Abstract
Heme binding and uptake are considered fundamental to the growth and virulence of the gram-negative periodontal pathogen Porphyromonas gingivalis. We therefore examined the potential role of the dominant P. gingivalis cysteine proteinases (gingipains) in the acquisition of heme from the environment. A recombinant hemoglobin-binding domain that is conserved between two predominant gingipains (domain HA2) demonstrated tight binding to hemin (Kd = 16 nM), and binding was inhibited by iron-free protoporphyrin IX (Ki = 2.5 μM). Hemoglobin binding to the gingipains and the recombinant HA2 (rHA2) domain (Kd = 2.1 nM) was also inhibited by protoporphyrin IX (Ki = 10 μM), demonstrating an essential interaction between the HA2 domain and the heme moiety in hemoglobin binding. Binding of rHA2 with either hemin, protoporphyrin IX, or hematoporphyrin was abolished by establishing covalent linkage of the protoporphyrin propionic acid side chains to fixed amines, demonstrating specific and directed binding of rHA2 to these protoporphyrins. A monoclonal antibody which recognizes a peptide epitope within the HA2 domain was employed to demonstrate that HA2-associated hemoglobin-binding activity was expressed and released by P. gingivalis cells in a batch culture, in parallel with proteinase activity. Cysteine proteinases from P. gingivalis appear to be multidomain proteins with functions for hemagglutination, erythrocyte lysis, proteolysis, and heme binding, as demonstrated here. Detailed understanding of the biochemical pathways for heme acquisition in P. gingivalis may allow precise targeting of this critical metabolic aspect for periodontal disease prevention.
Evidence for the potential importance of cysteine proteinases from Porphyromonas gingivalis in periodontal disease pathology is increasing. Periodontal disease affects the majority of adults to some degree and may be associated with significant systemic morbidity (2, 46), including dental infection and loss of teeth (36). P. gingivalis is implicated as an important periodontal pathogen by its high incidence and relative levels in human disease (1, 11) and by its virulence in monoinfected animals (14, 15). Virulence of P. gingivalis has been attributed to several components of the microorganism, including fimbriae (25, 37), short-chain volatile acids (12, 65), lipopolysaccharide (26, 58), collagenase activity (3, 39), and noncollagenolytic cysteine proteinase activity (8, 10, 54).
Cysteine proteinase activity may affect the remodeling of matrix proteins and disrupt the immune response by stimulating the collagen-degrading activity of host cells (8, 10, 62), degrading fibronectin (34), inactivating gamma interferon (68) and interleukins (6, 17), interfering with the complement cascade (63, 67), and degrading immunoglobulins (16, 52). Also, clotting and vascular permeability mechanisms may be disturbed (27, 28, 54), fibrinogen may be degraded (33, 54), and erythrocytes may be agglutinated and lysed (44, 56) by cysteine proteinase activity, possibly for the acquisition of metabolically necessary iron, heme, or porphyrin from hemoglobin. Numerous different P. gingivalis cysteine proteinases described in several reports have been demonstrated to be antigenically related (9, 47, 48) and the products of three related genes (41, 51). This unique family of enzymes, named gingipains, has two major gene products, Arg-gingipain-1 (RGP-1) and Lys-gingipain (KGP) (41), which prefer proteinacious substrates with an arginine or lysine in the P1 position, respectively.
Bacterial cysteine proteinase activity has been demonstrated within diseased periodontal pockets (13, 20), and epitopes of gingipains are detectable in clinical plaque samples from patients with adult periodontitis (unpublished data), so the gingipains are likely to be clinically relevant. The gingipains are expressed on the outer membrane of P. gingivalis and may also be released with vesicles or as soluble proteins (9, 18, 24). Gingipains have been suggested to account for up to 85% of trypsin-like proteolytic activity in a P. gingivalis culture (49), and under certain growth conditions in vitro, these enzymes can accumulate to become the most abundant P. gingivalis proteins in a culture (9).
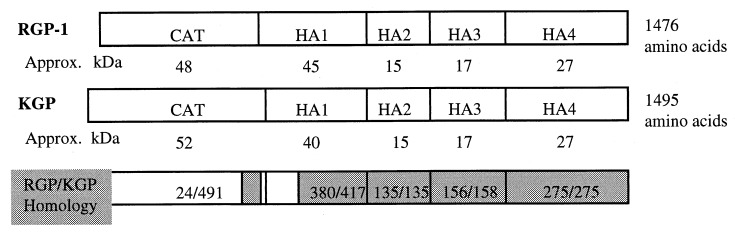
The catalytic domains of RGP-1 and KGP constitute approximately one-third of the translated protein products. The remaining two-thirds of these two gingipain molecules consist of four COOH-terminal domains (HA1 to HA4) which are highly homologous between these two predominant gingipains (Fig. 1). These noncatalytic COOH-terminal domains were originally named hemagglutinin (HA) domains because at least one was thought to participate in hemagglutination (47). They may each be separated posttranslationally from the catalytic domain and from one another, presumably through autolysis some time after logarithmic growth in vitro (9, 59). The functions of the first, third, and fourth HA domains are unknown. The second HA domain (HA2) has recently been implicated in hemoglobin binding (19, 43). Because all of the domains of the gingipains are found together predominately in loose, noncovalent associations with one another after hydrolytic separation (9, 59), the gingipains appear to be multifunctional proteins for aggregation of erythrocytes and then lysing of these cells to obtain hemoglobin for the acquisition of iron, heme, or porphyrin.
FIG. 1.
Domain structure and homologies between the gingipains RGP-1 and KGP. CAT represents the putative catalytic domain. Shaded areas represent regions of >98% amino acid identity between the two gingipains. Each fraction represents the degree of identity for each RGP-1 domain. Approx. kDa, approximate molecular mass in kilodaltons.
P. gingivalis (formerly Bacteroides sp.) can utilize inorganic iron, free or protein-associated heme, or organic iron sources such as transferrin (5). Several investigators have previously shown that P. gingivalis binds to and internalizes hemin with various affinities and at various rates (4, 21, 53, 57, 60, 64). These earlier reports suggest that there are at least two heme-binding proteins of P. gingivalis with different affinities for hemin which may respond to environmental changes by rapidly changing their position or associations within the outer membrane.
Hemin binding and uptake appear to be related to the regulation of proteinase and fimbriae expression and to vesicle formation (7, 38, 40) and were recently proposed to establish an antioxidative shield for protection from oxidative radicals (61). Binding of protoporphyrin IX in P. gingivalis was also implied by competition with labelled hemin (4, 64), and protoporphyrin IX was reported to support growth (53). Protoporphyrin IX limitation was shown to be coordinated with phenotypic expression of proteinase activity (42). Hemin binding by P. gingivalis may therefore represent a capacity for protoporphyrin binding.
Recently, Nakayama et al. have isolated a hemoglobin-binding protein associated with the outer membrane of P. gingivalis and identified this protein as one homologous with the HA2 domain of the gingipains (43). In that report, adsorption of hemoglobin to whole P. gingivalis cells was associated with the presence of the HA2 domain. Also, hemin accumulation within the P. gingivalis cells was shown to be dependent on functional expression of KGP (45). The HA2 gingipain domain may therefore function as a hemoglobin-binding domain in P. gingivalis.
Understanding the molecular and biochemical mechanisms involved in key regulatory pathways is paramount in developing strategies for control of disease. In this study, we obtained evidence, by using a monoclonal antibody (MAb) which recognizes the hemoglobin-binding (HA2) domain of P. gingivalis cysteine proteinases, that the HA2 domain can bind to hemoglobin primarily and specifically through a portion of the heme moiety that is surface exposed in the hemoglobin structure. We also found that the unique epitope of MAb 5A1 within this heme-binding domain was expressed in parallel with hemoglobin-binding activity and proteinase activity in cellular and cell-free culture fractions of P. gingivalis.
MATERIALS AND METHODS
RGP-1 and KGP isolation.
Polydomain RGP-1 and KGP were isolated and characterized as previously described (68) by arginine-Sepharose affinity chromatography of detergent-extracted P. gingivalis ATCC 33277 cells. Alternatively, polydomain RGP-1 and KGP were isolated as previously described (9) by arginine-Sepharose affinity chromatography from cell-free supernatant of a 10-day P. gingivalis batch culture.
Enzyme activity assays.
The proteinase activities of P. gingivalis culture fractions were measured by using the substrates N-tert-butoxycarbonyl-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin or N-tert-butoxycarbonyl-Glu-Lys-Lys-7-amido-4-methylcoumarin at 30°C in Tris buffer without added reducing agents. Substrate hydrolysis was monitored over time by measuring A460 with a 380-nm excitation beam on a Perkin-Elmer LS 50B luminescence spectrophotometer.
Development of MAbs 5A1 and IIB2.
Antigingipain MAbs 5A1 and IIB2 were prepared in mice as previously described (9).
Expression and purification of recombinant HA2 (rHA2).
Forward and reverse primers (AACCTGCAGCGCGCAGACTTCACGG and GGAAGCCAATGGCGCCAAAAGATCTAGT) were designed to amplify the HA2 domain from the P. gingivalis RGP-1 proteinase gene (accession no. U15282). Restriction sites for PstI and BglII were designed into the 5′ ends of the primers to facilitate cloning. The digested PCR product was ligated into the QIAexpressionist type III construct providing a six-His tag at the COOH terminus (Qiagen Corp.). Transformation of the ligated construct was performed by electroporation into Escherichia coli NM522 cells. E. coli cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 and then induced by incubation with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h. Cells were harvested and resuspended to 5 ml/g (wet weight) in buffer A (8 M urea, 0.1 mM NaH2PO4, 0.01 mM Tris-HCl, pH 7.9). The cells were stirred for 2 h at room temperature, taking care to avoid foaming. This cell lysate was subjected to centrifugation at 31,000 × g for 30 min at room temperature to pellet the cellular debris, and then the supernatant was subjected to ultracentrifugation at 130,000 × g for 2 h. The clarified lysate was loaded onto a nickel-nitrilotriacetic acid column (Qiagen Corp.), pre-equilibrated with buffer A. The nickel-nitrilotriacetic acid column was washed with buffer A until the baseline was reached. The protein was refolded on this column by running a linear gradient of urea from 8 to 0 M in 20 mM Tris-HCl–500 mM NaCl–10% glycerol (pH 7.9). The protein was then eluted with 50 mM Tris-HCl–500 mM NaCl–10% glycerol–250 mM imidazole (pH 7.9). The eluant was diluted 100-fold in 50 mM sodium acetate buffer (pH 5.5) and applied to a hemoglobin-agarose column pre-equilibrated with the dilution buffer. After loading, the column was washed with the same buffer until the baseline was reached and then the hemoglobin-binding protein was eluted with 50 mM Tris-HCl (pH 9). Protein concentrations were determined by Coomassie dye binding using bovine serum albumin as the standard.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed by using 12% gels with 4% stackers by the method of Laemmli (32). All samples were diluted with SDS sample buffer before electrophoresis with (reducing) or without 2-mercaptoethanol. Western blotting was performed by the method of Towbin et al. (66), and proteins were transferred from the gels to polyvinylidene difluoride (PVDF) paper (Bio-Rad) with 300 mA for 1 h. Blots were blocked with 0.1% bovine serum albumin in 20 mM Tris-HCl with 500 mM NaCl containing 0.1% Tween 20 (TBS/Tween). An alkaline phosphatase (AP) conjugate of rabbit anti-mouse immunoglobulin G (Dako Corp.) was used as a secondary antibody. Blots were washed with TBS/Tween between antibody applications. The substrate for AP was nitroblue tetrazolium in excess with 5-bromo-4-chloro-3-indolylphosphate (Bio-Rad), and color was developed in 5 mM Tris (pH 9.5).
NH2-terminal amino acid sequencing of proteins resolved by SDS-PAGE was performed as previously described (9).
ELISA.
Enzyme-linked immunosorbant assays (ELISA) were performed in polystyrene microtiter wells. Proteins were used to coat the surfaces in 2.7 mM KCl–1.5 mM KH2PO4–137 mM NaCl–8.1 mM Na2HPO4 (PBS) with 10 mM sodium azide (PBS/N3). All wells were blocked and washed in PBS with 0.1% Tween 20 (PBS/Tween). Primary murine antibodies were applied in PBS/Tween at a concentration of 0.5 μg/ml for at least 1 h. Secondary goat anti-mouse antibodies conjugated with AP (Dako Corp.) were applied at a concentration of 1.1 μg/ml for 30 min, and then AP activity was monitored at 414 nm by hydrolysis of the substrate 4-nitrophenylphosphate (Boehringer GmbH, Mannheim, Germany) in 5 mM Tris (pH 9.5) by using a Titertek Twinreader PLUS photometer (absorbance maximum of 3.0 ELISA units). Mean apparent dissociation constants (Kds) were derived by solid-phase ELISA as previously described (50) and are accompanied by standard errors of the means.
Ligand-binding assay.
The ligand-binding assay was a variant of the ELISA in which the ligand (i.e., hemin or hemoglobin) that had been used to coat the wells in PBS/N3 was subsequently allowed to bind to a second ligand-binding protein (i.e., rHA2 or gingipains) in PBS/Tween. The ligand-binding protein was then detected with MAb 5A1 or IIB2, followed by a rabbit anti-mouse AP conjugate, and developed as already described for ELISA. Bovine hemoglobin was used in these experiments. Hemin was from stock solutions dissolved in 0.1 N NaOH, and although the NaOH would replace the chloride ion of hemin with a hydroxylate ion (hematin), the term hemin will be used for this compound throughout this report. The Kd and apparent inhibition constant (Ki) for ligand binding were derived as previously described (50) in these assays by using serial dilutions of the ligand-binding protein or competitor, respectively, with even amounts of coated ligand. The reported results are means accompanied by the standard errors of the means.
Peptide synthesis.
Peptides were synthesized by Chiron Mimotopes with terminal amines and carboxylic acids. The peptide 1 sequence was ALNPDNYLISKDVTG, and the peptide 2 sequence was GEAPAEWTTIDADGDGQGWL.
Materials.
All chemicals and compounds were purchased from Sigma unless otherwise specified.
Statistics.
Statistical differences between measurements of the gingipains and rHA2 were determined with one-tailed Student t tests.
RESULTS
The polydomain RGP-1 and KGP isolated from 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)-extracted P. gingivalis cells possessed SDS-PAGE profiles, NH2-terminal sequences, proteolytic activities, and inhibition profiles characteristic of gingipain-like molecules previously described by us (9, 68) and others (3, 47, 54) (data not shown).
The HA2 domain was cloned, expressed, and purified as a six-His tag fusion. Nucleic acid and NH2-terminal amino acid sequencing verified the identities of the clone and the expressed protein, respectively, as the HA2 domain of RGP-1 (data not shown).
Hemoglobin is bound by rHA2 and by native but not denatured RGP-1 and KGP.
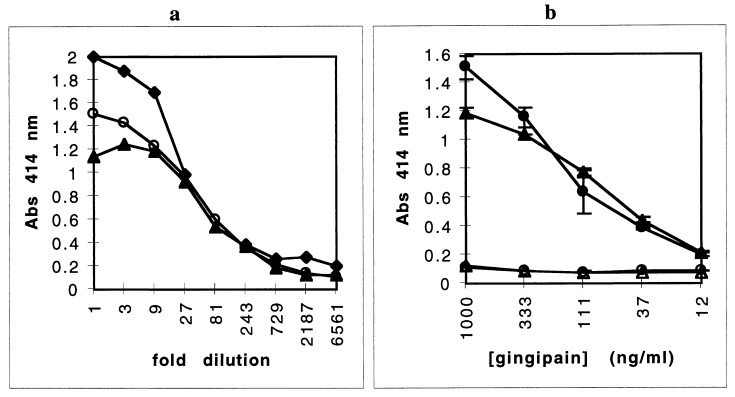
In the solid-phase ligand-binding assay, rHA2, RGP-1, and KGP each bound to hemoglobin (Fig. 2a). As MAb 5A1 was used to detect rHA2 bound to hemoglobin and did not interfere with this binding, it was evident that the epitope for MAb 5A1 within the HA2 domain was separate from the hemoglobin-binding site of HA2. The hemoglobin-binding affinities of rHA2, RGP-1, and KGP (Kd = 2.1 ± 0.6 nM) were similar (P = 0.24), and the binding curves of neither rHA2 nor the gingipains were indicative of multisite binding (Fig. 2a). High-affinity binding to hemoglobin at a single site within only the HA2 domain of both native RGP-1 and KGP is sufficient to account for these observations. The binding site for hemoglobin within the gingipains appeared to be associated with a higher-order protein structure, since denaturation of RGP-1 and KGP by boiling effectively eliminated their ability to bind hemoglobin (Fig. 2b).
FIG. 2.
Hemoglobin binding by rHA2, RGP-1, and KGP. (a) Microtiter wells were coated with hemoglobin and then incubated with threefold dilutions of purified rHA2 at 2.5 μg/ml (⧫), RGP-1 at 5 μg/ml (○), or KGP at 5 μg/ml (▴). Association of rHA2 with hemoglobin was measured with MAb 5A1, followed by substrate development after binding of a secondary anti-mouse AP-conjugated antibody. (b) Hemoglobin binding by native, but not denatured, gingipains. Wells were coated with hemoglobin and then incubated overnight with dilutions of either RGP-1 (●), KGP (▴), RGP-1 denatured by boiling (○), or KGP denatured by boiling (▵). For this experiment, native or denatured gingipains that bound to hemoglobin were recognized by MAb IIB2, which specifically detects both native and denatured gingipains. Primary antibody IIB2 was followed by substrate development after binding of a secondary anti-mouse AP-conjugated antibody. These data are representative of three separate experiments. Abs, absorbance.
Hemoglobin binding of the HA2 domain is mediated through the heme moiety.
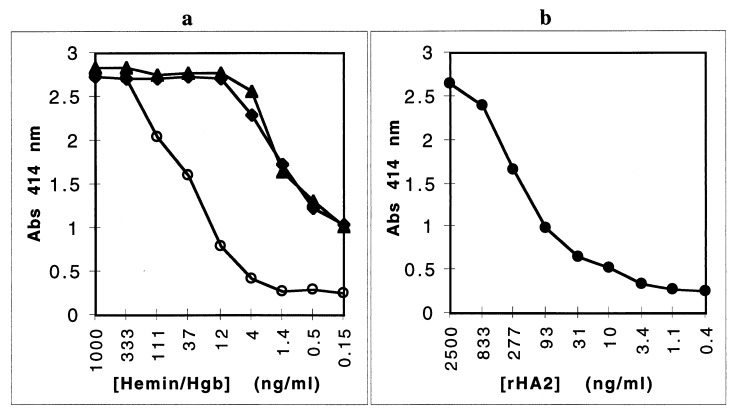
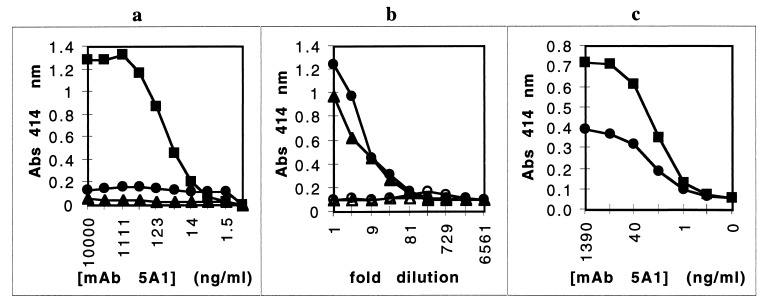
To begin characterizing the binding between rHA2 and hemoglobin, we examined the binding between rHA2 and hemin, as well as binding to hemoglobin degraded by proteinase K. rHA2 bound not only to wells coated with hemoglobin but also to wells coated with hemin or with proteolytically degraded hemoglobin (Fig. 3a). Binding of the rHA2 domain to hemin-coated wells was approximately eightfold weaker than binding to hemoglobin in solid-phase assays (Kd = 16 ± 1 nM) (Fig. 3b).
FIG. 3.
Binding of the HA2 domain to the heme moiety. (a) Binding of rHA2 to dilutions of hemin (⧫), hemoglobin (Hgb) (○), or hemoglobin degraded by proteinase K (▴). Microtiter wells were coated with dilutions of samples, and then overnight binding of rHA2 to coated wells was detected with MAb 5A1, followed by substrate development after binding of a secondary anti-mouse AP-conjugated antibody. The absence of contaminating protein within 90 μg of the hemin preparation and the absence of nondegraded subunits of hemoglobin remaining after proteinase K treatment were verified by SDS-PAGE (data not shown). (b) Binding of rHA2 to hemin. Microtiter wells were coated with hemin, and overnight binding of rHA2 dilutions was detected with MAb 5A1 as described above. These data are representative of two separate experiments. Abs, absorbance.
The HA2 domain binds the porphyrin ring structure.
To dissect the binding of the rHA2 domain to hemin, the Kis of iron-free protoporphyrin IX in solution phase competition assays were determined. By using the standard ligand-binding assay described herein, rHA2 or the gingipains were preincubated with dilutions of protoporphyrin IX and then allowed to bind to the hemin-coated wells. Binding of the gingipains or rHA2 to hemin was inhibited by the addition of protoporphyrin IX (Ki = 2.5 ± 0.3 μM) (Fig. 4a). The apparent Ki values of rHA2 and the gingipains were similar (P = 0.42). These data indicated that binding of rHA2 or the gingipains to hemin was specific for some aspect of the protoporphyrin ring. Importantly, binding of rHA2 or the gingipains to hemoglobin was also inhibited by protoporphyrin IX (Fig. 4b) (Ki = 10 ± 2 μM) and preincubation with the protoporphyrin effectively eliminated binding to hemoglobin.
FIG. 4.
Inhibition of hemin or hemoglobin binding. Microtiter wells were coated overnight with hemin (a) or hemoglobin (b). rHA2 in E. coli lysate (100-fold dilution) (×), RGP-1 at 65 ng/ml (●) or KGP at 65 ng/ml (▴) was preincubated with dilutions of 300 μM protoporphyrin IX for 1 h and then transferred to the ligand-coated plates for overnight incubation. Binding of rHA2 or the gingipains to coated wells was detected with MAb 5A1 or IIB2, respectively, followed by substrate development after binding of a secondary anti-mouse AP-conjugated antibody. These data are representative of two separate experiments. The absence of contaminating protein in a 90-μg protoporphyrin IX preparation was verified by SDS-PAGE and Coomassie dye binding (data not shown). ABS, absorbance.
Directed protoporphyrin binding by rHA2.
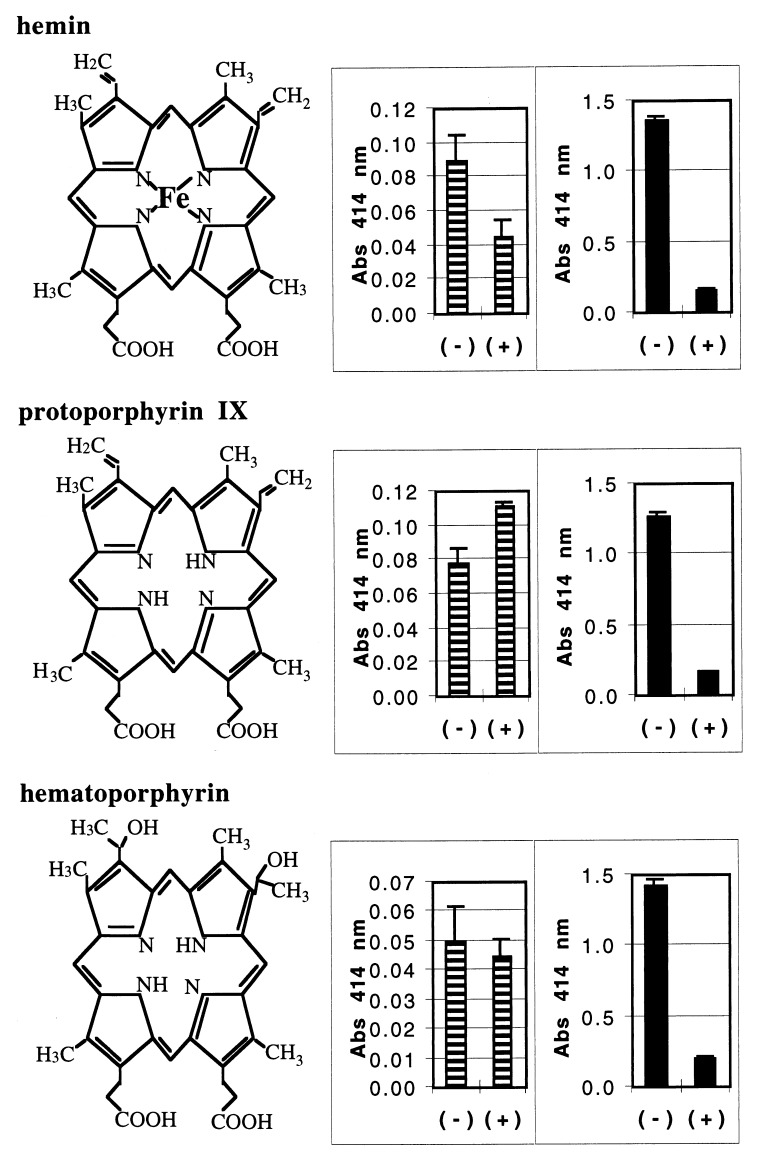
Examination of the hemoglobin crystal structure indicated that only the region of the heme moiety possessing the propionate functional groups (Fig. 5) would be exposed for possible protein-protein contact. We therefore reasoned that blocking access to the acidic region of protoporphyrin molecules would have an effect on rHA2 binding and allow more specific characterization of binding between the HA2 domain and the porphyrin ring. In a modification of the ligand-binding assay system described above, surfaces were first coated with ethylene diamine to provide fixed, free, primary amines for carbodiimide linkage of carboxylic acid groups. Hemin, protoporphyrin IX, and hematoporphyrin bound to wells coated with ethylene diamine with or without carbodiimide treatment, as determined by A414 measurement (Fig. 5, striped bars). rHA2 binding to the carbodiimide-treated porphyrins in the wells was almost eliminated, however, compared to the relatively greater association of rHA2 with the nonderivatized porphyrins (Fig. 5, solid bars). These data indicated that the rHA2 domain specifically recognized the three porphyrin compounds in the region of the propionic acid groups, as we were able to block rHA2 binding by directionally attaching the carboxylic acids of hemin, protoporphyrin IX, or hematoporphyrin to fixed amines. Since the heme moiety within hemoglobin is almost identical to these porphyrin molecules, the data suggested that the heme moiety of hemoglobin was bound by rHA2 and by the HA2 domain of the gingipains in a similar, directed, high-affinity manner.
FIG. 5.
Directed porphyrin binding by rHA2. Microtiter wells were coated with 100 mM ethylene diamine (pH 4.7) and then incubated with hemin, protoporphyrin IX, or hematoporphyrin at 90 μg/ml overnight in 50% dimethyl formamide in the presence (+) or absence (−) of 10 mM carbodiimide. Wells were washed four times with water, and then the amount of porphyrin bound to the wells was determined by measuring absorbance (Abs) at 414 nm (striped bars). Wells were blocked with PBS/Tween and then incubated with rHA2 at 125 ng/ml overnight. Binding of rHA2 to coated wells was detected with MAb 5A1, followed by substrate development after binding of a secondary anti-mouse AP-conjugated antibody (solid bars). Error bars represent standard deviations of absorbance measurements. Diagrams of the chemical structures of hemin, protoporphyrin IX, and hematoporphyrin are presented adjacent to the corresponding data.
The MAb 5A1 epitope is recognized in the rHA2 domain and in denatured, but not native, RGP-1 and KGP.
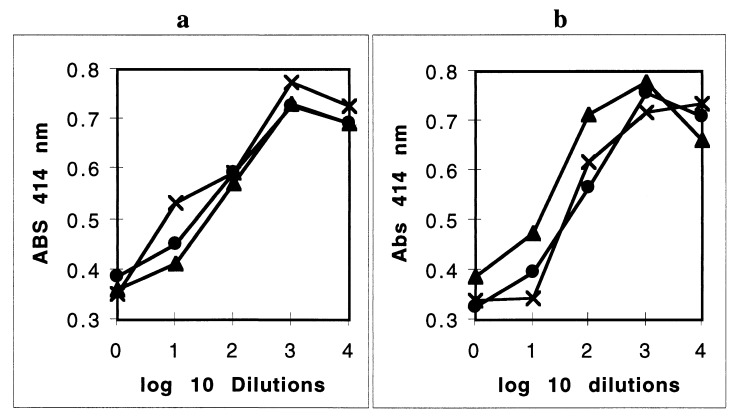
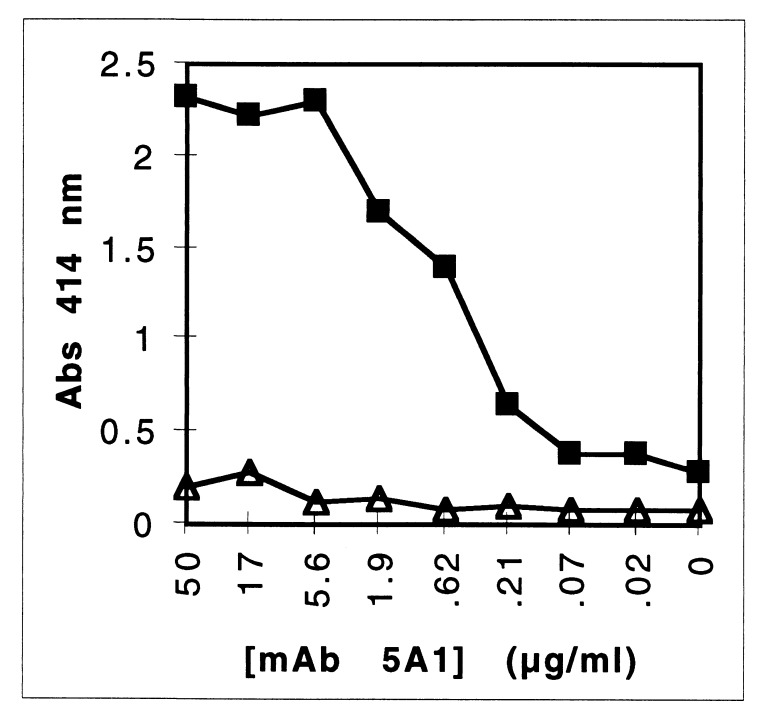
In ELISA, MAb 5A1 bound to rHA2 with high affinity (Kd = 2.2 × 10−10 ± 0.5 × 10−10 M) (Fig. 6a). MAb 5A1 also bound to denatured RGP-1 and KGP but did not bind to the native gingipains isolated from CHAPS-extracted P. gingivalis cells (Fig. 6b). Soluble high-molecular-weight aggregates of gingipain domains isolated from the cell-free fraction of a P. gingivalis batch culture by arginine-Sepharose affinity chromatography (9) were, however, recognized by MAb 5A1 (Kd = 1.7 × 10−10 ± 0.6 × 10−10 M) (Fig. 6c). The similarity of the dissociation constants (P = 0.36) and binding curves suggested that MAb 5A1 recognized the same HA2 epitope in these polydomain gingipains as in rHA2.
FIG. 6.
Measurement of high-affinity binding of MAb 5A1 with rHA2, denatured but not native gingipains, and gingipains from the culture supernatant. (a) RGP-1 (●), KGP (▴), or rHA2 in crude E. coli lysate (■) was used to coat microtiter wells and incubated with serial dilutions of MAb 5A1. (b) Dilutions of RGP-1 (○), KGP (▵), or heat-denatured RGP-1 (●) or KGP (▴) were used to coat microtiter wells with threefold dilutions from 10 μg/ml and then incubated with MAb 5A1. (c) Purified rHA2 (■) or purified high-molecular-weight aggregates of gingipain domains isolated from culture supernatant (●) were used to coat microtiter wells and incubated with serial dilutions of MAb 5A1. These data are representative of three separate experiments. Abs, absorbance.
The MAb 5A1 epitope is represented by an amino acid sequence within the HA2 gingipain domain.
By use of linear synthetic peptides, the epitope of MAb 5A1 was determined to be associated with the peptide ALNPDNYLISKDVTG (Kd = 3.8 nM), which represents amino acids 1215 to 1229 of translated KGP within the HA2 domain (Fig. 7, peptide 1). Dot blot analysis on a PVDF membrane confirmed the unique immunoreactivity of this peptide with MAb 5A1 (data not shown). A search of the SwissProt database for the linear sequence of peptide 1 or of the GenBank database by using the deduced nucleic acid sequence of this epitope resulted in no molecules with perfect homology to the peptide other than the gingipains and HagA, a large HA with regions of identity to the entire HA2 domain.
FIG. 7.
Immunoreactivity of synthetic peptides with MAb 5A1. ELISA demonstrating selective immunoreactivity of MAb 5A1 with peptide 1. Peptide 1 (■) or 2 (▵) was used to coat microtiter plates at a concentration of 5 μg/ml, incubated overnight, and then incubated with dilutions of MAb 5A1. These data are representative of two separate experiments. Abs, absorbance.
Correlation of HA2 domain immunoreactivity with hemoglobin binding in a P. gingivalis culture.
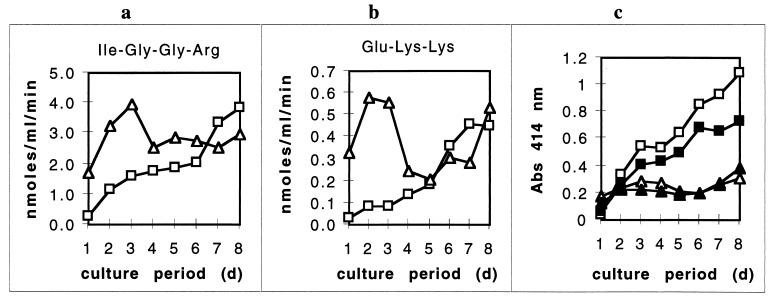
Detection of the HA2 epitope with MAb 5A1 in unfractionated P. gingivalis samples was correlated with hemoglobin binding. Because proteinase activity and gingipain expression have been shown to progressively change during the course of an extended P. gingivalis batch culture (9), we examined cell-associated and extracellular fractions during 8 days of culture. Both Arg- and Lys-specific proteinase activities of the P. gingivalis cells peaked near day 3 of culture (Fig. 8a and b, triangles). Proteinase activities of the cell-free culture supernatants steadily rose throughout the culture period (Fig. 8a and b, squares).
FIG. 8.
Expression of HA2-related immunoreactive hemoglobin-binding protein from P. gingivalis. Aliquots of P. gingivalis culture medium were removed daily during a period of 8 days (d), immediately separated into a cell pellet and culture supernatant, and then frozen until use. The OD660 and purity of the culture were measured daily. The cell pellets were dispersed evenly into 1 ml of PBS/N3. (a and b) Arg- and Lys-specific proteinase activities, respectively, of the cell-free culture supernatant (□) and cellular fraction (▵) were measured as described in Materials and Methods. Measurements of the cellular fractions were normalized to the culture densities (OD660) recorded daily. (c) The HA2 domain (1/243 dilution; □) and the HA2 domain associated with hemoglobin binding (1/81 dilution; ■) in culture supernatants were measured by ELISA and a ligand-binding assay, respectively, as described in Materials and Methods. In P. gingivalis whole-cell fractions, the HA2 domain (1/243 dilution; ▵) and the HA2 domain associated with hemoglobin binding (1/9 dilution; ▴) were measured by ELISA and a ligand-binding assay, respectively, as described in Materials and Methods. Measurements of the cell-associated fractions were normalized to the culture densities (OD660) recorded daily. The corresponding background immunoreactivity with murine anti-human CD-19 immunoglobulin G was subtracted from each measurement. These data are representative of two separate experiments in which the patterns of expression were similar. Abs, absorbance.
Immunoreactive protein in the cell-free conditioned culture medium detected with MAb 5A1 steadily accumulated throughout the 8-day culture period, similar to proteolytic activity (Fig. 8c, open squares). Immunoreactive protein associated with hemoglobin binding in this supernatant fraction also increased steadily throughout the extended culture in a parallel manner (Fig. 8c, closed squares). In the cellular fraction of the P. gingivalis culture, expression of immunoreactive protein increased early during the culture period with a peak near day 3 followed by a slight decrease and then an increase to peak levels again by day 7, similar to the proteolytic activity of this fraction (Fig. 8c, open triangles). Immunoreactive protein associated with hemoglobin binding in the cellular fraction followed a parallel pattern of expression (Fig. 8c, closed triangles). These data demonstrated that detection of protein immunoreactive with MAb 5A1 in crude cellular and extracellular fractions of a P. gingivalis culture was directly associated with hemoglobin binding, suggesting that MAb 5A1 specifically recognized the hemoglobin-binding HA2 domain within the P. gingivalis culture. Also, the data demonstrated a profile of HA2 domain expression and hemoglobin-binding activity similar to the profile of cellular and extracellular proteolytic activity expressed by P. gingivalis.
DISCUSSION
Control of P. gingivalis growth to prevent periodontal pathology might be achieved by interference with one or more pathways for obtaining heme. To this end, we have reported on a MAb which recognizes an epitope within the hemoglobin-binding domain of the abundant P. gingivalis cysteine proteinases, named gingipains, and demonstrated increasing levels of this HA2 domain associated with hemoglobin binding and proteinase activity in an extended P. gingivalis culture. Further, we have characterized the binding between the HA2 domain and hemoglobin, suggesting that binding is mediated in large part by specific recognition of the porphyrin ring of the heme moiety within hemoglobin.
The hemoglobin-binding affinities of RGP-1, KGP, and the HA2 domain measured in our experiments were similar. Also, binding curves for these interactions were typical of single-site binding, which is consistent with the idea that the HA2 domain of the cell-derived gingipains is solely responsible for hemoglobin binding. The similarity of the inhibition profiles for the gingipains to that of rHA2 further suggested that mediation of gingipain binding to heme was through only the HA2 domain. These data do not, however, rule out other possible heme-binding sites in the gingipains with affinity identical to that of HA2.
Hemoglobin binding by the separated catalytic domain of KGP was recently demonstrated (31). Our data, obtained by using polydomain gingipains, did not provide evidence for this second hemoglobin-binding site. It is likely, however, that separated domains of the gingipains behave differently than when associated either noncovalently or within a single polydomain polypeptide. The inability of MAbs which recognize either isolated gingipain domains or peptides to recognize the larger polydomain gingipains of cells exemplifies this potential (this report and reference 22).
Apparent dissociation constants in the nanomolar range represented significantly tighter binding of the HA2 domain to hemoglobin than previously reported (43). Further, this relatively tight binding in our experiments was measured at a nearly neutral pH and not at the pH maximum for binding of 5.5 reported earlier. Differences in experimental systems for measuring binding may account for this discrepancy.
Protoporphyrin IX inhibited binding of rHA2 to hemin. Also, protoporphyrin IX and hemin did not differ statistically in the ability to inhibit the binding of rHA2 to hemin (data not shown). This indicated that the sequestering of porphyrin by HA2 functioned independently of iron. The side chain groups of the porphyrin also did not appear to determine HA2 binding. Hematoporphyrin differs from protoporphyrin IX only by the hydroxylation of the two side chain ethylene groups. These groups are located opposite the positions of propionate groups across the plane of the porphyrin. As the binding to HA2 of these two porphyrins was comparably strong (Fig. 5), it can be concluded that HA2 binding was insensitive to the nature of the chemical groups attached at these positions. This contrasts with the blocking of rHA2 binding in both hematoporphyrin and protoporphyrin IX by directional attachment through chemical modification of the propionate groups.
The iron chelator 2,2′-dipyridal at a concentration of 2 mM also inhibited the binding of rHA2 to hemin, although the Ki of the dipyridal was 200-fold higher than the Ki of protoporphyrin IX (data not shown). This may indicate that rHA2 also had some weak interaction with the iron, but direct steric interference by the dipyridal in the absence of direct iron binding by rHA2 could also be considered.
Binding of hemin by the rHA2 domain was eightfold weaker than that of hemoglobin, although it would be expected to be similar if binding of the HA2 domain to hemoglobin occurred solely through the porphyrin ring of the heme ligand. Competition experiments demonstrated that protoporphyrin IX also inhibited hemoglobin binding, although it was approximately fourfold less competitive than in hemin-binding assays. A portion of the hemoglobin polypeptide may, therefore, contribute to the interaction of HA2 with hemoglobin in a cooperative manner. Because protoporphyrin IX alone completely blocked the interactions between rHA2 or the gingipains and hemoglobin, however, binding between the HA2 domain and the heme moiety must have been essential for the maintenance of this cooperative hemoglobin binding. Alternatively, the weaker binding of rHA2 with hemin in these experiments might also be due to the possibility that iron-protoporphyrins in solution can dimerize, ruffle, or associate differently than when bound to hemoglobin (23, 29, 55). Further, the HA2-binding region of the relatively smaller hemin ligand when bound directly to a surface may be less sterically accessible to the HA2 domain than when heme is presented and supported as part of a large globular protein where the propionate groups and the adjoining rim of the porphyrin ring protrude slightly beyond the surface of the protein (35).
Gingipains recovered from the culture supernatant subsequent to the first day of growth were previously shown to consist of noncovalently aggregated lower-molecular-weight domain fragments of the gingipains (9, 59). Although MAb 5A1 did not recognize native gingipains purified from solubilized P. gingivalis cells, MAb 5A1 did detect gingipain domain aggregates purified from the culture supernatant. This is not surprising, considering that the antibodies were made against the domain fragments of these gingipains (9), and it demonstrates potential differences between high-molecular-weight gingipains recovered by various means.
It is not known whether the HA2 domain was recognized in our cultures as a separate domain, as implicated by the isolation of the separate HA2 domain from envelope fractions (19), or whether the HA2 domain was part of a polydomain complex of gingipain fragments or derived from the hagA gene product. Since the gingipains would be required for hydrolytic release of the HA2 domain from the hagA gene product, as well as from the gingipains themselves (43, 45), analysis of porphyrin binding in hagA knockout strains of P. gingivalis is needed to address this question. Our data demonstrated that the presence of the HA2 domain released by the cells paralleled proteinase activity, as well as hemoglobin-binding activity, suggesting that the hemoglobin-binding HA2 domain was derived from the gingipains. Although these data do not directly implicate the HA2 domain in iron, heme, or porphyrin acquisition by the P. gingivalis organism, the HA2 domain was associated with hemoglobin binding and could be considered a specific target for interference with heme acquisition by P. gingivalis. An HA2-specific antibody which blocks HA2 binding to heme or hemoglobin might be useful in dissecting the role of this porphyrin-binding domain in whole-cell metabolism and virulence.
Hemagglutination was the original function ascribed to the four COOH-terminal domains of the gingipains (47). Although the HA2 domain functions as a porphyrin-binding domain, it might, in addition, participate in hemagglutination. The separate rHA2 domain, at a concentration of 2 μg/ml, did not agglutinate erythrocytes, however, and MAb 5A1, which bound to the HA2 domain, did not inhibit the hemagglutination capacity of whole P. gingivalis cells (data not shown). We are currently investigating the functions of each gingipain HA domain.
Sequence analysis and trypsin susceptibility make the hemin-binding Omp26 described by Bramanti and Holt clearly different from the HA2 domain (4, 30). We have therefore identified a second hemin-binding protein in P. gingivalis. Interestingly, a recent independent analysis of hemin binding by whole cells of P. gingivalis described two different affinities (64). Now we have demonstrated that hemin- or hemoglobin-binding activity is also released by P. gingivalis in batch cultures. It is not immediately clear what advantage P. gingivalis would gain by releasing heme-binding activity, but it may be speculated, considering the recovery of the separate HA2 domain from the outer membrane (43), that soluble HA2 might reassociate with other gingipain domains on the P. gingivalis cells after scavenging and binding to heme or hemoglobin. A specific association of the HA2 domain with an active catalytic domain may be required for removal of the heme moiety from hemoglobin.
Characterization of the binding between the rHA2 domain and porphyrins should allow design of efficient affinity ligands for purification of HA2 and allow structure-based design of inhibitors of heme or hemoglobin binding. Heme acquisition is considered to be fundamental to the growth of P. gingivalis, and intervention with specific agents to disrupt pathways for heme binding or uptake may allow the eventual control or prevention of periodontal disease.
ACKNOWLEDGMENT
Funding for this work was provided by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Albandar J M, Brown L J, Löe H. Putative periodontal pathogens in subgingival plaque of young adults with and without early-onset periodontitis. J Periodontol. 1997;68:973–981. doi: 10.1902/jop.1997.68.10.973. [DOI] [PubMed] [Google Scholar]
- 2.Beck J D, Offenbacher S, Williams R, Gibbs P, Garcia R. Periodontitis: a risk factor for coronary heart disease? Ann Periodontol. 1998;3:127–141. doi: 10.1902/annals.1998.3.1.127. [DOI] [PubMed] [Google Scholar]
- 3.Bedi G S, Williams T. Purification and characterization of a collagen-degrading protease from Porphyromonas gingivalis. J Biol Chem. 1994;269:599–606. [PubMed] [Google Scholar]
- 4.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalisW50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calkins C C, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis: implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 7.Carman R J, Ramakrishnan M D, Harper F H. Hemin levels in culture medium of Porphyromonas (Bacteroides) gingivalisregulate both hemin binding and trypsinlike protease production. Infect Immun. 1990;58:4016–4019. doi: 10.1128/iai.58.12.4016-4019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeCarlo A A, Grenett H E, Harber G J, Windsor L J, Bodden M K, Birkedal-Hansen B, Birkedal-Hansen H. Induction of matrix metalloproteinases and a collagen-degrading phenotype in gingival fibroblasts and mucosal epithelial cells by secreted proteinase from Porphyromonas gingivalis. J Periodontal Res. 1998;33:408–420. doi: 10.1111/j.1600-0765.1998.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 9.DeCarlo A A, Harber G J. Hemagglutinin activity and heterogeneity of related Porphyromonas gingivalisproteinases. Oral Microbiol Immunol. 1997;12:47–56. doi: 10.1111/j.1399-302x.1997.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 10.DeCarlo A A, Windsor L J, Bodden M K, Harber G J, Birkedal-Hansen B, Birkedal-Hansen H. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J Dent Res. 1997;76:1260–1270. doi: 10.1177/00220345970760060501. [DOI] [PubMed] [Google Scholar]
- 11.Dzink J L, Socransky S S, Haffajee A D. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 12.Eke P I, Laughon B E, Rotimi V O. Cytotoxic activity of crude extracts of Bacteroides gingivalis. J Med Microbiol. 1989;28:5–8. doi: 10.1099/00222615-28-1-5. [DOI] [PubMed] [Google Scholar]
- 13.Eley B M, Cox S W. Correlation between gingivain/gingipain and bacterial dipeptidyl peptidase activity in gingival crevicular fluid and periodontal attachment loss in chronic periodontitis patients. A 2-year longitudinal study. J Periodontol. 1996;67:703–716. doi: 10.1902/jop.1996.67.7.703. [DOI] [PubMed] [Google Scholar]
- 14.Evans R T, Klausen B, Ramamurthy N S, Golub L M, Sfintescu C, Genco R J. Periodontopathic potential of two strains of Porphyromonas gingivalisin gnotobiotic rats. Arch Oral Biol. 1992;37:813–819. doi: 10.1016/0003-9969(92)90115-o. [DOI] [PubMed] [Google Scholar]
- 15.Fiehn N E, Klausen B, Evans R T. Periodontal bone loss in Porphyromonas gingivalis-infected specific pathogen-free rats after preinoculation with endogenous Streptococcus sanguis. J Periodontol Res. 1992;27:609–614. doi: 10.1111/j.1600-0765.1992.tb01744.x. [DOI] [PubMed] [Google Scholar]
- 16.Fishburn C S, Slaney J M, Carman R J, Curtis M A. Degradation of plasma proteins by the trypsin-like enzyme of Porphyromonas gingivalisand inhibition of protease activity by a serine protease inhibitor of human plasma. Oral Microbiol Immunol. 1991;6:209–215. doi: 10.1111/j.1399-302x.1991.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher J, Reddi K, Poole S, Nair S, Henderson B, Tabona P, Wilson M. Interactions between periodontopathic bacteria and cytokines. J Periodontal Res. 1997;32:200–205. doi: 10.1111/j.1600-0765.1997.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 18.Fujimura S, Nakamura T. Multiple forms of proteases of Bacteroides gingivalisand their cellular location. Oral Microbiol Immunol. 1989;4:227–229. doi: 10.1111/j.1399-302x.1989.tb00257.x. [DOI] [PubMed] [Google Scholar]
- 19.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalisand isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazi M I, Cox S W, Clark D T, Eley B M. A comparison of cysteine and serine proteinases in human gingival crevicular fluid with tissue, saliva and bacterial enzymes by analytical isoelectric focussing. Arch Oral Biol. 1996;41:393–400. doi: 10.1016/0003-9969(96)00007-6. [DOI] [PubMed] [Google Scholar]
- 21.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalisare induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genco C A, Odusanya B M, Potempa J, Mikolajczyk-Pawlinska J, Travis J. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalisinfection in mice. Infect Immun. 1998;66:4108–4114. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goff H M, Morgan L O. Carbon-13 and proton nuclear magnetic resonance spectroscopy of water-soluble porphyrins and metalloporphyrins. Bioinorg Chem. 1978;9:61–79. doi: 10.1016/s0006-3061(00)82006-2. [DOI] [PubMed] [Google Scholar]
- 24.Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. 1987;55:111–117. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanazawa S, Murakami Y, Hirose K, Amano S, Ohmori Y, Higuchi H, Kitano S. Bacteroides (Porphyromonas) gingivalisfimbriae activate mouse peritoneal macrophages and induce gene expression and production of interleukin-1. Infect Immun. 1991;59:1972–1977. doi: 10.1128/iai.59.6.1972-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanazawa S, Nakada K, Ohmori Y, Miyoshi T, Amano S, Kitano S. Functional role of interleukin 1 in periodontal disease: induction of interleukin 1 production by Bacteroides gingivalislipopolysaccharide in peritoneal macrophages from C3H/HeN and C3H/HeJ mice. Infect Immun. 1985;50:262–270. doi: 10.1128/iai.50.1.262-270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura T, Pike R N, Potempa J, Travis J. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalisinduces vascular permeability enhancement through activation of the kallikrein/kinin pathway. J Clin Investig. 1994;94:361–367. doi: 10.1172/JCI117330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura T, Potempa J, Tanase S, Travis J. Activation of blood coagulation factor X by arginine-specific cysteine proteinases (gingipain-Rs) from Porphyromonas gingivalis. J Biol Chem. 1997;272:16062–16067. doi: 10.1074/jbc.272.25.16062. [DOI] [PubMed] [Google Scholar]
- 29.Jentzen W, Ma J G, Shelnutt J A. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S-J, Chu L, Holt S C. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 31.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalisis identical to lysine-specific cysteine proteinase (Lys-gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Lantz M S, Allen R D, Duck L W, Blume J L, Switalski L M, Hook M. Identification of Porphyromonas gingivaliscomponents that mediate its interactions with fibronectin. J Bacteriol. 1991;173:4263–4270. doi: 10.1128/jb.173.14.4263-4270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larjava H, Uitto V J, Haapasalo M, Heino J, Vuento M. Fibronectin fragmentation induced by dental plaque and Bacteroides gingivalis. Scand J Dent Res. 1987;95:308–314. doi: 10.1111/j.1600-0722.1987.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 35.Liddington R, Derewenda Z, Dodson E, Hubbard R, Dodson G. High resolution crystal structures and comparisons of T-state deoxyhaemoglobin and two liganded T-state haemoglobins: T(alphas-oxy)haemoglobin and T(met)haemoglobin. J Mol Biol. 1992;228:551–579. doi: 10.1016/0022-2836(92)90842-8. [DOI] [PubMed] [Google Scholar]
- 36.Löe H, Anerud A, Boysen H, Morrison E. Natural history of periodontal disease in man. Rapid, moderate and no loss of attachment in Sri Lankan laborers 14–46 years of age. J Clin Periodontol. 1986;13:431–440. doi: 10.1111/j.1600-051x.1986.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 37.Malek R, Fisher J G, Caleca A, Stinson M, van Oss C J, Lee J-Y, Cho M-I, Genco R J, Evans R T, Dyer D W. Inactivation of the Porphyromonas gingivalis fimAgene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsh P D, McDermid A S, McKee A S, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalisW50. Microbiology. 1994;140:861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 39.McDermid A S, McKee A S, Marsh P D. Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalisW50. Infect Immun. 1988;56:1096–1100. doi: 10.1128/iai.56.5.1096-1100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKee A S, McDermid A S, Baskerville A, Dowsett A B, Ellwood D C, Marsh P D. Effect of hemin on the physiology and virulence of Bacteroides gingivalisW50. Infect Immun. 1986;52:349–355. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton P A, Chen W-C A, Travis J, Potempa J. Genetic variation of Porphyromonas gingivalisgenes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 42.Minhas T, Greenman J, Schaffer A G. The influence of haemin levels on growth and enzyme production by Porphyromonas gingivalisin continuous culture. Microbios. 1993;76:105–114. [PubMed] [Google Scholar]
- 43.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 44.Nishikata M, Yoshimura F. Characterization of Porphyromonas (Bacteroides) gingivalishemagglutinin as a protease. Biochem Biophys Res Commun. 1991;178:336–342. doi: 10.1016/0006-291x(91)91819-x. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 46.Page R C. The pathobiology of periodontal diseases may affect systemic diseases: inversion of a paradigm. Ann Periodontol. 1998;3:108–120. doi: 10.1902/annals.1998.3.1.108. [DOI] [PubMed] [Google Scholar]
- 47.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 48.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalisare due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis(gingipains) using peptidyl chlormethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 50.Qiu X, Schroeder P, Bridon D. Identification and characterization of a C(K/R)TC motif as a common epitope present in all subtypes of hepatitis B surface antigen. J Immunol. 1996;156:3350–3356. [PubMed] [Google Scholar]
- 51.Rangarajan M, Aduse-Opoku J, Slaney J, Young K A, Curtis M A. The prpR1 and prR1 arginine-specific protease genes of Porphyromonas gingivalisW50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 52.Sato M, Otsuka M, Maehara R, Endo J, Nakamura R. Degradation of human secretory immunoglobulin A by protease isolated from the anaerobic periodontopathogenic bacterium Bacteroides gingivalis. Arch Oral Biol. 1987;32:235–238. doi: 10.1016/0003-9969(87)90016-1. [DOI] [PubMed] [Google Scholar]
- 53.Schifferle R E, Shostad S A, Bayers-Thering M T, Dyer D W, Neiders M E. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J Endod. 1996;22:352–355. doi: 10.1016/S0099-2399(96)80216-0. [DOI] [PubMed] [Google Scholar]
- 54.Scott C F, Whitaker E J, Hammond B F, Colman R W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalisthat cleaves kininogens and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 55.Shaanan B. Structure of human oxyhaemoglobin at 2.1 A resolution. J Mol Biol. 1983;171:31–59. doi: 10.1016/s0022-2836(83)80313-1. [DOI] [PubMed] [Google Scholar]
- 56.Shah H N, Gharbia S E. Lysis of erythrocytes by the secreted cysteine proteinase of Porphyromonas gingivalisW83. FEMS Microbiol Lett. 1989;52:213–217. doi: 10.1016/0378-1097(89)90199-7. [DOI] [PubMed] [Google Scholar]
- 57.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;131:313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 58.Sismey-Durrant H J, Hopps R M. Effect of lipopolysaccharide from Porphyromonas gingivalis on prostaglandin E2 and interleukin-1-beta release from rat periosteal and human gingival fibroblasts in vitro. Oral Microbiol Immunol. 1991;6:378–380. doi: 10.1111/j.1399-302x.1991.tb00510.x. [DOI] [PubMed] [Google Scholar]
- 59.Slakeski N, Bhogal P S, O’Brien-Simpson N M, Reynolds E C. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalisand identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology. 1998;144:1583–1592. doi: 10.1099/00221287-144-6-1583. [DOI] [PubMed] [Google Scholar]
- 60.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin binding as a factor in the virulence of Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;141:65–70. doi: 10.1111/j.1574-6968.1996.tb08364.x. [DOI] [PubMed] [Google Scholar]
- 61.Smalley J W, Silver J, Marsh P J, Birss A J. The periodontopathogen Porphyromonas gingivalisbinds iron protoporphyrin IX in the mu-oxo dimeric form—an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sorsa T, Ingman T, Suomalainen K, Haapasalo M, Konttinen Y, Lindy O, Saari H, Uitto V-J. Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect Immun. 1992;60:4491–4495. doi: 10.1128/iai.60.11.4491-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sundqvist G, Bengtson A, Carlsson J. Generation and degradation of the complement fragment C5a in human serum by Bacteroides gingivalis. Oral Microbiol Immunol. 1988;3:103–107. doi: 10.1111/j.1399-302x.1988.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 64.Tompkins G R, Wood D P, Birchmeier K R. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J Bacteriol. 1997;179:620–626. doi: 10.1128/jb.179.3.620-626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Touw J J, van Steenbergen T J, De Graaff J. Butyrate: a cytotoxin for Vero cells produced by Bacteroides gingivalis and Bacteroides asaccharolyticus. Antonie Leeuwenhoek. 1982;48:315–325. doi: 10.1007/BF00418285. [DOI] [PubMed] [Google Scholar]
- 66.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wingrove J A, DiScipio R G, Chen Z, Potempa J, Travis J, Hugli T E. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 68.Yun P L W, DeCarlo A A, Hunter N. Modulation of major histocompatability complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect Immun. 1999;67:2986–2995. doi: 10.1128/iai.67.6.2986-2995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]