Summary
Tetraspanins (TSPANs) are a class of four-transmembrane segmented proteins. The precise functions of TSPANs and their roles in pan-cancer are unclear. In this work, we analyzed TSPAN1, TSPAN10, TSPAN11, TSPAN12, TSPAN13, TSPAN14, TSPAN15, TSPAN16, TSPAN17, TSPAN18, TSPAN18-AS1, TSPAN19, TSPAN2, TSPAN3, TSPAN31, TSPAN32, TSPAN33, TSPAN4, TSPAN5, TSPAN6, TSPAN7, TSPAN8, TSPAN9, TSPAN9-IT1 (24 TSPAN family genes) in relation to tumor characteristics from 11,057 TCGA samples across 33 cancer types. On 24 TSPAN family genes, multidimensional studies were conducted, including gene differential expression, immunological subtype analysis, clinical analysis, stemness indices analysis, drug sensitivity analysis, alteration analysis, and multi-omics validation (including ATAC-seq validation, single-cell sequencing validation, and other external validation). Genes were differentially expressed in 33 cancers, and several of them showed high consistency in terms of tumor characteristics. In particular, the potential roles of TSPAN15 and TSPAN1 in cancer deserve further attention.
Subject areas: Cancer systems biology, Cancer, Transcriptomics
Graphical abstract
Highlights
-
•
A first pan-cancer analysis of the TSPAN family
-
•
Differential expression of TSPAN15 in HNSC may be associated with ADAM10
-
•
Tetraspanins may participate in tumor activity by complex transmembrane properties
Cancer systems biology; Cancer; Transcriptomics.
Introduction
Tetraspanins (TSPANs) are a protein family that consists of four transmembrane segments, a tiny extracellular structural domain, and a large extracellular loop (LEL) (Charrin et al., 2014; Florin and Lang, 2018). Intracellular domains are modest in size and contain palmitoylated cysteines and the N- and C-terminal tails. And, with the exception of a tiny variable domain located inside the LEL, the homology between isoforms is highly conserved (Seigneuret et al., 2001). Tetraspanin proteins are likewise highly conserved across species, with 33 of the 34 four-transmembrane proteins found in mammals also being found in humans (Beckwith et al., 2015). The tetraspanin transmembrane proteins have been implicated in a variety of activities including cancer, the immune system, fertility, and infectious disease. In the oncogenesis field, the TSPAN family of genes may contribute to tumor growth by influencing angiogenesis, immunological function, platelet coagulation, and infection (Hemler, 2008).
Therefore, it’s essential to investigate TSPANs expression patterns in pan-cancer and fully exert TSPAN-targeted drugs’ potential by matching targeted drugs to the specific TSPAN-expressed tumors.
The expression signatures of 24 TSPAN family genes in pan-cancer were addressed in this study. We discovered relationships between TSPAN gene expression levels and clinical correlation, immunological subtype, tumor purity, and drug sensitivity in multiple tumor types using multidimensional correlation analysis.
Results
Differential co-expression analysis of key genes
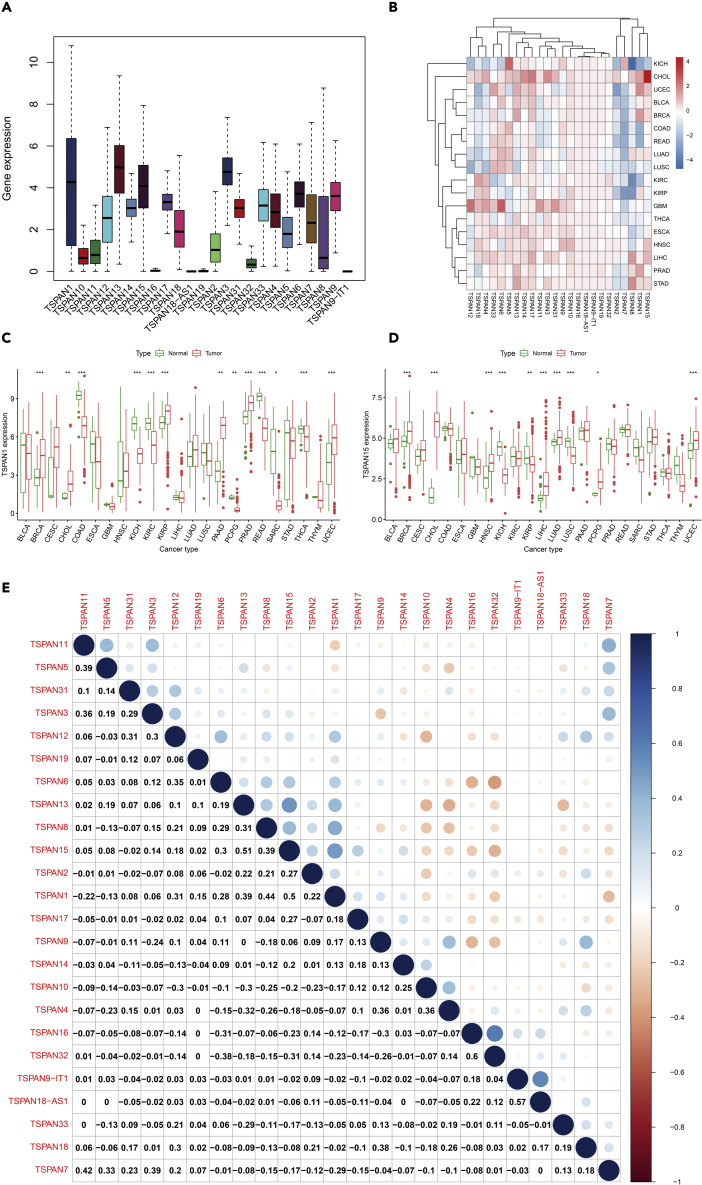
Flow diagrams outlining our data analysis process are in the graphical abstract. The expression of 24 TSPAN family genes showed in Figure 1A. The Wilcox test was used to determine the differential expression of 24 TSPAN family genes between tumor and adjacent normal samples (Figure 1B). Gene expression differed between tumors. TSPAN3-6, TSPAN9-11, TSPAN13-15, TSPAN17, TSPAN31, TSPAN33 are mainly upregulated, and TSPAN2, TSPAN7 are mainly downregulated. Contradictory results exist about the expression of TSPAN1, TSPAN8, TSPAN12, TSPAN18. TSPAN16, TSPAN18-AS1, TSPAN19, TSPAN32 remain low expression level (Figure S1).
Figure 1.
Differential expression and co-expression analysis of 24 TSPAN genes
(A) Gene expression in pan-cancer.
(B) The heatmap shows expression levels of TSPAN genes among 33 cancer types; the gradation of color represents log2 Fold Change (log FC) value.
(C and D) The differential expression of TSPAN1 and TSPAN15 between tumor and adjacent normal tissue in 23 cancer types. (Triple “∗” represent p < 0.001, double “∗” represent p < 0.01 and single “∗” represent p < 0.05).
(E) Co-expression heatmap demonstrates co-expression relations of TSPAN genes. The correlation coefficients are shown in the boxes.
TSPAN1 was observed significantly high expression in BCRA, CHOL, KIRP, PAAD, UCEC. TSPAN15 was observed significantly high expression in BCRA, CHOL, HNSC, LIHC, LUAD, UCEC, especially in CHOL (Figures 1B–1D).
The co-expression relationship could be observed in Figure 1E, including TSPAN15 and TSPAN13 (correlation coefficient = 0.51), TSPAN15 and TSPAN1 (correlation coefficient = 0.50), TSPAN15 and TSPAN32 (correlation coefficient = −0.31). The co-expression analyses of TSPAN15 showed the potential correlation network of TSPAN15, which was consistent with the external validation from the STRING Database (Figure S3).
Clinical correlation analysis
By performing the R package, Kaplan-Meier analyses were performed on 24 TSPAN family genes in 33 TCGA tumors (Figures 2A–2F, Figure S2). The patients’ cohorts were separated into high/low groups according to median gene expression. High expression of TSPAN15 was significantly associated with better prognosis in SARC (p = 0.006, Figure 2D), TGCT (p = 0.048, Figure 2E), and with poorer prognosis in HNSC (p = 0.015, Figure 2B), LIHC (p = 0.011, Figure 2C), PAAD (p = 0.007, Figure 2F). As shown in the KM plot in Figure 2C and a poor prognosis in patients with LIHC corresponded to the low gene expression of TSPAN15. Consistent results appear in Figure 1D, where the expression level of TSPAN15 was higher in tumor tissues than in adjacent normal tissues in LIHC (p < 0.001).
Figure 2.
Survival analysis and Cox proportional model analysis of TSPAN genes in 33 cancer types
(A-F) Kaplan-Meier survival curve of TSPAN1 and TSPAN15 in 33 cancer types.
(G) Cox proportional hazard analyses of TSPANs in TCGA cancers.
Similarly, the two sets of results for TSPAN1 showed consistency. As shown in the KM plot (Figure 2A), a poor prognosis in patients with PAAD corresponded to the high gene expression of TSPAN1 (p = 0.011). Consistent results appear in Figure 1C, where the expression level of Tspan1 was higher in tumor tissues than in adjacent normal tissues in PAAD (p < 0.01).
We performed a Cox regression analysis of the collected data. As shown in Figure 2G, the prognostic factor was characterized by hazard ratio (HR > 1) and p-value (p < 0.05) in the forest plot. We found TSPAN4, TSPAN12, TSPAN15, and TSPAN17 have pan-cancer significantly with HR > 1 in most cancer types.
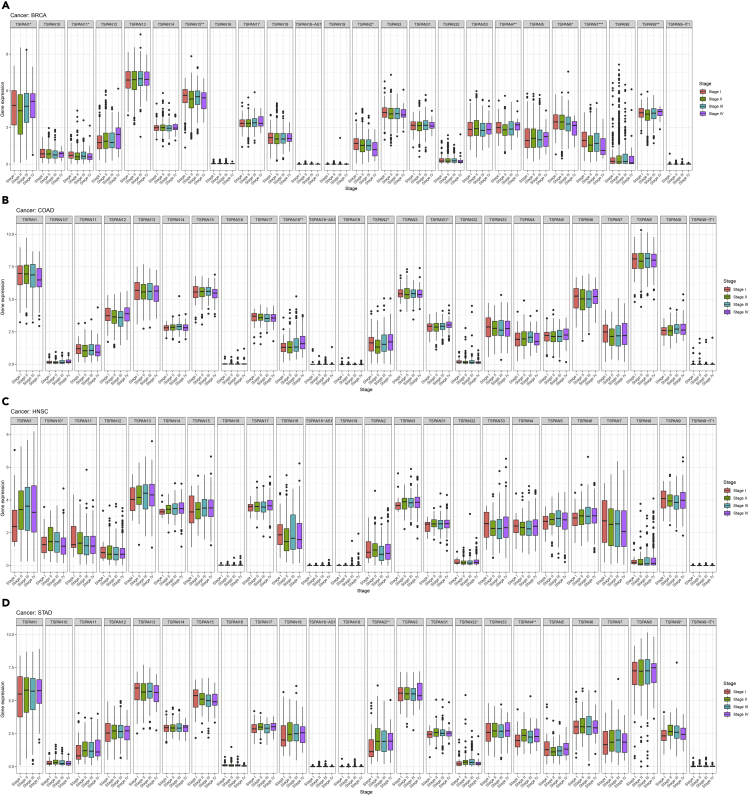
In particular, we found TSPAN15 expression was correlated with TNM stages (p < 0.01) in BRCA. The same expression situation occurs on TSPAN2, TSPAN32 in STAD, and TSPAN18 in COAD (Figure 3). TSPAN gene expression in different TNM stages might predict the prognosis of tumors.
Figure 3.
Correlation analysis between 24 TSPAN family genes and four stages in BRCA (A), COAD (B), HNSC (C), and STAD (D)
Correlation between tetraspanin family and tumor mutation burden in pan-cancer
Tumor mutation burden (TMB) in 33 cancer types was presented in the form of log10 (TMB+1). Different types of cancer are ordered by the median value of mutation frequency (Figure 4A). SKCM, LUSC, LUAD, and so forth showed relatively high TMB.
Figure 4.
Correlation analysis between TSPAN family and tumor mutation burden
(A) TMB of 33 cancer types was shown in the chat of log10 (TMB+1). Different types of cancer are ordered by the median value of mutation frequency.
(B) Correlation analysis between TSPAN family and TMB in 33 cancer types. (TMB: tumor mutation burden. Positive and negative correlations are indicated by red and blue dots, respectively).
Spearman correlation analysis was performed based on gene expression and TMB in 33 cancer types (Figure 4B). THYM showed a widely positive association with TSPAN3 (Pearson coefficient = 0.49), TSPAN6 (Pearson coefficient = 0.67), TSPAN9 (Pearson coefficient = 0.52), TSPAN12 (Pearson coefficient = 0.46), TSPAN17 (Pearson coefficient = 0.45), TSPAN31 (Pearson coefficient = 0.46) and so forth, suggesting a higher mutational heterogeneity from other cancers.
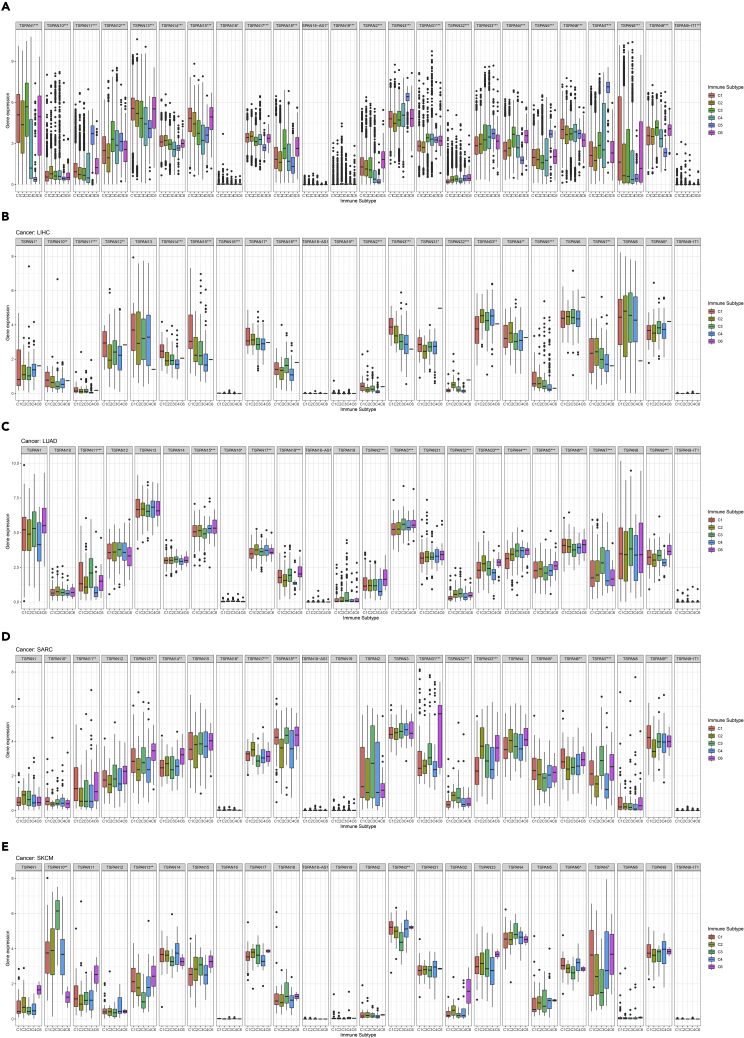
Immune subtype analysis
Among 33 TCGA cancer types, the mRNA differential expression levels of the 24 TSPAN family genes in six immune subtypes were analyzed with the Kruskal-Wallis test (Figure 5A).
Figure 5.
Results of 24TSPAN gene co-expression analysis with six immune subtypes
(A) The differential expression of the 24TSPAN gene in all tumor types.
(B-E) The correlation analysis between 24 TSPAN gene in LIHC, LUAD, SARC, and SKCM and immune subtypes. (Triple “∗” represent p < 0.001, double “∗” represent p < 0.01 and single “∗” represent p < 0.05).
The expression of TSPAN1-10, TSPAN13-15, TSPAN17-19, TSPAN31, TSPAN32, TSPAN9-IT1 (p < 0.001) exist significant differences among six immune subtypes (Figure 5A).
Samples of different tumor types exhibited variable results in immune subtype analysis. In LIHC, LUAD, SARC, and SKCM, the expression of TSPAN family genes differed significantly in six immune subtypes. TSPAN15 shows significant expression (p < 0.001) in LIHC and LUAD, and with the highest C1 expression in LIHC (Figures 5B and 5C). Likewise, it is worth noting that the expression level of TSPAN31 (p < 0.001) was the highest in C6 immune subtypes in SARC, and the expression level of TSPAN10 (p < 0.001) was the highest in C3 immune subtypes in SKCM, which may, therefore, be considered as a potential target to identify immune subtypes (Figures 5D and 5E).
Stemness indices and tumor microenvironment analysis in pan-cancer
In TCGA tumors, the correlation between the two stemness indices and the expression levels of TSPANs revealed the difference. For DNAss, TSPAN family genes has strong correlations with TGCT, OV and CHOL with positive correlations of TSPAN13 (p < 0.001), TSPAN18 (p < 0.001) in CHOL, negative correlations of TSPAN11 (p < 0.001), TSPAN2 (p < 0.001), TSPAN5 (p < 0.001) in OV and negative correlations of TSPAN11 (p < 0.001), TSPAN2 (correlation coefficient = −0.65, p < 0.001), TSPAN5 (p < 0.001) in TGCT (Figure 6A).
Figure 6.
Correlation analysis of 24TSPAN gene and tumor stemness score and tumor microenvironment score
(A and B) The two heatmaps show the correlation of expression level of TSPAN1-9, TSPAN9-IT1, TSPAN10-18, TSPAN18-AS1, TSPAN19, TSPAN31-33, and stemness indices (DNAss and RNAss) in 33 TCGA cancer types. (DNAss: DNA methylation-based stemness score; RNAss: RNA-based stemness score).
(C) Correlation analysis between 24 TSPAN genes and stromal score in 33 cancer types. (Positive and negative correlations are indicated by red and blue dots, respectively).
For RNAss, the expression of TSPAN2, TSPAN4, TSPAN9, TSPAN11, TSPAN18 was negatively correlated with several tumor types, including BRCA, ESCA, COAD, PCPG, PRAD, PAAD, TGCT, and THYM (Figure 6B).
The stromal score of TCGA cancer samples was calculated by applying the ESTIMATE algorithm (Yoshihara et al., 2013).
We describe the correlation between TSPAN family gene expression and pan-cancer stroma score by applying Spearman correlation analysis in Figure 6C expression of TSPAN2, TSPAN4, TSPAN9, TSPAN11, and TSPAN18 was positively correlated with several tumor types, including BLCA, BRCA, CHOL, COAD, LIHC, PRAD READ, STAD, and TGCT, suggesting that changes in TSPAN family gene expression are associated with tumor purity.
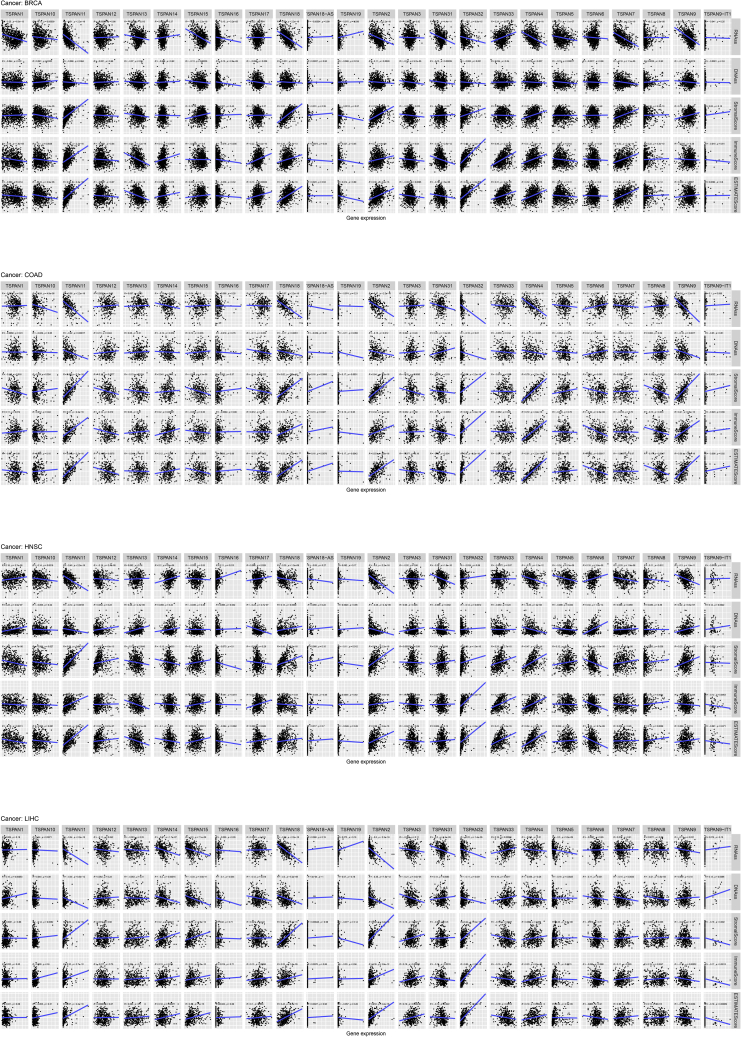
Figure 7 shows the relevance of TSPAN family genes and stromal score, immune score, ESTIMATE score, stemness indices in BRCA, COAD, HNSC, and LIHC.
Figure 7.
Correlation analysis between 24 TSPAN genes and stemness scores (RNAss and DNAss), stromal scores, immune scores, and ESTIMATE scores in BRCA, COAD, HNSC, LIHC
Figure 7A demonstrates the correlation of key TSPAN genes with multiple scores in BRCA. Four genes (TSPAN2, TSPAN11, TSPAN18, and TSPAN32) expression were positively correlated with ESTIMATE score, stromal score, immune score, and seven genes (TSPAN2, TSPAN4, TSPAN7, TSPAN9, TSPAN11, TSPAN15, and TSPAN18) expression was negatively correlated with DNAss and RNAss.
In COAD (Figure 7B), five genes (TSPAN2, TSPAN4, TSPAN9, TSPAN11, TSPAN32) expression was positively correlated with ESTIMATE score, stromal score, immune score. Three genes (TSPAN9, TSPAN11, TSPAN32) expression was negatively correlated with DNAss and RNAss.
In HNSC (Figure 7C), two genes (TSPAN2 and TSPAN11) expression was positively correlated with ESTIMATE score, stromal score, immune score. Three genes (TSPAN2, TSPAN4, TSPAN17) expression was negatively correlated with DNAss and RNAss.
In LIHC (Figure 7D), four genes (TSPAN2, TSPAN11, TSPAN18, TSPAN32) expression was positively correlated with ESTIMATE score, stromal score, immune score. Five genes (TSPAN2, TSPAN11, TSPAN15, TSPAN18, TSPAN32) expression was negatively correlated with DNAss and RNAss.
Drug sensitivity analysis in pan-cancer
By applying Pearson correlation analysis, we retrieved 263 antitumor drugs from the Cellminer database and correlated TSPAN gene expression with 263 drug activities in NCI-60 cancer cell lines (Puszkiel et al., 2019).
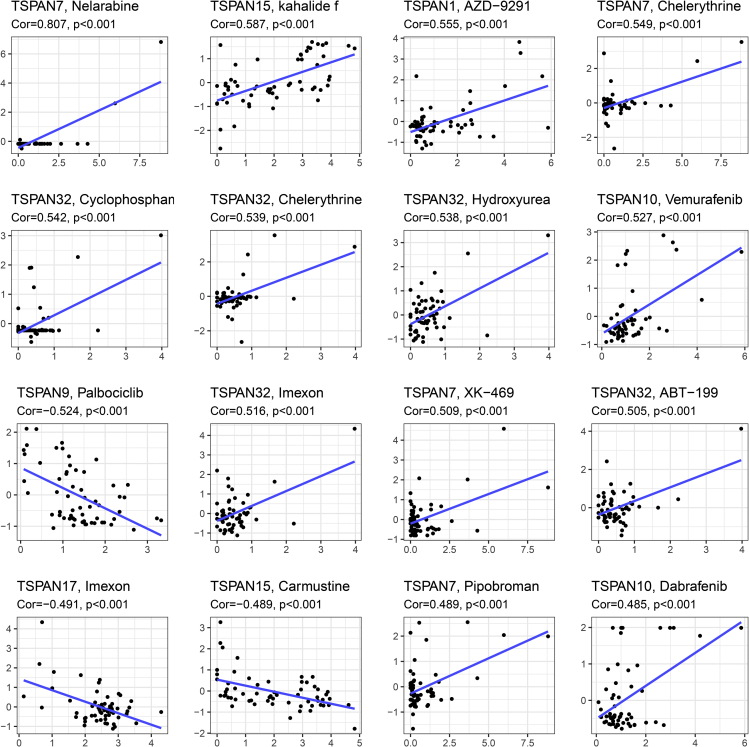
The results of the correlation analysis are presented in a scatterplot in Figure 8 (p < 0.05), which is ordered by the size of the p value. TSPAN1 was positively correlated with the sensitivity of AZD-9291 (p < 0.001). TSPAN15 was positively correlated with the sensitivity of kahalalide f (p < 0.001) and the resistance of Carmustine (p < 0.001).
Figure 8.
Drug response analysis
Correlation analysis between 24 TSPAN genes and drug sensitivity in 33 cancer types. The scatterplots are ranked by p value.
Tetraspanin family-related alteration of signaling pathways in pan-cancer
Numerous studies have shown TSPAN genes showed intense dependency on the WNT pathway to implement their function, including TSPAN12, TSPAN20, TSPAN24, and TSPAN27. So, we demonstrated an alteration plot of the WNT pathway in Figure 10. Data and design for Figure 10 were provided by Francisco, and so forth (Sanchez-Vega et al., 2018).
Figure 10.
ATAC-seq verifying that the chromosome region in the positions of TSPAN15 was open in multiple cell lines
Alteration in the WNT path showed that the CN-low subtype of UCEC took up the top of the list and CTNNB1 was serve as the most frequent gene to mutate in tumors (Figure 9).
Figure 9.
Extensive alteration of WNT pathway in most cancer types
CTNNB1 was the most frequent gene to mutate.
ATAC-seq validation
The transcriptional regulation of TSPAN15 and other TSPAN family members were further validated by Assay for Transposase-Accessible Chromatin using Sequencing (ATAC-seq). The results of ATAC-seq revealed that the chromatin regions were open in the chromosomal positions of these biomarkers in patients with multiple cell lines (Figure 10). The expression of other key biomarkers was also detected by using ATAC-seq validation (Figure S6).
Single-cell sequencing validation
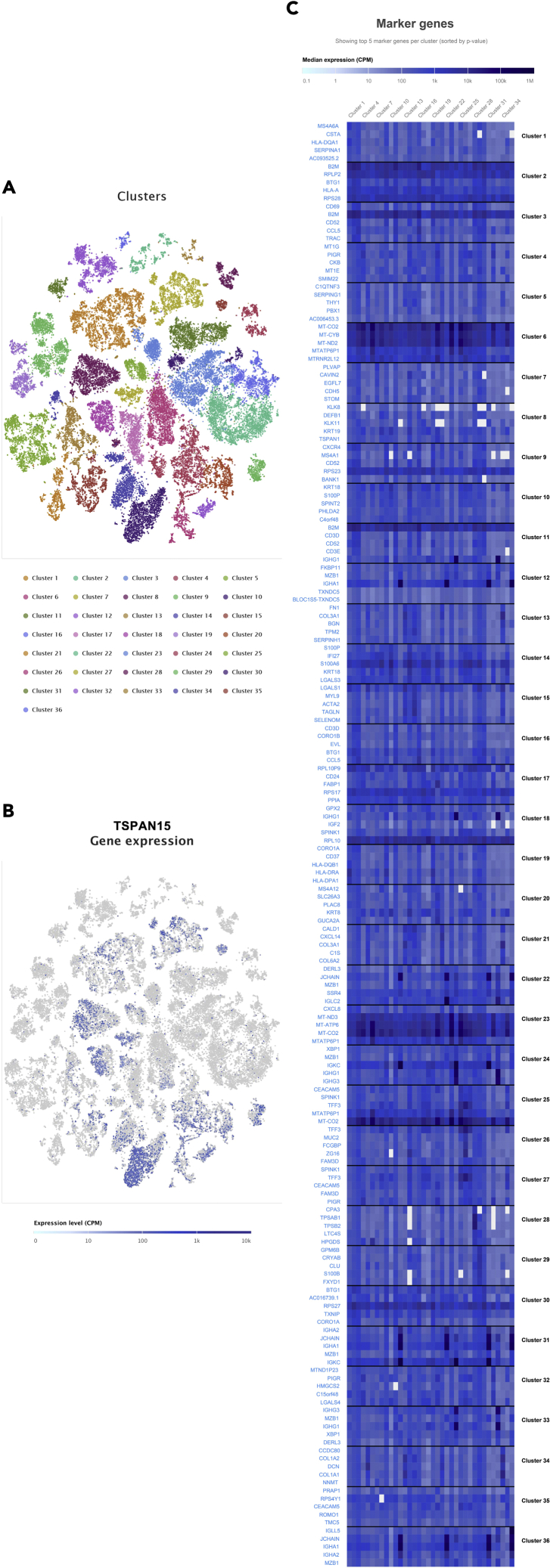
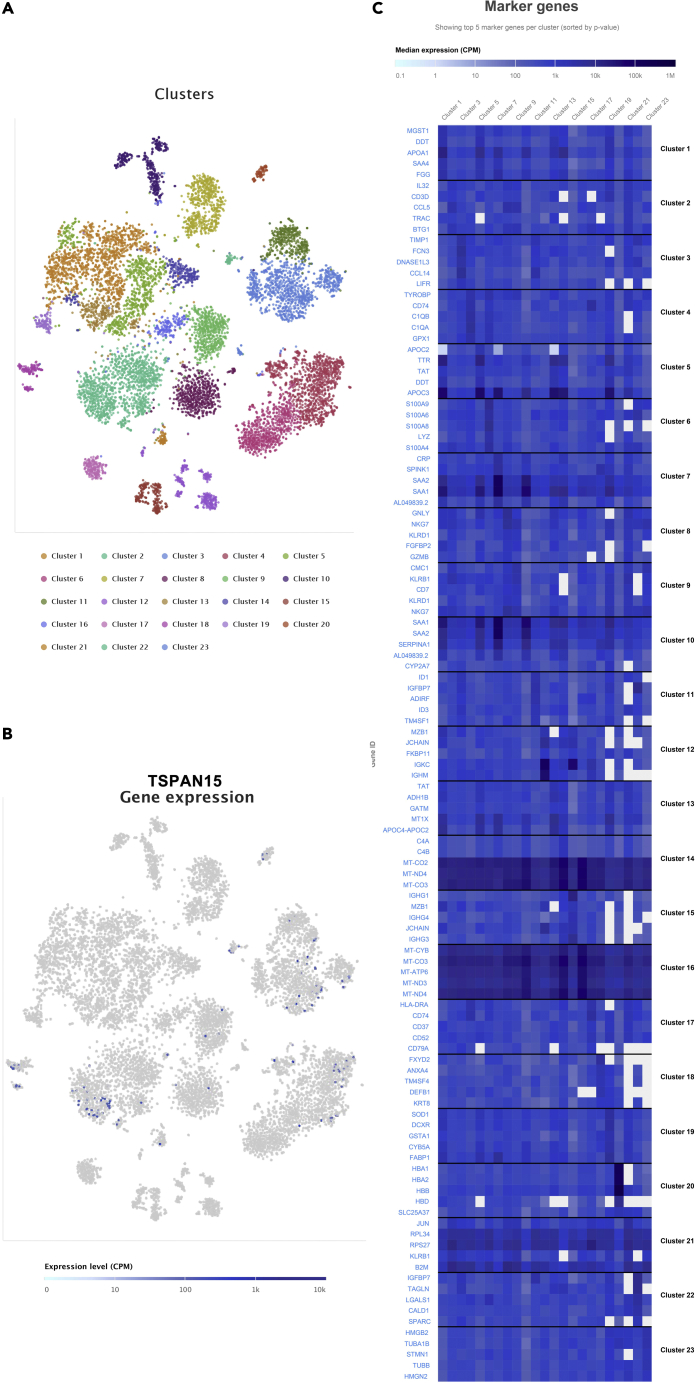
Based on the single-cell sequencing data of COAD and LIHC acquired from Single Cell Expression Atlas, we analyzed the expression of these biomarkers in a single cell in COAD and LIHC samples (Figures 11 and 12). The results indicated that TSPAN15 was highly expressed in B cells, CD8+ T cells, CD4+ T cells, alpha-beta T cell, gamma-delta T cell.
Figure 11.
The t-distributed stochastic neighbor embedding (t-SNE) plot of cell clusters in colorectal cancer in the Single Cell Expression Atlas (A).
The expression level and distribution of TSPAN15 in different clusters at single cellular level in COAD samples (B-C).
Figure 12.
The t-distributed stochastic neighbor embedding (t-SNE) plot of cell clusters in liver hepatocellular carcinoma in the Single Cell Expression Atlas (A).
The expression level and distribution of TSPAN15 in different clusters at single cellular level in LIHC samples (B-C).
External validation
By using the STRING database, the protein-protein interaction (PPI) network was constructed among TSPAN1-19, TSPAN31-33, and their top related genes. The protein-protein interaction network shows potential interactions between TSPANs, which may provide new ideas for further research (Figure S3).
Considering close correlations between the TSPAN family and clinical features, we explored the genetic alteration of the TSPAN family in pan-cancer for finding potential treatment targets by using cbioportal (http://www.cbioportal.org). The oncoprint in Figure S4 illustrated alteration breadth and frequency of the TSPAN family across 33 cancer types of which TSPAN2 and TSPAN12 were on the top roll with a high alteration frequency of 1.8 and 1.5%, respectively, mainly missense mutation. The result suggests that mutations in the TSPAN family may play a key role in oncogenic transformations (Figure S4).
In the human protein atlas, TSPAN15 protein expression in five types of cancer and normal tissues was performed in the form of immunohistochemical staining. The expression of TSPAN15 is lower in COAD and PRAD than normal tissue but is opposite in PAAD (Figure S5).
To reduce bias from a single database, we obtained a total of 544 samples of four cancer types (renal cancer, liver cancer, oral cancer, pancreatic cancer) from four regions (Japan, India, Canada, and European Union) from the ICGC database (https://dcc.icgc.org) to validate the expression of 24 genes of the TSPAN family in the four tumors. As shown in Figures S7–S10, the TSPAN family genes showed differential survival outcomes among the four tumors.
Meanwhile, we performed a single gene pan-cancer analysis for TSPAN1, TSPAN15 genes with good results in the analysis, as shown in Figures S9 and S10. We analyzed the expression of TSPAN1 in 33 tumor types species (Figure S11A) and performed km survival analysis and COX survival regression analysis (Figures S11B and S11C), and Figures S11D and S11E demonstrate the radar plot of TSPAN1 in each tumor for TMB and MSI indices. Figures S11G–S11I demonstrate the immune infiltration of TSPAN1. Similarly, the results of but genetic analysis of TSPAN15 are shown in Figure S12. From gene expression to survival analysis results, the single gene analysis was similar to our multi-gene analysis.
To increase the accuracy of the clinical analysis, we applied the PROgnoscan database for survival analysis of the TSPAN family genes and present the KM plots of the survival analysis results consistent with our result from the TCGA database in Figure S13.
Discussion
We sought to discuss the correlation between the transcriptional expression of TSPANs and TCGA tumor characteristics in this study, which contains clinical significance, immune subtype, drug sensitivity, stemness indices, and microtumor environment. Tetraspanins are a family of membrane proteins with four transmembrane domains. Because the proteins can be found in both plasma membranes and exosomes, tetraspanins are important actors in cell communication (Zöller, 2009). In terms of the results of the present study, TSPAN family genes exist a significant impact on cancer development. Multidimensional analysis of mass data was performed with 11,057 samples across 33 TCGA cancer types in differential expression analysis. The results showed that TSPAN gene expression levels differed in 33 tumors, while high and low TSPAN gene expression groups showed different survival outcomes in survival analysis. Broad TSPAN family genes expression in pan-cancer research across 33 cancer types promoted TSPAN family genes has been extensively implicated in tumor progression. Some research suggests that tetraspanins can control the functions of several cell surface proteins that may be involved in cell-cell and cell-matrix interactions (Charrin et al., 2014), which indicated that tetraspanins might demonstrate a significant role in cancers (Wang et al., 2016). Five members of the TSPAN family, TSPAN29, TSPAN26, TSPAN27, TSPAN24, and TSPAN8, TSPAN13, have been reported to be differentially expressed in cancer (Zöller, 2009; Hemler, 2014; Jiang et al., 2019). This clinical significance of TSPANs may provide clues for the diagnosis and treatment of specific cancers.
Tumor microenvironment (TME) is composed of immune characteristics, extracellular matrix, tumor vessels, and tumor cells. Tumor microenvironment heterogeneity has a great impact on the therapeutic effects and clinical prognosis of tumors (Junttila and de Sauvage, 2013). A classification was achieved for all tumor samples based on the C1-C6 immune subtypes (Thorsson et al., 2018). Expression levels of RNA-seq of 24 TSPAN family genes were examined, and all of them were identified as differentially expressed. In addition, to get more TME-related information among TCGA tumor samples, we also calculated the stromal score, immune score, and ESTIMATE scores. And the results show a statistically significant correlation between these scores and TSPAN family genes. Stemness is a definition that responds to tumor stem cell characteristics that include self-renewal and dedifferentiation (Friedmann-Morvinski and Verma, 2014). We calculated the RNAss score and DNAss score by using the OCLE approach to evaluate stemness characteristics. And the association was found between TSPANs and stemness, which suggested the potential stemness maintenance relevance in TSPANs. Evaluation indicators of tumor microenvironment and stemness characteristics show a correlation with TSPAN gene expression. Thus, we speculate that TSPAN family genes may be widely involved in tumor progression and metastasis.
In this study, we found that TSPAN1, TSPAN7, TSPAN9, TSPAN10, TSPAN15, TSPAN17, TSPAN32 gene expression levels also related to drug sensitivity. It is noteworthy that high expression of TSPAN15 will elevate the sensitivity of kahalidef, and high expression of TSPAN1 will elevate the sensitivity of AZD-9291.
The findings of TSPAN15 from the present analysis are noteworthy. In LIHC, TSPAN15 showed relatively high expression in tumor tissue than in adjacent normal tissues (Figure 1D). Survival analysis revealed a worse outcome in the high TSPAN15 expression group in LIHC (Figure 2C). Meanwhile, TSPAN15 displayed statistical significance in stemness features (Figure 7D) and cytotoxic drug sensitivity analysis (Figure 8). Sidahmed-Adrar et al. found that TSPAN15 may regulate cell proliferation through the secretion of soluble factors such as CTGF/CCN2 or other proteins that produce proliferation-promoting effects on HCC cells (Sidahmed-Adrar et al., 2019). This study also found that TSPAN15 expression was associated with certain stemness markers (epithelial cell adhesion molecule (EpCAM) and cytokeratin-19 (CK19), suggesting that it has some stem cell properties. This is consistent with our analysis.
Multiple research articles have reported the close relationship between TSPAN15 and ADAM10 (Eschenbrenner et al., 2020; Koo et al., 2020; Matthews et al., 2018; Saint-Pol et al., 2017). Chek Ziu Koo et al. posed and empirically validated four hypotheses: 1) TSPAN15 and ADAM10 were co-localized on the cell surface; 2) There is an interactive relationship between TSPAN15 and ADAM10 proteins; 3) Tspan15 protein expression requires ADAM10; 4) The combination of TSPAN15 and ADAM10 forms a functional complex, which performs scissor function. From this, we propose a new hypothesis. ADAM10 has now been shown to exert a vital part in tumorigenesis and tumor development in HNSC (Huang et al., 2014; Loganathan et al., 2020; Mitsuda et al., 2021). In our study, consistent with LIHC, TSPAN15 was relatively highly expressed in HNSC (Figures 2B and 2D), with poorer survival outcomes (Figure 3C) and high correlation with stromal and immune scores (Figure 7C), suggesting that TSPAN15 may also play a role in HNSC. There is no research report on the TSPAN15 gene in HNSC. But the potential role of TSPAN15 in HNSC deserves further attention based on the close relationship between TSPAN15 and ADAM10 and the findings of ADAM10 in HNSC.
Kahalalide F is a partially cyclic depsipeptide with a short chain fatty acid amide on the amino-terminal residue (Hamann and Scheuer, 1993; López-Macià et al., 2001). Though the specific biological mechanisms of KF remain unclear, there has been some speculation, alterations plasma membrane permeability (Suárez et al., 2003; Sewell et al., 2005), alterations in the lysosome morphology (García-Rocha et al., 1996; Shilabin et al., 2007), blocked expression of the ErbB3 signaling pathway (Shilabin et al., 2007; Janmaat et al., 2005; Piggott and Karuso, 2008), and other cytotoxic effects (Suárez et al., 2003). Meanwhile, clinical trials of the drug for prostate cancer are underway. We, therefore, speculate that the coherence shown in Figure 7 (TSPAN15, Kahalalide F) may be owing to cell proliferation promoting characteristics of TSPAN15 and the cytotoxicity mediated by necrotic cell death.
The results of TSPAN1 in this study are also noteworthy. In PAAD, TSPAN1 showed relatively high expression in tumor tissue than in adjacent normal tissues (Figure 1C). Survival analysis revealed a worse outcome in the high TSPAN1 expression group in PAAD (Figure 2A). And TSPAN1 was positively correlated with the sensitivity of AZD-9291 (p < 0.001) The role of TSPAN1 in tumors has been mainly summarized as related to proliferation, metastasis, apoptosis, and autophagy (Garcia-Mayea et al., 2021). Also, TSPAN1 has been reported to increase treatment resistance and act as a prognostic factor in tumors (Garcia-Mayea et al., 2021). Among the pan-cancer studies, there are more studies related to pancreatic cancer. In pancreatic cancer studies of TSPAN1, it was demonstrated that TSPAN1 is involved in nearly all the mechanisms mentioned above, including proliferation (Wang et al., 2020b), metastasis (Wang et al., 2020a; Zhang et al., 2019), apoptosis (Hou et al., 2015; Tian et al., 2018), and autophagy (Zhou et al., 2021). TSPAN1 promotes cell proliferation by activating AKT to upregulate CDK1 expression (Wang et al., 2020b); miR-216a/TSPAN1/ITGA2 axis is involved in pancreatic metastasis (Wang et al., 2020a); TSPAN1 deletion induces apoptosis in pancreatic cancer (Hou et al., 2015; Tian et al., 2018); TSPAN1 regulate the autophagic process by altering the protein expression of SQSTM1/p62 and LC3-II in pancreatic cancer, and LC-3 protein promotes autophagic maturation by binding to TSPAN1 (Zhou et al., 2021).
In our study, TSPAN genes show conflicting results on expression in different tumor types. Though studies of TSPAN family genes are few, similar conflicting results can be found in studies about CD9, which is a member of the TSPAN family. It has been shown that CD9 inhibits cell motility such as migration and invasion of tumor cell lines in vitro, including lung, breast, skin, gastric, pancreatic, and bladder tumors (Funakoshi et al., 2003; Ikeyama et al., 1993; Mitsuzuka et al., 2005; Ono et al., 2001; Sauer et al., 2003). However, the promotion effect of CD9 on cell motility has also been reported. Over-expression of CD9 in cell line SbCl2 and B16F1 resulted in the gain of Matrigel invasion activity in melanoma (Fan et al., 2010). Another two studies show simultaneous knockdown of CD9 and CD81 decreases MT1-MMP proteolytic functions in cancer cells (Lafleur et al., 2009) and silencing of MT1-MMP decreases invasive and migratory properties of breast cancer cells (Jiang et al., 2006). According to M. M. Richardson et al., this may be owing to differences in the lipid composition of different cells, such as differences in GM3 content in the plasma membrane of different tumor cells. (Richardson et al., 2011; Ono et al., 2001; Mitsuzuka et al., 2005). As transmembrane proteins, the regulatory mechanism of tetraspanins is highly complex (Richardson et al., 2011). The contradicting results among different tumor types suggest TSPAN genes may participate in tumor progression by different mechanisms.
Limitations of the study
Although this study is the first to perform multidimensional analyses on Tetraspanins (TSPANs) in pan-cancer, there were still a few limitations to the present study. To begin, all data were obtained from publicly available sources such as TCGA, GEO, Single Cell Expression Atlas, human protein atlas, string, and cbioportal. So, the clinicopathological parameters of patients in our data may not be comprehensive enough. Second, all data series used for analysis and processing were from publicly available databases, with the majority of the data sample coming from Western countries, so attention should be paid to potential data bias owing to ethnicity when extending the scope of the study. Third, the present study was not designed to elucidate the mechanism of TSPAN family genes. And research in the field of TSPAN is still in its infancy and needs further progress. We, therefore, made only limited inferences across groups. TSPANs and their co-activators have the potential to be pharmacological targets for cancer, but additional research is needed to determine this, making our findings even more valuable as a contribution to the expression signature analysis of 24 TSPAN family genes.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Bulk RNA-seq and clinical data for TCGA cohort | TCGA | https://gdc.cancer.gov/about-data/publications/pancanatlas |
| Bulk RNA-seq and clinical data for ICGC cohort | ICGC | https://dcc.icgc.org |
| Liver hepatocellular carcinoma ATAC-seq data | Grubert et al., 2020 | GSE139099 |
| Colorectal cancer ATAC-seq data | Lee et al., 2020 | EGAS00001003779,EGAS00001003769 |
| LIHC and COAD scRNA-seq data | Expression Atlas | https://www.ebi.ac.uk/gxa/home |
| Software and algorithms | ||
| R version 4.0.3 | https://www.r-project.org/ | https://www.r-project.org/ |
| ggpubr_0.4.0 | github | https://github.com/kassambara/ggpubr |
| ggcorrplot_0.1.3 | github | https://github.com/kassambara/ggcorrplot |
| survival_3.2-7 | github | https://github.com/therneau/survival |
| limma_3.46.0 | github | https://github.com/cran/limma |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zongqiang Huang (gzhuangzq@163.com).
Materials availability
This study did not generate new unique reagents.
Method details
Data collection and processing
The multimodal data of 24 TSPAN family genes about 11,057 samples across 33 cancer types were downloaded from TCGA. And we performed the read count assignment and analyzed transcriptional levels by calculating Fragments Per Kilobase per Million (FPKM) and HTSeq-counts. Demographics, follow-up information, and tumor characteristics were also obtained from TCGA. A part of data and datasets was obtained from Gene Expression Omnibus (GEO) (Accession no. GSE139099) (Grubert et al., 2020) and Single Cell Expression Atlas (Accession no. E-HCAD-14) (Papatheodorou et al., 2020). All data and code are provided as supplemental information.
Differential co-expression analysis of key genes
By applying the "ggpubr" R package, we performed Wilcox tests in 33 tumor types for differential analysis of tumor and normal tissue gene expression, where cancer types with fewer than three normal samples were excluded. Differential gene levels showed in the heatmap in a form of log2 fold change (log2 FC).
Co-expression analysis of the 24 TSPAN family genes was also performed at the transcriptional level using the R package "corrplot" to investigate the probable expression pattern between each pair of TSPAN genes.
Clinical correlation analysis
By performing the R package, we analyzed the survival differences of high and low expression groups to generate Kaplan-Meier plots. GDC TCGA sets in the UCSC Xena database (http://xena.ucsc.edu/) were used to download survival data for 33 TCGA cancer types. Patients were classified as either high- or low-expression (more or less than the median expression level) groups based on the median expression level of the 24 TSPAN family genes, respectively.
Other than that, hazard ratios of the 24 TSPAN family genes in all TCGA tumor types were calculated by using Cox regression. Differential expression analysis assessed differences of the clinical stage in 33 TCGA cancer types. Significance threshold was set at p < 0.05 (two-tailed).
Correlation between TSPAN family and TMB (tumor mutation burden) in pan-cancer
TMB can characterize mutation-driven tumors and help in prediction of immune therapy in most types of cancer (Samstein et al., 2019). Lawrence et al. performed a comprehensive investigation of TMB in 27 cancer types by calculating non-synonymous mutations per mega base (Lawrence et al., 2013). The same process was applied for describing the correlation between TSPANs and TMB level with Pearson correlation analysis.
Immune subtype analysis
TME has both therapeutic and prognostic value in antitumor therapy. Thorsson Vésteinn et al. has identified six immune subtypes based on the immune signature of TCGA tumor types. Difference in subtype distribution in TCGA tumors give them unique tumor characteristics that are associated with prognosis, genetic, and immune modulatory alterations, reflecting some extent the important role of TME in therapeutic and prediction of clinical outcomes, etc (Thorsson et al., 2018). Among 33 TCGA cancer types, the mRNA differential expression levels of the 24 TSPAN family genes in six immune subtypes (wound healing (C1), IFN-γ dominant (C2), inflammatory (C3), lymphocyte depleted (C4), immunologically quiet (C5), TGF-β dominant (C6)) were analyzed with the Kruskal–Wallis test (Thorsson et al., 2018).
Stemness indices and TME analysis in pan-cancer
The importance of the tumor microenvironment in tumor research is increasingly accepted. TME includes components such as stromal cells, immune cells, and blood vessels that play an important role in tumor development. By using the ESTIMATE score (Estimation of stromal and immune cells in malignant tumors using expression data) (Yoshihara et al., 2013), we quantified the proportion of stromal and immune cells in the tumor microenvironment and performed co-expression analysis with TSPAN family genes.
With the help of the one-class logistic regression (OCLR) algorithm, stemness indices are created as a novel model to reflect tumor pathology and clinical outcome. The stemness indices describe two basic characteristics of tumor cells: self-renewal and dedifferentiation, which may promote distant metastasis and tumorigenesis (Malta et al., 2018). We performed stemness analysis on TCGA tumor samples and calculated two stemness index characteristics called DNAss and RNAss (DNA methylation-based stemness index and mRNA expression-based stemness index) to reflect the tumor stemness of the samples, and performed Spearman correlation analysis on gene expression and stemness index.
Correlation analysis between gene expression and drug activity
With the help of CellMiner database (https://discover.nci.nih.gov/cellminer/) and the Impute package of Bioconductor (http://www.bioconductor.org/packages/release/bioc/html/impute.html), we acquired RNA-seq profiles and drug activity data and performed data pre-processing. Pearson correlation analysis was applied for correlation analysis between the 24 TSPAN genes expression and drug sensitivity (p < 0.05).
Landscape of alteration in PI3K and TGF-beta pathways in pan-cancer
Former studies pointed out that TSPAN genes showed widely depends on the WNT pathway to implement their function, including TSPAN12, TSPAN20, TSPAN24, and TSPAN27. So, a landscape of PI3K and TGF-beta pathway alteration in pan-cancer was painted, using Genomic Alteration Matrices and Curated pathway templates offered by Francisco, etc (Sanchez-Vega et al., 2018).
Chromatin accessibility analysis
The transcriptional regulation of TSPAN15 and other TSPAN family members were further validated by ATAC-seq methods. Initially, we downloaded the ATAC-seq data of COAD (Lee et al., 2020) and LIHC patients from the Gene Expression Omnibus (GEO) (Accession no. GSE139099) and detected the chromatin accessibility in the location of these biomarker genes.
Single cell sequencing
With the help of the single-cell expression atlas, we downloaded single-cell sequencing data to better analyze the expression of key biomarkers such as TSPAN15 at the single-cell level and hence to validate the cellular location of these biomarker expressions.
External validation
Validation results from multiple databases showed the consistency among multi-omics methods, including the human protein atlas (Uhlén et al., 2015), string (Snel et al., 2000), cbioportal (http://www.cbioportal.org), ICGC (https://dcc.icgc.org) and Prognoscan (Mizuno et al., 2009).
Statistical analysis
All statistical analysis was applied by R version 4.0.3 (Institute for Statistics and Mathematics, Vienna, Austria; www.r-project.org) (Package: impute, UpSetR, ggplot2, rms, glmnet, preprocessCore, forestplot, survminer, survivalROC, limma et al.). All data and code are provided as supplemental information. Except where noted, two-tailed p < 0.05 was regarded statistically significant.
Acknowledgments
We thank The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) team for using their data.
This study was supported in part by the National Natural Science Foundation of China (Grant No. 82173168, 81772856, 82073207; 81801620); National Natural Science Foundation of China, joint fund cultivation project (Grant No. U1504822); Shanghai Rising-Star Program (No. 21QA1407500); Henan medical science and technology research project (No. 201602031). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Author contributions
Conception/design: Runzhi Huang, Hanlin Sun, Ruoyi Lin, Jie Zhang, Huabin Yin, Shuyuan Xian, Man Li, Siqiao Wang, Zhenyu Li, Yannan Qiao, Meiyun Jiang, Penghui Yan, Tong Meng, and Zongqiang Huang.
Provision of study material: Runzhi Huang, Hanlin Sun, Ruoyi Lin, Jie Zhang, Huabin Yin, Shuyuan Xian, Man Li, Siqiao Wang, Zhenyu Li, Yannan Qiao, Meiyun Jiang, Penghui Yan, Tong Meng, and Zongqiang Huang.
Collection and/or assembly of data: Runzhi Huang, Hanlin Sun, Ruoyi Lin, Jie Zhang, Huabin Yin, Shuyuan Xian, Man Li, Siqiao Wang, Zhenyu Li, Yannan Qiao, Meiyun Jiang, Penghui Yan, Tong Meng, and Zongqiang Huang.
Manuscript writing: Runzhi Huang, Hanlin Sun, Ruoyi Lin, Jie Zhang, Huabin Yin, Shuyuan Xian, Man Li, Siqiao Wang, Zhenyu Li, Yannan Qiao, Meiyun Jiang, Penghui Yan, Tong Meng, and Zongqiang Huang.
Final approval of article: Runzhi Huang, Hanlin Sun, Ruoyi Lin, Jie Zhang, Huabin Yin, Shuyuan Xian, Man Li, Siqiao Wang, Zhenyu Li, Yannan Qiao, Meiyun Jiang, Penghui Yan, Tong Meng, and Zongqiang Huang.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104777.
Contributor Information
Penghui Yan, Email: 877492185@qq.com.
Tong Meng, Email: mengtong@medmail.com.cn.
Zongqiang Huang, Email: gzhuangzq@163.com.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All original code is available in supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Beckwith K.A., Byrd J.C., Muthusamy N. Tetraspanins as therapeutic targets in hematological malignancy: a concise review. Front. Physiol. 2015;6:91. doi: 10.3389/fphys.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S., Jouannet S., Boucheix C., Rubinstein E. Tetraspanins at A glance. J. Cell Sci. 2014;127:3641–3648. doi: 10.1242/jcs.154906. [DOI] [PubMed] [Google Scholar]
- Eschenbrenner E., Jouannet S., Clay D., Chaker J., Boucheix C., Brou C., Tomlinson M.G., Charrin S., Rubinstein E. Tspanc8 tetraspanins differentially regulate Adam10 endocytosis and half-life. Life Sci. Alliance. 2020;3:e201900444. doi: 10.26508/lsa.201900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhu G.Z., Niles R.M. Expression and function of Cd9 in melanoma cells. Mol. Carcinog. 2010;49:85–93. doi: 10.1002/mc.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L., Lang T. Tetraspanin assemblies in virus infection. Front. Immunol. 2018;9:1140. doi: 10.3389/fimmu.2018.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D., Verma I.M. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T., Tachibana I., Hoshida Y., Kimura H., Takeda Y., Kijima T., Nishino K., Goto H., Yoneda T., Kumagai T., et al. Expression of tetraspanins in human lung cancer cells: frequent downregulation of Cd9 and its contribution to cell motility in small cell lung cancer. Oncogene. 2003;22:674–687. doi: 10.1038/sj.onc.1206106. [DOI] [PubMed] [Google Scholar]
- Garcia-Mayea Y., Mir C., Carballo L., Sánchez-García A., Bataller M., Me L.L. Tspan1, A novel tetraspanin member highly involved in carcinogenesis and chemoresistance. Biochim. Biophys. Acta Rev. Canc. 2021;1877:188674. doi: 10.1016/j.bbcan.2021.188674. [DOI] [PubMed] [Google Scholar]
- García-Rocha M., Bonay P., Avila J. The antitumoral compound kahalalide F acts on cell lysosomes. Cancer Lett. 1996;99:43–50. doi: 10.1016/0304-3835(95)04036-6. [DOI] [PubMed] [Google Scholar]
- Grubert F., Srivas R., Spacek D.V., Kasowski M., Ruiz-Velasco M., Sinnott-Armstrong N., Greenside P., Narasimha A., Liu Q., Geller B., et al. Landscape of cohesin-mediated chromatin loops in the human Genome. Nature. 2020;583:737–743. doi: 10.1038/s41586-020-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann M.T., Scheuer P.J. Kahalide F: a bioactive depsipeptide from the sacoglossan mollusk elysia rufescens and the green alga bryopsis sp. J. Am. Chem. Soc. 1993;115:5825–5826. [Google Scholar]
- Hemler M.E. Targeting of tetraspanin proteins--potential benefits and strategies. Nat. Rev. Drug Discov. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler M.E. Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- Hou F.Q., Lei X.F., Yao J.L., Wang Y.J., Zhang W. Tetraspanin 1 is involved in survival, proliferation and carcinogenesis of pancreatic cancer. Oncol. Rep. 2015;34:3068–3076. doi: 10.3892/or.2015.4272. [DOI] [PubMed] [Google Scholar]
- Huang Y., Benaich N., Tape C., Kwok H.F., Murphy G. Targeting the sheddase activity of adam17 By an anti-adam17 antibody D1(A12) inhibits Head and Neck squamous cell carcinoma cell proliferation and motility via blockage of bradykinin induced hers transactivation. Int. J. Biol. Sci. 2014;10:702–714. doi: 10.7150/ijbs.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeyama S., Koyama M., Yamaoko M., Sasada R., Miyake M. Suppression of cell motility and metastasis by transfection with human motility-related protein (Mrp-1/Cd9) dna. J. Exp. Med. 1993;177:1231–1237. doi: 10.1084/jem.177.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmaat M.L., Rodriguez J.A., Jimeno J., Kruyt F.A.E., Giaccone G. Kahalalide F induces necrosis-like cell death that involves depletion of Erbb3 and inhibition of akt signaling. Mol. Pharmacol. 2005;68:502–510. doi: 10.1124/mol.105.011361. [DOI] [PubMed] [Google Scholar]
- Jiang L., Zhang X., Geradts J., Wei Q., Hochwald S., Xu H., Huang H. Expression of tetraspanins net-6 and Cd151 in breast cancer as a potential tumor biomarker. Clin. Exp. Med. 2019;19:377–384. doi: 10.1007/s10238-019-00554-x. [DOI] [PubMed] [Google Scholar]
- Jiang W.G., Davies G., Martin T.A., Parr C., Watkins G., Mason M.D., Mansel R.E. Expression of membrane type-1 matrix metalloproteinase, Mt1-mmp in human breast cancer and its impact on invasiveness of breast cancer cells. Int. J. Mol. Med. 2006;17:583–590. [PubMed] [Google Scholar]
- Junttila M.R., de Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- Koo C.Z., Harrison N., Noy P.J., Szyroka J., Matthews A.L., Hsia H.E., Müller S.A., Tüshaus J., Goulding J., Willis K., et al. The tetraspanin Tspan15 is an essential subunit of an Adam10 scissor complex. J. Biol. Chem. 2020;295:12822–12839. doi: 10.1074/jbc.RA120.012601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur M.A., Xu D., Hemler M.E. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol. Biol. Cell. 2009;20:2030–2040. doi: 10.1091/mbc.E08-11-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-O., Hong Y., Etlioglu H.E., Cho Y.B., Pomella V., Van Den Bosch B., Vanhecke J., Verbandt S., Hong H., Min J.-W., et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020;52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- Loganathan S.K., Schleicher K., Malik A., Quevedo R., Langille E., Teng K., Oh R.H., Rathod B., Tsai R., Samavarchi-Tehrani P., et al. Rare driver mutations in Head and Neck squamous cell carcinomas converge on notch signaling. Science. 2020;367:1264–1269. doi: 10.1126/science.aax0902. [DOI] [PubMed] [Google Scholar]
- López-Macià A., Jiménez J.C., Royo M., Giralt E., Albericio F. Synthesis and structure determination of kahalalide F (1, 2) J. Am. Chem. Soc. 2001;123:11398–11401. doi: 10.1021/ja0116728. [DOI] [PubMed] [Google Scholar]
- Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., Kamińska B., Huelsken J., Omberg L., Gevaert O., et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.L., Koo C.Z., Szyroka J., Harrison N., Kanhere A., Tomlinson M.G. Regulation of leukocytes by Tspanc8 tetraspanins and the "molecular scissor" Adam10. Front. Immunol. 2018;9:1451. doi: 10.3389/fimmu.2018.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda J., Tsujikawa T., Yoshimura K., Saburi S., Suetsugu M., Kitamoto K., Takenaka M., Ohmura G., Arai A., Ogi H., et al. A 14-marker multiplexed imaging panel for prognostic biomarkers and tumor heterogeneity in Head and Neck squamous cell carcinoma. Front. Oncol. 2021;11:713561. doi: 10.3389/fonc.2021.713561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzuka K., Handa K., Satoh M., Arai Y., Hakomori S. A specific microdomain ("Glycosynapse 3") controls phenotypic conversion and reversion of bladder cancer cells through Gm3-mediated interaction of Alpha3beta1 integrin with Cd9. J. Biol. Chem. 2005;280:35545–35553. doi: 10.1074/jbc.M505630200. [DOI] [PubMed] [Google Scholar]
- Mizuno H., Kitada K., Nakai K., Sarai A. Prognoscan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Handa K., Sonnino S., Withers D.A., Nagai H., Hakomori S. Gm3 Ganglioside inhibits Cd9-facilitated haptotactic cell motility: coexpression of Gm3 and Cd9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry. 2001;40:6414–6421. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- Papatheodorou I., Moreno P., Manning J., Fuentes A.M.-P., George N., Fexova S., Fonseca N.A., Füllgrabe A., Green M., Huang N., et al. Expression atlas update: from tissues to single cells. Nucleic Acids Res. 2020;48:D77–D83. doi: 10.1093/nar/gkz947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggott A.M., Karuso P. Rapid identification of A protein binding partner for the marine natural product kahalalide F by using reverse chemical proteomics. Chembiochem. 2008;9:524–530. doi: 10.1002/cbic.200700608. [DOI] [PubMed] [Google Scholar]
- Puszkiel A., Noé G., Bellesoeur A., Kramkimel N., Paludetto M.N., Thomas-Schoemann A., Vidal M., Goldwasser F., Chatelut E., Blanchet B. Clinical pharmacokinetics and pharmacodynamics of dabrafenib. Clin. Pharmacokinet. 2019;58:451–467. doi: 10.1007/s40262-018-0703-0. [DOI] [PubMed] [Google Scholar]
- Richardson M.M., Jennings L.K., Zhang X.A. Tetraspanins and tumor progression. Clin. Exp. Metastasis. 2011;28:261–270. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- Saint-Pol J., Eschenbrenner E., Dornier E., Boucheix C., Charrin S., Rubinstein E. Regulation of the trafficking and the function of the metalloprotease adam10 by tetraspanins. Biochem. Soc. Trans. 2017;45:937–944. doi: 10.1042/BST20160296. [DOI] [PubMed] [Google Scholar]
- Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer Genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G., Kurzeder C., Grundmann R., Kreienberg R., Zeillinger R., Deissler H. Expression of tetraspanin adaptor proteins below defined threshold values is associated with in vitro invasiveness of mammary carcinoma cells. Oncol. Rep. 2003;10:405–410. [PubMed] [Google Scholar]
- Seigneuret M., Delaguillaumie A., Lagaudrière-Gesbert C., Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with A structurally variable domain insertion. J. Biol. Chem. 2001;276:40055–40064. doi: 10.1074/jbc.M105557200. [DOI] [PubMed] [Google Scholar]
- Sewell J.M., Mayer I., Langdon S.P., Smyth J.F., Jodrell D.I., Guichard S.M. The mechanism of action of kahalalide F: variable cell permeability in human hepatoma cell lines. Eur. J. Cancer. 2005;41:1637–1644. doi: 10.1016/j.ejca.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Shilabin A.G., Kasanah N., Wedge D.E., Hamann M.T. Lysosome and Her3 (Erbb3) selective anticancer agent kahalalide F: semisynthetic modifications and antifungal lead-exploration studies. J. Med. Chem. 2007;50:4340–4350. doi: 10.1021/jm061288r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidahmed-Adrar N., Ottavi J.F., Benzoubir N., Ait Saadi T., Bou Saleh M., Mauduit P., Guettier C., Desterke C., Le Naour F. Tspan15 is A new stemness-related marker in hepatocellular carcinoma. Proteomics. 2019;19:E1900025. doi: 10.1002/pmic.201900025. [DOI] [PubMed] [Google Scholar]
- Snel B., Lehmann G., Bork P., Huynen M.A. String: a web-server to retrieve and display the repeatedly occurring neighbourhood of A gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez Y., González L., Cuadrado A., Berciano M., Lafarga M., Muñoz A. Kahalalide F, A new marine-derived compound, induces oncosis in human prostate and breast cancer cells. Mol. Cancer Ther. 2003;2:863–872. [PubMed] [Google Scholar]
- Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Zhang R., Piao H., Li X., Sheng W., Zhou J., Dong M., Zhang X., Yan X., Shang W., et al. Silencing Tspan1 inhibits migration and invasion, and induces the apoptosis of human pancreatic cancer cells. Mol. Med. Rep. 2018;18:3280–3288. doi: 10.3892/mmr.2018.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wang S., Liu X., Khan A.A., Li H., Tahir M., Yan X., Wang J., Huang H. Mir-216a-Mediated upregulation of Tspan1 contributes to pancreatic cancer progression via transcriptional regulation of Itga2. Am. J. Cancer Res. 2020;10:1115–1129. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gao X., Tian J., Zhang R., Qiao Y., Hua X., Shi G. Tspan1 promotes human pancreatic cancer cells proliferation by modulating Cdk1 via akt. bioRxiv. 2020 doi: 10.1101/2020.05.29.123091. Preprint at. [DOI] [Google Scholar]
- Wang Z., Von Au A., Schnölzer M., Hackert T., Zöller M. Cd44v6-Competent tumor exosomes promote motility, invasion and cancer-initiating cell marker expression in pancreatic and colorectal cancer cells. Oncotarget. 2016;7:55409–55436. doi: 10.18632/oncotarget.10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi G., Gao F., Liu P., Wang H., Tan X. Tspan1 upregulates Mmp2 to promote pancreatic cancer cell migration and invasion via plcγ. Oncol. Rep. 2019;41:2117–2125. doi: 10.3892/or.2019.6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Liang Y., Zhou L., Yan Y., Liu N., Zhang R., Huang Y., Wang M., Tang Y., Ali D.W., et al. Tspan1 promotes autophagy flux and mediates cooperation between wnt-ctnnb1 signaling and autophagy via the mir454-fam83a-tspan1 Axis in pancreatic cancer. Autophagy. 2021;17:3175–3195. doi: 10.1080/15548627.2020.1826689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All original code is available in supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.