Abstract
Various RNA-binding proteins (RBPs) are key components in RNA metabolism and contribute to several neurodevelopmental disorders. To date, only a few of such RBPs have been characterized for their roles in neocortex development. Here, we show that the RBP, Rbms1, is required for radial migration, polarization and differentiation of neuronal progenitors to neurons in the neocortex development. Rbms1 expression is highest in the early development in the developing cortex, with its expression gradually diminishing from embryonic day 13.5 (E13.5) to postnatal day 0 (P0). From in utero electroporation (IUE) experiments when Rbms1 levels are knocked down in neuronal progenitors, their transition from multipolar to bipolar state is delayed and this is accompanied by a delay in radial migration of these cells. Reduced Rbms1 levels in vivo also reduces differentiation as evidenced by a decrease in levels of several differentiation markers, meanwhile having no significant effects on proliferation and cell cycle rates of these cells. As an RNA binding protein, we profiled the RNA binders of Rbms1 by a cross-linked-RIP sequencing assay, followed by quantitative real-time polymerase chain reaction verification and showed that Rbms1 binds and stabilizes the mRNA for Efr3a, a signaling adapter protein. We also demonstrate that ectopic Efr3a can recover the cells from the migration defects due to loss of Rbms1, both in vivo and in vitro migration assays with cultured cells. These imply that one of the functions of Rbms1 involves the stabilization of Efr3a RNA message, required for migration and maturation of neuronal progenitors in radial migration in the developing neocortex.
Keywords: Efr3a, neurogenesis, radial migration, RNA binding motif single stranded interacting protein 1
INTRODUCTION
Neurogenesis in the developing brain involves cells that develop from neural progenitor cells (NPCs). The ventricular zone is the home of NPCs where they divide to become neurons and radial glial cells (RGCs) (Ji et al., 2017; Noctor et al., 2004; Pilaz and Silver, 2015). Neurons then start an RGC glia-guided migration, called the radial migration, from the sub-ventricular zone to the cortical plate. For radial migration, key signaling pathways are activated and fine-tuned and in proper orchestration for optimal growth of the developing brain (Huttner and Kosodo, 2005). This radial migration results in neurons moving to their proper cortical layers and in correct orientations. Flaws in radial migration lead to developmental brain malformation, resulting in various documented developmental disorders, with functional defects such as cognitive dysfunction and autism and anatomical defects such subcortical band heterotopia and lissencephaly (Bryant and Yazdani, 2016; Moffat et al., 2015; Pilaz and Silver, 2015).
Among the signaling entities required for proper execution of radial migration for the neurons, RNA-binding proteins (RBPs) are thought to play critical roles; an expression study of RBPs in the developing brain also found almost 323 putative RBPs being expressed (Cha et al., 2020; McKee et al., 2005). A number of these RBPs have been proven to be functionally important for neurogenesis, having critical roles in cell proliferation, differentiation and migration with specific tasks such as regulating alternative splicing, RNA transport, alternative polyadenylation, RNA localization and RNA stability (Pilaz and Silver, 2015; Yeh and Yong, 2016).
RNA binding motif single stranded interacting protein 1 (Rbms1), also known as c-Myc single strand binding protein (MSSP-1), is expressed in almost all human tissues, although in varying levels (Taira et al., 1998). For adult mouse, it is also expressed in the brain (https://www.proteinatlas.org/ENSG00000153250-RBMS1/brain). Rbms1 has been shown to bind to both RNA and DNA via its two consensus RRM motifs for RNA/DNA binding (Bandziulis et al., 1989; Dreyfuss et al., 1988; Niki et al., 2000; Schwer et al., 2009). Rbms1 also has one conserved C-terminal domain (Schwer et al., 2009). The sequence comparison of Rbms1 from different vertebrate species reveals a high degree of conservation, implying important roles in their biology. As Rbms1 has a shorter linker region between its RRM domains, it is implied that it provides stronger binding to its RNA/DNA targets (Lunde et al., 2007).
On the functions of Rbms1, it can act as a transcription suppressor of alpha-smooth muscle actin (Kimura et al., 1998). It can also suppress the transcription activity of c-Myc by binding to the E box region of c-Myc and forming a suppressive ternary complex of Rbms1/Myc/Max (Niki et al., 2000). Rbms1 can also induce apoptosis in Hela cells when overexpressed (Iida et al., 1997). Homozygous deletion of Rbms1 in mice leads to placental defects in null embryos with the embryos having reduced levels of progesterone, leading to a smaller percentage of littermates being born (Fujimoto et al., 2001). Rbms1 also acts as a post transcriptional regulator of RNA stability of its target regulon in a cancer study (Yu et al., 2020).
To date it is not known if Rbms1 affects a developing brain. In this study, we show that Rbms1 is differentially expressed in the mouse developing brain. We use in utero electroporation (IUE) to manipulate endogenous Rbms1 levels in the embryos and study the role of Rbms1 in radial migration of newly born neurons. We found that knockdown of Rbms1 causes defective radial migration and suppresses neuronal differentiation. Using an RNA binding and sequencing technique, we addressed the role of Rbms1 in the developing brain as a post transcriptional RNA regulator and discovered a role for Efr3a, as the RNA substrate for Rbms1. Efr3a is also expressed in the mouse brain (https://www.proteinatlas.org/ENSG00000132294-EFR3A/brain) and it is a plasma membrane associated protein and studies have identified a signaling role in keeping active pools of phosphatidylinositol kinase PI4KA at the plasma membrane (Baird et al., 2008). Efr3a has also been reported to have a role in the nervous system in adult neurogenesis in the hippocampus (Qian et al., 2017). A set of mutations of human Efr3a is also associated with onset of autism spectrum disorder (ASD) (Gupta et al., 2014). In our study, Rbms1 was shown to bind and stabilize the Efr3a RNA message and demonstrated a 3’ -UTR region of Efr3a transcript that would bind to Rbms1. Functionally, the Rbms1 migration defect was corrected when Efr3a was overexpressed in the same cells, both in utero and in migration assays. In summary, we ascribe a functional link between Rbms1 as an RBP for regulating neuronal migration and differentiation in the neocortex via binding and stabilizing the RNA message for Efr3a.
MATERIALS AND METHODS
Animals and IUE
All the animal studies and experimental protocols were approved by Hallym University Institutional Animal Care and Use Committee (IACUC) (approval No. Hallym2020-19). In this study we used pregnant mice (C57BL/6J strain) purchased from Orient Bio (Korea). According to the company’s mating schedule, the vaginal plug at noon of the day was defined as the embryonic day 0.5 (E0.5). The mice were kept in the optimum 12-h dark/12-h light cycle, 22°C ± 2°C temperature and 50% ± 10% humidity with ad libitum access to food and water. Mice feeds were purchased from commercial food company Purina (Korea). IUE was performed according to (Saito, 2006). Briefly, pregnant mice of targeted stages were anesthetized by 4% isoflurane. Intact uterine horn was then manifested by seizure of the abdominal wall and it was kept moist with sterilized phosphate buffer saline (PBS). Into the lateral ventricles of the target embryo 2 mg/ml plasmid DNA in 2 µl TE buffer containing 0.01% fast green dye (Sigma-Aldrich, USA) was injected through the uterine wall of the mouse. Electroporation across the brain was performed by fixing 5-mm diameter tweezertrodes (BTX [Harvard Biosciences], USA) during electroporation by square-wave pulse generator (ECM830; BTX) of five 45 V and 50 ms-long pulses with 950 ms intervals been discharged. The intact uterus was then returned back to the abdomen of the pregnant mouse and the abdominal cut was carefully and aseptically sutured. After the IUE procedure, the pregnant mouse was then kept in a pre-warmed cage to recover; after the operation, the embryos were allowed to develop to the desired stages in the pregnant mouse.
DNA construction
Various DNA constructs were prepared by the subcloning of either the open reading frame (ORF) or knockdown oligomers in the corresponding expression plasmid vectors in (Table 1). The expression vector was produced by subcloning of the ORF of mouse Rbms1 (NCBI accession No. NM_001141931) into the base vector pCAGIG (Connie Cepko [plasmid #11159]; Addgene, USA) and that for the knockdown vector by inserting 19-mer shRNA oligonucleotide of 5’ GGAGACGTCTAATGACCATTC 3’ into pGPH1/GFP/Neo vector (Bicistronic GFP shRNA vector; GenePharma, China). FLAG-tagged Rbms1 expression vector were prepared by subcloning of Rbms1 ORF into the N-terminal FLAG vector, pCMV3-N-FLAG. The mouse Rbms1 addback vector was generated by mutating four nucleotides of shRbms1 targeting sequence in complete ORF of mouse Rbms1; the sequence changes were as 5’ G/AGAG/AACGTCT/GAATGACCAT/CTC 3’, without changing their coding amino acids. The mutant subcloned into pCAGIG by Enzynomics (Korea).
Table 1.
List of vectors
| No. | Plasmid | Insert | Remarks |
|---|---|---|---|
| 1 | pCAGIG (GFP) | Control | Bicistronic GFP control expression vector |
| 2 | pCAGIG (GFP) | Mouse Rbms1 (NM_001141931) ORF | Bicistronic GFP and mouse Rbms1 expression vector |
| 3 | pCAGIG (GFP) | Mouse Rbms1 (NM_001141931) ORF (G/AGAG/AACGTCT/GAATGACCAT/CTC silent point mutations) | Addback expression vector, bicistronic GFP and Rbms1 shRNA binding region silent mutation |
| 4 | pGPH1/GFP/Neo | Control | Bicistronic GFP containing control shRNA vector |
| 5 | pGPH1/GFP/Neo | shRbms1 (shRNA oligonucleotide of 5´GGAGACGTCTAATGACCATTC-3´) | Bicistronic GFP containing mouse Rbms1 silencing vector |
| 6 | pCMV3-N-FLAG | Control N-FLAG vector | N-terminal FLAG tagged control expression vector |
| 7 | pCMV3-N-FLAG | Mouse Rbms1 (NM_001141931) ORF | N-terminal FLAG tagged mouse Rbms1 expression vector |
| 8 | pCAGIG (GFP) | Mouse Efr3a (NM_133766.3) ORF | Bicistronic GFP and mouse Efr3a expression vector |
Primary culture
Electroporated/transfected cerebral cortices were manually dissected and chopped into small pieces. The tissue was dissociated and digested with 4 unit/ml DNase I (Sigma-Aldrich) with TrypLE (Gibco, USA) at 37°C for 30 min. Digested tissue was then dissociated by pipetting, washed and cultured in Neurobasal Medium (Invitrogen, USA) supplemented with 1% each of 100X penicillin-streptomycin, GlutaMAX and B27 supplement solutions (Thermo Fisher Scientific, USA). Dissociated neurons were plated in 24-well culture dishes previously coated with poly-L-Lysine (Sigma-Aldrich) at a concentration of 100,000 cells and were cultured at 37°C in humidified, 5% CO2 incubator.
Immunofluorescence
The IUE brain samples were fixed in 4% paraformaldehyde in PBS at 4°C overnight and the samples were soaked in 30% sucrose solution in PBS overnight at 4°C. Cortices were sliced into 15 µm thickness coronal sections and directly transferred onto precoated glass slides (Superfrost-20; Matsunami, Japan). The sectioned samples were then stored at –20°C. Presectioned, preserved samples were then removed from the freezer and dried for 10 min. Immunostaining was then performed using a standard protocol. Briefly, the sectioned brain samples were permeabilized by immerging the slides in PBS with 0.1% Triton X-100 for 10 min. The samples were then blocked in 3% bovine serum albumin in PBS with 0.1% Triton X-100 for 40 min at room temperature. This was followed by primary antibody (Table 2) incubation for either 3 h at room temperature or overnight at 4°C. The samples were then washed with PBS for 3 times for 5 min each and then incubated with secondary antibody Alexa 488 anti-chicken, Abcam or anti-rabbit immunoglobulin G (IgG) (1:1,000; Abcam, UK) and 4′,6-diamidino-2-phenylindole (DAPI) (1:1,000) were incubated for 1 h in room temperature. After washing with PBS for 3 times for 5 min each, the samples were mounted in mounting solution (Choi et al., 2019). The fluorescence images were captured by a Zeiss LSM710 confocal microscope (Zeiss, Germany).
Table 2.
List of antibody used in experiment
| Serial No. | Antibody | Catalogue | Primary antibody | Secondary antibody | Experiments |
|---|---|---|---|---|---|
| 1 | GFP | AB13970 | Abcam, UK; 1:1,000 | Alexa 488 | ICC |
| 2 | Alexa Fluor® 647 Anti-SATB2 antibody | ab196536 | Abcam, UK; 1:200 | Conjugated | ICC |
| 3 | Tbr2 | #14-4875-82 | Abcam, UK; 1: 200 | Anti-mouse, Alexa 594 | ICC |
| 4 | Pax6 | #901302 | Bio Legend, USA; 1:200 | Rabbit, Alexa 594 | ICC |
| 5 | Tuj1 | #4466 | Cell Signaling Technology, USA; 1:1,000 | Anti-mouse, Alexa 488 Anti-mouse, Alexa 555 |
ICC |
| 6 | Rbms1 | ab150353 | Abcam, UK; 1:500 | Anti-rabbit, Alexa 488 Anti-rabbit, Alexa 594 |
Western blot, ICC |
| 7 | GFAP | #3670 | Abcam, UK; 1:500 | Anti-mouse, Alexa 555 | ICC |
| 8 | Map2 | ab32454 | Abcam, UK; 1:1,000 | Anti-rabbit, Alexa 647 | ICC |
| 9 | Actin | #4967 | Cell Signaling Technology, USA; 1:5,000 | Rabbit | Western blot |
| 10 | Gapdh | #2118S | Cell Signaling Technology, USA; 1:5,000 | Rabbit | Western blot |
| 11 | N cadherin | ab98952 | Abcam, UK; 1:1,000 | Mouse | Western blot |
RNA immunoprecipitation
RNA immunoprecipitation with chemical crosslinking were performed according to (Keene et al., 2006; Nilsen, 2014). Briefly, mouse E15.5 brains cortices were collected in cold Hanks’ Balanced Salt Solution (HBSS) and manually dissociated into smaller sizes by pipetting and vortexing and they were then washed 3 times with ice cold PBS for 5 min, spinning them at 1,000 × g for 5 min before each wash. The brain tissues were re-suspended in 1 ml crosslinking buffer (20 mM HEPES [pH 7.0], 10 mM KCl, 1.5 mM MgCl2) and were crosslinked by adding from a 10% formaldehyde stock solution in PBS to a final concentration of 1% formaldehyde and mixing (AR grade, 37%). Crosslinking was allowed to proceed for 10 min at room temperature, and was stopped by adding 1 M glycine in PBS to a final concentration of 0.25 M glycine and incubating for 5 min at room temperature. The cells were then suspended in freshly prepared ice-cold polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.0], 0.5%, NP40, 1 mM DTT, 100 U/ml RNaseOUT, 400 µM vanadyl ribonucleoside complexes [VRC] [Sigma-Aldrich], 1X Protease inhibitor cocktail [Cell Signaling Technology, USA], RNase inhibitors [50 units, Invitrogen RNaseOUT]) and incubated for 5 min on ice. The samples were then sonicated using a microprobe in 3 ml total volume on ice for 15 s at 8% amplitude (Vibro-cell VCX-600; Artisan, USA). The lysates from above were centrifuged at 13,000 rpm for 20 min in a 4°C chilled centrifuge to obtain the supernatant. The cleared supernatant was precleared with Protein A/G agarose beads (Santa Cruz Biotechnology, USA). From 1 ml of lysates, the capture of mouse Rbms1 with antibody (3 µg of anti-Rbms1) in parallel with isotype matched control antibody via 20 µl of Protein A/G beads was in presence of 200 units of RNaseOUT, 400 µM VRC, 10 µl of 100 mM DTT and 20 mM EDTA in PBS for 6 h at 4˚C on a rotor. The beads were washed 5 times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP40) and followed by 30 µg proteinase K treatment for 30 min at 55°C. Immunoprecipitated RNAs were extracted with Trizol (Invitrogen) and RNA samples were then send for sequencing in a custom service at eBiogen (Korea), or cDNA was prepared using either ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Japan) or miScript II RT Kit (Qiagen, Germany) according to the manufacturer’s protocol. Targets RNA message were detected and quantified with the SYBR Green RT-PCR kit (Qiagen).
Western blotting
Brain tissue, N2A, HEK293T and primary neurons were cultured in humidified 37°C, 5% CO2 incubator. Cells were collected by PBS and washed two times at 500 g for 5 min. Then lysed the cells in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 1 mM Na3VO4, 5 mM NaF, and added protease inhibitor cocktail). Protein concentration were determined by Bradford assay (Bio-Rad, USA). Then equal amount of proteins was separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes and blocked by 5% skimmed milk in TBST. Transferred membranes were probed with primary antibody at 4°C for overnight. After washing with TBST, the blots were incubated with appropriate primary antibody and the protein bands were visualized by Western HRP in Fusion FX (Vilber, France).
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA were extracted from brain sample, and RIP samples by qiazen miscript mini RNasy kits, and cDNA were prepared either ReverTra AceTM qPCR RT Master Mix with gDNA Remover (Toyobo) or miScript II RT Kit (Qiagen) with standard procedure supplied by the manufacturer. Expression levels of the targets mRNA were quantified by SYBER Green RT-PCR kits either (Qiagen [Germany] or Bioneer [Korea]). The primers used in the qPCR are listed in (Table 3). The relative expression of the messenger RNA was calculated by the expression of Gapdh for normalization. The formula used in the calculation of delta Ct were 2 – [(Ct of target gene) – (Ct of Gapdh)] where Ct is threshold (Ct).
Table 3.
List of primers
| Serial No. | Gene name | Primer |
|---|---|---|
| 1 | Gapdh | Forward: GAGTCTACTGGTGTCTTCAC |
| Reverse: GTTCACACCCATCACAAAC | ||
| 2 | Rbms1 | Forward: CACTTATGACCCGACTACAG |
| Reverse: CAGGAGATGCAATATAGGGTG | ||
| 3 | Efr3a | Forward: TAGAAGAAGTTGACAGTCGC |
| Reverse: AGTTCTCTGAAACAGCTCTC | ||
| 4 | Zbtb46 | Forward: ACTCAAATGTAGACCTGACG |
| Reverse: CAAGTAGCTGGTTTCTCCTC | ||
| 5 | Zcchc24 | Forward: CCCTGCACTCCAGTTATC |
| Reverse: GTTATTGAGGGAACCGTAGG | ||
| 6 | Wwtr1 | Forward: GATTAGGATGCGTCAAGAGG |
| Reverse: TCTCATATCTGTGCTCATGG | ||
| 7 | Twist1 | Forward: AGCTGAGCAAGATTCAGAC |
| Reverse: CAGCTTGCCATCTTGGAG | ||
| 8 | Stat6 | Forward: TGGTTATCTGGCTCATGTTG |
| Reverse: ACAGTCCATCCACTCATTTC | ||
| 9 | Klf4 | Forward: CACCCACACTTGTGACTATG |
| Reverse: TCACAGTGGTAAGGTTTCTC | ||
| 10 | Emb | Forward: CCAGTCAGTACAGGTTCATC |
| Reverse: TGGGTACGTGGATGTTAAAG | ||
| 11 | Dnaaf4 | Forward: ATGCTGACGTATTCTGTGG |
| Reverse: CAATCTTGGCTTTGCTCTTC | ||
| 12 | Itga8 | Forward: CTCCAGCTTCCTATGATGAC |
| Reverse: TATCCAAAGTTCTGTGCTCC | ||
| 13 | Drd2 | Forward: CTTTGGACTCAACAACACAG |
| Reverse: GATATAGACCAGCAGGGTG | ||
| 14 | Adm | Forward: TATTGGGTTCACTCGCTTTC |
| Reverse: ATGCTTGTAGTTCCCTCTTC |
Lentivirus production and primary neuron transduction
Vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped lentivirus particles were produced and used for transduction of primary culture neurons. We parched shRbms1 and corresponding control vector from Vectorbuilder (USA). To make Over-expression Rbms1 lenti viral vector we inserted ORF of Rbms1 into pLJM1-EGFP (plasmid #19319; Addgene) vector by cutting EGFP region. The viral particles were produced by co-transfection of human embryonic kidney cells (HEK293T) with a mixture of plasmids through Lentifectin transfection reagent (abm, Canada). The ratio of plasmids and transfection reagent were measured first and it was 5 to 4. The plasmids use in the transfection were either shRbms1 or corresponding control or (pLJM1-Rbms1) or corresponding control vector and packaging mix (abm), and the ratio of transfer vector and packaging mix (packaging plasmid & envelop plasmid) used for transfection were 2 to 3. Transfection was in serum free conditions, then after 4 h of transfection 10% fetal bovine serum (FBS) was added and incubated at 37°C for overnight. The lentivirus particles in the medium were harvested for 48 h after transfection. The medium was then collected and centrifuged by 500 × g for 7 min to remove cell debris. The lenti particles were incubated overnight at 4°C in the ice cold lenti concentrator solution (OriGene Technologies, USA). Next day, virus particle containing media was centrifuged at 3,500 × g for 25 min at 4°C and resuspended in lentivirus stabilizer (OriGene Technologies) and aliquots were stored at –80°C. The lentivirus particles were titrating in HEK293T cells by FACS and Western blot. Appropriate titers of lenti particles were used for transduction of DIV (days in vitro)1 primary cultured cortical neurons and harvested at DIV3.
RNA half-life analysis
To stop transcription freshly prepared Actinomycin D (5 µg/ml; Sigma-Aldrich) was added in cultured cells. Then, cells were collected at different time point (0, 2, 4, and 8 h). RNA was extracted using RNeasy Mini RNA extraction kit (Qiagen) according to manufacturer’s protocol. Then reverse transcription was performed to change mRNA to cDNA by miScript II RT Kit (Qiagen) according to manufacturer’s guideline. Mouse Efr3a and Klf4 mRNA were determined by qRT-PCR and normalized by Gapdh. The expression of the Efr3a and Klf4 mRNA at the starting point was taken as 1 and t1/2 was calculated from RNA level trend plot.
Synthetic RNA binding competition assay
Brain samples were collected in ice-cold HBSS and washed with ice-cold PBS for three times at 500 g. The samples were resuspended in Polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.0], 0.5%, NP40, 1 mM DTT, 100 U/ml RNaseOUT, 400 M VRC, Protease inhibitor cocktail) and sonicated in 3 ml total volume on ice for 15 s at 8% amplitude (Vibro-cell VCX-600; Artisan). The lysates were centrifuged at 13,000 rpm for 20 min and the supernatants were saved and then incubated with 150 µl Protein A/G plus-agarose beads for preclearing. One milliliter of this lysate solution was incubated with 150 µl beads (A/G plus-agarose; Santa Cruz Biotechnology) and appropriate amount of antibody (3 µg), and incubated at 4˚C overnight. Beads were next washed 5 times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP40) at 2,500 rpm for 6 min at 4°C and were resuspended in 20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 plus added protease inhibitors and RNaseOUT at 100 Units/ml, and then divided into 6 aliquots with each tube having 100 µl of the bead suspension. The candidate competition RNA oligo at 1.5 µM was added to each tube plus 200 µl of binding buffer. The suspension was incubated on a rotator for 5-6 h at 4°C. The beads were washed 5 times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP40) and followed by 30 µg proteinase K treatment for 30 min at 55°C. Immunoprecipitated RNAs were extracted with Trizol (Invitrogen) and cDNA was prepared using either ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) or miScript II RT Kit (Qiagen) according to the manufacturer’s protocol. Targets RNA message were detected and quantified with the SYBR Green RT-PCR kit (Qiagen).
Biotinylated RNA-protein capture assay
Biotinylated RNA pulldown and immunoblotting was performed to address RNA-protein interactions. Biotin labeled RNA oligoes were purchased from Bioneer. RNA oligoes at 100 pmol were immobilized on 5 µl of streptavidin agarose beads. E15.5 brain samples were collected in ice cold HBSS and homogenized by pipetting. They were then twice washed with chilled HBSS and gently resuspended in hypotonic buffer solution (20 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2). Cells were lysed by rigorous vortexing in NP40 (10%) solution with protease phosphatase inhibitor and RNase I solution (100 U/ml). Nuclear extracts were collected as a pellet by centrifuging the mixture at 3,000 rpm at 4°C and resuspended in NP 40 and quantified. Conjugated RNA and beads were then incubated in nuclear extracts at 4°C overnight with rotation. Next day washed them and boiled them with 4X loading dye and detected the Rbms1 level by Western blotting (Bishayee et al., 2022). Biotinylated RNA oligo sequences were as follows Efr3a_3 5´- Biotin-GUGUGCUUUUGUAUGGAAAA, Efr3a_4 5´- Biotin-CUAUUCUUUUGUAUGACUCC, nonspecific oligo GAA AGG ACU CCU UUG ACA GGC AUC GG.
Statistical analysis
Statistical analyses were via GraphPad Prism 5 (GraphPad Software, USA). Data are presented as mean ± SEM with the two-tailed Student’s t-test for comparing the means of 2 groups. For data P < 0.05, they were considered as being statistically significant.
RESULTS
Rbms1 is expressed in the neocortical mouse brain and its protein levels are regulated in a temporal fashion
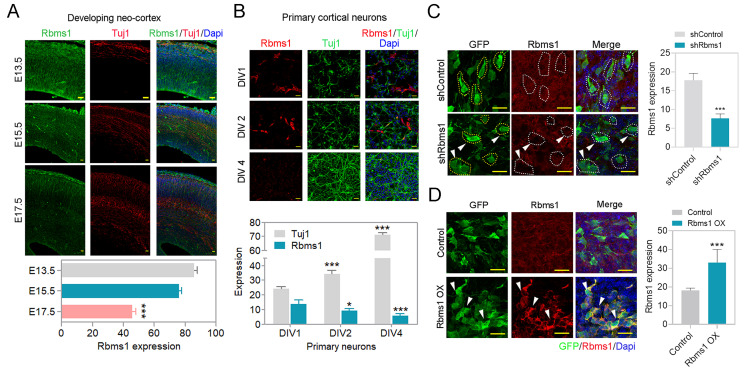
To address the function of Rbms1 in the developing mouse brain, we evaluated the expression of Rbms1 during different stages of mouse brain development. To assay the protein levels of Rbms1 during the progenitor pool expansion to postnatal day 0 (P0), we collected brain samples from embryonic mouse brains from E13.5, E15.5, E17.5, E18.5, and P0. By Western blot, we found the expression of Rbms1 to be most prominent in the pre-mitotic stage; however, it was detectable throughout the developmental period (Supplementary Fig. S1A). This data suggested that Rbms1 may be developmentally relevant during the neuronal migration and differentiation periods. For spatial expression patterns of Rbms1 during development, we performed immunostaining of cortical brain slices for Rbms1 protein expression at different embryonic stages of mouse brain (E13.5, E15.5, and E17.5) with an anti-Rbms1 antibody (Fig. 1A). The result was Rbms1 being expressed throughout the developing cortical layers and being most prominent in the early stages (E13.5). At E13.5, Rbms1 expression was higher in the sub ventricular zone (SVZ) layers, and in later stages, it was primarily found in marginal zone area. The specificity of the anti-Rbms1 antibody was checked by Western blots and ICC of Neuro-2a (N2a) cells with knockdown (KD) endogenous Rbms1 or overexpression (OX) of recombinant Rbms1 in those cells (Supplementary Figs. S1B and S1C). With the in vivo samples, the same changes were seen with the KD and OX samples in the IUE samples (Figs. 1C and 1D).
Fig. 1. Rbms1 is expressed in the developing cortex.
(A) Developmental expression pattern of Rbms1. Immunostaining for Tuj1 (red), Dapi (blue), and Rbms1 (green) in coronal sections of the E13.5, E15.5, and E17.5 mouse cortices. Scale bars = 20 µm. Data presented as mean ± SEM; Student’s t-test, ***P < 0.001. (B) Rbms1 expression from in vitro primary cortical neuron, Immunostaining for Tuj1 (green), Rbms1 (red), and Dapi (blue), at DIV1 (days in vitro), DIV2, and DIV4. Scale bars = 20 µm. Data presented as mean ± SEM; Student’s t-test, *P < 0.05 and ***P < 0.001. (C) Knockdown Rbms1 vector with corresponding control vector introduced by IUE at E15.5 and 48 h after harvested and immunostained against Rbms1 specific antibody, respective GFP labeled cells marked with yellow dots, and GFP and Rbms1 labeled cells marked with white dots and white arrowheads. Scale bars = 10 µm; their relative quantification in the right panel. Data presented as mean ± SEM; Student’s t-test, ***P < 0.001 (n = 3). (D) Overexpression Rbms1 vector with corresponding control vector introduced by IUE at E15.5 and 48 h after harvested and immunostained against Rbms1 specific antibody. Scale bars = 10 µm; their relative quantification in the right panel. Data presented as mean ± SEM; Student’s t-test, ***P < 0.001 (n = 3).
Reduced Rbms1 levels lead to defective neuronal migration during corticogenesis
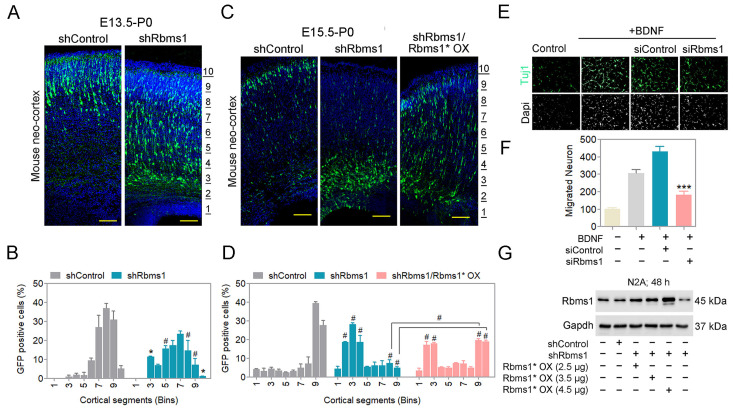
Rbms1 shRNA could efficiently downregulate the expression of Rbms1 in vivo. The shRNA vector co-expressed the green fluorescent protein (GFP) in a bicistronic arrangement (shRbms1) and the template control vector served as the negative control in various experiments. With IUE at E15.5 with samples harvested at E17.5, the Rbms1 protein levels were reduced by more than 60% compared with the control vector (Fig. 1C). For knockdown of Rbms1 via IUE at E15.5 with samples harvested at E17.5, there was little effect on radial migration (Supplementary Fig. S2A). At a longer time point of 5 and 7 days’ post IUE, the effect of radial migration with shRbms1 was significant with radial migration of transfected cells being delayed; these were for early-born neuron at E13.5 to P0 and late-born-neuron at E15.5 to P0 (Figs. 2A and 2B). When we observed the knockdown effect from E15.5 to post-natal 5 days, we found that Rbms1 knockdown cells also migrated to the intermediate zone. The migration defects due to shRbms1 were also seen in vitro with the Boyden chamber transwell migration assay for siRNA negative control and siRbms1 transfected neurons; the cells were made to migrate for the chemoattractant BDNF in the lower chamber and knockdown of Rbms1 delayed the chemoattractant mediated migration in compare to control (Figs. 2E and 2F).
Fig. 2. Knockdown of Rbms1 delayed radial migration during neurogenesis.
(A) Immunostaining of GFP (green) and Dapi (blue) mouse cortices at postnatal days 0, seven days after electroporation at E13.5 of Rbms1 knockdown vector (shRbms1) and backbone control vector. The area from the outer layer of the CP to the inner layer of the VZ was divided into ten different bins. Scale bars = 100 µM (n = 3 brain from three different injections). (B) Quantify the percentage of GFP positive cells in the ten different cortical segments (Bins) areas. Data presented as mean ± SEM; *P < 0.05, #P < 0.001 (n = 3, two-way ANOVA). (C) Immunostaining of GFP (green) and Dapi (blue) mouse cortices at postnatal days 0, five days after electroporation at E15.5 of Rbms1 knockdown vector (shRbms1) and backbone control vector and addback vector with knockdown Rbms1 vector. The area from the outer layer of the CP to the inner layer of the VZ was divided into ten different bins. Scale bars = 100 µM (n = 3 brain from three different injections). (D) Quantify the percentage of GFP positive cells in the ten different cortical segments (Bins) areas. Data presented as mean ± SEM; #P < 0.001 (n = 3, two-way ANOVA). (E) siRbms1 and si negative control transfected primary neuron’s migration were measured by Boyden transwell migration assay and Immunocytochemistry. Vehicle or BDNF (25 ng/ml) was present in the bottom chamber. Tuj1 (green) and Dapi (gray) (n = 3). Scale bars = 50 µM. (F) Quantification of the migrated neurons. Data presented as mean ± SEM; ***P < 0.001 (n = 3, Student’s t-test). (G) Western blot analysis of control, shRbms1, and shRbms1 with add-back vector-transfected N2A cells for 48 h.
We also prepared an Rbms1 overexpression vector (Rbms1* OX) with silent point mutation of the shRNA targeting sequence for addback studies. This design would allow the addback Rbms1 protein to become expressed even in presence of shRbms1 in the same cell and be a true addback. The efficiency of silently mutated addback vector was confirmed by transfection and Western blotting in N2a cells (Fig. 2G). Co-injection of the shRbms1 vector and the Rbms1 addback vector partially rescued the knockdown effect of Rbms1 on migration delays, verifying that the majority of changes in the phenotype of shRbms1 transfected cells were on-target (Figs. 2C and 2D). The partial recovery may also be due to Rbms1 OX also causing a partial migration defect when singly transfected, thus offering the partial recovery of the addback.
Loss of Rbms1 leads to maturation defects for the developing neurons
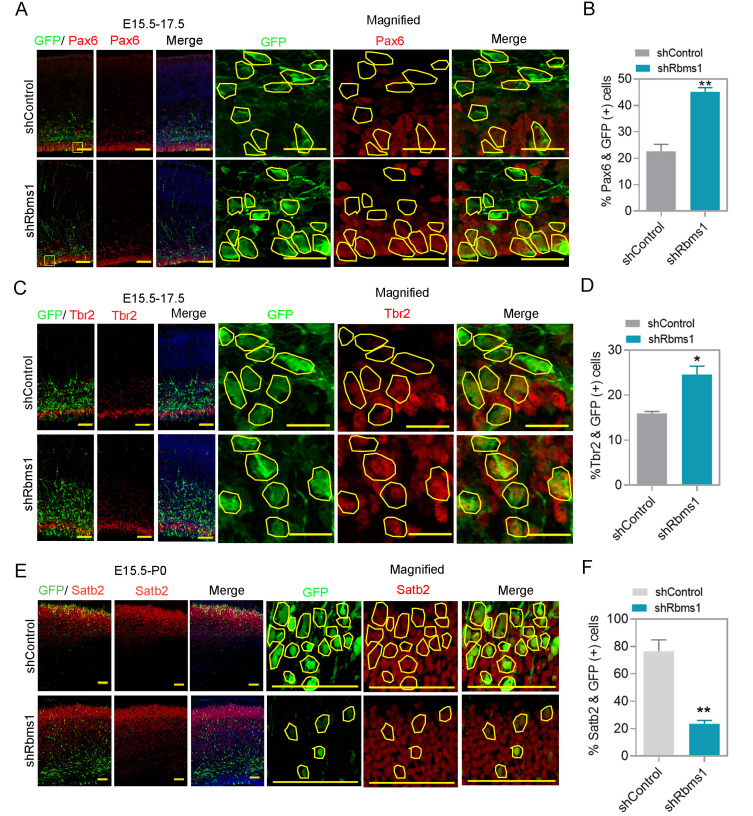
For cultured primary neurons collected from E13.5 brain, the levels of Rbms1 and the differentiation marker Tuj1 inversely correlated, similar to the IHC and Western data from the cortical slices for the various developmental time points suggested that Rbms1 might also have a role in neuronal differentiation (Fig. 1B). We then performed an IUE with the knockdown vector targeting Rbms1 of E15.5 mouse brain with harvest at E17.5 and immunostained brain slices with antibody against Pax6, the marker for neocortical/basal progenitors. We found that knockdown Rbms1 led to a higher percentage (45%) versus control (22%) for GFP-labeled Pax6 positive cells in the VZ and SVZ (Figs. 3A and 3B). For intermediate progenitor cells, knockdown Rbms1 increased Tbr2 positive GFP labeled cells (25%) compared with control (15%) in the SVZ to intermediate zone for undifferentiated and intermediate progenitors (Figs. 3C and 3D). With these observations, we conclude that knockdown of Rbms1 increases the proportion of basal progenitor cells in the VZ and SVZ. For long term effects of knockdown Rbms1 in the developing brain, we immunostained for Satb2, a marker for layer II-V colossal axon projecting neuron, in the cortices of E15.5 injected and P0 harvested samples and found that Rbms1 knockdown samples had reduced (20%) compared with the control samples (75%) for the number of GFP-labeled Satb2 positive cells (Figs. 3E and 3F).
Fig. 3. Rbms1 regulates cortical progenitor differentiation in vivo.
(A) Immunostaining for GFP (green) and Pax6 (red) at the SVZ in E17.5 mouse cortices two days after electroporation with shcontrol and Rbms1 knockdown vector, respective GFP labeled cells marked with yellow lines. Scale bars = 50 µm (normal image), 20 µM (magnified image) (n = 3). (B) Quantify the percentage of GFP-labeled Pax6+ cells in the VZ and SVZ (VZ/SVZ). Data presented as mean ± SEM; **P < 0.005 (n = 3, Student’s t-test). (C) Immunostaining for GFP (green) and Tbr2 (red) at the SVZ in E17.5 mouse cortices two days after electroporation with shcontrol and Rbms1 knockdown vector, respective GFP labeled cells marked with yellow lines. Scale bars = 50 µM (normal image), 20 µM (magnified image) (n = 3). (D) Quantification of GFP-labeled Tbr2+ cells in the VZ and SVZ (VZ/SVZ). Data presented as mean ± SEM, *P < 0.05 (n = 3, Student’s t-test). (E) Immunostaining for GFP (green) and Satb2 (red) at the upper-layer of postnatal day 0 mouse cortices five days after electroporation of shcontrol and Rbms1 knockdown vector, respective GFP labeled cells marked with yellow lines. Scale bars = 100 µM (normal image), 50 µM (magnified image) (n = 3, Student’s t-test). (F) Quantification of GFP-labeled Satb2+ cells in the upper-layer cortex. Data presented as mean ± SEM; **P < 0.05 (n = 3, Student’s t-test).
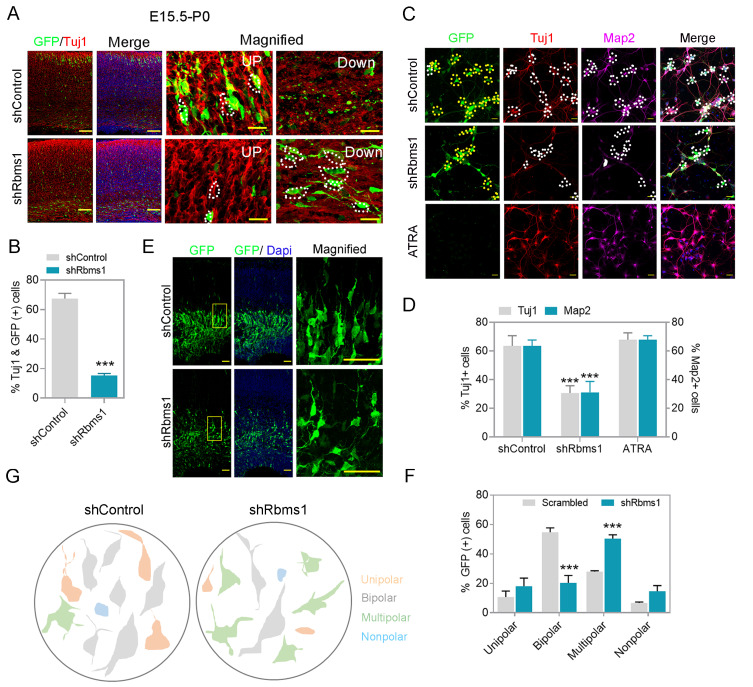
On whether knockdown of Rbms1 prevented or delayed the progenitor to neuron transition, we injected the Rbms1 knockdown vector at E15.5 and harvested at P0 and immunostained with Tuj1, an immature neuronal marker. We found that knockdown of Rbms1 significantly decreased the percentage of Tuj1 positive GFP labeled cells (18%) compared with the control vector (65%) (Figs. 4A and 4B). This data confirmed a retardation of migrating neurons maturing to Tuj1 positivity with the knockdown of Rbms1. This motivated us to figure out on whether the Rbms1 knockdown also converted a neuronal to astrocytes conversion. To test this, we immunostained E15.5 transfected and P0 harvested brain cortices with for the astrocytes marker, and did not found any astrocyte conversion due to knockdown of Rbms1. In vitro experiments with cultured neurons also had reduced differentiation markers (Tuj1 and Map2) with the knockdown of Rbms1 compared with control vector transfection; all-trans retinoic acid (ATRA) served as a positive control (Figs. 4C and 4D). In N2A cells, knockdown of Rbms1 also blunted the percentage of Tuj1 positive cells (Figs. 5A-5C). We conclude that optimal levels of Rbms1 are required for both migration and maturation of various neuronal cells.
Fig. 4. Knockdown of Rbms1 decreased differentiation in vivo and in vitro, and delayed multipolar, bipolar transition.
(A) Immunostaining for GFP (green) and Tuj1 (red) in the upper and lower layer of the postnatal days 0 mouse cortices five days after electroporation with shcontol (control) or knockdown Rbms1 vector (shRbms1) GFP and Tuj1 positive cells marked with white dots. Scale bars = 100 µM (normal image), 20 µM (magnified image). (B) Quantification of the percentage of GFP-labeled Tuj1+ cells in the upper and lower level of the mouse cortices. Data presented mean ± SEM; ***P < 0.001 (n = 3, Student’s t-test). (C) Knockdown Rbms1 and corresponding control lenti particle were infected with primary neuron at DIV1 and measured the Tuj1 (red), GFP, and Tuj1 positive cells marked with white dots in control, and shRbms1 white dots represent Tuj1 negative and GFP positive cells, and Map2 (purple) positive neurons by immunocytochemistry at DIV3, map2 and GFP positive cells marked with white dots in control and shRbms1 white dots represent GFP positive and Map2 negative cells, yellow dots represent GFP positive cells both in control and shRbms1, ATRA was a positive control. Scale bars = 100 µM. (D) Quantification of the percentage of Tuj1+ and cells in the three different experiments. Data presented mean ± SEM, ***P < 0.001 (n = 3, Student’s t-test). (E) Immunostaining for GFP (green) and Dapi (blue) at the SVZ in E17.5 mouse cortices two days after electroporation with shcontrol and Rbms1 knockdown vector. Scale bars = 50 µM (n = 4). (F) Quantification of the percentage of GFP-labeled unipolar, bipolar, multipolar, and nonpolar cells in the SVZ and IZ. Data presented mean ± SEM; ***P < 0.001 (n = 3, Student’s t-test). (G) Trace of GFP positive cells was redrawn by Paint software (Microsoft, USA), representing shcontrol and shRbms1.
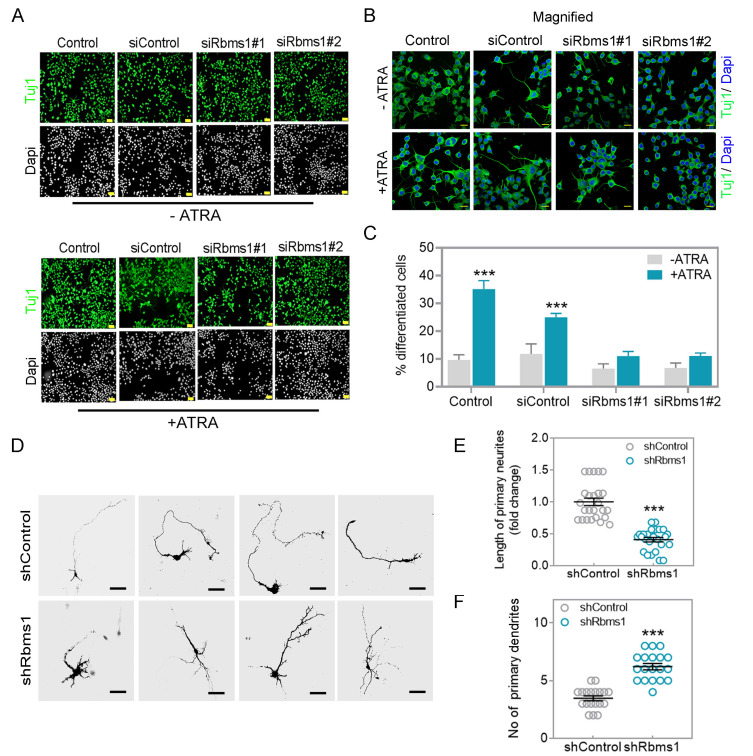
Fig. 5. Knockdown of Rbms1 suppressed ATRA mediated differentiation in N2A cells and decreased primary neurite length in vitro.
(A) Two siRbms1 and respective si negative control transfected N2A cells were subjected to 5 µM of ATRA (lower panel) or without ATRA (upper panel) in low serum conditions. After 48 h, immunostained with Tuj1 (green) and Dapi (gray). Scale bars = 100 µM. (B) Magnification of Fig. 5A Tuj1 green and Dapi blue. Scale bars = 20 µM. (C) Quantify the percentage of differentiated cells from three different experiments; ***P < 0.001 (Student’s t-test). (D) In vivo transfected GFP labeled shRbms1 and shcontrol primary neurons were cultured until DIV4 and immunostained with anti-GFP. Scale bar = 50 µM. (E) Length of primary neurite measured by Image J Fiji. ***P < 0.001 (Student’s t-test). (F) No of primary dendrites, data presented as mean ± SEM; ***P < 0.001 (n = 15, Student’s t-test).
Rbms1 is required for multipolar to bipolar transition of post mitotic neurons
During the development of the cerebral cortex, newborn neurons produced in VZ transition from a multipolar phenotype to a bipolar one. Upon coming into close proximity to the RGCs, they established a bipolar morphology with one elongated process/ neurite and start migration toward the cortical plate (CP) using the RGC long processes as guide for their radial migration towards the neocortical mantle. To identify any neuronal polarization defects related to the levels of Rbms1, the knockdown of Rbms1 was performed via IUE at E15.5 and then soon afterwards at E17.5, the morphology of the transfected newborn neurons was analyzed. Around 60% of control cells in intermediate zone (IZ) had bipolar shape but in comparison only about 15% of shRbms1 treated cells (Figs. 4E-4G). For the multipolar cells, there were about 25% in the control samples and 55% in the shRbms1 group (Figs. 4E-4G). To further quantitate the differences in the multipolar to bipolar ratios, we cultured the in utero transfected primary neurons and cultivated them until DIV4. They were then immunostained with anti-GFP antibody to visualize their shape. From ImageJ software analysis for single neurite tracing, knockdown of Rbms1 also decreased the neurite length and increase the number dendrites in the transfected cells (Figs. 5D-5F). Together these data support optimal Rbms1 levels being required for multipolar to bipolar transition of neurons during radial migration.
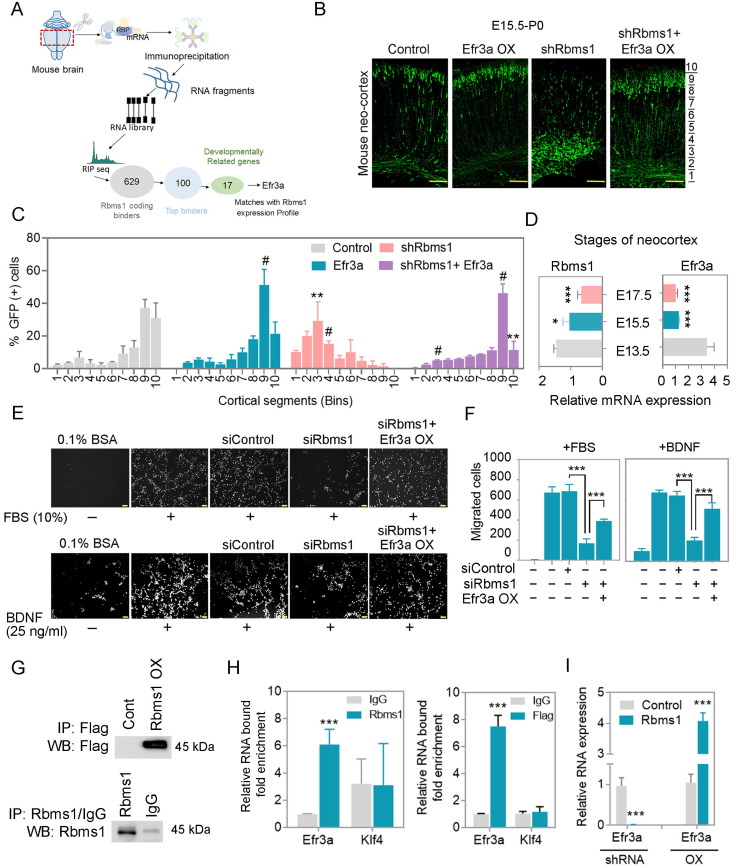
Identification of Efr3a as target RNA for Rbms1 in the developing mouse brain
We profiled the targets of Rbms1 as an RBP in the mouse embryonic brain via RIP-seq (Fig. 6A). This involved anti-Rbms1/isotype control immunoprecipitation, followed by crosslinking and sequencing of the associated RNA transcripts from E15.5 whole brain lysates (Supplementary Figs. S3A-S3C). Efficiency of the immunoprecipitation of Rbms1 was judged by immunoblotting with the anti Rbms1 antibody. The immunoprecipitated RNA was subjected to high-throughput RNA sequencing (RNA-seq) using an Illumina HiSeq platform. From RIP-seq reads, we found 629 coding transcripts, 35 lncRNA and 15 snoRNA species (Supplementary Fig. S3A). Gene ontology analysis revealed these binders being involved in RNA splicing, PI3K-AKT pathway, mTOR pathway, differentiation, migration and neurogenesis (Supplementary Fig. S3C). Among these targets, we selected 100 coding genes as being top binders; from those, 17 were known to be developmentally related from a scan of Mouse Genome Informatics database (www.informatics.jax.org/) and being involved in neurogenesis. We validated the binders by RIP-qPCR. A further narrowing of the list was by selecting the targets whose expression in the neocortex correlated with the levels of Rbms1 during neurogenesis (Fig. 6D). From these targets, Efr3a was selected for further study. We then used lentivirus knockdown (KD) and overexpression (OX) versions of Rbms1 to test for correlative changes with respect to Efr3a in cultured cortical neurons with qPCR. Efficiency of lentivirus vector mediated KD and OX of Rbms1 in primary neurons was showed by immunoblotting (Supplementary Fig. S3E). qPCR experiments showed Efr3a RNA levels to significantly and positive correlate with recombinant changes of Rbms1 protein levels (Fig. 6I, Supplementary Fig. S3F). Overexpression of a Flag-tagged version of Rbms1 in N2a cells also led to increased Efr3a RNA message levels by qPCR, similar to the previous findings with untagged Rbms1 (Figs. 6G and 6H). The non-binder RNA message for Klf4 did not correlate. These results support Rbms1 protein as an RBP binding and promoting the levels of Efr3a RNA message.
Fig. 6. Rbms1 regulates migration through Efr3a.
(A) Schematic diagram of Efr3a selection from the RIP seq. (B) IUE of Efr3a and knockdown Rbms1 along with control vector and Efr3a and knockdown vector at E15.5 and harvested at P0 and immunostained with anti-GFP (green). The area from the outer layer of the CP to the inner layer of the VZ was divided into ten different bins. Scale bars = 100 µM (n = 3). (C) Quantification of the percentage of GFP positive cells in the ten different cortical segments (Bins) areas. Data presented as mean ± SEM; Student’s t-test; **P < 0.01 #P < 0.001 (n = 3). (D) Relative mRNA expression of Rbms1 and Efr3a from different stages of developing brain by qPCR. Data presented as mean ± SEM; Student’s t-test; *P < 0.05, ***P < 0.001 (n = 3). (E) siRbms1 and si negative control, and siRbms1 and Efr3a transfected TR cells migration were measured by Boyden transwell migration assay and Immunocytochemistry against Dapi after 24-h migration. Vehicle or 10% FBS was present (upper panel), and Vehicle or BDNF (25 ng/ml) was present (lower panel). Scale bars = 100 µm. (F) Quantification of Fig. 6E, left panel with FBS (10%) and right panel with BDNF (25 ng/ml). Data presented as mean ± SEM; Student’s t-test; ***P < 0.001 (n = 3). (G) In the upper panel, over-expressed Flag Rbms1 and respective control vectors were transfected in N2A cells and harvested after 48 h; the lysate was immunoprecipitated (IP) with anti-Flag and immunoblot by anti-flag antibody. In the down panel, overexpressed pCAGIG-Rbms1 vector and pCAGIG vector were transfected in N2A cells, and then lysates were immunoprecipitated with anti Rbms1 and IgG and immunoblot with anti Rbms1. (H) N2a cells were transfected with Rbms1 expression vector and 48 h after transfection perform RIP and checked the binding by qPCR (left panel), and N2a cells were transfected with Flag-tagged Rbms1 expression vector and 48 h after transfection perform RIP and checked the binding by qPCR (right panel), and klf4 kept as a negative control. Data presented as mean ± SEM; Student’s t-test; ***P < 0.001. (I) shRbms1 and overexpressed Rbms1 lentivirus particles and their corresponding control lentivirus particles were infected with primary neurons and measured the relative mRNA expression of Efr3a and other targets in the supplementary figure. Data presented as mean ± SEM; Student’s t-test; ***P < 0.001.
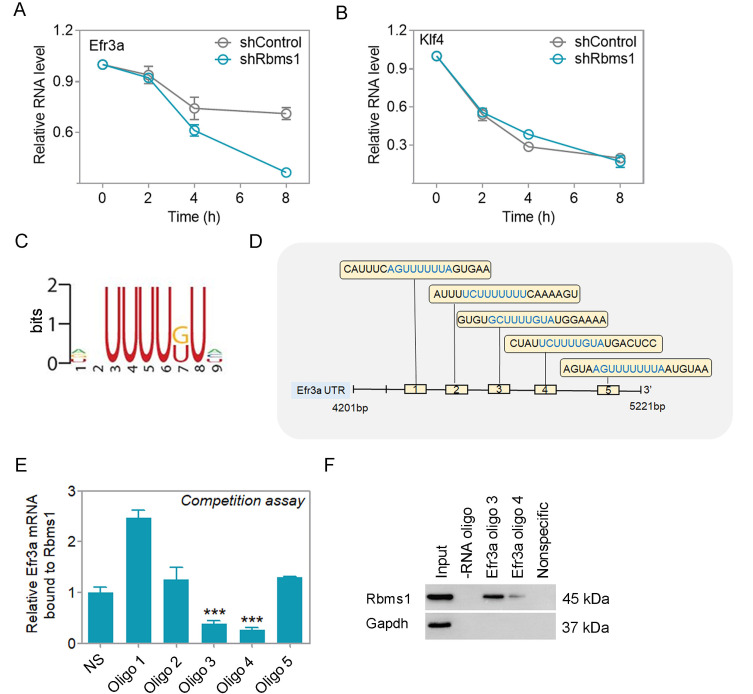
Rbms1 increases the stability of Efr3a RNA message
N2a cells were transfected to test whether Rbms1 promotes the stability of Efr3a RNA message. N2a cells were transfected with control vector or vector for knockdown Rbms1 followed by transcriptional inhibitor Actinomycin D block of the cells and their cellular RNA harvest after 0, 2, 4, and 8 h post block. The levels of Efr3a and the negative control and non-binder Klf4 RNA were quantitated by qPCR. In these cells, knockdown of Rbms1 led to a more rapid, time-dependent decrease in Efr3a cellular RNA levels and not for the negative control Klf4 (Figs. 7A and 7B). This implied that cellular Rbms1 levels lead to stabilization of Efr3a RNA message.
Fig. 7. Rbms1 stabilizes Efr3a by binding with its 3' UTR.
(A) N2a cells were transfected with control, and knockdown Rbms1 vector 48 h after adding 5 µg/ml Actinomycin D and harvested after 0, 2, 4, and 8 h after and checked the stability of Efr3a mRNA by qPCR and (B) klf4 as a negative control. (C) The binding motif of Rbms1 was reported by Yu et al. (2020). (D) Schematic Diagram of 5 RNA oligoes in the UTR of Efr3a. (E) Binding competition of Rbms1 and different RNA oligoes from 3’ UTR of Efr3a and nonspecific RNA oligoes. Data presented as mean ± SEM; Student’s t-test, ***P < 0.001. (F) Biotinylated RNA oligoes specific to Efr3a were used to pulldown Rbms1 protein by RNA pulldown assay in brain lysate and checked the pull-down by immunoblot anti Rbms1. Nonspecific oligo and beads not coated with any oligoes served as negative controls in the capture.
We found that there are five potential Rbms1 consensus binding sequences (Fig. 7C) in the 3’ UTR region of Efr3a (Fig. 7D, Supplementary Fig. S3H). To identify the optimal potential binding site of Efr3a RNA on Rbms1, we performed RIP of Rbms1, followed by incubation with excess amounts RNA oligoes from Efr3a 3’ UTR to outcompete the binding of native Efr3a RNA to the immunoprecipitated Rbms1. We found that oligoes 3 and 4 were best able to outcompete the native Efr3a RNA compared with the nonspecific oligo (NS) and oligoes 1, 2, and 5 (Fig. 7E). To verify that oligoes 3 and 4 could bind to Rbms1 protein, the oligoes were 5’-end biotin labelled, immobilized on streptavidin agarose beads and were used to reverse capture native Rbms1 protein from cell lysates. Compared with the nonspecific labeled oligo, oligo 4, oligo 3 was best able to capture native Rbms1 protein and oligo 4 was least capable (Fig. 7F). Around the region for oligo 3, the sequences were conserved between human, mouse and rat (Supplementary Fig. S3G). From these results, we propose that as an RBP, Rbms1 binds and stabilizes the RNA message for Efr3a at the cellular level.
Rbms1 promotes neuronal migration through Efr3a
Efr3a mutation is related to the incidence of ASD as it is also expressed in adult human brain tissue (Gupta et al., 2014). In the developing mouse embryos, we compared the RNA expression of Rbms1 and Efr3a by qPCR at stages E13.5, E15.5, and E17.5 (Fig. 6D) and found that they followed similar patterns. Efr3a and Rbms1 expressions in developing neocortex were high at E13.5 and gradually decreased in the later developmental stages. To ascribe a role for Efr3a in the developing neocortex, we performed an IUE at E15.5 with Efr3a overexpressing vector (Efr3a OX), and compared any effects on radial migration compared the control vector injected samples at P0 (Figs. 6B and 6C). It was found that Efr3a ectopic overexpression did not disrupt or slow radial migration, and in fact, it promoted it when compared with control (Figs. 6B and 6C). In the same experiment, the knockdown of Rbms1 (Rbms1 KD) had the opposite effect, as also shown previously, and it retarded the radial migration of the newly born neurons. When Efr3a OX and Rbms1 KD vectors were co-transfected, the net result was similar to the control vector samples, with Efr3a overexpression correcting the defect from loss of Rbms1 (Figs. 6B and 6C). Recovery from Rbms1 knockdown by Efr3a was also tested in a trans-well migration assay using mouse TR cells in a Boyden chamber with migration induced either with BDNF or fetal bovine serum as chemoattractants. We found that Efr3a overexpression could also nearly repair the migration defect due to Rbms1 knockdown in these cells (Figs. 6E and 6F). These data suggest that Efr3a as an RNA substrate of Rbms1 can functionally mediate the radial migration promotion properties of Rbms1 in the developing mouse neocortex.
DISCUSSION
RBPs promote multiple functions with respect to their RNA substrates and in the brain, these affect development in the brain (La Fata et al., 2014; Xia et al., 2018). In this study, we studied the role of one such RBP, namely Rbms1, with respect to being required for radial migration of maturing neurons in the developing neocortex. Previously, homozygous deletion of this protein leads to defective implantation of the embryos and reduced rates of implantation; however, to our knowledge, the brain development was not studied in the null homozygous litters (Fujimoto et al., 2001). A role for Rbms1 in the developing neocortex has not been described and by using recombinant techniques of in utero knockdown and overexpression of Rbms1, we demonstrate a requirement for Rbms1 during neocortical brain development, as it is also notably expressed in the VZ/SVZ in the early embryonic brain (Fig. 1A). Knockdown of Rbms1 impairs radial migration, differentiation and cell polarization (Figs. 2-4), but it did not change the conversion of cell type from neuron to astrocytes (Supplementary Fig. S4A). However, the staining of neuronal progenitors with Ki67 marker of proliferation was not affected for the E15.5 to E17.5 window (Supplementary Fig. S2C). Interestingly, Rbms1 increases cell proliferation and transformation for colony formation in NIH3T3 cells only when the cells co-express recombinant c-myc and H-ras. Rbms1 overexpression by itself (without added c-myc and H-ras) had no effect in the colony formation assay, implying that there are additional factors required for an Rbms1 proliferative response. There may also be differences in the comparison of different cell types and for the differences in the in vivo versus in vitro settings (Niki et al., 2000).
IUE of Rbms1 knockdown vector at E15.5 and assay at E17.5 revealed a major delayed transformation from multipolar to bipolar morphology of neurons in the SVZ and IZ compared with control (Figs. 4E and 4F). As N-cadherin levels are a known regulator in this multipolar to bipolar transition stage, we checked the expression of N-cadherin after knockdown of Rbms1 but those levels were only changed slightly in the primary neurons from E15.5 brain samples, transfected for 48 h (Supplementary Fig. S2E). As Efr3a was able to compensate for loss of Rbms1 in migration and differentiation defects, we assume that the differentiation defect lies downstream of Efr3a signaling and this requires further analysis.
Efr3a is a plasma membrane associated protein and studies have identified a role in keeping an active pool of PI4KA at the plasma membrane (Baird et al., 2008; Nakatsu et al., 2012). PI4KA is a lipid kinase, required to synthesize phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P] that is hydrolyzed by phospholipase C (PLC) to generate the secondary messenger molecules, Ins(1,4,5)P3 and diacylglycerol (Qian et al., 2017). Both Efr3a and the related Efr3b are highly expressed in the brain, suggesting that they play essential roles in the nervous system (Bojjireddy et al., 2015; Nakatsu et al., 2012). Rare mutations of the human EFR3A gene have been found in a number of ASD cases; however, we do not know if these affect the functioning of Efr3a as in the study, functional assays were not performed, although a comparison with a known crystal structure of the portion of the molecule was made (Gupta et al., 2014). Interestingly, brain-specific ablation of adult mouse Efr3a in the hippocampus did affect the neural stem cell/NPC differentiation or proliferation; thus, there may be differences in the role of Efr3a in the embryonic versus the adult mouse brain and roles for radial migration of neurons in the neocortex compared with proliferation/differentiation in the adult hippocampus (Qian et al., 2017).
In this study, we validated the regulation of Efr3a RNA cellular levels being dependent on Rbms1 protein levels and Efr3a RNA being one of the top binders to Rbms1. Using synthetic RNA oligoes specific to a conserved region of 3’UTR of Efr3a, we also demonstrated specific capture of cellular Rbms1 protein (Fig. 7F). Finally, over expression of Efr3a reversed the migration defects due by the knockdown of Rbms1, implying that the Rbms1 effect on radial migration may be mediated by Rbms1 binding and stabilizing the RNA message for Efr3a and thus allowing the cell to maintain sufficient levels of Efr3a to promote radial migration of the newly born neurons in the neocortex from the ventricular zone to cortical plate. In this study, we could not rule out the involvement of additional roles for Efr3a, besides radial migration and whether there are additional modulators of Efr3a, besides Rbms1, in the scenarios described. Together, this study describes a novel role for Rbms1 and its substrate Efr3a underlying neuronal migration and cortical formation.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Foundation of Korea (NRF) grant funded by the Korean Government (2019M3C7A1032601, 2021R1A2C1005395, 2021M3E5D9021902) to S.-O.H.
Footnotes
AUTHOR CONTRIBUTIONS
S.-O.H. and A.S. concept and study design. K.H., K.B., and J.K. acquisition of data and analysis. K.H. and K.B. preparation of figures and sketches. K.H. and A.S. writing. A.S. review and editing. S.-O.H. funding acquisition. All authors have reviewed and agreed to this submission.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Baird D., Stefan C., Audhya A., Weys S., Emr S.D. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis R.J., Swanson M.S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Bishayee K., Habib K., Nazim U.M., Kang J., Szabo A., Huh S.O., Sadra A. RNA binding protein HuD promotes autophagy and tumor stress survival by suppressing mTORC1 activity and augmenting ARL6IP1 levels. J. Exp. Clin. Cancer Res. 2022;41:18. doi: 10.1186/s13046-021-02203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojjireddy N., Guzman-Hernandez M.L., Reinhard N.R., Jovic M., Balla T. EFR3s are palmitoylated plasma membrane proteins that control responsiveness to G-protein-coupled receptors. J. Cell Sci. 2015;128:118–128. doi: 10.1242/jcs.157495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C.D., Yazdani N. RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 2016;15:169–186. doi: 10.1111/gbb.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha I.J., Lee D., Park S.S., Chung C.G., Kim S.Y., Jo M.G., Kim S.Y., Lee B.H., Lee Y.S., Lee S.B. Ataxin-2 dysregulation triggers a compensatory fragile X mental retardation protein decrease in Drosophila C4da neurons. Mol. Cells. 2020;43:870–879. doi: 10.14348/molcells.2020.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Sadra A., Kang J., Ryu J.R., Kim J.H., Sun W., Huh S.O. Farnesylation-defective Rheb increases axonal length independently of mTORC1 activity in embryonic primary neurons. Exp. Neurobiol. 2019;28:172–182. doi: 10.5607/en.2019.28.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M.S., Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Matsumoto K., Iguchi-Ariga S.M., Ariga H. Disruption of MSSP, c-myc single-strand binding protein, leads to embryonic lethality in some homozygous mice. Genes Cells. 2001;6:1067–1075. doi: 10.1046/j.1365-2443.2001.00488.x. [DOI] [PubMed] [Google Scholar]
- Gupta A.R., Pirruccello M., Cheng F., Kang H.J., Fernandez T.V., Baskin J.M., Choi M., Liu L., Ercan-Sencicek A.G., Murdoch J.D., et al. Rare deleterious mutations of the gene EFR3A in autism spectrum disorders. Mol. Autism. 2014;5:31. doi: 10.1186/2040-2392-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner W.B., Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Iida M., Taira T., Ariga H., Iguchi-Ariga S.M. Induction of apoptosis in HeLa cells by MSSP, c-myc binding proteins. Biol. Pharm. Bull. 1997;20:10–14. doi: 10.1248/bpb.20.10. [DOI] [PubMed] [Google Scholar]
- Ji L., Bishayee K., Sadra A., Choi S., Choi W., Moon S., Jho E.H., Huh S.O. Defective neuronal migration and inhibition of bipolar to multipolar transition of migrating neural cells by Mesoderm-Specific Transcript, Mest, in the developing mouse neocortex. Neuroscience. 2017;355:126–140. doi: 10.1016/j.neuroscience.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Keene J.D., Komisarow J.M., Friedersdorf M.B. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kimura K., Saga H., Hayashi K., Obata H., Chimori Y., Ariga H., Sobue K. c-Myc gene single-strand binding protein-1, MSSP-1, suppresses transcription of alpha-smooth muscle actin gene in chicken visceral smooth muscle cells. Nucleic Acids Res. 1998;26:2420–2425. doi: 10.1093/nar/26.10.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fata G., Gartner A., Dominguez-Iturza N., Dresselaers T., Dawitz J., Poorthuis R.B., Averna M., Himmelreich U., Meredith R.M., Achsel T., et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci. 2014;17:1693–1700. doi: 10.1038/nn.3870. [DOI] [PubMed] [Google Scholar]
- Lunde B.M., Moore C., Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A.E., Minet E., Stern C., Riahi S., Stiles C.D., Silver P.A. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.J., Ka M., Jung E.M., Kim W.Y. Genes and brain malformations associated with abnormal neuron positioning. Mol. Brain. 2015;8:72. doi: 10.1186/s13041-015-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Baskin J.M., Chung J., Tanner L.B., Shui G., Lee S.Y., Pirruccello M., Hao M., Ingolia N.T., Wenk M.R., et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki T., Izumi S., Saegusa Y., Taira T., Takai T., Iguchi-Ariga S.M., Ariga H. MSSP promotes ras/myc cooperative cell transforming activity by binding to c-Myc. Genes Cells. 2000;5:127–141. doi: 10.1046/j.1365-2443.2000.00311.x. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. Preparation of cross-linked cellular extracts with formaldehyde. Cold Spring Harb. Protoc. 2014;2014:1001–1003. doi: 10.1101/pdb.prot080879. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Martinez-Cerdeno V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Pilaz L.J., Silver D.L. Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. Wiley Interdiscip. Rev. RNA. 2015;6:501–515. doi: 10.1002/wrna.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q., Liu Q., Zhou D., Pan H., Liu Z., He F., Ji S., Wang D., Bao W., Liu X., et al. Brain-specific ablation of Efr3a promotes adult hippocampal neurogenesis via the brain-derived neurotrophic factor pathway. FASEB J. 2017;31:2104–2113. doi: 10.1096/fj.201601207R. [DOI] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Schwer B., Schneider S., Pei Y., Aronova A., Shuman S. Characterization of the Schizosaccharomyces pombe Spt5-Spt4 complex. RNA. 2009;15:1241–1250. doi: 10.1261/rna.1572709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira T., Maeda J., Onishi T., Kitaura H., Yoshida S., Kato H., Ikeda M., Tamai K., Iguchi-Ariga S.M., Ariga H. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells. 1998;3:549–565. doi: 10.1046/j.1365-2443.1998.00206.x. [DOI] [PubMed] [Google Scholar]
- Xia W., Su L., Jiao J. Cold-induced protein RBM3 orchestrates neurogenesis via modulating Yap mRNA stability in cold stress. J. Cell Biol. 2018;217:3464–3479. doi: 10.1083/jcb.201801143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H.S., Yong J. Alternative polyadenylation of mRNAs: 3'-untranslated region matters in gene expression. Mol. Cells. 2016;39:281–285. doi: 10.14348/molcells.2016.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Navickas A., Asgharian H., Culbertson B., Fish L., Garcia K., Olegario J.P., Dermit M., Dodel M., Hanisch B., et al. RBMS1 suppresses colon cancer metastasis through targeted stabilization of its mRNA regulon. Cancer Discov. 2020;10:1410–1423. doi: 10.1158/2159-8290.CD-19-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.