Abstract
Mutants of Escherichia coli that lack cytoplasmic superoxide dismutase (SOD) exhibit auxotrophies for sulfur-containing, branched-chain, and aromatic amino acids and cannot catabolize nonfermentable carbon sources. A secondary-site mutation substantially relieved all of these growth defects. The requirement for fermentable carbon and the branched-chain auxotrophy occur because superoxide (O2−) leaches iron from the [4Fe-4S] clusters of a family of dehydratases, thereby inactivating them; the suppression of these phenotypes was mediated by the restoration of activity to these dehydratases, evidently without changing the intracellular concentration of O2−. Cloning, complementation, and sequence analysis identified the suppressor mutation to be in dapD, which encodes tetrahydrodipicolinate succinylase, an enzyme involved in diaminopimelate and lysine biosynthesis. A block in dapB, which encodes dihydrodipicolinate reductase in the same pathway, conferred similar protection. Genetic analysis indicated that the protection stems from the intracellular accumulation of tetrahydro- or dihydrodipicolinate. Heterologous expression in the SOD mutants of the dipicolinate synthase of Bacillus subtilis generated dipicolinate and similarly protected them. Dipicolinates are excellent iron chelators, and their accumulation in the cell triggered derepression of the Fur regulon and a large increase in the intracellular pool of free iron, presumably as a dipicolinate chelate. A fur mutation only partially relieved the auxotrophies, indicating that Fur derepression assists but is not sufficient for suppression. It seems plausible that the abundant internal iron permits efficient reactivation of superoxide-damaged iron-sulfur clusters. This result provides circumstantial evidence that the sulfur and aromatic auxotrophies of SOD mutants are also directly or indirectly linked to iron metabolism.
The discovery of superoxide dismutase (SOD) by McCord and Fridovich introduced biology to the field of free radical chemistry (37). The presence of this enzyme in virtually all aerobic organisms (38) forced the conclusion that superoxide (O2−) must be formed as a by-product of aerobic metabolism and, if not scavenged, must damage critical biomolecules. Left unknown were the identities of those target biomolecules. This point was controversial, since chemical studies indicated that amino acids, nucleotides, and sugars are essentially not reactive with O2− (6, 7, 15, 43).
In 1986 Carlioz and Touati reported the construction of mutants of Escherichia coli that lacked both the manganese- and iron-containing isozymes of SOD (10). These mutants were capable of aerobic growth only if branched-chain, aromatic, and sulfurous amino acids were provided in the medium, and if a fermentable carbon source was available. They also displayed a high rate of spontaneous mutagenesis (14). Similar phenotypes were reported for wild-type cells during exposure to hyperbaric oxygen, presumably because of the accelerated pace of O2− formation (8). The requirement for branched-chain amino acids was traced to the inactivation by O2− of dihydroxyacid dehydratase, an enzyme in the common biosynthetic pathway (31). This enzyme utilizes a [4Fe-4S]2+ cluster as a Lewis acid to catalyze substrate dehydration. The solvent exposure of the cluster allows it to be accessible to O2−, which can univalently oxidize it. The resultant [4Fe-4S]3+ cluster is unstable and disintegrates to the [3Fe-4S]+ form, which is catalytically inactive, with loss of a ferrous iron atom to the cytosol (16, 17). Several reports following this discovery showed that other dehydratases in the cell, including the tricarboxylic acid (TCA) cycle enzymes aconitase and fumarase, contain [4Fe-4S]2+ clusters that suffer the same damage when exposed to superoxide (20, 21, 34). E. coli can repair these enzymes by a process that is presently undefined but which must entail the reduction of the cluster and replacement of the lost iron atom. The amounts of SOD and of repair enzymes in E. coli appear to be calibrated to precisely balance the rate of dehydratase damage by endogenous O2−, so that the dehydratases retain near-maximal activity during aerobic growth (22).
Interestingly, the hypermutagenesis of SOD mutants arises from the iron that is released by the damaged clusters. This iron accumulates in the cytosol, where it catalyzes the oxidation of DNA (28, 30, 35). Thus, several phenotypes of SOD mutants have been traced to oxidation by O2− of iron-sulfur clusters.
However, the auxotrophies for sulfur-containing and aromatic amino acids and the slow growth in rich medium are not directly attributable to cluster damage. It appears that neither amino acid biosynthetic pathway contains labile dehydratases. The sulfur requirement has been linked to the apparent leakage of sulfur from SOD mutants (5), although the specific lesion that causes this phenomenon is unknown. The aromatic amino acid biosynthetic defect has recently been ascribed to the oxidation of the intermediate 1,2-dihydroxyethyl thiamine pyrophosphate of transketolase (3). In vitro, superoxide oxidized and released this compound with progressive inactivation of the enzyme, and catabolic processes that require transketolase function were shown to be defective in SOD mutants. A resultant deficiency in erythrose-4-phosphate production could plausibly diminish aromatic amino acid biosynthesis.
A pleiotropic suppressor which relieves all the auxotrophies of SOD mutants—those for sulfurous and aromatic as well as branched-chain amino acids—was isolated and characterized by Imlay and Fridovich (25, 26). It was originally hoped that this suppressor might protect multiple targets by curbing the rate of endogenous O2− production. However, the original analysis indicated that the mutation did not act to change the internal O2− concentration (26). The goal of the present study was to decipher the mechanism of suppression and thereby gain further understanding of the nature of O2− toxicity.
MATERIALS AND METHODS
Chemicals and enzymes.
o-Nitrophenyl-β-d-galactopyranoside, fluorocitrate, deferoxamine mesylate (desferrioxamine), isopropyl-β-d-thiogalactopyranoside, 6-phosphogluconate, NADP+, NADH, diethylenetriaminepentaacetic acid (DTPA), hydrogen peroxide, dipicolinate, diaminopimelic acid, lactate, succinate, glutathione, ATP, horse heart cytochrome c, xanthine, xanthine oxidase, porcine heart isocitrate dehydrogenase, bovine erythrocyte Cu,ZnSOD, and rabbit muscle lactic dehydrogenase were purchased from Sigma. Coomassie protein reagent was obtained from Pierce. Magnesium sulfate heptahydrate, ferrous sulfate heptahydrate, sodium nitrite, and manganese chloride were obtained from Aldrich. β-Mercaptoethanol and sodium citrate dihydrate were obtained from Fisher Scientific. Restriction enzymes were purchased from Gibco BRL. Ready-to-go PCR beads were obtained from Pharmacia Biotech. Shrimp alkaline phosphatase was obtained from Boehringer Mannheim. Water was purified from a Labconco Water Pro PS system by using house deionized water as the feedstock.
Growth media.
Luria-Bertani (LB) medium contained (per liter) 10 g of Bacto Tryptone, 5 g of yeast extract, 10 g of sodium chloride, and 2 g of glucose. Minimal medium consisted of minimal A or E salts medium (39) with 1 mM MgSO4 · 7H2O and with 5 mg of thiamine and 2 g of glucose per liter. Casamino Acids medium was additionally supplemented with 0.2% Casamino Acids. l-Amino acid supplements were used at a final concentration of 0.5 mM, and necessary vitamin supplements were used at 3 μg/ml. Spectinomycin, tetracycline, and ampicillin were used at 150, 12, and 100 μg/ml, respectively. Where indicated, diaminopimelic acid was added to a final concentration of 50 μg/ml.
Growth studies.
Aerobic cultures were routinely grown in flasks at 37°C in a shaking water bath. Anaerobic cultures were grown in a Coy chamber (Coy Laboratory Products, Inc.) under 85% N2–10% H2–5% CO2. Optical densities (OD) of cultures were measured at 600 nm. For studies of auxotrophies, cells were grown anaerobically in minimal A medium supplemented with all amino acids except sulfurous amino acids (cysteine and methionine) or aromatic amino acids (tyrosine, phenylalanine, and tryptophan). Cultures were grown for at least four generations to reach an OD at 600 nm (OD600) of 0.2 before they were diluted in an aerobic medium of the same composition to an OD600 of 0.01. The ability to grow on amino acid-supplemented minimal medium lacking the sulfurous amino acids was used in general as a diagnostic feature to identify the suppressor strains. When the cells were grown on minimal medium alone, they were routinely supplemented with arginine, histidine, leucine, proline, and threonine in order to meet the amino acid requirement of strain AB1157. Where specifically indicated, cells were grown to log phase in LB medium, centrifuged, and washed stringently with minimal salts before they were diluted to an OD600 of 0.01 in minimal medium supplemented with all but the sulfurous amino acids. To test aerobic growth on nonfermentable carbon sources, cells were pregrown in anaerobic minimal medium that contained 20 amino acids, 0.4% lactate or succinate, and 40 mM sodium nitrate.
Strain construction.
All strains were K-12 derivatives (Table 1). The dapD and dapB mutations were transduced (39) into sodA sodB strains with selection for the linked fhuA468::Tn10 and thr::Tn10, respectively, on LB plates containing tetracycline and diaminopimelic acid. Transductants were screened for diaminopimelate auxotrophy on LB plates. In general, in order to avoid suppressor mutations, transductions were performed under anaerobic conditions. Strain AS237 was rendered chloramphenicol sensitive by penicillin enrichment in the presence of chloramphenicol. The fur mutation was transduced by linkage to zbf-507::Tn10. Strain SM1106 was constructed by excising the Tn10 (36) from SM1093, thereby generating the tetracycline-sensitive derivative SM1104, and then transducing the fur null allele with the linked Tn10. To verify that the fur mutation was inherited with the Tn10, the Tn10 was transduced back into AN387, and transductants were screened for coinheritance of the kanamycin marker from the fur::Tn5 allele. tonB mutants were screened by their ability to confer resistance to infection by bacteriophage φ80.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| AB1157 | F−thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1 galk2 rpsL supE44 are-14 xyl-15 mtl-1 tsx-33 | 27 |
| JI132 | Same as AB1157 plus (sodA::MudPR13)25 (sodB::kan)1-Δ2 | 27 |
| JI200 | Same as JI132 plus ssa-1 | 25 |
| JI210 | Same as AT986 plus fhuA468::Tn10 pan-6 | 25 |
| AN387 | F−rpsL gal | Bob Gennis |
| AS237 | Same as AN387 plus (sodA::MudPR13)25 (sodB::kan)1-Δ2; chloramphenicol sensitive | Lab collection |
| AT986 | HfrPO45 of KL16 relA1 spoT1 thi-1 dapD8 | 9 |
| CAG18442 | As MG1655 plus thr-34::Tn10 | John Cronan |
| UM122 | rpoS::Tn10 | John Cronan |
| BN407 | As AB1157 plus ΔlacU169 and ColV-K30 (iucC::lacZ) | 2 |
| KP1032 | W3110 plus tonB::kan | 1 |
| β224 | ΔdapA::cat | Steve Farrand |
| β161 | dapB::kan | Paul Shaw |
| SM1004 | As AS237 plus dapD8 linked to fhuA468∷Tn10 | This study |
| SM1005 | As AS237 plus dapA::cat | This study |
| SM1006 | As SM1004 plus dapA::cat | This study |
| SM1008 | As AS237 plus dapB17::Mu thr-34∷Tn10 | This study |
| SM1009 | As SM1008 plus dapA::cat | This study |
| SM1024 | As AS237 plus vector pMTL21 | This study |
| SM1025 | As AS237 plus pSM912 | This study |
| SM1077 | As AN387 plus dapA::cat | This study |
| SM1093 | As AS237 plus dapB::kan linked to thr-34∷Tn10 | This study |
| SM1095 | As AN387 plus dapB::kan | This study |
| SM1104 | As SM1093; tetracycline sensitive | This study |
| SM1105 | As AS237 plus fur::Tn5 linked to zbf-507::Tn10 | This study |
| SM1109 | As SM1104 plus rpoS::Tn10 | This study |
| SM1116 | As SM1104 plus fur::Tn5 linked to zbf-507::Tn10 | This study |
| SM1117 | As SM1022 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1110 | As SM1095 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1111 | As AS237 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1112 | As SM1104 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1113 | As SM1103 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1114 | As SM1005 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1119 | As SM1025 plus ColV-K30 (iucC∷lacZ) | This study |
| SM1022 | As AN387 plus pMTL21 | This study |
| SM1103 | As SM1004 but tetracycline sensitive | This study |
| SM1120 | As AN387 plus tonB::kan | This study |
| SM1122 | As SM1120 plus (sodA::mudPR13)25 (sodB::kan)1-Δ2 zdg299∷Tn10 | This study |
| SM1123 | As SM1116 plus pSM915 | This study |
| SM1127 | As SM1095 plus pSM912 | This study |
| SM1128 | As SM1077 plus pSM912 | This study |
| KK210 | As AB1157 plus fur::Tn5 and zbf-507::Tn10 | This study |
| Plasmids | ||
| pSM900 | pCL1921 derivative carrying ORFs for tsf, rpsB, map, glnD, dapD, and yaeI | This study |
| pSM902 | pCL1921 derivative carrying ORFs for dapD and yaeI | This study |
| pSM903 | pCL1921 derivative carrying ORF for yaeI | This study |
| pSM904 | pCL1921 derivative carrying ORF for dapD | This study |
| pSM912 | pMTL21 derivative carrying ORF encoding dipicolinate synthase | This study |
| pSM915 | Wild-type dapB cloned in pCR 2.1 | This study |
| pMTL21 | pUC18 derivative | 10a |
| pDB2 | pACYC-based plasmid with a 20-kb insert from the 4-min region | Christian Raetz |
| pASD6 | pUC18 derivative carrying part of the dap operon from B. subtilis | 11 |
Recombinant DNA techniques.
Standard cloning techniques were used (42). Oligonucleotide synthesis and sequencing were performed at the genetic engineering facility of the University of Illinois. PCR was performed directly on overnight cultures or on purified DNA by using Ready-to-Go PCR beads from Pharmacia. The primers for dapD, dapB, and the open reading frame (ORF) encoding dipicolinate synthase were designed by using the sequence available from GenBank and are as follows: dapD 5′ end, GAT GGA TCC CGA ATT ACA ACC ATT; dapD 3′ end, AAC CGA ATT CTG AGC TCG TGG; dapB 5′ end, GGA TCC ATG CAT GAT GCA AAC ATCC G; dapB 3′ end, AAG CTT TTA CAA ATT ATT GAG ATCA A; dipicolinate synthase ORF 5′ end, GTT TAC CAT GGT AAC CGG ATT; dipicolinate synthase ORF 3′ end, GAC CGG ATC CTT TAG TTT GGG. The dapD8 allele was sequenced and found to contain the missense mutations R164→G, G167→S, and M178→I within the dapD locus. It was not determined whether additional mutations lie outside the gene.
Biochemical assays.
β-Galactosidase assays (39) were performed in triplicate. For 6-phosphogluconate dehydratase assays, the cells were grown in Casamino Acids medium with 0.2% gluconate as a carbon source. Cultures (250 ml) were grown to an OD of 0.1, centrifuged, and resuspended in 1 ml of ice-cold Tris-HCl (pH 7.65). The cells were lysed by passage through a French press. The lysates were immediately centrifuged in a microcentrifuge for 1 min at 12,000 rpm, and the supernatants were frozen immediately in a dry-ice–ethanol bath to prevent the inactivation of these labile enzymes by air. The enzyme activity of rapidly thawed extract was determined by the two-step method of Fraenkel and Horecker (18). A 100-μl reaction mixture containing extract, 8 mM 6-phosphogluconate, and 10 mM MgCl2 was incubated at room temperature for 5 min. The reaction mixture was then diluted into 2 ml of 50 mM Tris (pH 7.65) and boiled for 2 min. The tubes were centrifuged to remove the particulates, and the supernatant was then assayed for pyruvate at 340 nm with lactate dehydrogenase and 0.2 mM NADH. To show that the active 6-phosphogluconate dehydratase recovered from suppressor strains is still superoxide sensitive, the extracts were exposed to superoxide that was generated by xanthine and xanthine oxidase (22). Extracts for aconitase assays were prepared in a similar way except that the cultures were grown in Casamino Acids medium containing 0.2% glucose. The lysis buffer contained 50 mM Tris (pH 7.4), 0.6 mM MnCl2, and 20 μM fluorocitrate to block damage to iron-sulfur clusters (19). Aconitase activity was assayed as described previously (20). One milliliter of assay mixture consisted of extract, 30 mM citrate, 0.2 mM NADP+, 0.6 mM MnCl2, 50 mM Tris-HCl (pH 7.4), and 1 U of isocitrate dehydrogenase. The production of NADPH was monitored at 340 nm. Protein was measured by the Bradford dye-binding assay by using ovalbumin as a standard.
Sensitivity to hydrogen peroxide.
Overnight cultures grown in LB medium were diluted into fresh medium and were grown for at least four generations to reach an OD600 of 0.2. One-milliliter aliquots were exposed to 2.5 mM H2O2, and the cultures were shaken at 37°C for 10 min. Killing was stopped by diluting the cultures 625-fold into LB medium containing 130 U of catalase/ml. Cells were plated onto LB plates in 2 ml of LB top agar, and the plates were incubated at 37°C overnight. Diaminopimelic acid auxotrophs were plated on LB plates containing 5 μg of diaminopimelic acid/ml.
Miscellaneous methods.
Electron paramagnetic resonance (EPR) measurements were performed as described previously (30). Peak areas were normalized to those of iron standards, and intracellular iron concentrations were calculated by correlating OD to intracellular volumes (24). For iron starvation experiments, cells were grown anaerobically to log phase in minimal A medium supplemented with all amino acids. Cells were then diluted into an aerobic medium of the same composition containing various concentrations of DTPA, and growth was monitored. Measurement of expression of the SoxRS regulon was made possible by the introduction of a λ bacteriophage carrying a soxS::lacZ fusion (22) into control and suppressor strains. λ infections and screens for lysogens were carried out as described previously (44). Lysogens were identified by their ability to lyse a λ-sensitive strain after brief exposure to UV light. β-Galactosidase assays were performed on the lysogens to determine the expression of soxS. RpoS induction was tested by introducing a λ construct carrying a bolA::lacZ fusion into the control and suppressor strains. The dapB null mutation was also transduced into a strain containing a marOII::lacZ fusion (12), and β-galactosidase activity was measured.
HPLC quantitation of intracellular dipicolinic acid.
Cultures were grown overnight in minimal E salts medium containing 0.2% glucose and 0.05% Casamino Acids. They were then subcultured into 2.5 ml of minimal medium containing all amino acids except cysteine and containing 10 μM [14C]aspartate (Amersham). Cysteine was omitted from the medium because it inhibits the growth of dap mutants, presumably by competing with diaminopimelic acid for its transport. At an OD600 of 0.8 the cells were pelleted by centrifugation, immediately resuspended in 5% trichloroacetic acid, and incubated on ice for 10 min. The precipitated material was separated by centrifugation, and the clear supernatant was lyophilized to dryness. The dried sample was resuspended in 0.2 ml of buffer A (5 mM Tris-HCl [pH 7]). The sample was injected onto a Beckman system gold high-pressure liquid chromatograph (HPLC) using a Waters SAX column with a constant flow rate of 1 ml of buffer A/min. The column was washed with buffer A for 5 min and then eluted on a linear gradient to 100% buffer B (Tris-HCl [pH 7], 5 mM; NaCl, 1 M) over 20 min. The column was then washed with 100% buffer B for an additional 5 min. Eluate from the column was passed through a Beckman 168 UV/visible spectrophotometer set at 270 nm and an in-line scintillation counter (Beckman detector 171).
SOD assays of dipicolinate-metal chelates.
In vitro solution assays for SOD (37) were performed with few modifications. Reaction mixtures included 100 μM dipicolinate and either 100 μM FeSO4 · 7H2O, 20 μM CuSO4, or 1 to 4 μM MnCl2. EDTA was omitted in order to avoid metal binding.
Measurements of dipicolinate-iron binding and oxidation rates.
Binding, titration, and oxidation studies of the ferrous iron-dipicolinate complex relied upon an ɛ of 1,510 M−1 cm−1 at 484 nm. The ferric complex does not absorb at this wavelength. In these studies water was used without buffers, since buffering anions displace equilibria by competing for free iron; instead, reagents were independently adjusted to pH 7 before mixing, and postreaction pH measurements verified that the pH did not change. The binding constant of the complex was determined by titrating 10.0 μM ferrous iron with dipicolinate. The 2:1 stoichiometry of the dipicolinate-Fe2+ complex was demonstrated by similar titrations of 100 and 400 μM ferrous iron. Binding competition experiments between dipicolinate and citrate, ATP, and reduced glutathione were conducted by adding 50 μM FeSO4 · 7H2O to a solution containing 200 μM dipicolinate and either 1 to 150 mM citrate, 0.167 to 42 mM ATP, or 0.5 to 86 mM glutathione. Time courses of A484 were observed to ensure that equilibrium concentrations of the ferrous iron-dipicolinate complex had been obtained. Solutions were assembled from anaerobic stock solutions and monitored in sealed anaerobic cuvettes to prevent any oxidation of the ferrous complexes by molecular oxygen. The rates of oxidation of the ferrous iron-dipicolinate, -citrate, and -ATP complexes by dissolved oxygen in air-saturated water were determined by monitoring the disappearance of the 484-nm peak (for dipicolinate) or the appearance of the 330-nm absorbance of the ferric chelates (for citrate and ATP). The absorbance maxima of authentic ferric chelates were demonstrated by adding H2O2.
The rate constant for the oxidation of ferrous iron-dipicolinate chelate by H2O2 was measured by monitoring the disappearance of the 484-nm absorbance peak. Reaction mixtures contained 25 μM FeSO4 · 7H2O, 1 mM dipicolinate, and 15.6 to 125 μM H2O2. The absorbance of chelate declined exponentially with time at rates proportional to the H2O2 concentration, justifying the calculation of a second-order rate constant. The hydroxyl radical that is generated by iron oxidation could plausibly perturb the apparent reaction rate either by oxidizing ferrous chelate or by producing O2−; the O2− could, in theory, rereduce ferric iron. Therefore, control reactions included 10 mM ethanol or 20 U of copper, zinc SOD to scavenge hydroxyl radical and O2−, respectively. No effect on the reaction kinetics was observed. This is reasonable, since the excess dipicolinate is likely to be the primary target of the hydroxyl radical.
RESULTS
The suppressor mutation creates a stop codon in dapD.
An earlier study showed that the ssa-1 mutation suppressed the auxotrophies for branched-chain, aromatic, and sulfurous amino acids that are otherwise imposed by O2− stress. Mapping experiments had placed the ssa-1 mutation in the 4-min region of the chromosome but had not identified the altered gene (25).
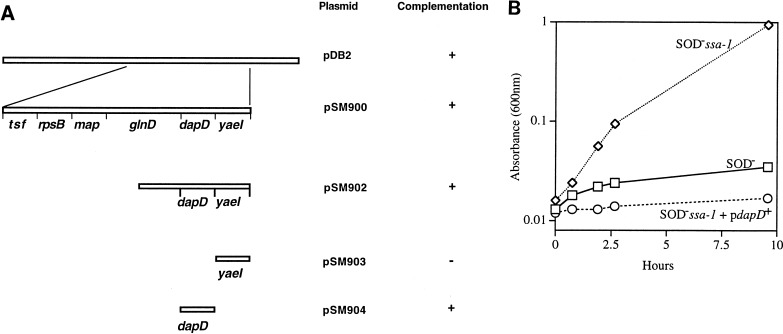
The insert from pDB2, a pACYC-based plasmid carrying part of the 4-min region, was subcloned into the spectinomycin-resistant low-copy-number vector pCL1921 (33) and transformed into JI200 (Fig. 1A). Subclones pSM900 and pSM902 complemented the ssa-1 mutation of strain JI200 and inhibited growth in minimal medium under aerobic conditions, while the parental strain JI200 continued to grow (Fig. 1B). Sequencing demonstrated that pSM902 carries the 3′ end of glnD, all of dapD, and an uncharacterized ORF (yaeI). Further subcloning showed that a minimal clone carrying only the dapD gene (pSM904) complemented the ssa-1 mutation in JI200 (data not shown). The dapD gene encodes tetrahydrodipicolinate succinylase, an enzyme midway in the pathway of diaminopimelate and lysine biosynthesis.
FIG. 1.
Complementation of the suppressor mutation with dapD. (A) Subclones of pDB2 are represented. pSM904, carrying dapD alone, complements the ssa-1 mutation. (B) Aerobic growth of JI200 carrying pSM902. Anaerobic log-phase cultures in minimal medium containing all amino acids except Met and Cys were shifted to an aerobic medium of same composition. Symbols: diamonds, JI200 (SOD− ssa-1); rectangles, JI132 (SOD−); circles, SM506 (SOD− ssa-1/pSM904 [dapD+]).
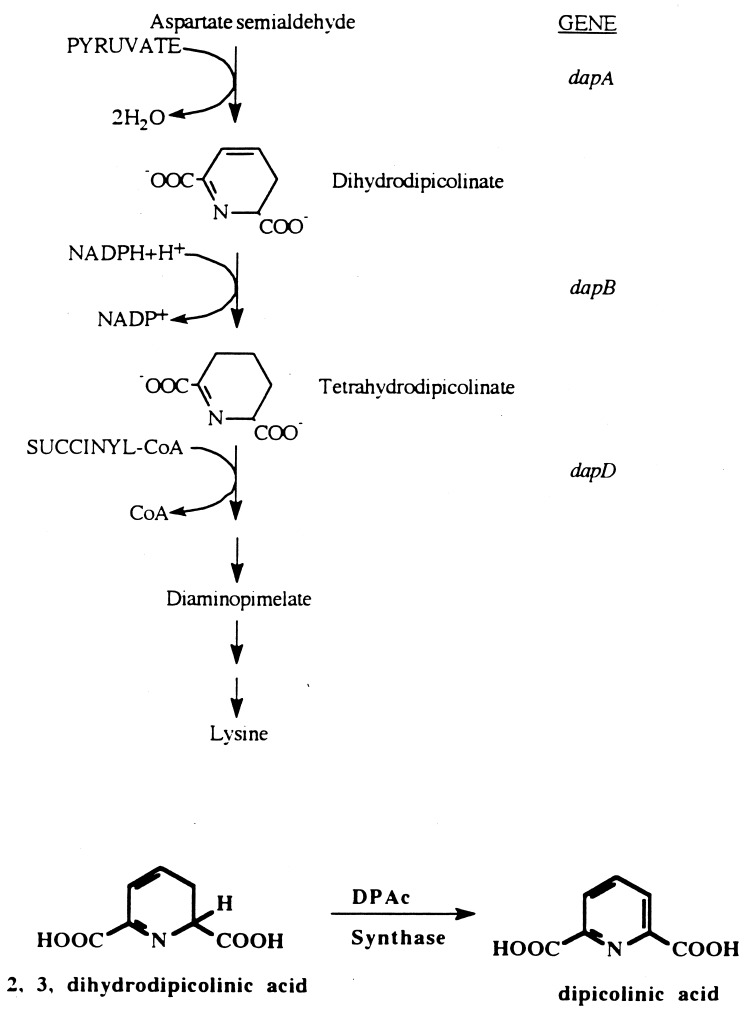
The dapD alleles from strains JI200 and AB1157 (wild type for dapD) were amplified by PCR. Three independent PCR products of dapD from JI200 and two products from AB1157 were cloned into pCL1921 and sequenced. Comparison of the dapD DNA sequence from JI200 with that from AB1157 showed a G→A base substitution at position 428 of the ORF. This results in the conversion of a tryptophan codon at position 143 of the 274-codon gene to an amber codon. Theoretically, this should lead to diaminopimelate and lysine auxotrophy (Fig. 2), which would have prevented its recovery in the selection that was originally used to isolate suppressors. However, the supE mutation of AB1157 partially suppresses amber mutations, allowing low-level expression of dapD. This level is evidently sufficient to prevent auxotrophy for diaminopimelate and lysine in this genetic background.
FIG. 2.
Pathway of diaminopimelate and lysine biosynthesis. The conversion of dihydrodipicolinate to dipicolinate by dipicolinate synthase is represented at the bottom.
Accumulation of the intermediates upstream of dapD is important for protection.
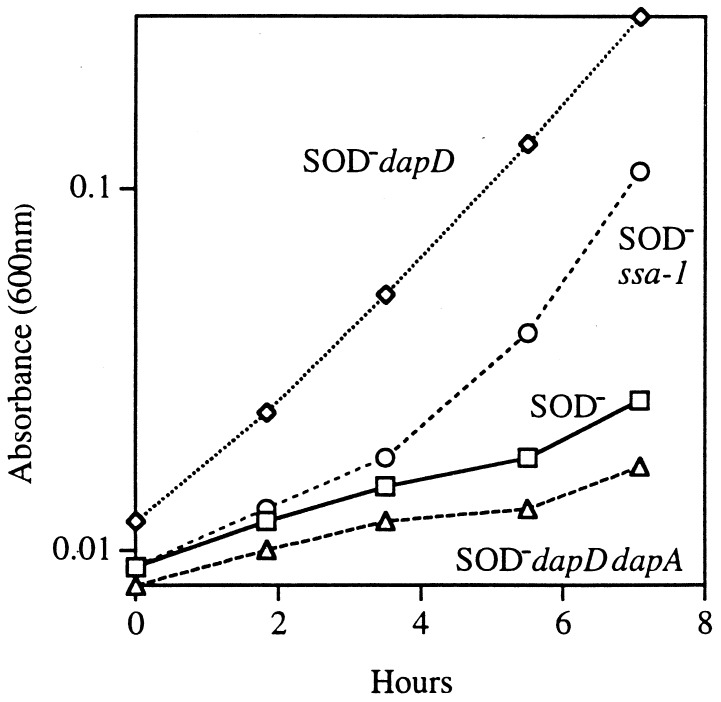
The sequence data from the PCR products suggested that lowering the amount of tetrahydrodipicolinate synthetase in the cell is useful in a SOD mutant. To verify this, we introduced a null mutation of dapD into a SOD mutant. This mutation also relieved the auxotrophies (Fig. 3). The suppression of auxotrophies was reproduced in each genetic background that we examined (AB1157, GC4468, and AN387).
FIG. 3.
The dapD mutation allows SOD− strains to grow without branched-chain, aromatic, and sulfurous amino acids. Cultures were grown to log phase in anaerobic minimal medium supplemented with five amino acids (see Materials and Methods) and were diluted into an aerobic medium of same composition. Dap mutants were supplemented with diaminopimelic acid. Symbols: rectangles, AS237 (SOD−); circles, JI200 (SOD− ssa-1); diamonds, SM1004 (SOD− dapD); triangles, SM1006 (SOD− dapD dapA).
To check whether suppression resulted from a loss of pathway function per se, a sodA sodB dapA strain was constructed (SM1005). The dapA gene encodes the first enzyme in the lysine and diaminopimelate pathway (Fig. 2), and a null mutation in this gene renders a strain auxotrophic for both of these products. Unlike the dapD mutation, the dapA mutation failed to suppress the O2− imposed auxotrophies (data not shown). Moreover, when the dapA mutation was introduced into a sodA sodB dapD strain, it reversed the suppression that had been achieved by the dapD allele (Fig. 3). This result indicated that the initial steps in the Dap pathway were essential for minimal tolerance in the sodA sodB dapD strain.
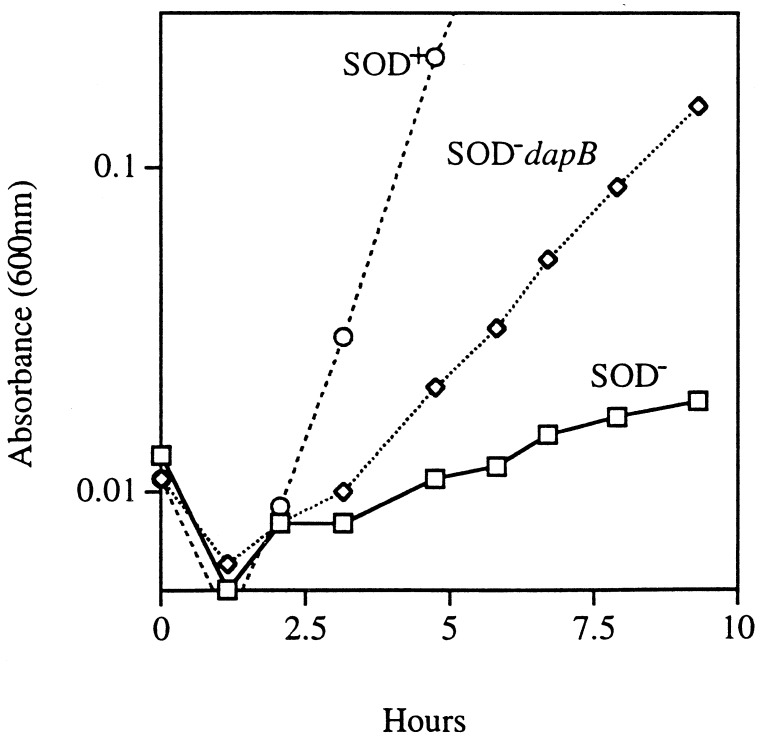
A block in dapD should result in the accumulation of one or both of the upstream intermediates, tetrahydrodipicolinate and dihydrodipicolinate. Since a functional dapA allele was essential for protection against the auxotrophies caused by the lack of SOD, it seemed reasonable that the accumulation of these intermediates might have a role in suppression. In order to test this hypothesis, a strain with a dapB mutation was constructed in the sodA sodB background. A block at dapB should cause the accumulation of dihydrodipicolinate (Fig. 2). In fact, the sodA sodB dapB strain grew well without amino acid supplements (data not shown). The introduction of a dapA mutation into the sodA sodB dapB strain once again restored auxotrophic behavior, confirming that pathway intermediates must be synthesized to confer the protection (data not shown).
The suppressors exhibit an increase in the activities of labile dehydratases.
Superoxide destroys the iron-sulfur clusters of a subfamily of dehydratases, thereby eliminating the function of the pathways to which they belong. The relaxed requirement in suppressed strains for branched-chain amino acid supplements suggested that the activity of dihydroxyacid dehydratase might be increased. The suppressed strains also showed better growth on nonfermentable carbon sources such as lactate (Fig. 4) and succinate (data not shown). This growth requires functional aconitase and fumarase and suggested a general improvement in dehydratase activities. In fact, while SOD-deficient strains usually show very low aconitase activity in air—about 10% of wild-type levels—the suppressor strains exhibited about 70% of the wild-type level (Table 2). This was also true for 6-phosphogluconate dehydratase, another labile dehydratase. The residual 6-phosphogluconate dehydratase activity in the cell extracts remained fully vulnerable to O2−, confirming that the increased activity was due to stabilization of the labile Fe-S dehydratase rather than its replacement by a O2−-resistant isozyme (data not shown).
FIG. 4.
Growth on a nonfermentable carbon source. Cultures were grown to log phase in an anaerobic medium containing all amino acids with lactic acid as a carbon source. They were then diluted into an aerobic medium of the same composition. Symbols: circles, AN387 (SOD+); rectangles, AS237 (SOD−); diamonds, SM1008 (SOD− dapB).
TABLE 2.
Suppressors exhibit increased iron-sulfur cluster enzyme activity
| Strain | Concna (% activity)
of:
|
|
|---|---|---|
| Aconitase | 6-P-gluconate dehydratase | |
| AN387 (SOD+) | 25.4 (100) | 0.274 (100) |
| AS237 (SOD−) | 1.7 (6.6) | 0.034 (12.4) |
| SM1004 (SOD−dapD) | 16.8 (66) | NDb |
| SM1093 (SOD−dapB) | 8.15 (32) | 0.192 (70) |
In units per milligram.
ND, not determined.
The reductive properties of the intermediates are not important for suppression.
The structures of tetrahydrodipicolinate and dihydrodipicolinate suggested two possible scenarios by which their accumulation could benefit SOD mutants. The first possibility was that these compounds could act as reductants in the cell. Superoxide destabilizes 4Fe-4S clusters by univalently oxidizing them; a reductant might prevent their inactivation by rereducing the cluster before its iron atom is lost. Tetrahydrodipicolinic acid, in particular, is expected to be a good reducing agent, since its oxidation product is aromatic (32). In fact, tetrahydrodipicolinic acid spontaneously oxidizes in solution. Spontaneous disproportionation of dihydrodipicolinic acid creates tetrahydrodipicolinic acid, leading to the same result.
A second possibility was that these dicarboxylate compounds might chelate metal atoms in the cell. Superoxide-stressed cells contain increased amounts of free iron due to the disintegration of their iron-sulfur clusters. It seemed plausible that the accumulated dipicolinates might bind this iron and present it to the cluster-rebuilding apparatus in a “usable” form. Alternatively, iron chelation might perturb metal metabolism in an unanticipated but fortuitous way.
The following experiment was designed to distinguish the action of dipicolinates as potential reductants and chelators. Dipicolinate, the oxidation product of tetrahydrodipicolinate, should retain its putative chelating ability but not its reductive capacity, so if metal binding were the essential aspect of suppression, then dipicolinate might also protect a sodA sodB strain. Dipicolinate is a charged molecule, unlikely to cross cell membranes, and in fact exogenous dipicolinate did not suppress the auxotrophies of SOD mutants. We anticipated, however, that dipicolinate might accumulate in E. coli if we expressed dipicolinate synthase, an enzyme from Bacillus subtilis that during sporulation converts dihydrodipicolinate to dipicolinate in a single step. We hoped that high titers of this enzyme might divert sufficient dihydrodipicolinate from the diaminopimelate and lysine pathway to generate substantial dipicolinate. For this purpose the dipicolinate synthase gene from B. subtilis was amplified from pASD6 and cloned into a high-copy-number vector, pMTL21, thereby generating pSM912. This plasmid was transformed into SOD-proficient and -deficient strains.
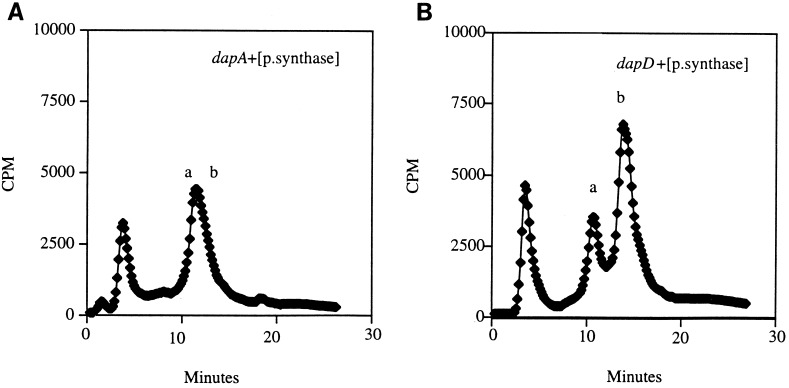
A standard colorimetric method was too insensitive to detect dipicolinate (see Materials and Methods). However, we detected substantial intracellular [14C]dipicolinate when [14C]aspartate was fed to an SOD-proficient strain carrying the dapB mutation and a plasmid overexpressing dipicolinate synthase (Fig. 5). This peak was absent from the dapA control strain (SM1128), which cannot convert aspartate to dihydrodipicolinate. The intracellular dipicolinate concentration was approximately 3 mM.
FIG. 5.
Accumulation of dipicolinate. Fifty microliters of the sample (see Materials and Methods) was analyzed on an HPLC anion-exchange column. An unlabelled internal standard of dipicolinate was monitored at 270 nm, and the 14C-labelled dipicolinate was counted on a flowthrough scintillation counter attached in series to the HPLC. (A) SM1128 (SOD+ dapA/SM912 [encoding dipicolinate synthase]). (B) SM1127 (SOD+ dapB/SM912). Peak a represents [14C]aspartate, and peak b represents 14C-labelled dipicolinate. Authentic dipicolinate and aspartate standards coeluted with these peaks (data not shown).
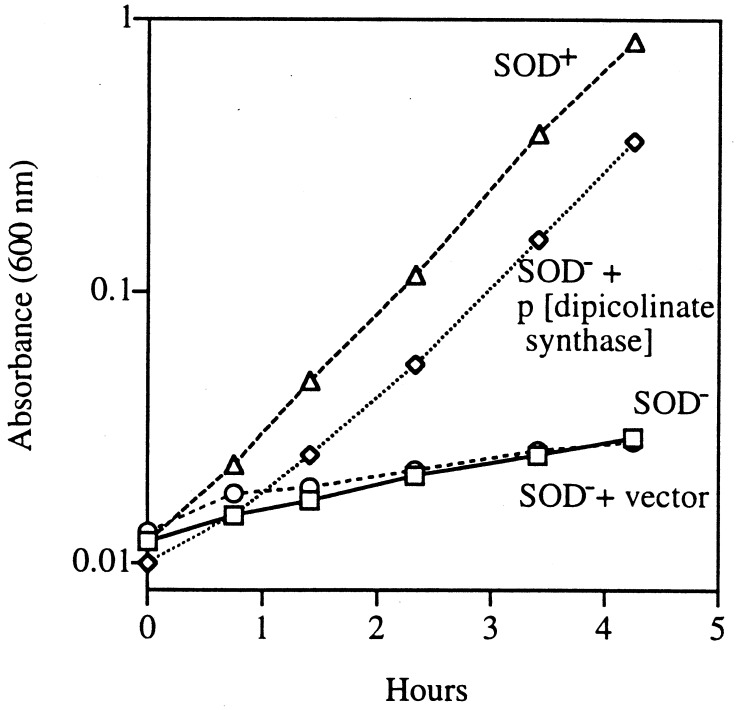
When it was transformed into a sodA sodB strain, this plasmid suppressed its amino acid auxotrophies (Fig. 6), exactly like the dapD and dapB mutations. Again, the introduction of dapA into this strain restored the auxotrophies, presumably because the diaminopimelate pathway must be functional in order for dipicolinate accumulation to occur (data not shown). Thus, the possibility that the dipicolinates restored dehydratase activities through cluster reduction was eliminated.
FIG. 6.
Dipicolinate synthase overexpression in a SOD-deficient strain relieves the auxotrophies. Cultures were grown to log phase in minimal medium supplemented with all but the sulfurous amino acids and were diluted into an aerobic medium of same composition. Symbols: circles, AS237 (SOD−); rectangles, SM1024 (SOD− plus vector); triangles, AN387 (SOD+); diamonds, SM1025 (SOD−/pSM912 [encoding dipicolinate synthase]).
Dipicolinate binds and stabilizes ferrous iron in vitro.
The structure of dipicolinate should predispose it to be a cation chelator, and its iron-binding ability has been used in assays of the dipicolinate content of spores. However, its function in spores is thought to arise from an ability to bind calcium rather than iron. In order to serve as an effective iron chelator in vivo, dipicolinate must outcompete other suspected intracellular iron chelators, including citrate, ATP, and glutathione.
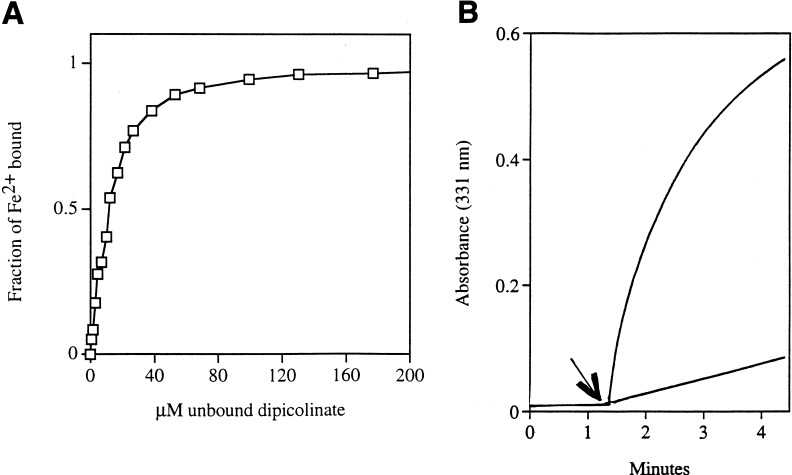
The avidity with which dipicolinate binds iron was demonstrated by binding studies (Fig. 7A), which demonstrated that 10 μM dipicolinate is sufficient for half-maximal iron binding. Titrations showed that the stoichiometry was 2 dipicolinate atoms per iron atom; it is likely that they form a sandwich, with pi-orbital interactions contributing to the iron binding. A 67-fold excess of ATP or a 280-fold excess of citrate was necessary to compete with 200 μM dipicolinate (Fig. 7B). Reduced glutathione was ineffective even at a 500-fold excess. Therefore, the millimolar concentrations of dipicolinate in suppressed cells should be sufficient to sequester iron from these potential carriers.
FIG. 7.
Dipicolinate binds ferrous iron with greater avidity than does citrate. (A) Titration of 10.0 μM ferrous iron with dipicolinate. Half of all ferrous iron is in the Fe2+–dipicolinate complex in 11 μM free dipicolinate. The amount of free dipicolinate was calculated based on the stoichiometry of 2 dipicolinate atoms/complex. (B) Citrate-stimulated Fe2+ oxidation is inhibited by the addition of dipicolinate. Top curve, 330 μM Fe2+ in water. Citrate (33 mM) was added at the time indicated by the arrow, and the appearance of Fe3+ was monitored at 331 nm. The reaction slows due to depletion of Fe2+. For the bottom curve, a mixture of 33 mM citrate and 1.7 mM dipicolinate was added to 330 μM Fe2+ at the time indicated by the arrow.
In addition to binding ferrous iron, dipicolinate stabilizes it in the ferrous form, with only about 1% being oxidized per min in air-saturated solutions. In contrast, many chelators bind ferric iron with higher avidity than ferrous iron, and they therefore lower its reduction potential. Among those chelators that we tested, citrate strongly accelerated Fe2+ oxidation. In saturating citrate, ferrous iron oxidized at a rate of 25% per min. Therefore, dipicolinate could plausibly recover the free ferrous iron released by oxidized clusters and, by maintaining it in its reduced form, more effectively recycle it into the enzyme repair process.
The accumulation of dipicolinates causes the derepression of the Fur regulon in suppressor strains.
The chelation of intracellular iron was tested by measurement of the expression of genes in the Fur regulon. In iron-replete cells, the Fur regulatory protein binds ferrous iron and gains DNA-binding capacity. Iron starvation, through either exhaustion of extracellular iron or the presence of extracellular chelators, prevents DNA binding by Fur, apparently by diminishing the pool of iron available for it to bind. The occasional appearance of extracellular chromophores similar to enterochelin suggested that the Fur regulon might be induced in the suppressed strains.
To directly test whether the Fur regulon was induced in the suppressed strains, a reporter plasmid containing the Fur-regulated iucC::lacZ allele (2) was introduced into both control and suppressor strains. The expression level was monitored by the measurement of β-galactosidase activity (Table 3). SOD-deficient strains exhibited approximately the same level of activity as SOD-proficient cells. However, the presence of either dapD or dapB mutations, or of the dipicolinate synthase-expressing plasmid, caused substantial iucC expression. These data confirm that the dipicolinates effectively chelate iron in vivo as well as in vitro.
TABLE 3.
The Fur regulon is derepressed in suppressed strains
| Straina | β-Galactosidase activity (Miller units)b |
|---|---|
| SM1117 (SOD+) | 500 (30) |
| SM1110 (SOD+dapB) | 1,140 (18) |
| SM1111 (SOD−) | 530 (15) |
| SM1112 (SOD−dapB) | 1,860 (49) |
| SM1113 (SOD−dapD) | 1,370 (23) |
| SM1114 (SOD−dapA) | 410 (14) |
| SM1119 (SOD−/pSM912)c | 1,800 (15) |
All the strains carry iucC::lacZ reporter plasmid.
The average of three assays is reported, with the standard deviation in parentheses.
pSM912 encodes dipicolinate synthase.
It seemed possible that, by binding metal atoms such as iron, copper, and manganese, dipicolinate might form a complex capable of scavenging intracellular O2−. However, in vitro assays demonstrated that dipicolinate, by itself or in combination with metals, cannot prevent O2− from reducing cytochrome c (data not shown). In fact, dipicolinate inhibited manganese cations from dismuting superoxide. Further, a previous study indicated that suppression does not occur by reducing the intracellular O2− concentration (26).
Suppressed strains accumulate high levels of intracellular iron.
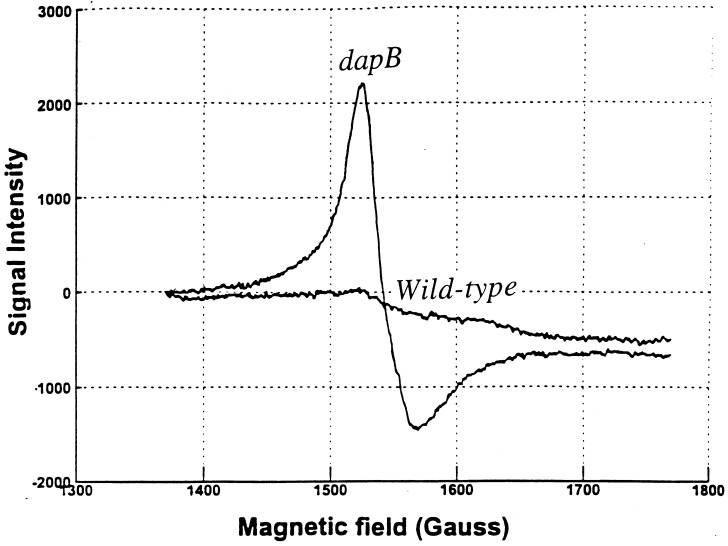
The dapD and dapB mutations and dipicolinate synthase overproduction did not suppress the characteristic sensitivity of SOD mutants to hydrogen peroxide; in fact, sensitivity was somewhat increased (data not shown). This effect was more obvious in SOD-proficient cells: exposure of wild-type cells to 2.5 mM H2O2 for 15 min typically permitted survival of 80% of the cells, whereas only 2% of dapB mutants survived. This suggested that dapB mutants might accumulate high levels of free intracellular iron that could catalyze oxidative DNA damage. EPR spectroscopy of whole cells confirmed that dapB mutants contained 300 μM free iron, in contrast to the 30 μM iron detected in their dapB+ parents (Fig. 8).
FIG. 8.
The dapB mutation increases the intracellular iron concentration. EPR scans of AN387 (SOD+) (wild type) and SM1095 (SOD+ dapB) are shown. Scans are normalized to the intracellular volume.
Although derepression of the Fur regulon induces the synthesis of iron transporters and causes accumulation of excess iron (30, 46), the amount of accumulated iron is not as high in fur mutants as was seen in the dapD strain. Furthermore, the addition of a tonB mutation did not diminish the sensitivity of the dapD mutants to hydrogen peroxide (data not shown), indicating that high-affinity iron transport was not necessary for the high iron levels of the dapD mutants. Therefore the accumulated dipicolinates captured iron that was either imported through constitutive processes or released from proteins.
The continued susceptibility to oxidative DNA damage of dipicolinate synthase-expressing cells would require that any putative iron chelates still react rapidly with H2O2. It might seem that chelators would inhibit this reaction by sterically obstructing it. However, the rate constant for the reaction of dipicolinate-chelated iron with H2O2 (260 M−1 s−1 [data not shown]) was determined to be even higher than that of unchelated ferrous iron (76 M−1 s−1). EDTA, notably, also forms a hexacoordinate ferrous complex yet enhances the rate of inner-sphere electron transfer to H2O2.
SOD-deficient strains are starved for iron.
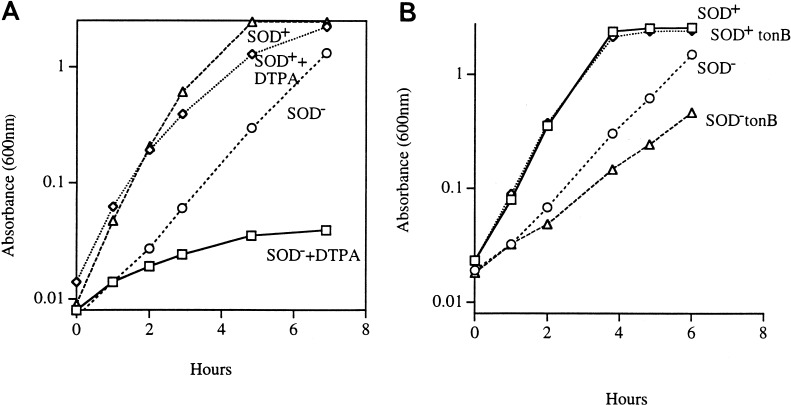
The presence of high concentrations of iron chelates in the suppressed strains suggested that in unsuppressed SOD mutants the rate of cluster repair might be limited by the availability of intracellular iron. Although SOD-deficient cells are replete with free iron which is released from the iron-sulfur clusters by O2−, this iron may not be available to repair the damage to the iron-sulfur clusters; in fact, it appears to be rapidly removed from the cytosol by an undefined process (29). Thus, the constant turnover of iron-sulfur clusters could plausibly lead to iron starvation in SOD-deficient strains. To test whether this was the case, cells were exposed to the cell-impermeant chelator DTPA in order to abruptly terminate iron uptake. When 10 mM DTPA was added to cultures of SOD-proficient cells in amino acid-supplemented medium, a sevenfold increase in cell mass occurred before growth stopped. Under the same conditions SOD− mutants stopped growing sooner, after only a 1.6-fold increase. This result suggested that either SOD− mutants had lower levels of iron reserves or they had a higher iron demand. Subsaturating concentrations of DTPA were also far more inhibitory to SOD mutants than to wild-type strains (Fig. 9A).
FIG. 9.
SOD− mutants are sensitive to iron limitation. (A) Subsaturating concentrations of DTPA can inhibit the growth of a SOD-deficient strain. Log-phase cultures grown under anaerobic conditions in minimal medium supplemented with all amino acids were diluted into an aerobic medium with or without 50 μM DTPA. Growth in 50 μM DTPA is represented. Symbols: triangles, AN387 (SOD+); circles, AS237 (SOD−); diamonds, AN387 plus DTPA; rectangles, AS237 plus DTPA. (B) A tonB mutation slows the growth of a SOD-deficient strain. Log-phase cultures grown under anaerobic conditions in minimal medium supplemented with all amino acids were diluted into an aerobic medium of same composition. Symbols: diamonds, AN387 (SOD+); rectangles, SM1120 (SOD+ tonB); circles, AS237 (SOD−); triangles, SM1122 (SOD− tonB).
In the absence of chelators, our attempts to supplement a SOD-deficient strain with iron did not result in any significant improvement in growth rates or suppression of amino acid auxotrophies (data not shown). Most lab media may contain enough iron to saturate iron transporters, so that the increased demand may be satisfied only by increasing the number of transporters.
The addition of a tonB mutation to wild-type strains did not slow their growth, but it did slow the growth of SOD− mutants (Fig. 9B). These results support the idea that the rate of iron uptake limits the growth of O2−-stressed cells.
Fur derepression alone is not enough for suppression.
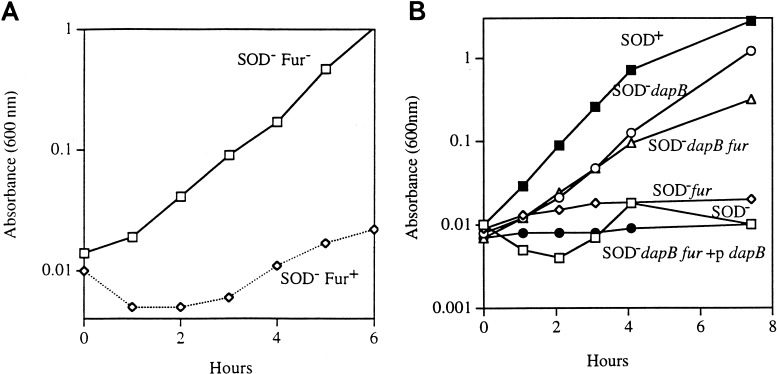
Fur derepression and consequent rapid iron uptake can be achieved by introducing a null mutation in the Fur repressor protein. A fur mutation significantly relieved the aromatic amino acid auxotrophy of the SOD− strain (Fig. 10A), but the sulfur-containing and branched-chain amino acid auxotrophies persisted.
FIG. 10.
(A) A fur mutation relieves the aromatic amino acid auxotrophy of a SOD-deficient strain. Log-phase cultures grown under anaerobic conditions in minimal medium supplemented with all but the aromatic amino acids were diluted into an aerobic medium of the same composition. Symbols: rectangles, SM1105 (SOD− fur); diamonds, AS237 (SOD−). (B) Fur derepression alone is not sufficient for suppression. Cultures growing logarithmically in LB medium were washed and resuspended in minimal medium containing all but the sulfurous amino acids. Symbols: solid rectangles, AN387 (SOD+); solid circles, AS237 (SOD−); open circles, SM1093 (SOD− dapB); triangles, SM1116 (SOD− dapB fur); diamonds, SM1105 (SOD− fur); open rectangles, SM1123 (SOD− dapB fur/pSM915 [dapB+]).
Those data indicated that Fur derepression must not be the sole mechanism of suppression by dap mutations. To test this more directly, a SOD− dapB fur strain (SM1116) was constructed and found to grow without any amino acid supplements. When a plasmid carrying the wild-type dapB allele was introduced into the strain, growth was once again inhibited (Fig. 10B). Thus, although Fur derepression alone partially suppresses some aspects of the SOD− phenotype, the dap mutations exert some additional effect that more fully suppresses damage. This effect could conceivably be the capture and recycling of iron released from clusters. That putative effect has not yet been demonstrated in vitro.
Suppression is not mediated by induction of antioxidant defenses.
The SoxR sensor protein, which activates the response to superoxide-generating agents, may be activated in vivo by removal of iron from its [2Fe-2S] cluster. However, assays of soxS::lacZ expression determined that this regulon was not activated in the suppressor background, despite the accumulation of dipicolinates. Further, neither bolA::lacZ nor marOII::lacZ alleles were induced, indicating that the RpoS and Mar responses also remained inactive (data not shown).
DISCUSSION
SOD deficiency imposes upon E. coli an inability to use nonfermentable carbon sources, auxotrophies for several families of amino acids, and slow growth even in glucose- and amino acid-supplemented medium. It is remarkable that pleiotropic suppression of these disparate phenotypes is achieved by the accumulation of a single metabolite, dipicolinate, or its reduced derivatives. This observation is of interest not only with respect to cell physiology but also because of its possible utility in pharmaceutical research.
The original report of the isolation of these mutants mapped the suppressor locus near but outside dapD. The data presented here show that that placement was erroneous. In retrospect, it is apparent that some of the mapping analyses were confounded by the fact that the suppressor allele was a nonsense mutation in a gene that, in the media used, was essential. Partial suppression of the mutation was achieved by the supE44 allele of the strain in which the mutation was isolated but was variable in the genetic backgrounds used for mapping. This resulted in incorrect phenotyping, since too little suppression of the nonsense allele would prohibit growth of the mutants, and too much would preclude suppression of the SOD auxotrophies. Furthermore, we have observed that growth of the dapD mutants in cysteine-supplemented media is also strain dependent, presumably manifesting some aspect of the competition between cysteine and diaminopimelate for the cysteine transporter. In any case, the sequence and complementation data reported here definitively establish that the original suppression occurred by mutation of dapD.
We do not yet understand the details of the connection between dipicolinate accumulation and suppression of SOD− phenotypes. The restoration of branched-chain amino acid synthesis and of TCA cycle function is clearly due to the increased activity of the labile dehydratases in these pathways. Because dipicolinate chelates iron both in vitro and in vivo and maintains it in a reduced form, we have suggested that it may capture iron lost from the clusters and recycle it into a cluster repair process. This idea is speculative and, since the repair process has not been biochemically defined, has not been easily testable yet in vitro. However, previous work demonstrated that in nonsuppressed strains the iron lost from clusters is not available to rebuild them (29). A predictable consequence is that the cycles of enzyme damage and remetallation catalytically deplete the cells of usable iron, until there is little iron left to remetallate them. This would explain the hypersensitivity of SOD− mutants to tonB mutations and to extracellular chelators, both of which reduce the rate of new iron import. Thus, iron recycling mediated by dipicolinates would benefit SOD− cells. This concurs with Benov and Fridovich’s observation that by providing additional iron in the growth medium they could increase the activities of the dehydratases and the cell growth rate in SOD mutants (4). While we were unable to see a similar effect (data not shown), that discrepancy could simply reflect differences in the abundance of trace metals in the salts used in growth media.
It is intriguing that an intracellular chelator which restores the function of the dehydratases also suppresses the sulfur and aromatic auxotrophies. This seems to comprise circumstantial evidence that these phenotypes as well are somehow connected to cluster damage. This idea is not easily reconciled with the recent demonstration that the aromatic auxotrophy may derive from the inactivation of transketolase (3). It is not clear how accumulation of iron-dipicolinate might enhance transketolase activity. However, some association of the aromatic auxotrophy with iron metabolism is implied by the observations that the auxotrophy is suppressed both by excessive iron supplements and by fur mutations. There is no evidence that either of the E. coli tkt genes is regulated by fur, and we do not recognize any Fur binding site in their promoter regions.
Although the connections between these phenotypes and cluster damage remain to be unravelled, this idea also dovetails with observations made in other studies. First, the amount of intracellular O2− needed to confer sulfur and aromatic auxotrophies is about the same as that necessary to inactivate the dehydratases (22). Second, the ability of Mycoplasma pneumoniae (23) to thrive in an aerobic habitat without any SOD correlates with the absence of any of the known labile dehydratases. Finally, Culotta and colleagues discovered that mutations in genes suspected of being involved in cluster assembly can suppress the sulfur auxotrophy of SOD− yeast (45).
That SOD-deficient strains are iron starved, despite the fact that their cytosol is replete with iron released from the iron-sulfur cluster enzymes, seems odd. However, one can imagine that a pipeline must deliver iron from transporters and storage proteins to the cluster assembly process and that iron adrift in the cytosol might not easily reenter that pipeline. The pipeline idea has an appeal, both because free iron is so sticky that it would seem unwise to rely upon its random diffusion through the cell and because of its extreme toxicity. Indeed, it has been found that other, less-toxic metals such as copper and nickel are carried through cells by chaperones (13, 40, 41). No such system has yet been uncovered for iron, but mutants would presumably have escaped simple genetic screens because of their inviability.
These experiments suggested that the Fur protein perceives a different pool of iron than do the dehydratases. For example, while the dehydratases were demetallated in the SOD mutants, the Fur protein remained metallated and active as a repressor. In contrast, the accumulation of the intracellular chelator caused Fur demetallation at the same time that it improved the assembly of dehydratase clusters. One explanation might be that Fur monitors the abundance of free iron in the medium and perceives SOD mutants as being iron rich. This could seem surprising: one might think that Fur would measure the iron content of the putative pipeline that delivers iron to the clusters. However, Touati et al. (46) showed that the import of iron in excess of the cellular iron storage capacity causes free iron to spill into the cytosol. Therefore, under normal conditions the presence of free iron would signal that iron import systems should be shut down, and Fur would respond accordingly. Only when free iron arises from cluster disintegration would this sensing system be inappropriate.
ACKNOWLEDGMENTS
We are grateful to Kathleen Postle, Bruce Demple, Stuart Levy, and Henry Paulus for generously providing the strains and plasmids used in this study. We thank Alex Smirnov, Tatyana Smirnova, and R. Linn Belford for technical assistance with the EPR experiments conducted at the Illinois EPR Research Center, a National Institutes of Health Biomedical Research Technology Resource (P41-RR01811). We are also thankful to Irwin Fridovich for providing useful suggestions during the course of this study, Craig Kuettner for assistance in growth studies, and Kay Keyer for assistance in EPR experiments.
This work was supported by a National Institutes of Health grant (GM49640).
REFERENCES
- 1.Ahmer B M M, Thomas M G, Larsen R A, Postle K. Characterization of the exbBD operon of Escherichia coliand the role of ExbB and ExbD in TonB function and stability. J Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 3.Benov L. Why superoxide imposes an aromatic amino acid auxotrophy in Escherichia coli. J Biol Chem. 1999;274:4202–4206. doi: 10.1074/jbc.274.7.4202. [DOI] [PubMed] [Google Scholar]
- 4.Benov L, Fridovich I. Growth in iron-enriched medium partially compensates Escherichia colifor the lack of manganese and iron superoxide dismutase. J Biol Chem. 1998;273:10313–10316. doi: 10.1074/jbc.273.17.10313. [DOI] [PubMed] [Google Scholar]
- 5.Benov L, Kredich N M, Fridovich I. The mechanism of the auxotrophy for sulfur-containing amino acids imposed upon Escherichia coliby superoxide. J Biol Chem. 1996;271:21037–21040. doi: 10.1074/jbc.271.35.21037. [DOI] [PubMed] [Google Scholar]
- 6.Bielski B H J, Cabelli D E, Arudi R L. Reactivity of HO2/O2−radicals in aqueous solution. J Phys Chem Ref Data. 1985;14:1041–1062. [Google Scholar]
- 7.Bielski B H J, Richter H W. A study of the superoxide radical chemistry by stopped-flow radiolysis and radiation-induced oxygen consumption. J Am Chem Soc. 1977;99:3019. [Google Scholar]
- 8.Boehme D E, Vincent K, Brown O R. Oxygen and toxicity: inhibition of amino acid biosynthesis. Nature. 1976;262:418–420. doi: 10.1038/262418a0. [DOI] [PubMed] [Google Scholar]
- 8a.Bradley T M, Hidalgo E, Leautand V, Ding H, Demple B. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucleic Acids Res. 1997;25:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukhari A I, Taylor A L. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J Bacteriol. 1971;105:844–854. doi: 10.1128/jb.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Chambers S P, Prior S E, Barstow D A, Minton N P. The pMTL nic−cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 11.Chen N Y, Jiang S Q, Klein D A, Paulus H. Organization and nucleotide sequence of the Bacillus subtilisdiaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268:9448–9465. [PubMed] [Google Scholar]
- 12.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culotta V C, Klomp L W, Strain J, Casareno R L, Krems B, Gitlin J D. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 14.Farr S B, D’Ari R, Touati D. Oxygen-dependent mutagenesis in Escherichia colilacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fee J A. Is superoxide important in oxygen poisoning? Trends Biochem Sci. 1982;7:84–86. [Google Scholar]
- 16.Flint D H, Emptage M H. Dihydroxyacid dehydratase: isolation, characterization as Fe-S proteins, and sensitivity to inactivation by oxygen radicals. In: Barak Z, Chipman D, Schloss J V, editors. Biosynthesis of branched chain amino acids. New York, N.Y: VCH Publishers; 1990. pp. 285–314. [Google Scholar]
- 17.Flint D H, Smyk-Randall E, Tuminello J F, Draczynska-Lusiak B, Brown O R. The inactivation of dihydroxyacid dehydratase in Escherichia colitreated with hyperbaric oxygen occurs because of the destruction of its Fe-S cluster, but the enzyme remains in the cell in a form that can be reactivated. J Biol Chem. 1993;268:25547–25552. [PubMed] [Google Scholar]
- 18.Fraenkel D G, Horecker B L. Pathways of d-glucose metabolism in Salmonella typhimurium. A study of a mutant lacking phosphoglucose isomerase. J Biol Chem. 1964;239:2765–2771. [PubMed] [Google Scholar]
- 19.Gardner P R, Fridovich I. Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J Biol Chem. 1992;267:8757–8763. [PubMed] [Google Scholar]
- 20.Gardner P R, Fridovich I. Superoxide sensitivity of the Escherichia coli6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 21.Gardner P R, Fridovich I. Superoxide sensitivity of the Escherichia coliaconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 22.Gort A, Imlay J. Balance between endogenous superoxide stress and antioxidant defenses. J Bacteriol. 1998;180:1402–1410. doi: 10.1128/jb.180.6.1402-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imlay J A, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 25.Imlay J A, Fridovich I. Isolation and genetic analysis of a mutation that suppresses the auxotrophies of superoxide dismutase-deficient Escherichia coliK12. Mol Gen Genet. 1991;228:410–416. doi: 10.1007/BF00260634. [DOI] [PubMed] [Google Scholar]
- 26.Imlay J A, Fridovich I. Suppression of oxidative envelope damage by pseudoreversion of a superoxide dismutase-deficient mutant of Escherichia coli. J Bacteriol. 1992;174:953–961. doi: 10.1128/jb.174.3.953-961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imlay J A, Linn S. Mutagenesis and stress responses induced in Escherichia coliby hydrogen peroxide. J Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyer K, Gort A S, Imlay J A. Superoxide and the production of oxidative DNA damage. J Bacteriol. 1995;177:6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keyer K, Imlay J A. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J Biol Chem. 1997;272:27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- 30.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo C F, Mashino T, Fridovich I. α,β-Dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 32.Kuthan J, Kurfurst A. Developments in dihydropyridine chemistry. Ind Eng Chem Prod Res Dev. 1982;21:191–261. [Google Scholar]
- 33.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coliwith blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liochev S I, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liochev S I, Fridovich I. The role of superoxide in the production of hydroxyl radical: in vitro and in vivo. Free Radic Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 36.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCord J M, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 38.McCord J M, Keele B B, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Park I S, Carr M B, Hausinger R P. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pufahl R A, Singer C P, Peariso K L, Lin S, Schmidt P J, Fahrni C J, Culotta V C, Penner-Hahn J E. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Sawyer D T, Valentine J S. How super is superoxide? Accounts Chem Res. 1981;14:393–400. [Google Scholar]
- 44.Silhavy T, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 45.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron-sulfur cluster assembly. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 46.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]