Abstract
Background
Human leukocyte antigen (HLA) genes play critical roles in immune surveillance, an important defence against tumors. Imputing HLA genotypes from existing single-nucleotide polymorphism datasets is low-cost and efficient. We investigate the relevance of the major histocompatibility complex region in breast cancer susceptibility, using imputed class I and II HLA alleles, in 25,484 women of Asian ancestry.
Methods
A total of 12,901 breast cancer cases and 12,583 controls from 12 case–control studies were included in our pooled analysis. HLA imputation was performed using SNP2HLA on 10,886 quality-controlled variants within the 15–55 Mb region on chromosome 6. HLA alleles (n = 175) with info scores greater than 0.8 and frequencies greater than 0.01 were included (resolution at two-digit level: 71; four-digit level: 104). We studied the associations between HLA alleles and breast cancer risk using logistic regression, adjusting for population structure and age. Associations between HLA alleles and the risk of subtypes of breast cancer (ER-positive, ER-negative, HER2-positive, HER2-negative, early-stage, and late-stage) were examined.
Results
We did not observe associations between any HLA allele and breast cancer risk at P < 5e−8; the smallest p value was observed for HLA-C*12:03 (OR = 1.29, P = 1.08e−3). Ninety-five percent of the effect sizes (OR) observed were between 0.90 and 1.23. Similar results were observed when different subtypes of breast cancer were studied (95% of ORs were between 0.85 and 1.18).
Conclusions
No imputed HLA allele was associated with breast cancer risk in our large Asian study. Direct measurement of HLA gene expressions may be required to further explore the associations between HLA genes and breast cancer risk.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12282-022-01366-w.
Keywords: HLA, Breast cancer risk, Breast cancer subtypes
Introduction
The major histocompatibility complex (MHC) region in the human genome is gene dense, highly polymorphic, and known as a hotspot for disease associations (mainly autoimmune and infectious). Mapping to the short arm of chromosome 6, the MHC region includes human leukocyte antigen (HLA) genes and plays various critical roles in regulating immune response. One such function is immune surveillance, which is an important defense against tumors [1].
Many studies have examined HLA genotypes in the MHC region in relation to breast cancer susceptibility [2–4]. However, low statistical power due to the limited sample sizes in these studies has made it difficult to obtain consistent and convincing results. Nonetheless, in a large-scale GWAS study comprising 62,533 breast cancer cases and 60,976 controls of European ancestry, a variant within the MHC region (rs9257408, chr6: 28,926,220) was found to be significantly associated with the risk of breast malignancy [5]. Pathway analyses of a larger, follow-up GWAS study comprising over 200,000 individuals revealed that immune-related processes may underlie some of the observed associations with breast cancer susceptibility [6].
Traditional GWAS analyses do not go beyond the interrogation of individual single-nucleotide polymorphisms (SNPs). The MHC region is unique in that beyond SNPs, there is an additional layer of functional polymorphisms in the form of HLA genotypes [7]. Transforming SNP-based associations into HLA-allele level associations may thus yield more information and biological meaning behind the associations [8].
Probe-based genotyping commonly used in GWAS is suboptimal for querying variation in the MHC region due to its extensive polymorphic nature [9]. However, with the recent development of new computational tools and large reference panels, HLA genotypes can be inferred from SNPs that are in close proximity to classical HLA loci. Compared to serological typing and direct sequencing approaches, imputation is a less accurate method of obtaining HLA genotype calls. However, determining HLA genotypes in silico using readily available SNP datasets has the benefits of being low-cost and efficient in both time and labor. We aim to investigate the relevance of the MHC region in breast cancer susceptibility, using imputed HLA alleles, in 25,484 women of Asian ancestry.
Methods
Study population
We studied 25,484 women from 12 case–control studies participating in the Breast Cancer Association Consortium (BCAC). These women were genetically Asian with known breast cancer status. Our study includes a total of 12,901 breast cancer patients and 12,583 controls (Supplementary Table 1 and Supplementary Fig. 1). This study was approved by the ethics board of the Agency for Science, Technology and Research (ASTAR IRB Ref: 2020-154). Informed consent was obtained by the individual studies which contributed to BCAC.
Single nucleotide variants (SNPs)
DNA was genotyped using OncoArray (Illumina Infinium array) [6, 10]. A data request application was made to BCAC to obtain all variants within the 15–55 Mb region (build 37) on chromosome 6 (concept 686); of which 15,412 variants were received. A total of 10,886 variants remained after excluding 181 variants with missing genotype data and 4345 variants with minor allele frequency (MAF) below 0.01. The total genotyping rate was 0.999.
Imputation of human leukocyte antigen (HLA) alleles
We imputed 273 HLA alleles (in dosage format; 94 classical 2-digit alleles and 179 classical 4-digit alleles.) using the SNP2HLA package (v1.0.3) with using a pre-built reference panel (available as part of the software) based on the data of pan-Asian subjects (including Han Chinese, Southeast Asian Malay, Tamil Indian ancestries, and Japanese) [11, 12]. Imputation was done in batches of 100 individuals. The info score for each individual was calculated from all individuals, based on the ratio of empirical and expected variance in dosage. The frequency of HLA alleles present ranges from 0.0005 to 0.489 (Supplementary Table 2). HLA alleles (n = 175) with info scores greater than 0.8 and frequency greater than 0.01 were selected for association analysis [9].
Breast cancer risk factors
Information on breast cancer risk factors was collected at enrollment for nested case–control studies, or post breast cancer diagnosis for cases in case–control studies. Risk factors used in our study: number of first-degree family members with breast cancer (none, 1, ≥ 2), body mass index (BMI), age at menarche (years), parity, menopausal status (post-menopausal, pre-/peri-menopausal), oral contraceptive use (≥ 4 months, < 4 months), and hormone replacement therapy use (HRT; ≥ 3 months, < 3 months). Women were considered post-menopausal if their date of last menstruation was > 12 months prior to the date of breast cancer diagnosis for cases or enrollment for controls. Ethnicity was self-reported with the exception of Korean and Thai, where they were coded based on the study’s country.
Statistical analysis
Pooled analysis was performed as the mean and standard deviation of the HLA alleles were similar across studies (Supplementary Figs. 2 and 3). To study the associations of SNPs and HLA alleles with breast cancer status, logistic regression was used with adjustment for age (at recruitment for controls, and at diagnosis for cases) and the first 15 principal components (PC) (i.e. population structure). The PCs were based on 22 chromosomes. Further adjustment for breast cancer risk factors (study, family history, BMI, parity, age at menarche, menopausal status, contraceptive use, and hormone replacement therapy use) was done for the study of HLA alleles with breast cancer status. To compare our results with published literature, we did a systematic review of studies on HLA and breast cancer risk in Asian patients (Supplementary Methods).
Breast cancer tumor characteristics were obtained from medical records. The association analysis, using logistic regression, was repeated with all controls and subgroups of cases, (1) estrogen receptor (ER) positive, (2) ER-negative, (3) human epidermal growth factor receptor 2 (HER2) positive, (4) HER2-negative, (5) early-stage (stages 0 or I), and (6) late-stage (stage III or IV).
Results
Ethnicity distribution in our study
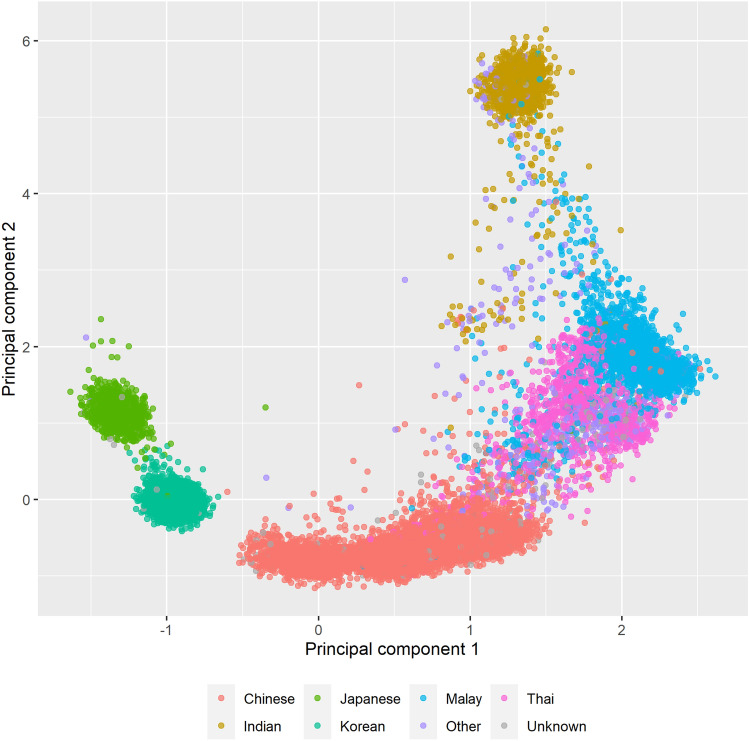
A PC analysis (PCA) plot of genotyping data shows the distinction between the six major ethnicities present in our study (Fig. 1). Over half of all women were Chinese (n = 14,048), and were from seven of the twelve studies (Table 1, Supplementary Table 3). The same PCA plot, but colored by study, showed that the spread seen in the Chinese is due to the country of recruitment (Supplementary Fig. 1). Indians were from multi-ethnic Singapore and Malaysia (Supplementary Table 3).
Fig. 1.
Principal components plot plotting principal component 1 and 2 colored by ethnicity
Table 1.
Characteristics of Asian breast cancer cases and controls from 12 studies (n = 25,484)
| Controls | Cases | |
|---|---|---|
| n = 12,583 | n = 12,901 | |
| Age, years (at recruitment for controls, at diagnosis for cases) | 51 (44–58) | 52 (45–60) |
| Unknown | 234 | 34 |
| Ethnicity | ||
| Chinese | 6,809 (54) | 7239 (56) |
| Indian | 1019 (8) | 577 (4) |
| Japanese | 657 (5) | 661 (5) |
| Koreana | 1772 (14) | 2394 (19) |
| Malay | 1335 (11) | 1061 (8) |
| Thaia | 642 (5) | 448 (3) |
| Other | 161 (1) | 243 (2) |
| Unknown | 188 (1) | 278 (2) |
| Family history of breast cancer | ||
| None | 6753 (54) | 8885 (69) |
| 1 | 607 (5) | 1,265 (10) |
| ≥ 2 | 89 (1) | 192 (1) |
| Unknown | 5134 (41) | 2559 (20) |
| Body mass index, kg/m2 | ||
| < 18.5 | 566 (4) | 584 (5) |
| 18.5–24.9 | 7226 (57) | 5458 (42) |
| 25.0–29.9 | 3292 (26) | 2189 (17) |
| ≥ 30 | 1140 (9) | 705 (5) |
| Unknown | 359 (3) | 3965 (31) |
| Age at menarche, years | ||
| ≤ 12 | 3708 (29) | 3567 (28) |
| 13–14 | 5024 (40) | 4320 (33) |
| > 14 | 3087 (25) | 2356 (18) |
| Unknown | 764 (6) | 2658 (21) |
| Parity | ||
| Nulliparous | 1823 (14) | 1940 (15) |
| 1 child | 1719 (14) | 1926 (15) |
| 2 children | 4276 (34) | 3434 (27) |
| ≥ 3 children | 4168 (33) | 3320 (26) |
| Unknown | 597 (5) | 2281 (18) |
| Menopausal status | ||
| Postmenopausal | 5718 (45) | 5603 (43) |
| Premenopausal or peri-menopausal | 6028 (48) | 3989 (31) |
| Unknown | 837 (7) | 3309 (26) |
| Oral contraceptive | ||
| Never | 4227 (34) | 4828 (37) |
| Ever | 1125 (9) | 1841 (14) |
| Unknown | 7231 (57) | 6232 (48) |
| Hormone replacement therapy | ||
| Never | 5088 (40) | 8199 (64) |
| Ever | 682 (5) | 749 (6) |
| Unknown | 6813 (54) | 3953 (31) |
aWe re-coded Korean and Thai from other ethnicities based on the country (Korea and Thailand, respectively) the study was from
Characteristics of cases and controls
The median age at breast cancer diagnosis for cases (n = 12,901) was 52 years (interquartile range [IQR]: 45–60) and the median age at enrollment for controls (n = 12,583) was 51 years (IQR: 44–58) (Table 1). The majority of all women were of normal BMI (18.5–24.9 kg/m2, 42% in cases, and 57% in controls). The majority of cases (68%) and controls (81%) had at least one child. Forty-three percent of cases were post-menopausal at the time of diagnosis, a similar proportion of post-menopausal controls (45%) were recruited. Older age at menarche was observed in some studies (Asia Cancer Program [ACP], Korean Hereditary Breast Cancer Study [KOHBRA], Shanghai Breast Cancer Genetic Study [SBCGS], Seoul Breast Cancer Study [SEBCS], and Taiwanese Breast Cancer Study [TWBCS]) (Supplementary Table 4). The majority of breast cancer cases were ER-positive (n = 8558, 66%), HER2-negative (n = 3777, 45%), and stage II (n = 4597, 36%) (Supplementary Table 1).
Associations between SNPs and breast cancer risk
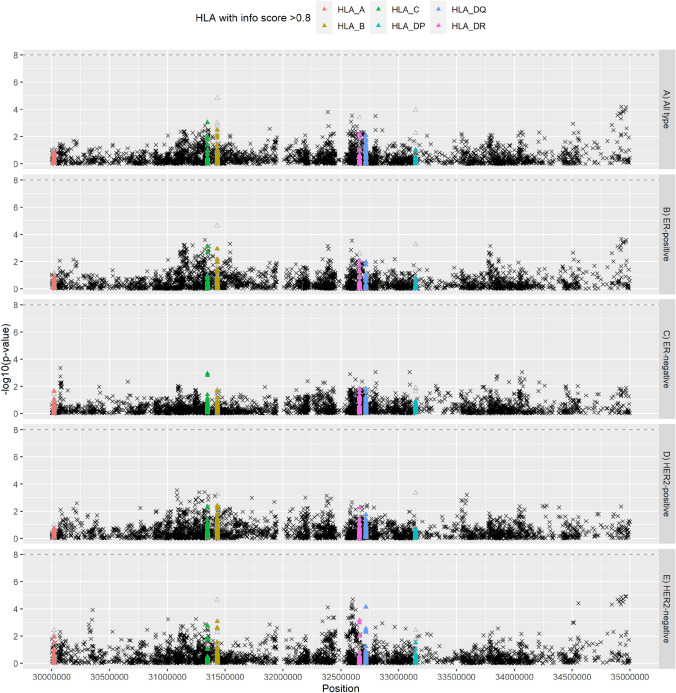
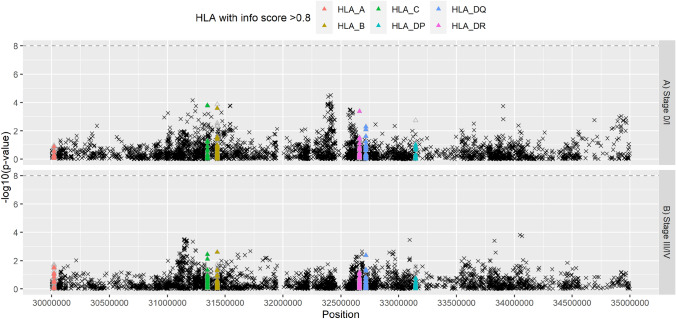
None of the associations between the 10,886 SNPs chromosome 6 (15–55 Mb) and breast cancer risk reached genome-wide significance (P < 5e−8) (Fig. 2A). The smallest p-value observed was for SNP rs12663096 (odds ratio [OR] = 0.92, P = 5.01e−05). Effect sizes (odds ratios, OR) observed ranged from 0.77 to 1.25, where 95% of the effect sizes observed were between 0.92 and 1.07. Similar non-significant results were observed when subgroups of breast cancer were studied, with the smallest p-value observed for rs9784889 and HER2-negative breast cancer (OR = 0.889, P = 1.19e−5) (Figs. 2B–E and Fig. 3).
Fig. 2.
Manhattan plot of the associations between imputed human leukocyte antigen alleles (HLA, n = 273; triangles; as predicted by SNP2HLA), single nucleotide polymorphisms (SNP) on chromosome 6 (30–35 Mb, build 37) and the risk of developing breast cancer as compared to controls (n = 12,583)—a all types (n = 12,901), b estrogen receptor (ER)-positive (n = 8558), c ER-negative (n = 3722), d HER2-positive (n = 3722), and e HER2-negative (n = 5848). Filled triangles denote associations with HLA (n = 175) with high (≥ 0.8) info score and frequency > 0.01; unfilled triangles denote HLA class. Logistic regression models were used and adjusted for the first 15 principal components for population stratification and age (at recruitment for controls; at diagnosis for cases). Info score: info score from HLA prediction by SNP2HLA
Fig. 3.
Manhattan plot of the associations between imputed human leukocyte antigen alleles (HLA, n = 273; triangles; as predicted by SNP2HLA), single nucleotide polymorphisms (SNP) on chromosome 6 (30–35 Mb, build 37) and the risk of developing breast cancer as compared to controls (n = 12,583)—a stage 0 or I (n = 3896), and b stage III or IV (n = 1972). Filled triangles denote associations with HLA (n = 175) with high (≥ 0.8) info score and frequency > 0.01; unfilled triangles denote HLA class. Logistic regression models were used and adjusted for the first 15 principle components for population stratification and age (at recruitment for controls; at diagnosis for cases). Info score: info score from HLA prediction by SNP2HLA
Associations between HLA alleles and breast cancer risk
Of the 273 imputed HLA alleles, 175 had info scores greater than 0.8 and frequencies greater than 0.01; seventy-one were classic 2-digits HLA alleles and 104 were classic 4-digits HLA alleles (Supplementary Table 5). Heatmap of the 71 classic 2-digits HLA alleles by study participants did not show obvious clustering of cases and controls (Supplementary Fig. 4).
Of the 175 HLA alleles, the association between HLA-C*12:03 and breast cancer risk attained the smallest p-value (OR = 1.29, P = 1.08e-3) (Fig. 2A). Effect sizes (OR) observed ranged from 0.87 to 1.29, where 95% of the effect sizes (OR) observed were between 0.90 and 1.23 (Supplementary Table 5). No appreciable change was observed after adjustments for study and breast cancer risk factors (age, body mass index number of first-degree family members with breast cancer, number of children, age at menarche, menopausal status, oral contraceptive use, and hormone replacement therapy use) (Supplementary Fig. 5A and Supplementary Table 56).
Three of 14 articles identified in the literature reported associations between classic HLA class I or class II alleles and breast cancer risk (Supplementary Methods). All studies used a case–control design and HLA ascertained from blood samples. Only Leong et al. reported significant associations between HLA-A*31 and breast cancer risk (Fisher’s exact test P = 0.020) (Table 2). Two other HLA alleles (HLA-A*26 and HLA-A*36) were reported to be correlated with metastasis (Table 2). We did not observe the same associations in our study; the associations between HLA-A*31and breast cancer risk (OR = 1.05, P = 0.324) and between HLA-A*26 and late-stage breast cancer (OR = 1.11, P = 0.260) were not significant. HLA-A*36 alleles were not imputed.
Table 2.
Findings from the three articles that were identified from our systematic review (Supplementary Methods: a systematic review of studies on HLA in Asian breast cancer patients)
| First author (year of publication) | Country | Sample size | HLA measurement | Results from literature | Our resultsa |
|---|---|---|---|---|---|
| Leong PP (2011) | Malaysia | 59 cases; 77 controls | HLA-A Sequence-Specific primers PCR | HLA-A*31 (cases vs controls; Fisher's exact test, P = 0.020) | HLA-A*31 and breast cancer risk (Odds ratio [OR] = 1.05, P = 0.324) |
| HLA-A*26 (Metastasis; Spearman's rank test, r = − 0.430, P = 0.001) | HLA-A*26 and breast cancer risk (OR = 0.96, P = 0.454) | ||||
| HLA-A*36 (Metastasis; Spearman's rank test, r = − 0.430, P = 0.001) | HLA-A*26 and late-stage breast cancer risk (OR = 1.11, P = 0.260) | ||||
| HLA-A*36 was not imputed | |||||
| Yang XX (2011) | China | 216 cases; 216 controls | 16 variants in HLA class II region were genotyped | HLA class II variants studied were not associated with breast cancer risk | Associations for class II variants did not reach genome-wide significance (P < 5e−8) |
| Chen PC (2007) | Taiwan | 101 cases; 115 controls | Genotyping was performed by Sequence-Specific primers PCR | No significant differences in phenotype frequencies of HLA-DQA1 and -DQB1 between patients with breast cancer and matched control subjects | Associations did not reach genome-wide significance (P < 5e−8), for HLA-DQA1 (imputed six 2-digits HLA alleles) and HLA-DQB1 (imputed five 2-digits HLA alleles) |
The studies applied a case–control design and used blood samples
aResults on the association of HLA alleles and breast cancer risk, using logistic regression, adjusted for age and the first 15 principal components (Supplementary Table 4)
Subgroup analysis on the associations between HLA alleles and breast cancer risk
Similar results were observed when different subtypes of breast cancer were studied (ER-positive, ER-negative, HER2-positive, HER2-negative, early-stage, and late-stage) (Figs. 2B–E and 3; Supplementary Figs. 5B-E and 6; and Supplementary Table 5). Effect sizes (OR, adjusted for age and the first 15 PCs) observed ranged from 0.75 to 1.47, where 95% of the effect sizes observed were between 0.85 and 1.18 (Supplementary Table 5). The association between HLA-B*35:03 and early-stage breast cancer showed the largest effect size after additional adjustments for study and breast cancer risk factors (OR = 0.60, P = 0.040) (Supplementary Table 6).
Discussion
It was suggested that the MHC region is associated with breast cancer susceptibility studies of European populations [5, 13]. In contrast to the European genome-wide association study, we observed no significant association (P < 5e−8) between variants in the MHC region (imputed 175 classic class I and II HLA alleles) and breast cancer risk in our Asian study of 12,901 breast cancer cases and 12,583 controls (smallest P = 1.08e−3, HLA-C*12:03) [5].
Gourley et al. [14] reported that while studies showed no consistent association between HLA class I type (HLA-A, HLA-B and HLA-C) and breast cancer risk, certain HLA class I types may be more common in specific disease subclasses. Zhao et al. [15] reported down-regulation of HLA class I genes (which comprise HLA-A, HLA-B, or HLA-C) on CD4(+) and on CD8(+) T lymphocytes in breast cancer patients when as compared with healthy controls. However, we did not observe any significant association between any of the inferred HLA class I alleles and breast cancer risk, nor with any of the breast cancer subtypes in our Asian study population. Compared to other cancers, MHC class I mutations are less associated with the expression of killer lymphocyte effector genes in breast cancer [13]. Hence, possible differences in expression may be missed when studying HLA alleles (not expression).
Our systematic review identified one Asian study examining the relationship between HLA class I alleles and breast cancer risk [16]. Leong et al. [17] identified HLA-A*31 (6.8% in cases and 0% in controls; Fisher's exact test, P = 0.020) as a risk allele for breast cancer in 59 invasive ductal carcinoma breast cancer patients and 77 controls without breast cancer in Malaysia. In a subset of Malaysian women in our study, HLA-A*31 was not associated with breast cancer risk (adjusted OR = 0.77, P = 0.166; data not shown). It should be noted that the Malaysia study determined HLA-A expression using the Biotest HLA-A Sequence-Specific primers (SSP), while only imputed HLA alleles were interrogated in our study [17].
While it is conceivable that HLA genes play critical roles in immune surveillance against breast cancers, there are many other studies that show no consistent link between HLA and breast cancer [18]. Results from our study are in agreement with the two Asian studies on HLA class II alleles in the systematic review which reported the lack of associations between HLA alleles and breast cancer risk [16, 19]. Contrary to this, alleles from MHC class II sub-region were found to be associated with breast cancer risk in studies on populations of European ancestry (HLA-DQB1*03 [20], HLA-DRB1*10:01 [21] HLA-DRB1*11:01 [21], and HLA-DRB1*13 [20]) and Middle-Eastern populations (HLA-DQB1*02 [14, 22, 23], HLA-DRB1*03 [22], HLA-DQB1*06 [22], HLA-DRB1*07 [14], HLA-DRB1*12 [24], HLA-DRB1*13 [22], and HLA-DRB1*18:01 [25]) populations. It should be noted that previously published results have small sample sizes (largest sample size of 216 cases and 216 controls), which makes validation of the results challenging [16, 17, 19]. It is noteworthy that HLA alleles are exceedingly diverse, and may be confined to certain ethnic groups and geographical locations [26]. Indeed, we observed that by accounting for study populations, significant associations were attenuated and no longer significant, suggesting that correction for population stratification is essential in the testing of the associations.
This is the largest study to date to examine the associations between the MHC region and breast cancer risk in Asians (12,901 breast cancer cases and 12,583 controls). However, there are several limitations worth noting. Validation of the imputed HLA loci was not performed by the direct typing of HLA loci. An average error rate of between three to six percent has been reported for the prediction of high-resolution (four-digit) HLA alleles using imputation software [27]. Due to weaker linkage disequilibrium between rare variants, the imputation of rare HLA alleles is challenging. Variants with MAF > 1% in were filtered out prior to HLA imputation, and only imputed HLA alleles with MAF > 1% were retained in our analyses, which resulted in the exclusion of many informative alleles. In addition, non-classical HLA alleles were not studied, as the imputation software used imputes only classical HLA alleles (class I and class II). The accuracy of HLA imputation is dependent on the reference panel used [12]. The ethnic representativeness of the reference panel used for HLA loci imputation may limit the generalizability of our findings. While the chosen Pan-Asian reference panel is large, it may not represent the ethnic groups of our study population fully [27]. As there were no genome-wide significant results, we did not perform orthogonal experiments to verify the findings.
In conclusion, imputed class I and II HLA alleles were not associated with breast cancer risk in our Asian sample. Direct measurement of HLA gene expressions may be required to further explore the associations between HLA genes and breast cancer risk.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. The ACP study wishes to thank the participants in the Thai Breast Cancer study. Special thanks also go to the Thai Ministry of Public Health (MOPH), doctors and nurses who helped with the data collection process. Finally, the study would like to thank Dr Prat Boonyawongviroj, the former Permanent Secretary of MOPH and Dr Pornthep Siriwanarungsan, the former Department Director-General of Disease Control who have supported the study throughout. CBCS thanks study participants, co-investigators, collaborators and staff of the Canadian Breast Cancer Study, and project coordinators Agnes Lai and Celine Morissette. HKBCS thanks Hong Kong Sanatorium and Hospital, Dr Ellen Li Charitable Foundation, The Kerry Group Kuok Foundation, National Institute of Health 1R03CA130065 and the North California Cancer Center for support. We thank all investigators of the KOHBRA (Korean Hereditary Breast Cancer) Study. MYBRCA thanks study participants and research staff (particularly Patsy Ng, Nurhidayu Hassan, Yoon Sook-Yee, Daphne Lee, Lee Sheau Yee, Phuah Sze Yee and Norhashimah Hassan) for their contributions and commitment to this study. SBCGS thanks study participants and research staff for their contributions and commitment to the studies. SGBCC thanks the participants and all research coordinators for their excellent help with recruitment, data and sample collection.
Author contributions
Study design: JL, PJH; writing group: JL, PJH, MH; data management: MKB, AMD, DFE, PDPP, AJK; statistical analysis: PJH, JL; provided DNA samples and phenotypic data: MH, BKT, YET, ST, VKMT, GHL, KJA, JC, JD, WH, MH, HI, MI, EMJ, DK, SK, AWK, AK, AL,KM, NAMT, KM, RAM, SKP, CS, SHT, QW, TY, WZ. All authors read and approved the final version of the paper.
Funding
This study was supported by the National Research Foundation Singapore (NRF-NRFF2017-02, awarded to J Li) and the Agency for Science, Technology and Research (A*STAR, awarded to J Li). BCAC is funded by the European Union's Horizon 2020 Research and Innovation Programme (Grant nos. 634935 and 633784 for BRIDGES and B-CAST, respectively), and the PERSPECTIVE I&I project, funded by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministère de l’Économie et de l'Innovation du Québec through Genome Québec, the Quebec Breast Cancer Foundation. The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Additional funding for BCAC is provided via the Confluence project which is funded with intramural funds from the National Cancer Institute Intramural Research Program, National Institutes of Health. Genotyping of the OncoArray was funded by the NIH Grant U19 CA148065, and Cancer Research UK Grant C1287/A16563 and the PERSPECTIVE project supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (grant GPH-129344) and, the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec and the PSRSIIRI-701 grant, and the Quebec Breast Cancer Foundation. Funding for iCOGS came from: the European Community's Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, and Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The ACP study is funded by the Breast Cancer Research Trust, UK. KM and AL are supported by the NIHR Manchester Biomedical Research Centre, the Allan Turing Institute under the EPSRC grant EP/N510129/1. CBCS is funded by the Canadian Cancer Society (grant # 313404) and the Canadian Institutes of Health Research. The HERPACC was supported by MEXT Kakenhi (No. 170150181 and 26253041) from the Ministry of Education, Science, Sports, Culture and Technology of Japan, by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from Ministry Health, Labour and Welfare of Japan, by Health and Labour Sciences Research Grants for Research on Applying Health Technology from Ministry Health, Labour and Welfare of Japan, by National Cancer Center Research and Development Fund, and "Practical Research for Innovative Cancer Control (15ck0106177h0001 and 20ck0106553)" from Japan Agency for Medical Research and development, AMED, and Cancer Bio Bank Aichi. The KOHBRA study was partially supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (HI16C1127; 1020350; 1420190). MYBRCA is funded by research grants from the Wellcome Trust (v203477/Z/16/Z), the Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Malaysia. MYMAMMO is supported by research grants from Yayasan Sime Darby LPGA Tournament and Malaysian Ministry of Higher Education (RP046B-15HTM). The Northern California Breast Cancer Family Registry (NC-BCFR) was supported by grant U01CA164920 from the USA National Cancer Institute of the National Institutes of Health. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The NGOBCS was supported by the National Cancer Center Research and Development Fund (Japan). The SBCGS was supported primarily by NIH grants R01CA64277, R01CA148667, UMCA182910, and R37CA70867. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The scientific development and funding of this project were, in part, supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network U19 CA148065. SEBCS was supported by the BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0000347). SGBCC is funded by the National Research Foundation Singapore, NUS start-up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant, Breast Cancer Prevention Programme, Asian Breast Cancer Research Fund and the NMRC Clinician Scientist Award (SI Category). Population-based controls were from the Multi-Ethnic Cohort (MEC) funded by grants from the Ministry of Health, Singapore, National University of Singapore and National University Health System, Singapore. The TWBCS is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan.
Availability of data and materials
The genetic and clinical data used in this study were obtained via a data request (concept #686) to the Breast Cancer Association Consortium (BCAC). All data requests can be directed to the BCAC data access committee (http://bcac.ccge.medschl.cam.ac.uk/bcacdata/).
Declarations
Conflict of interests
The authors declare no potential conflicts of interest.
Ethics approval
All studies were performed in accordance with the Declaration of Helsinki and all participants provided written informed consent. This study was approved by the ethics board of the Agency for Science, Technology and Research (ASTAR IRB Ref: 2020-154).
Consent to participate
Informed consent was obtained by the individual studies which contributed to BCAC.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mikael Hartman and Jingmei Li equally contributed.
Contributor Information
Peh Joo Ho, Email: ho_peh_joo@gis.a-star.edu.sg.
Alexis Jiaying Khng, Email: khngja@gis.a-star.edu.sg.
Benita Kiat-Tee Tan, Email: benita.tan.k.t@singhealth.com.sg.
Ern Yu Tan, Email: ern_yu_tan@ttsh.com.sg.
Su-Ming Tan, Email: tan.su.ming@singhealth.com.sg.
Veronique Kiak Mien Tan, Email: veronique.tan.k.m@singhealth.com.sg.
Geok Hoon Lim, Email: lim.gh@singhealth.com.sg.
Kristan J. Aronson, Email: aronson@queensu.ca
Tsun L. Chan, Email: tlchan@hksh.com
Ji-Yeob Choi, Email: jiyeob.choi@gmail.com.
Joe Dennis, Email: jgd29@cam.ac.uk.
Weang-Kee Ho, Email: weangkee.ho@nottingham.edu.my.
Ming-Feng Hou, Email: mifeho@kmu.edu.tw.
Hidemi Ito, Email: hidemi@aichi-cc.jp.
Motoki Iwasaki, Email: moiwasak@ncc.go.jp.
Esther M. John, Email: emjohn@stanford.edu
Daehee Kang, Email: dhkang@snu.ac.kr.
Sung-Won Kim, Email: brcakorea@gmail.com.
Allison W. Kurian, Email: akurian@stanford.edu
Ava Kwong, Email: akwong@asiabreastregistry.com.
Artitaya Lophatananon, Email: artitaya.lophatananon@manchester.ac.uk.
Keitaro Matsuo, Email: kmatsuo@aichi-cc.jp.
Nur Aishah Mohd-Taib, Email: nuraish@gmail.com.
Kenneth Muir, Email: kenneth.muir@manchester.ac.uk.
Rachel A. Murphy, Email: Rachel.murphy@ubc.ca
Sue K. Park, Email: suepark@snu.ac.kr
Chen-Yang Shen, Email: bmcys@ibms.sinica.edu.tw.
Xiao-Ou Shu, Email: xiao-ou.shu@vanderbilt.edu.
Soo Hwang Teo, Email: soohwang.teo@cancerresearch.my.
Qin Wang, Email: qw232@medschl.cam.ac.uk.
Taiki Yamaji, Email: tyamaji@ncc.go.jp.
Wei Zheng, Email: wei.zheng@vumc.org.
Manjeet K. Bolla, Email: mkh39@medschl.cam.ac.uk
Alison M. Dunning, Email: amd24@medschl.cam.ac.uk
Douglas F. Easton, Email: dfe20@medschl.cam.ac.uk
Paul D. P. Pharoah, Email: pp10001@medschl.cam.ac.uk
Mikael Hartman, Email: ephbamh@nus.edu.sg.
Jingmei Li, Email: lijm1@gis.a-star.edu.sg.
References
- 1.Ribatti D. The concept of immune surveillance against tumors. The first theories. Oncotarget. 2017;8(4):7175–7180. doi: 10.18632/oncotarget.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Jong MM, et al. The HLA class III subregion is responsible for an increased breast cancer risk. Hum Mol Genet. 2003;12(18):2311–2319. doi: 10.1093/hmg/ddg245. [DOI] [PubMed] [Google Scholar]
- 3.Ouni N, et al. The Impact of HLA-G 3'UTR Polymorphisms in Breast Cancer in a Tunisian Population. Immunol Invest. 2019;48(5):521–532. doi: 10.1080/08820139.2019.1569043. [DOI] [PubMed] [Google Scholar]
- 4.Jobim MR, et al. Analysis of KIR gene frequencies and HLA class I genotypes in breast cancer and control group. Hum Immunol. 2013;74(9):1130–1133. doi: 10.1016/j.humimm.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Michailidou K, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy AE, Ozbek U, Dorak MT. What has GWAS done for HLA and disease associations? Int J Immunogenet. 2017;44(5):195–211. doi: 10.1111/iji.12332. [DOI] [PubMed] [Google Scholar]
- 8.Meyer D, Nunes K. HLA imputation, what is it good for? Hum Immunol. 2017;78(3):239–241. doi: 10.1016/j.humimm.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8(6):e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrubsole MJ, et al. MTR and MTRR polymorphisms, dietary intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15(3):586–588. doi: 10.1158/1055-9965.EPI-05-0576. [DOI] [PubMed] [Google Scholar]
- 11.Okada Y, et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum Mol Genet. 2014;23(25):6916–6926. doi: 10.1093/hmg/ddu387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillai NE, et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet. 2014;23(16):4443–4451. doi: 10.1093/hmg/ddu149. [DOI] [PubMed] [Google Scholar]
- 13.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33(11):1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavado R, et al. The HLA-B7 allele confers susceptibility to breast cancer in Spanish women. Immunol Lett. 2005;101(2):223–225. doi: 10.1016/j.imlet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhao S, et al. The amount of surface HLA-I on T lymphocytes decreases in breast infiltrating ductal carcinoma patients. J Int Med Res. 2011;39(2):508–513. doi: 10.1177/147323001103900218. [DOI] [PubMed] [Google Scholar]
- 16.Chen PC, et al. HLA-DQA1 and -DQB1 allele typing in southern Taiwanese women with breast cancer. Clin Chem Lab Med. 2007;45(5):611–614. doi: 10.1515/CCLM.2007.132. [DOI] [PubMed] [Google Scholar]
- 17.Leong PP, et al. HLA-A and breast cancer in West Peninsular Malaysia. Med Oncol. 2011;28(1):51–56. doi: 10.1007/s12032-009-9414-6. [DOI] [PubMed] [Google Scholar]
- 18.Gourley C. HLA in breast cancer: its relevance for immunotherapy. The Breast. 1998;7(6):308–313. doi: 10.1016/S0960-9776(98)90072-8. [DOI] [Google Scholar]
- 19.Yang XX, et al. HLA class II variants in Chinese breast cancer patients. Asian Pac J Cancer Prev. 2011;12(11):3075–3079. [PubMed] [Google Scholar]
- 20.Chaudhuri S, et al. Genetic susceptibility to breast cancer: HLA DQB*03032 and HLA DRB1*11 may represent protective alleles. Proc Natl Acad Sci U S A. 2000;97(21):11451–11454. doi: 10.1073/pnas.97.21.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aureli A, et al. Breast cancer is associated with increased HLA-DRB1*11:01 and HLA-DRB1*10:01 allele frequency in a population of patients from Central Italy. Immunol Invest. 2020;49(5):489–497. doi: 10.1080/08820139.2020.1737539. [DOI] [PubMed] [Google Scholar]
- 22.Gun FD, et al. HLA class-II allele frequencies in Turkish breast cancer patients. Med Oncol. 2012;29(2):466–471. doi: 10.1007/s12032-011-9873-4. [DOI] [PubMed] [Google Scholar]
- 23.Atoum MF, Tanashat RQ, Mahmoud SA. Negative association of the HLA-DQB1*02 allele with breast cancer development among Jordanians. Asian Pac J Cancer Prev. 2013;14(11):7007–7010. doi: 10.7314/APJCP.2013.14.11.7007. [DOI] [PubMed] [Google Scholar]
- 24.Ghaderi A, et al. HLA-DBR 1 alleles and the susceptibility of Iranian patients with breast cancer. Pathol Oncol Res. 2001;7(1):39–41. doi: 10.1007/BF03032603. [DOI] [PubMed] [Google Scholar]
- 25.Bayraktar B, et al. Distribution of HLA antigens in breast cancer. Bratisl Lek Listy. 2012;113(6):372–375. doi: 10.4149/bll_2012_084. [DOI] [PubMed] [Google Scholar]
- 26.Cantu de Leon D et al. High resolution human leukocyte antigen (HLA) class I and class II allele typing in Mexican mestizo women with sporadic breast cancer: case-control study. BMC Cancer 2009;9:48. [DOI] [PMC free article] [PubMed]
- 27.Brandt DY et al. Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 genomes project phase I data. G3 (Bethesda) 2015;5(5):931–41. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic and clinical data used in this study were obtained via a data request (concept #686) to the Breast Cancer Association Consortium (BCAC). All data requests can be directed to the BCAC data access committee (http://bcac.ccge.medschl.cam.ac.uk/bcacdata/).