky A., Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- Demirel Ö., Bangert I., Tampé R., Abele R. Tuning the cellular trafficking of the lysosomal peptide transporter TAPL by its N-terminal domain. Traffic. 2010;11:383–393. doi: 10.1111/j.1600-0854.2009.01021.x. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frank G.A., Shukla S., Rao P., Borgnia M.J., Bartesaghi A., Merk A., Mobin A., Esser L., Earl L.A., Gottesman M.M., et al. Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle. Mol. Pharmacol. 2016;90:35–41. doi: 10.1124/mol.116.104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Fukuda Y., Aguilar-Bryan L., Vaxillaire M., Dechaume A., Wang Y., Dean M., Moitra K., Bryan J., Schuetz J.D. Conserved intramolecular disulfide bond is critical to trafficking and fate of ATP-binding cassette (ABC) transporters ABCB6 and sulfonylurea receptor 1 (SUR1)/ABCC8. J. Biol. Chem. 2011;286:8481–8492. doi: 10.1074/jbc.M110.174516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Haffke M., Menzel A., Carius Y., Jahn D., Heinz D.W. Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides. Acta Crystallogr. D Biol. Crystallogr. 2010;66:979–987. doi: 10.1107/S0907444910028593. [DOI] [PubMed] [Google Scholar]
- Hansen S., Stüber J.C., Ernst P., Koch A., Bojar D., Batyuk A., Plückthun A. Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity. Sci. Rep. 2017;7:16292. doi: 10.1038/s41598-017-15711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimerl S., Bosserhoff A.K., Langmann T., Ecker J., Schmitz G. Mapping ATP-binding cassette transporter gene expression profiles in melanocytes and melanoma cells. Melanoma Res. 2007;17:265–273. doi: 10.1097/CMR.0b013e3282a7e0b9. [DOI] [PubMed] [Google Scholar]
- Helias V., Saison C., Ballif B.A., Peyrard T., Takahashi J., Takahashi H., Tanaka M., Deybach J.C., Puy H., Le Gall M., et al. ABCB6 is dispensable for erythropoiesis and specifies the new blood group system Langereis. Nat. Genet. 2012;44:170–173. doi: 10.1038/ng.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Sali A., Baker M.L., Carragher B., Devkota B., Downing K.H., Egelman E.H., Feng Z., Frank J., Grigorieff N., et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januchowski R., Zawierucha P., Andrzejewska M., Ruciński M., Zabel M. Microarray-based detection and expression analysis of ABC and SLC transporters in drug-resistant ovarian cancer cell lines. Biomed. Pharmacother. 2013;67:240–245. doi: 10.1016/j.biopha.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Kiss K., Brozik A., Kucsma N., Toth A., Gera M., Berry L., Vallentin A., Vial H., Vidal M., Szakacs G. Shifting the paradigm: the putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLoS One. 2012;7:e37378. doi: 10.1371/journal.pone.0037378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss K., Kucsma N., Brozik A., Tusnady G.E., Bergam P., van Niel G., Szakacs G. Role of the N-terminal transmembrane domain in the endo-lysosomal targeting and function of the human ABCB6 protein. Biochem. J. 2015;467:127–139. doi: 10.1042/BJ20141085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J., Guntrum R., Heintke S., Kyritsis C., Tampé R. Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP) J. Biol. Chem. 2004;279:10142–10147. doi: 10.1074/jbc.M312816200. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy P.C., Du G., Fukuda Y., Sun D., Sampath J., Mercer K.E., Wang J., Sosa-Pineda B., Murti K.G., Schuetz J.D. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443:586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- Krossa S., Scheidig A.J., Grötzinger J., Lorenzen I. Redundancy of protein disulfide isomerases in the catalysis of the inactivating disulfide switch in A Disintegrin and Metalloprotease 17. Sci. Rep. 2018;8:1103. doi: 10.1038/s41598-018-19429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Matsumura H., Lovell S., Yao H., Rodríguez J.C., Battaile K.P., Moënne-Loccoz P., Rivera M. Replacing the axial ligand tyrosine 75 or its hydrogen bond partner histidine 83 minimally affects hemin acquisition by the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 2014;53:2112–2125. doi: 10.1021/bi500030p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Yang J.G., Zhitnitsky D., Lewinson O., Rees D.C. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science. 2014;343:1133–1136. doi: 10.1126/science.1246489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O., Bibi E. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry. 2001;40:12612–12618. doi: 10.1021/bi011040y. [DOI] [PubMed] [Google Scholar]
- Loo T.W., Bartlett M.C., Clarke D.M. Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J. Biol. Chem. 2003;278:39706–39710. doi: 10.1074/jbc.M308559200. [DOI] [PubMed] [Google Scholar]
- Lynch J., Fukuda Y., Krishnamurthy P., Du G., Schuetz J.D. Cell survival under stress is enhanced by a mitochondrial ATP-binding cassette transporter that regulates hemoproteins. Cancer Res. 2009;69:5560–5567. doi: 10.1158/0008-5472.CAN-09-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolaridis I., Jackson S.M., Taylor N.M.I., Kowal J., Stahlberg H., Locher K.P. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018;563:426–430. doi: 10.1038/s41586-018-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattle D., Zeltina A., Woo J.S., Goetz B.A., Locher K.P. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J. Mol. Biol. 2010;404:220–231. doi: 10.1016/j.jmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Mehmood S., Domene C., Forest E., Jault J.M. Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10832–10836. doi: 10.1073/pnas.1204067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense S.M., Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- Minami K., Kamijo Y., Nishizawa Y., Tabata S., Horikuchi F., Yamamoto M., Kawahara K., Shinsato Y., Tachiwada T., Chen Z.S., et al. Expression of ABCB6 is related to resistance to 5-FU, SN-38 and vincristine. Anticancer Res. 2014;34:4767–4773. [PubMed] [Google Scholar]
- Mitsuhashi N., Miki T., Senbongi H., Yokoi N., Yano H., Miyazaki M., Nakajima N., Iwanaga T., Yokoyama Y., Shibata T., et al. MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J. Biol. Chem. 2000;275:17536–17540. doi: 10.1074/jbc.275.23.17536. [DOI] [PubMed] [Google Scholar]
- Park S., Shimizu C., Shimoyama T., Takeda M., Ando M., Kohno T., Katsumata N., Kang Y.K., Nishio K., Fujiwara Y. Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res. Treat. 2006;99:9–17. doi: 10.1007/s10549-006-9175-2. [DOI] [PubMed] [Google Scholar]
- Paterson J.K., Shukla S., Black C.M., Tachiwada T., Garfield S., Wincovitch S., Ernst D.N., Agadir A., Li X., Ambudkar S.V., et al. Human ABCB6 localizes to both the outer mitochondrial membrane and the plasma membrane. Biochemistry. 2007;46:9443–9452. doi: 10.1021/bi700015m. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polireddy K., Khan M.M., Chavan H., Young S., Ma X., Waller A., Garcia M., Perez D., Chavez S., Strouse J.J., et al. A novel flow cytometric HTS assay reveals functional modulators of ATP binding cassette transporter ABCB6. PLoS One. 2012;7:e40005. doi: 10.1371/journal.pone.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Putman M., Koole L.A., van Veen H.W., Konings W.N. The secondary multidrug transporter LmrP contains multiple drug interaction sites. Biochemistry. 1999;38:13900–13905. doi: 10.1021/bi991262k. [DOI] [PubMed] [Google Scholar]
- Rakvács Z., Kucsma N., Gera M., Igriczi B., Kiss K., Barna J., Kovács D., Vellai T., Bencs L., Reisecker J.M., et al. The human ABCB6 protein is the functional homologue of HMT-1 proteins mediating cadmium detoxification. Cell. Mol. Life Sci. 2019;76:4131–4144. doi: 10.1007/s00018-019-03105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt B.F., Keeffe E.B., Blankenship N.M., Ockner R.K. Validation of a recording spectrophotometric method for measurement of membrane-associated Mg- and NaK-ATPase activity. J. Lab. Clin. Med. 1979;93:790–799. [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soehnlein O. The ABC of thrombopoiesis. Arterioscler. Thromb. Vasc. Biol. 2014;34:700–701. doi: 10.1161/ATVBAHA.114.303365. [DOI] [PubMed] [Google Scholar]
- Song G., Zhang S., Tian M., Zhang L., Guo R., Zhuo W., Yang M. Molecular insights into the human ABCB6 transporter. Cell Discov. 2021;7:55. doi: 10.1038/s41421-021-00284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Aller S.G., Beis K., Carpenter E.P., Chang G., Chen L., Dassa E., Dean M., Duong Van Hoa F., Ekiert D., et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 2020;594:3767–3775. doi: 10.1002/1873-3468.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida M., Emi Y., Kida Y., Sakaguchi M. Human ABC transporter isoform B6 (ABCB6) localizes primarily in the Golgi apparatus. Biochem. Biophys. Res. Commun. 2008;369:369–375. doi: 10.1016/j.bbrc.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Ulrich D.L., Lynch J., Wang Y., Fukuda Y., Nachagari D., Du G., Sun D., Fan Y., Tsurkan L., Potter P.M., et al. ATP-dependent mitochondrial porphyrin importer ABCB6 protects against phenylhydrazine toxicity. J. Biol. Chem. 2012;287:12679–12690. doi: 10.1074/jbc.M111.336180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varatharajan S., Abraham A., Karathedath S., Ganesan S., Lakshmi K.M., Arthur N., Srivastava V.M., George B., Srivastava A., Mathews V., et al. ATP-binding casette transporter expression in acute myeloid leukemia: association with in vitro cytotoxicity and prognostic markers. Pharmacogenomics. 2017;18:235–244. doi: 10.2217/pgs-2016-0150. [DOI] [PubMed] [Google Scholar]
- Vasiliou V., Vasiliou K., Nebert D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genomics. 2009;3:281–290. doi: 10.1186/1479-7364-3-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Cao C., Wang N., Wang X., Wang X., Zhang X.C. Cryo-electron microscopy structure of human ABCB6 transporter. Protein Sci. 2020;29:2363–2374. doi: 10.1002/pro.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., He F., Bu J., Zhen Y., Liu X., Du W., Dong J., Cooney J.D., Dubey S.K., Shi Y., et al. ABCB6 mutations cause ocular coloboma. Am. J. Hum. Genet. 2012;90:40–48. doi: 10.1016/j.ajhg.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.B., Szewczyk P., Grimard V., Lee C.W., Martinez L., Doshi R., Caya A., Villaluz M., Pardon E., Cregger C., et al. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13386–13391. doi: 10.1073/pnas.1309275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K., Mihara S., Zhao C., Okamoto H., Saito-Ohara F., Tomida A., Funato T., Yokomizo A., Naito S., Imoto I., et al. Alteration in copy numbers of genes as a mechanism for cacquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.CAN-3263-2. [DOI] [PubMed] [Google Scholar]
- Zeytuni N., Dickey S.W., Hu J., Chou H.T., Worrall L.J., Alexander J.A.N., Carlson M.L., Nosella M., Duong F., Yu Z., et al. Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins. Sci. Adv. 2020;6:eabb8219. doi: 10.1126/sciadv.abb8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li D., Zhang J., Chen X., Huang M., Archacki S., Tian Y., Ren W., Mei A., Zhang Q., et al. Mutations in ABCB6 cause dyschromatosis universalis hereditaria. J. Invest. Dermatol. 2013;133:2221–2228. doi: 10.1038/jid.2013.145. [DOI] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P., Bortolus M., McHaourab H.S. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-30790 (CPIII-bound state) and EMD-30791 (hemin:GSH-bound state). The atomic coordinates have been deposited in the Protein Data Bank under accession codes 7DNY (CPIII-bound state) and 7DNZ (hemin:GSH-bound state).