Abstract
Hepatocellular carcinoma (HCC) is an aggressive and incurable cancer. Although understanding of the molecular pathogenesis of HCC has greatly advanced, therapeutic options for the disease remain limited. In this study, we demonstrated that SETD5 expression is positively associated with poor prognosis of HCC and that SETD5 depletion decreased HCC cell proliferation and invasion while inducing cell death. Transcriptome analysis revealed that SETD5 loss downregulated the interferon-mediated inflammatory response in HCC cells. In addition, SETD5 depletion downregulated the expression of a critical glycolysis gene, PKM (pyruvate kinase M1/2), and decreased glycolysis activity in HCC cells. Finally, SETD5 knockdown inhibited tumor growth in xenograft mouse models. These results collectively suggest that SETD5 is involved in the tumorigenic features of HCC cells and that targeting SETD5 may suppress HCC progression.
Keywords: epigenetics, glycolysis, hepatocellular carcinoma, histone lysine methyltransferase, interferon response, SETD5
INTRODUCTION
Liver cancer is a globally prevalent disease, and its incidence has increased annually (Qiu et al., 2019). Among different types of liver cancer, hepatocellular carcinoma (HCC) is the most common, accounting for approximately 90% of liver cancers (Llovet et al., 2021). Although extensive efforts have reduced the mortality of many cancers through early detection and treatment, HCC remains challenging and has a poor prognosis (Siegel et al., 2018). Along with the difficulty of diagnosing asymptomatic cancerous outgrowth in the liver, limited effective therapeutic options for liver cancer are obstacles to improving HCC prognosis. Patients with early-stage HCC tumors can be cured by resection, transplantation or transarterial chemoembolization and ablation (Llovet et al., 2021). However, patients with advanced HCC who are not eligible for surgical treatments require systemic therapies. In an effort to develop more efficient systemic therapies, more than 10 candidate drugs have been clinically tested in recent decades, but only a few of these drugs, such as sorafenib, lenvatinib, regorafenib, and cabozantinib, have been approved (Chen et al., 2020). Intriguingly, the approved drugs are small molecules that inhibit multiple protein kinases rather than single molecular targets, and the efficacies of these drugs have been quite small. The overall survival of sorafenib-treated patients increased ~3 months compared with control treatment (Llovet et al., 2008). The effective response rate for lenvatinib was reported to be only 24% (Kudo et al., 2018). The median survival time of regorafenib administered to sorafenib-resistant patients was only 6.4 months (Bruix et al., 2017). In addition, the median progression-free survival of cabozantinib increased 3.3 months compared to the placebo group (Abou-Alfa et al., 2018). Targeting the tumor microenvironment by inhibiting angiogenesis (bevacizumab, ramucirumab) or immune checkpoints (atezolizumab, nivolumab, ipilimumab) through monoclonal antibody-based drugs was also approved for the treatment of advanced HCC and displayed prominent efficacy in some cases (Saung et al., 2021). However, these immunotherapies have been beneficial for only a subset of patients, the biomarkers of which remain unclear (Feng et al., 2021). These low efficacies of current approved therapies highlight the necessity to explore more therapeutic targets to overcome HCC.
Epigenetic alterations linked to transcriptional reprogramming or genomic stability are highly implicated in cancer (Shen and Laird, 2013). Epigenetic instability often mediates the phenotypic plasticity of cancer cells and thus promotes aggressive progression via drug resistance and metastasis (Brown et al., 2014). Several epigenetic regulators, including SET domain-containing histone lysine methyltransferases (KMTs), which enable the catalysis of histone and/or nonhistone protein methylation at lysine residues (Dillon et al., 2005), have emerged as potent biomarkers and/or therapeutic targets in different cancers (Jones et al., 2016). For example, EZH2 and MLL are intimately associated with the tumorigenesis of glioblastoma and leukemia (Kim and Roberts, 2016; Liedtke and Cleary, 2009). Other KMTs, including Suv39H1 and SETD2, were reported to be involved in the drug resistance of colorectal cancer and non-small-cell lung cancer, respectively (Kim et al., 2019; Lu et al., 2020). Together, these findings raise the possibility that the identification of KMTs involved in HCC progression may aid in the development of more efficient targets for HCC.
The human genome encodes at least 48 different SET domain-containing KMTs. Some of these KMTs have been well characterized, whereas the others remain understudied (Albert and Helin, 2010). SETD5 is a putative KMT-containing SET domain. The biological function of SETD5 was initially characterized in mouse models. SETD5-haploinsufficient mice exhibit severe developmental defects, such as abnormal brain-to-body weight ratios (Deliu et al., 2018) and impairment in the proliferative dynamics of neural progenitors (Sessa et al., 2019). At the cellular level, SETD5 loss reduced embryonic stem cell proliferation by increasing apoptosis and aberrant cell cycle progression (Osipovich et al., 2016). The physiological importance of SETD5 in humans was reported by studies to show loss-of-function mutations of SETD5 in intellectual disability and autism disorders (Deliu et al., 2018; Fernandes et al., 2018; Moore et al., 2019). Molecular analyses proposed that SETD5 might function through its interaction with the HDAC3 and PAF1 complexes (Deliu et al., 2018; Osipovich et al., 2016). More recently, SETD5 was shown to be involved in different cancers (Piao et al., 2020; Sowalsky et al., 2015; Yu et al., 2019; Yang et al., 2021). These studies demonstrate that SETD5 is implicated in aggressive tumorigenic progression. However, the functional involvement of SETD5 in liver cancer remains unclear.
In this study, we explored the clinical significance of 48 different SET domain-containing human KMTs in liver cancer progression and showed that SETD5 is a novel HCC-related gene. In subsequent functional analyses, we demonstrated that SETD5 promotes glycolytic activity and survival of HCC. Together, this study suggests SETD5 as a possible target to treat HCC.
MATERIALS AND METHODS
Cell culture
The human HCC cell lines HLE, HepG2, HLF, and Huh1 were obtained from the American Type Culture Collection (ATCC, USA). The human HCC cell line Huh7 was purchased from the Japanese Collection of Research Bioresources Cell Bank (JCRB, Japan). HepG2, Huh1, and Huh7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM), and HLE and HLF cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI 1640). All of these media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% (v/v) antibiotic-antimycotic. Cells were cultured at 37°C with 5% CO2.
Transfection reagents and chemicals
Small interfering RNA (siRNA) transfection and DNA transfection were performed using RNAiMAX (Invitrogen, USA) and Lipofectamine 3000 transfection reagent (Invitrogen), respectively. 2-Deoxy-D-glucose (D6134) and doxycycline (D9891) were purchased from Sigma-Aldrich (USA). Doxycycline was added at 50 ng/ml for short hairpin RNA (shRNA) induction.
Western blot analysis
Whole cells were lysed in a chilled lysis buffer (150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris pH 8.0) with protease inhibitor cocktail (Roche, Switzerland). The protein concentration was determined using the Bradford assay. The same amount of protein was resolved by SDS-PAGE. After electrophoresis, the proteins were transferred onto nitrocellulose membranes (BioTrace NT Nitrocellulose membrane; PALL, USA). Then, the membrane was blocked for 1 h at room temperature using 5% skim milk in TBST buffer. The membrane was incubated overnight at 4°C with the following primary antibodies: anti-SETD5 (MBS8245298, 1:1,000; MyBioSource, USA), anti-alpha-tubulin (SC-23948, 1:10,000; Santa Cruz Biotechnology, USA), anti-PARP (9542, 1:1,000; Cell Signaling Technology, USA), anti-H3K27me3 (07-449, 1:1,000; Sigma-Aldrich), anti-H3K4me3 (39159, 1:1,000; Active Motif, Germany), anti-H3K9me2 (39375, 1:1,000; Active Motif), anti-H3K36me3 (ab9050, 1:1,000; Abcam, UK), anti-total H3 (ab1791, 1:1,000; Abcam), anti-phospho-STAT1 (9167, 1:1,000; Cell Signaling Technology), anti-STAT1 (9172, 1:1,000; Cell Signaling Technology), anti-MDA5 (5321, 1:1,000; Cell Signaling Technology), and anti-PKM (ab137791, 1:1,000; Abcam).
Cell proliferation assay
To examine the growth rate of SETD5-depleted cells, cells were directly transfected in 96-well plates (Day 0). After transfection, the cells were analyzed 1 and 3 days after transfection. At the experimental time point, MTT solution was added, and the sample was incubated for 2 h in a 37°C incubator. Then, the medium was removed by suctioning, and insoluble formazan was dissolved in DMSO. The absorbance was measured at 570 nm using a Luminoskan Ascent Microplate Luminometer (Thermo Fisher Scientific, USA). The value at Day 3 was normalized to the value at Day 1, and the value of the measurement was calculated relative to the control. All experiments were conducted independently thrice. To examine the sensitivity of SETD5-depleted HCC cells to 2-DG, cells were directly transfected in 96-well plates. Twenty-four hours after transfection, the cells were treated with 2.5 mM 2-DG for 48 h. Cell viability was determined by measuring luminescent signals from cells treated with CellTiter-Glo reagent (G7572; Promega, USA) using a multilabel plate reader (Victor X3; Perkin Elmer, USA). All experiments were conducted independently thrice.
Transfection of small-interfering RNAs
For transfection, cells were treated with a complex containing RNAi duplex and Lipofectamine. To achieve 60%-70% confluence 24 h after plating, cells were diluted in complete growth medium without antibiotics. Complex-made RNAi duplexes and Lipofectamine RNAiMAX were prepared according to the manufacturer’s protocol. The complex was added to the diluted cells. At 48 h, the assays were examined. The siRNA for transfection is described in Supplementary Table S1.
Invasion assay
An invasion assay was performed using a plate with a pore size of 8 µm (3422; Corning, USA). Transwell inserts were precoated with human collagen type IV and Matrigel (354234; Corning) for 2 h. Cells (1 × 104) were suspended in 200 µl of serum-free DMEM. As a source of chemoattractants, the lower chamber contained DMEM supplemented with 10% (v/v) calf serum. After incubation for 24 h at 37°C, cells that passed through the Matrigel-coated membrane were stained with 0.5% crystal violet solution.
Clonogenic assay
Using a lentiviral vector, a stable SETD5-depleted cell line was constructed. The sequences of shRNA are shown in Supplementary Table S1. shSETD5 or shControl cells were seeded in 6-well plates (1 × 105 cells) and incubated for 7 days. After 7 days of incubation, the cells were washed in phosphate-buffered saline (PBS) and stained with crystal violet solution.
Soft agar assay
To investigate the anchorage-independent growth ability in SETD5-depleted cells, we employed the doxycycline-inducible cell line. Agar was prepared at concentrations of 0.6% and 1.2% in PBS. Using 2× FBS medium, the bottom agar was generated by mixing 2× FBS medium and 1.2% agar, and the final concentration was 0.6% agar. Agar (0.6%) was poured into 6-well plates and incubated for 1 h at room temperature until the agar solidified. After the bottom agar was solidified, the upper layer of agar was prepared. The upper layer was made by mixing 2× FBS medium containing 1 × 105 cells and 0.6% agar. The final concentration of the upper agar was 0.3%.
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed using RevertAid Reverse Transcriptase (Thermo Fisher Scientific). RT-PCR was performed with AccuPower PCR Master Mix (Bioneer, Korea) and PCR machinery (MiniAmp Plus Thermal Cycler; Thermo Fisher Scientific). Quantitative reverse transcription-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems, USA) and a StepOnePlus Real-Time PCR System (Applied Biosystems). All experiments were performed independently in triplicate. Primer sequences used in quantitative real-time RT-PCR listed in Supplementary Table S2.
Flow cytometry analysis
For cell cycle analysis, the cells were trypsinized and collected using ice-cold PBS containing 2% FBS. The cells were fixed with ethanol and incubated for at least 1 h. After fixation, the cells were treated with RNase and propidium iodide (PI) and were then analyzed by flow cytometry.
For cell death analysis, the cells were collected, including cells in the supernatant. The cells were washed twice using ice-cold PBS containing 2% FBS. The cells were prepared in suspension using 1× Annexin V Binding Buffer (556547; BD Bioscience, USA) followed by Annexin V and PI staining. The cells were incubated for 15 min at room temperature in the dark. Within 1 h, the samples were analyzed by flow cytometry.
RNA sequencing (RNA-seq) analysis
To profile the transcriptome in cells, total RNA was extracted using an RNeasy micro kit (74004; Qiagen, Germany). The RNA-seq library was made with total RNA using the TruSeq Stranded mRNA LT Sample Prep Kit according to the manufacturer’s protocol (RS-122-9004DOC; Illumina, USA), and sequencing was performed using the Illumina HiSeq2000 platform to generate 100-bp paired-end reads. The reference genome of humans was obtained from the National Center for Biotechnology Information (NCBI) genome (https://www.ncbi.nlm.nih.gov/genome/), and genome indexing was performed using STAR (v.2.5.1) (Dobin et al., 2013). The sequenced reads were mapped to the human genome (mm10) STAR, and the gene expression levels were quantified with the count module in the STAR. The edgeR (v.3.12.1) (McCarthy et al., 2012) package was used to select differentially expressed genes from the RNA-seq count data between conditions (fold change > 1.5, FDR [false discovery rate] < 0.05). The heatmap was generated using MeV (Howe et al., 2011) and R (v.3.5.0; https://www.r-project.org/) pheatmap package (v.1.0.12; https://cran.r-project.org/web/packages/pheatmap/index.html). NGS (next generation sequencing) data were deposited in the NCBI Gene Expression Omnibus under accession number GSE193431 and Korean Nucleotide Archive (KoNA; https://www.kobic.re.kr/kona/) with the accession ID, PRJKA220167.
Histone extraction
Histones were extracted as previously described (Shechter et al., 2007). Briefly, cells were collected and washed with PBS. Cell pellets were lysed in hypotonic lysis buffer (10 mM Tris-Cl pH 8.0, 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and protease inhibitor [Roche]). After centrifugation, the supernatant containing histones was stored in a tube. To precipitate histones, trichloroacetic acid solution (TCA solution) was added dropwise, and the sample was inverted to mix the solutions. After centrifugation, histone pellets were obtained.
In vitro histone methyltransferase assays
For in vitro histone methyltransferase assays, the Set1B complex was prepared as previously described (Kwon et al., 2020). Flag-tagged SETD5 was purified from insect cells using the same purification process as that described for the Set1B complex (Jeon et al., 2018). In vitro histone methyltransferase assays were also performed as previously described (Jeon et al., 2018). Briefly, for histone H3 methyltransferase assays, reactions containing 200 ng of recombinant histone H3 and 50 ng of purified SETD5 in 15 µl of reaction buffer (25 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 10% glycerol) supplemented with 100 µM SAM (S-adenosyl methionine; NEB, USA) were incubated at 30°C for 1 h. Proteins were resolved by SDS-PAGE and subjected to immunoblotting.
Extracellular acidification rate (ECAR) measurement
To measure the ECAR, we used the Agilent Seahorse XF Glycolysis Stress Test Kit (103020-100; Agilent Technologies, USA). Prior to the assay, cells were plated in a Seahorse XF microplate using DMEM. On the following day, the appropriate compounds were loaded into the infection ports of the sensor cartridge: glucose (25 mM), oligomycin (1 µM), and 2-DG (50 mM). Glycolytic measurements were calculated using the Seahorse XF glycolysis stress test report generator. The experiment was performed independently thrice.
Mouse xenografts
Six-week-old mice were used for xenograft studies. Cells (each 5 × 106) resuspended in HBSS were subcutaneously injected into the mice. When the tumor volume reached ~150 mm3, the mice were divided into two groups: one group was fed a doxycycline diet, and the other group was fed a standard diet. The tumor volumes and weights of individual mice were measured every other day. After 13 days on a doxycycline diet, mice were sacrificed to excise primary tumors.
Immunohistochemistry
Samples were fixed in 10% formalin and embedded in paraffin. The paraffin blocks were sectioned at 3 μm. Sections on microslides were deparaffinized with xylene and hydrated using a diluted alcohol series. Sections were treated with sodium citrate buffer (pH 6.0) at 95°C for 30 min. To minimize nonspecific staining, each section was blocked with 10% goat serum at room temperature for 1 h. The sections were then incubated with anti-Ki67 (ab16667, 1:200; Abcam) in PBS with 1% bovine serum albumin overnight at 4°C. To quench endogenous peroxidase activity, sections were immersed in 3% H2O2 and incubated for 15 min. Biotinylated antibody was incubated with the cells for 30 min. Sections were then incubated with peroxidase substrate (NovaRED; LSBio, USA). Finally, slides were counterstained with hematoxylin.
Statistical analysis
The results are represented as the means of three independent experiments ± SEM. The statistical significance of the differences between the control and experimental groups was calculated with Student’s t-tests (two-tailed, unpaired). For multiple-group comparisons, one-way ANOVA followed by Tukey’s multiple comparison test was performed. Prism 9 software (GraphPad Software, USA) was used to calculate the statistical significance.
RESULTS
Elevated SETD5 expression is associated with liver cancer progression
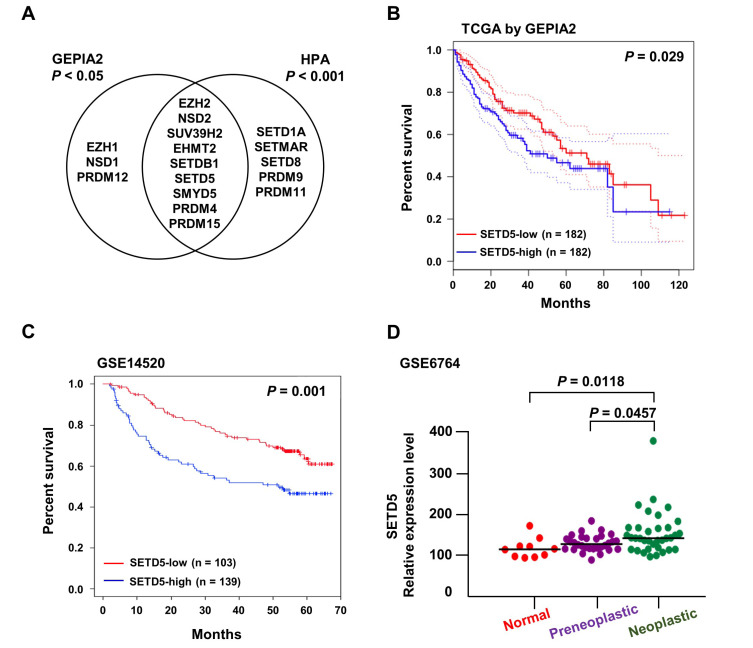
To identify liver cancer-related KMTs, we analyzed the clinical significance of KMT genes encoding SET domain-containing proteins (Albert and Helin, 2010). Gepia2 analysis of the TCGA database (Tang et al., 2019) revealed that high expression of 12 KMTs was associated with short survival of liver cancer patients (Figs. 1A and 1B, P < 0.05). In contrast, the expression levels of 14 KMTs were significantly correlated with poor prognosis of liver cancer in the Human Protein Atlas (Uhlen et al., 2017) (P < 0.001). Among the 9 genes identified by both analyses, we decided to further investigate the functional relevance of SETD5 in liver cancer, which has remained unclear to date. In an independent liver cancer cohort (GSE14520), high SETD5 expression displayed a significant correlation with poor patient survival compared with low SETD5 expression (Fig. 1C). In addition, SETD5 expression was highly associated with the carcinogenic process from normal to neoplastic liver (GSE6764, Fig. 1D). Together, these results suggest that upregulation of SETD5 in liver cancer is associated with unfavorable patient outcomes.
Fig. 1. Elevated SETD5 expression correlates with poor prognosis of HCC.
(A) Venn diagram displaying KMT genes, the high expression of which displays significant correlations with poor survival of liver cancer determined by GEPIA2 (http://gepia2.cancer-pku.cn/)- and Human Protein Atlas (HPA; https://www.proteinatlas.org/)-associated analyses. (B and C) Kaplan–Meier plots visualizing the significant association of SETD5 expression with low survival of liver cancer patients in TCGA (B) and GSE14520 (C) datasets. The red line depicts the SETD5-low expression group, and the blue line represents the SETD5-high expression group. (D) Correlation of SETD5 expression with HCC progression in GSE6764. HCC progression was divided into 3 stages: normal, preneoplastic, and neoplastic.
SETD5 depletion attenuates the tumorigenic features of liver cancer cells
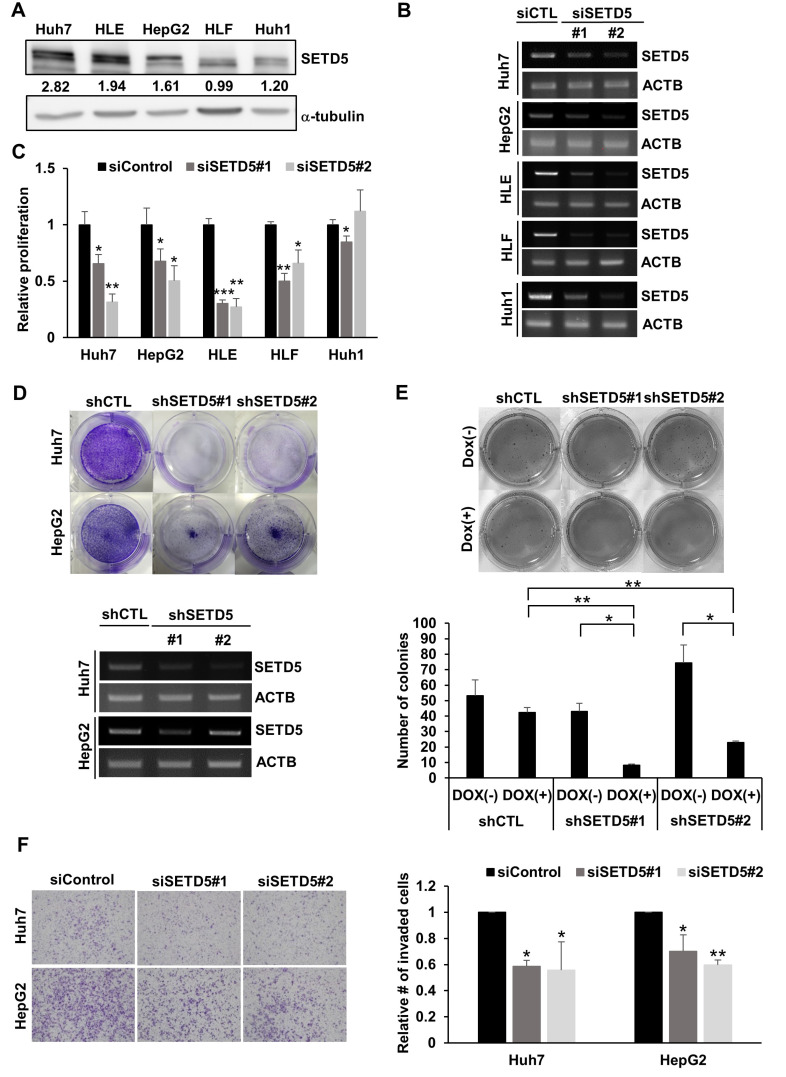
To assess SETD5 expression in liver cancer cell lines, we examined endogenous SETD5 protein levels in various HCC cell lines (Fig. 2A). All the tested cells displayed substantial amounts of SETD5 protein to different degrees. The highest level was noted in Huh7 cells, whereas the lowest was noted in Huh1 cells. To investigate whether SETD5 was involved in cancerous phenotypes, we genetically manipulated SETD5 expression. Knockdown (KD) of SETD5 by two independent siRNAs targeting SETD5 (Fig. 2B) impaired the growth and proliferation of all the cells examined except Huh1 cells (Fig. 2C). Clonogenic assays also showed that stable depletion of SETD5 via lentiviral shRNAs specific to SETD5 led to severe growth defects in Huh7 and HepG2 cells (Fig. 2D). We also observed that SETD5 KD significantly reduced sphere formation of Huh7 cells (Fig. 2E). Transwell invasion assays demonstrated that depletion of SETD5 impaired the motility of both Huh7 and HepG2 cells (Fig. 2F). Collectively, these findings indicate that SETD5 is involved in the tumorigenic features of HCC cells.
Fig. 2. SETD5 depletion impairs HCC cell proliferation and motility.
(A) Endogenous SETD5 protein levels in different HCC cell lines were determined by immunoblotting against SETD5. α-Tubulin served as the loading control. The intensities of immunoblotting signals were semiquantitatively measured by ImageJ. (B) RT-PCR to show siRNA-mediated SETD5 KD in different HCC cell lines. β-Actin served as the loading control. (C) SETD5 KD decreased proliferation in SETD5-depleted HCC cells, as determined by MTT assays (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA). (D) Upper: Clonogenic assay of Huh7 and HepG2 cells stably expressing shRNAs targeting SETD5 (shSETD5#1, shSETD5#2) and control shRNA (shCTL). Lower: SETD5 depletion was verified by RT-PCR visualized in agarose gel. ACTB was used as a loading control for RT-PCR. (E) Upper: Representative image of the soft agar assay to examine anchorage-independent growth of SETD5-depleted Huh7 cells. SETD5 KD was induced by doxycycline treatment. Dox(+) denotes cells treated with doxycycline, whereas Dox(–) indicates cells without doxycycline treatment. Lower: Quantification of colony numbers in soft agar assays counted by ImageJ (n = 3, *P < 0.05, **P < 0.01, one-way ANOVA). (F) Left: Transwell invasion assays showed that SETD5 KD in the indicated cells attenuated the invasion capacity. Right: The number of invaded cells was normalized to the control group (n = 3, *P < 0.05, **P < 0.01, one-way ANOVA).
SETD5 loss induces apoptotic cell death without affecting the cell cycle
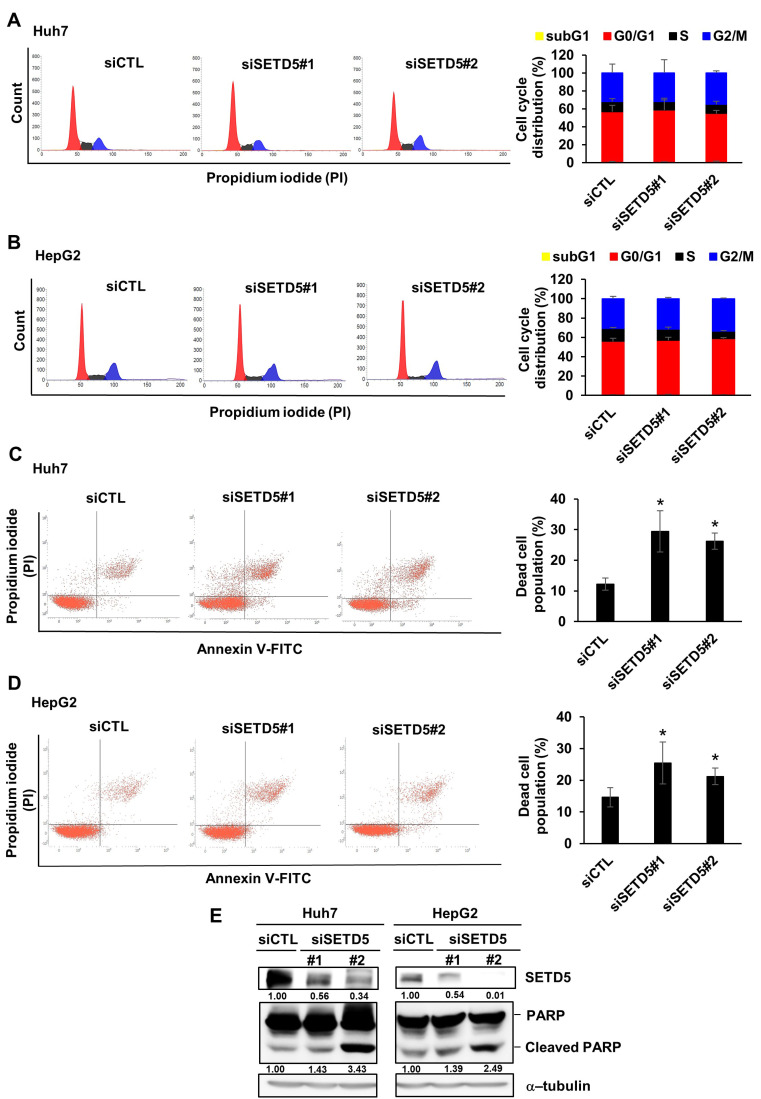
Then, we examined molecular pathways associated with defects in the growth and proliferation of SETD5 KD HCC cells. First, we analyzed the distribution of the cell cycle phase in Huh7 and HepG2 cells and found that SETD5 KD had little effect on the cell cycle (Figs. 3A and 3B). This finding demonstrates that SETD5 is not involved in regulating the cell cycle. In contrast, SETD5 KD increased the population of annexin V and PI double-stained cells, indicating that cells are dying via apoptosis (Figs. 3C and 3D). Moreover, cleaved PARP, a well-known molecular marker of apoptotic cell death, was elevated in SETD5-depleted Huh7 and HepG2 cells (Fig. 3E). These results indicate that SETD5 loss induced apoptotic death of liver cancer cells without affecting the cell cycle.
Fig. 3. SETD5 KD induces Huh7 and HepG2 cell apoptosis.
(A and B) Left: Effects of SETD5 KD in Huh7 and HepG2 cells on the cell cycle were examined by FACS analysis to detect propidium iodide signals. Right: Quantification of cell cycle distribution as determined using a flow cytometer. Values are the mean ± SEM of biological triplicate experiments. (C and D) Left: Effects of SETD5 KD in Huh7 and HepG2 cells on cell death were examined by FACS analysis based on annexin V and propidium iodide double staining. Right: Quantification of the dead cell population as determined using a flow cytometer. Values are the mean ± SEM of biological triplicate experiments (n = 3, *P < 0.05, one-way ANOVA). (E) Cleaved PARP in Huh7 and HepG2 cells was examined by immunoblotting with anti-PARP antibody. The intensities of immunoblotting signals were semiquantitatively measured by ImageJ.
SETD5 lacks intrinsic histone methyltransferase activity
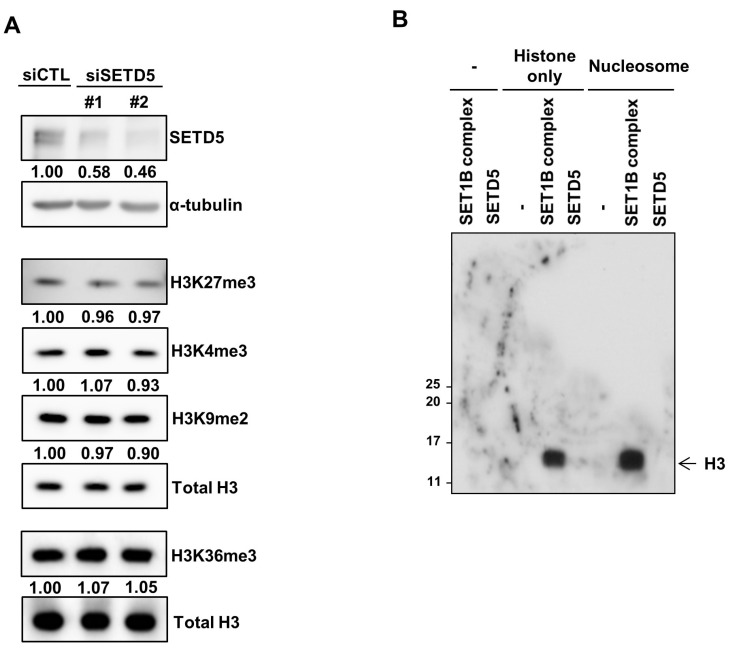
SETD5 contains a SET domain that generally exhibits lysine methyltransferase activity. We assessed whether potential KMT activity was involved in the function of SETD5 in HCC cells. To examine the effect of SETD5 KD on the methylation status of histones in HCC cells at the global level, we determined the levels of four well-known histone methylation markers: H3K27me3, H3K4me3, H3K9me2, and H3K36me3. Intriguingly, SETD5 KD in Huh7 cells had minimal effect on global levels of histone methylation marks (Fig. 4A). Then, we postulated that SETD5 might be involved in the methylation of only a subset of chromatin, a change that is subtle to detect at the global level. However, our in vitro histone methyltransferase assay showed that the SETD5 protein purified from insect cells failed to methylate any histone proteins in either the octamer or nucleosomal form (Fig. 4B). Of note, the SET1B complex used as a positive control for the assay (Lee et al., 2007) robustly generated methylated histones in both modes. This result suggests that unlike the SET1B complex, SETD5 lacks intrinsic histone methyltransferase activity. Therefore, it is unlikely that SETD5-mediated histone methylation underlies the functional involvement of SETD5 in HCC cells.
Fig. 4. SETD5 has no intrinsic histone methyltransferase activity.
(A) Global levels of four different histone methylation marks were determined using the indicated antibodies. Total H3 was used as the loading control. Immunoblots were semiquantitatively measured by ImageJ. (B) In vitro methyltransferase assays to examine the catalytic activity of SETD5 toward histones in either octamers (histone only) or nucleosomes. The SET1B complex was used as a positive control to display histone methyltransferase activity in this assay.
SETD5 depletion downregulates interferon (IFN)/inflammatory response pathways in HCC cells
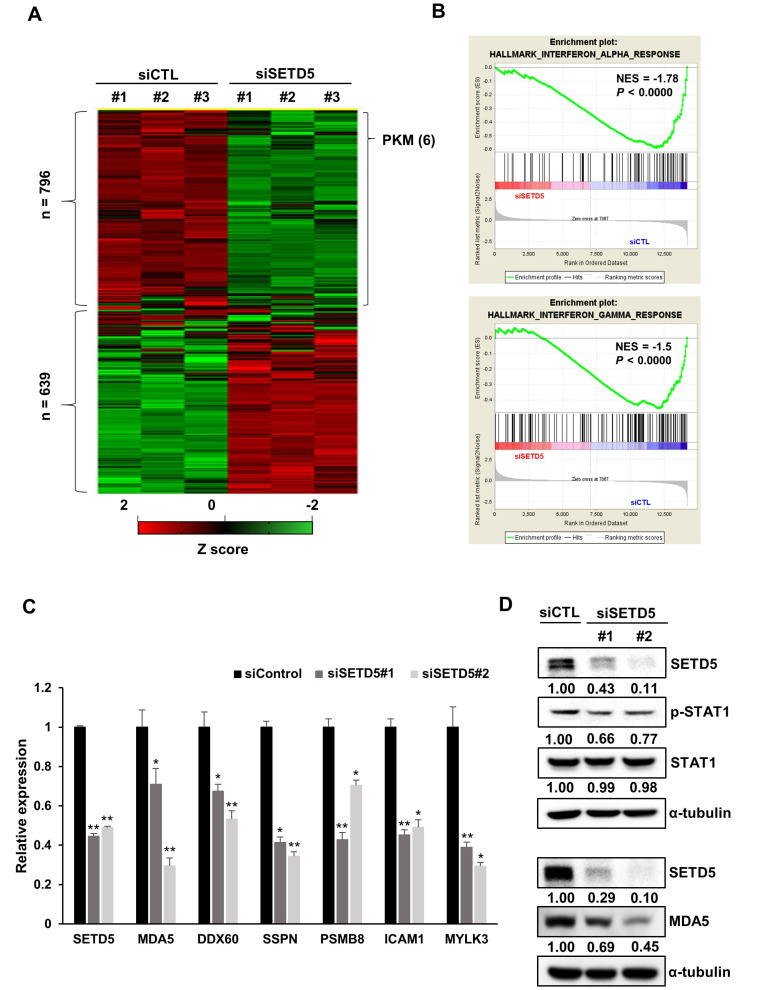
To identify the underlying mechanism by which SETD5 is involved in cancer cell survival, we analyzed transcriptomic changes in SETD5-depleted Huh7 cells by RNA-seq and found that 796 and 639 genes were differentially downregulated and upregulated, respectively (Fig. 5A). Gene set enrichment analysis showed that the inflammatory and IFN responses were significantly downregulated in SETD5 KD cells (Fig. 5B). The effect of SETD5 depletion on the expression of genes involved in the IFN/inflammatory pathways was validated by assessing the expression of individual representative genes, including MDA5 and DDX60, which are crucial for intrinsic inflammatory signals (Fig. 5C). Consistently, we observed that SETD5 KD decreased the MDA5 protein level as well as STAT1 phosphorylation, reflecting the activity of IFN and the inflammatory response pathways (Fig. 5D). These results imply that SETD5 is involved in the active state of IFN/inflammatory response pathways in HCC cells.
Fig. 5. SETD5 regulates the interferon-mediated inflammatory response in HCC cells.
(A) Heatmap showing the differentially expressed genes between control (siCTL) and SETD5 KD (siSETD5) Huh7 cells. (B) Gene set enrichment analysis displaying the downregulation of interferon responses in SETD5-depleted cells. (C) The mRNA levels of genes related to the interferon response in SETD5-depleted Huh7 cells were measured by quantitative real-time RT-PCR (n = 3, *P < 0.05, **P < 0.01, one-way ANOVA). (D) Immunoblots showed that SETD5 KD decreased the levels of MDA5 and phosphorylated STAT1.
SETD5 promotes glycolysis in HCC cells
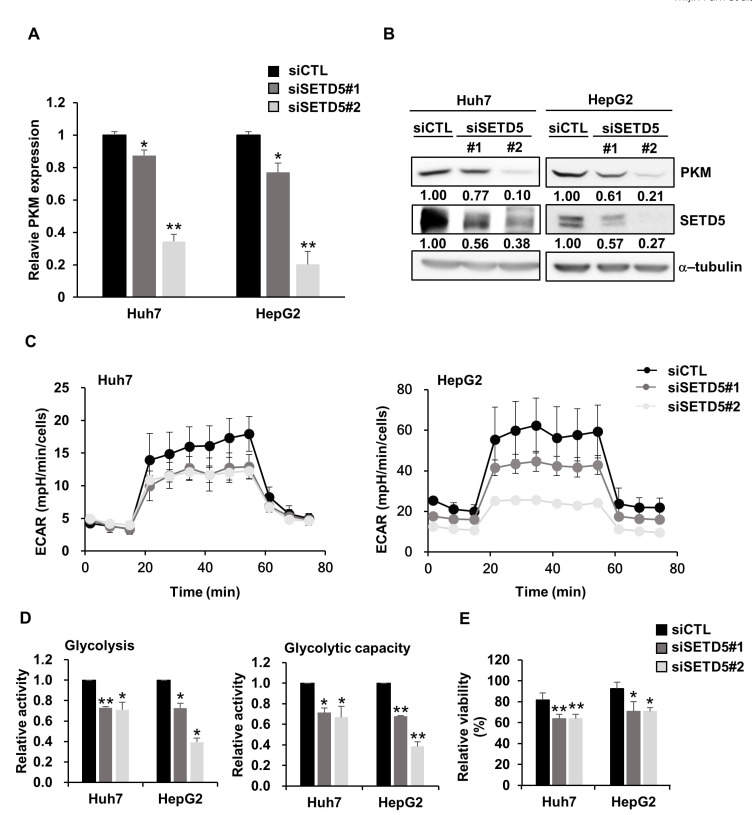
Given that IFN/inflammatory responses function as negative regulators of liver cancer cells (Grivennikov et al., 2010), the positive association of SETD5 expression with the IFN/inflammatory response pathways fails to explain SETD5 KD-mediated defects in liver cancer cell growth and proliferation. In the transcriptome data above, we found that a critical glycolysis gene encoding pyruvate kinase M1/2 (PKM) was significantly downregulated in SETD5 KD cells (Fig. 5A). The decrease in PKM expression by SETD5 depletion was further verified by individually examining the mRNA and protein levels (Figs. 6A and 6B). Given that PKM functions in the rate-limiting step of glycolysis, which is essential for cancer cell growth and proliferation, we examined the effect of SETD5 KD on glycolysis in liver cancer cells. The ECAR, which reflects glycolytic activity and capacity, was significantly attenuated in SETD5 KD cells (Figs. 6C and 6D). In addition, we observed that SETD5-depleted cells were significantly more sensitive to treatment with 2-Deoxy-D-glucose, a glycolysis inhibitor (Fig. 6E). These results imply that SETD5 promotes glycolysis by upregulating PKM expression, supporting liver cancer cell survival.
Fig. 6. SETD5 controls the glycolytic rate in HCC.
(A and B) In SETD5-depleted Huh7 and HepG2 cells, PKM expression was downregulated at both the mRNA (A) and protein levels (B). The numbers indicate the value of semiquantified immunoblotting bands by ImageJ. Statistical significance was determined by one-way ANOVA (*P < 0.05, **P < 0.01). (C) The ECAR in SETD5-depleted HCC cells upon the sequential addition of glucose (25 mM), oligomycin (1 µM), and 2-DG (50 mM). ECAR values were normalized to the number of cells. All experiments were performed independently in triplicate. (D) Glycolysis and glycolytic capacity were quantified in SETD5-depleted Huh7 and HepG2 cells. Relative activity was normalized to the values for siCTL. Values are the mean ± SEM of biological triplicate experiments. (n = 3, *P < 0.05, **P < 0.01, one-way ANOVA). (E) SETD5-depleted Huh7 and HepG2 cells were treated with 2.5 mM 2-DG for 48 h. Cell viability was measured using the Cell titer-Glo assay. Relative viability indicates the viability of cells treated with 2-DG normalized to those treated with vehicle only. Values are the mean ± SEM of biological triplicate experiments (n = 3, *P < 0.05, **P < 0.01, one-way ANOVA).
SETD5 is important for maintaining HCC tumor growth in vivo
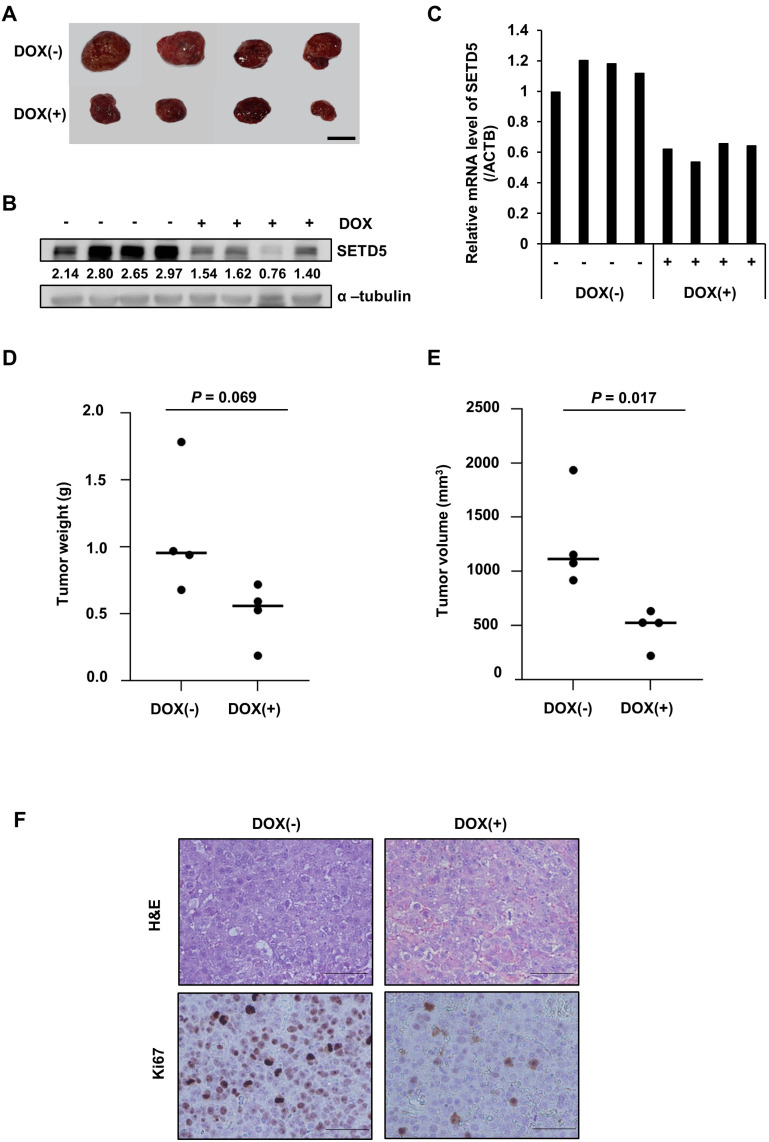
To determine whether SETD5 was relevant to tumor growth in vivo, we generated a xenograft mouse model harboring Huh7-derived tumors in which SETD5 expression was regulated by shSETD5 RNAs under the regulation of a doxycycline-responsive promoter (Figs. 7A-7C). Consistent with the defects in cancer cell growth in vitro, SETD5 depletion compromised tumor growth in vivo (Figs. 7D and 7E). In addition, the expression of Ki67, which indicates cancer cell proliferation, was largely decreased in SETD5 KD tumor tissues (Fig. 7F). Collectively, these results demonstrate that SETD5 functions are important for HCC tumor growth in vivo.
Fig. 7. SETD5 depletion inhibited tumorigenesis in vivo.
(A) Images of tumors taken from xenograft mice fed doxycycline (DOX(+)) or control diets (DOX(–)) for 13 days. (B and C) SETD5 protein (B) and mRNA levels (C) in the tumor tissues displayed in (A). The numbers indicate the value of semiquantified immunoblotting bands by ImageJ. (D and E) Tumor weight and size were measured at the time mice were sacrificed. Statistical significance was determined by Student’s t-test. (F) Representative images of H&E staining and immunohistochemistry for Ki67 in paraffin-embedded tumor sections. Scale bars = 200 µm.
DISCUSSION
Despite extensive efforts in recent decades, liver cancer remains one of the most challenging malignancies (Llovet et al., 2021). The findings that only a subset of liver cancer patients possess targetable oncogenic mutations (de Lope et al., 2012; Ji and Wang, 2012; Wörns and Galle, 2010) highlight the difficulty of developing molecular therapies to treat liver cancer. Targeting epigenetic regulators has emerged as a promising approach to treat cancers associated with less distinctive genomic aberrations (Cheng et al., 2019). KMTs containing well-conserved SET domains are among the druggable epigenetic regulators possibly exhibiting enzymatic activities (Vougiouklakis et al., 2020). In this study, by analyzing the association of different KMT expression levels with liver cancer patient prognosis, we identified SETD5 as a novel putative KMT involved in liver cancer progression. We found that SETD5 expression was highly correlated with low survival of liver cancer patients. In addition, functional analyses showed that SETD5 loss induced cell death without affecting the cell cycle. Intriguingly, SETD5 failed to methylate histones either in a free form or in a nucleosomal context. Nonetheless, our transcriptomic analyses revealed that SETD5 was involved in IFN-mediated inflammatory responses in HCC cells. In addition, we disclosed that SETD5 depletion compromised glycolytic activity by downregulating the expression of PKM, a rate-limiting enzyme of glycolysis. Finally, we demonstrated that SETD5 loss impairs tumor growth in mouse xenograft models.
Several previous studies have shown the functional implication of SETD5 in cancer. Consistent with our findings, these studies showed that SETD5 plays a tumorigenic role in different cancers. Wang et al. (2020) found that SETD5 promoted the resistance of pancreatic ductal adenocarcinoma (PDAC) to MEK1/2 inhibitors (MEKi). This group found that SETD5 lacked intrinsic methyltransferase activity toward nucleosomes. Instead, SETD5 formed a complex with G9a and thus mediated histone H3K9 methylation-associated MEKi resistance in PDAC cells. Nonetheless, it remains unclear whether SETD5 methylates nonhistone proteins linked to cancer progression. The involvement of SETD5 in the MEK-ERK pathway was also observed in non-small cell lung cancer (Yu et al., 2019). In addition, SETD5 promoted cancer stem cell properties and PI3K-Akt-mTOR axis activity in esophageal squamous cell carcinoma (ESCC) (Piao et al., 2020) and breast cancer (Yang et al., 2021). More interestingly, SETD5 expression in ESCC was positively correlated with HIF-1α levels, which regulate glycolytic activity (Piao et al., 2020). Our finding that SETD5 promoted glycolysis by upregulating PKM expression raises the possibility that SETD5 may cooperate with HIF-1α for metabolic reprogramming in cancer cells or cells exposed to hypoxic stress.
IFN-mediated inflammatory responses are pleiotropic, either tumor-suppressive or tumor-promoting, in different cellular and microenvironmental contexts (Martin-Hijano and Sainz, 2020). Our transcriptomic analyses demonstrated that SETD5 depletion downregulated IFN responses and inflammatory pathways in HCC cells. Double-stranded RNA (dsRNA) generated in cells for various reasons endogenously activates the IFN-inflammatory response without extracellular stimuli. We found that SETD5 loss reduced the expression of MDA5 and DDX60, which are critical for sensing dsRNAs (Cao et al., 2015; Miyashita et al., 2011; Oshiumi et al., 2015; Reikine et al., 2014). This finding implies that SETD5 may be involved in IFN-inflammatory signaling pathways by regulating the expression of inflammatory signal sensors. However, given that activation of innate IFN signaling pathways induces tumorigenic activities, we hypothesized that suppression of these pathways might not be directly related to SETD5 loss-induced HCC cell death in this study. Interestingly, IFN signaling is implicated in cancer cell dormancy, a major obstacle to efficient cancer therapy (Pietras et al., 2014). Our finding that SETD5 loss compromised IFN responses in liver cancer cells suggests that suppressing IFN-mediated signaling by SETD5 inactivation may awaken dormant cancer cells to be susceptible to agents targeting proliferative cancer cells. Future investigation may reveal a strategy to target SETD5 for the removal of dormant cancer cells.
In summary, this study proposes that SETD5 is a novel tumorigenic gene involved in HCC progression. We anticipate that further studies to characterize the molecular mechanisms underlying SETD5 function in liver HCC will shed light on the development of novel anticancer strategies against liver cancer.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by a grant (NRF-2019R1A2C 1086151 to J.-A.K.) from the National Research Foundation, Ministry of Science and ICT and Future Planning and by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (171134054 to J.-A.K. and KGM99922111 to J.-H.K. [Jeong-Hoon Kim] and J.-A.K.).
Footnotes
AUTHOR CONTRIBUTIONS
M.P. conceived the study, performed the experiments, analyzed the data, and wrote the manuscript. B.M., K.P., and J.K. performed the experiments and analyzed the data. J.-H.K. (Jong-Hwan Kim), S.-J.P., S.-K.K., and S.-Y.K. analyzed RNA-seq and public cancer data. J.-H.K. (Jeong-Hoon Kim) supervised the study. J.-A.K. conceived and supervised the study and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abou-Alfa G.K., Meyer T., Cheng A.L., El-Khoueiry A.B., Rimassa L., Ryoo B.Y., Cicin I., Merle P., Chen Y., Park J.W., et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N. Engl. J. Med. 2018;379:54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M., Helin K. Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Brown R., Curry E., Magnani L., Wilhelm-Benartzi C.S., Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat. Rev. Cancer. 2014;14:747–753. doi: 10.1038/nrc3819. [DOI] [PubMed] [Google Scholar]
- Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- Cao X., Ding Q., Lu J., Tao W., Huang B., Zhao Y., Niu J., Liu Y.J., Zhong J. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J. Hepatol. 2015;62:771–778. doi: 10.1016/j.jhep.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Chen Z., Xie H., Hu M., Huang T., Hu Y., Sang N., Zhao Y. Recent progress in treatment of hepatocellular carcinoma. Am. J. Cancer Res. 2020;10:2993–3036. [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lope C.R., Tremosini S., Forner A., Reig M., Bruix J. Management of HCC. J. Hepatol. 2012;56(Suppl 1):S75–S87. doi: 10.1016/S0168-8278(12)60009-9. [DOI] [PubMed] [Google Scholar]
- Deliu E., Arecco N., Morandell J., Dotter C.P., Contreras X., Girardot C., Kasper E.L., Kozlova A., Kishi K., Chiaradia I., et al. Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat. Neurosci. 2018;21:1717–1727. doi: 10.1038/s41593-018-0266-2. [DOI] [PubMed] [Google Scholar]
- Dillon S.C., Zhang X., Trievel R.C., Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G.S., Hanley K.L., Liang Y., Lin X. Improving the efficacy of liver cancer immunotherapy: the power of combined preclinical and clinical studies. Hepatology. 2021;73(Suppl 1):104–114. doi: 10.1002/hep.31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes I.R., Cruz A.C.P., Ferrasa A., Phan D., Herai R.H., Muotri A.R. Genetic variations on SETD5 underlying autistic conditions. Dev. Neurobiol. 2018;78:500–518. doi: 10.1002/dneu.22584. [DOI] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe E.A., Sinha R., Schlauch D., Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., McGinty R.K., Muir T.W., Kim J.A., Kim J. Crosstalk among Set1 complex subunits involved in H2B ubiquitylation-dependent H3K4 methylation. Nucleic Acids Res. 2018;46:11129–11143. doi: 10.1093/nar/gky920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Wang X.W. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin. Oncol. 2012;39:461–472. doi: 10.1053/j.seminoncol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- Kim I.K., McCutcheon J.N., Rao G., Liu S.V., Pommier Y., Skrzypski M., Zhang Y.W., Giaccone G. Acquired SETD2 mutation and impaired CREB1 activation confer cisplatin resistance in metastatic non-small cell lung cancer. Oncogene. 2019;38:180–193. doi: 10.1038/s41388-018-0429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M., Finn R.S., Qin S., Han K.H., Ikeda K., Piscaglia F., Baron A., Park J.W., Han G., Jassem J., et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- Kwon M., Park K., Hyun K., Lee J.H., Zhou L., Cho Y.W., Ge K., Skalnik D.G., Muir T.W., Kim J. H2B ubiquitylation enhances H3K4 methylation activities of human KMT2 family complexes. Nucleic Acids Res. 2020;48:5442–5456. doi: 10.1093/nar/gkaa317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Tate C.M., You J.S., Skalnik D.G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- Liedtke M., Cleary M.L. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Lu C., Klement J.D., Yang D., Albers T., Lebedyeva I.O., Waller J.L., Liu K. SUV39H1 regulates human colon carcinoma apoptosis and cell cycle to promote tumor growth. Cancer Lett. 2020;476:87–96. doi: 10.1016/j.canlet.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Hijano L., Sainz B., Jr. The interactions between cancer stem cells and the innate interferon signaling pathway. Front. Immunol. 2020;11:526. doi: 10.3389/fimmu.2020.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M., Oshiumi H., Matsumoto M., Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.M., Seidman J.S., Ellegood J., Gao R., Savchenko A., Troutman T.D., Abe Y., Stender J., Lee D., Wang S., et al. Setd5 haploinsufficiency alters neuronal network connectivity and leads to autistic-like behaviors in mice. Transl. Psychiatry. 2019;9:24. doi: 10.1038/s41398-018-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Miyashita M., Okamoto M., Morioka Y., Okabe M., Matsumoto M., Seya T. DDX60 is involved in RIG-I-dependent and independent antiviral responses, and its function is attenuated by virus-induced EGFR activation. Cell Rep. 2015;11:1193–1207. doi: 10.1016/j.celrep.2015.04.047. [DOI] [PubMed] [Google Scholar]
- Osipovich A.B., Gangula R., Vianna P.G., Magnuson M.A. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development. 2016;143:4595–4607. doi: 10.1242/dev.141465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao L., Li H., Feng Y., Yang Z., Kim S., Xuan Y. SET domain-containing 5 is a potential prognostic biomarker that promotes esophageal squamous cell carcinoma stemness. Exp. Cell Res. 2020;389:111861. doi: 10.1016/j.yexcr.2020.111861. [DOI] [PubMed] [Google Scholar]
- Pietras E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., Passegue E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 2014;211:245–262. doi: 10.1084/jem.20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Li H., Zhang Z., Zhu Z., He S., Wang X., Wang P., Qin J., Zhuang L., Wang W., et al. A pharmacogenomic landscape in human liver cancers. Cancer Cell. 2019;36:179–193.e11. doi: 10.1016/j.ccell.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikine S., Nguyen J.B., Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saung M.T., Pelosof L., Casak S., Donoghue M., Lemery S., Yuan M., Rodriguez L., Schotland P., Chuk M., Davis G., et al. FDA approval summary: nivolumab plus ipilimumab for the treatment of patients with hepatocellular carcinoma previously treated with sorafenib. Oncologist. 2021;26:797–806. doi: 10.1002/onco.13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A., Fagnocchi L., Mastrototaro G., Massimino L., Zaghi M., Indrigo M., Cattaneo S., Martini D., Gabellini C., Pucci C., et al. SETD5 regulates chromatin methylation state and preserves global transcriptional fidelity during brain development and neuronal wiring. Neuron. 2019;104:271–289.e13. doi: 10.1016/j.neuron.2019.07.013. [DOI] [PubMed] [Google Scholar]
- Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Sowalsky A.G., Xia Z., Wang L., Zhao H., Chen S., Bubley G.J., Balk S.P., Li W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol. Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- Vougiouklakis T., Bernard B.J., Nigam N., Burkitt K., Nakamura Y., Saloura V. Clinicopathologic significance of protein lysine methyltransferases in cancer. Clin. Epigenetics. 2020;12:146. doi: 10.1186/s13148-020-00897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hausmann S., Lyu R., Li T.M., Lofgren S.M., Flores N.M., Fuentes M.E., Caporicci M., Yang Z., Meiners M.J., et al. SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020;37:834–849.e13. doi: 10.1016/j.ccell.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörns M.A., Galle P.R. Future perspectives in hepatocellular carcinoma. Dig. Liver Dis. 2010;42(Suppl 3):S302–S309. doi: 10.1016/S1590-8658(10)60521-X. [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang C., Che N., Feng Y., Li C., Xuan Y. Su(var)3-9, Enhancer of zeste, and Trithorax domain-containing 5 facilitates tumor growth and pulmonary metastasis through up-regulation of AKT1 signaling in breast cancer. Am. J. Pathol. 2021;191:180–193. doi: 10.1016/j.ajpath.2020.10.005. [DOI] [PubMed] [Google Scholar]
- Yu H., Sun J., Zhao C., Wang H., Liu Y., Xiong J., Chang J., Wang M., Wang W., Ye D., et al. SET domain containing protein 5 (SETD5) enhances tumor cell invasion and is associated with a poor prognosis in non-small cell lung cancer patients. BMC Cancer. 2019;19:736. doi: 10.1186/s12885-019-5944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.