Abstract
Regulation of the purine biosynthetic gene purA was examined by using a transcriptional fusion to a luciferase reporter gene. Transcription was repressed about 10-fold by the addition of adenine and increased approximately 4.5-fold by the addition of guanosine. This regulation is mediated by a purine repressor (PurR). In a purR mutant, basal expression was increased 10-fold, and there was no further stimulation by guanosine or repression by adenine. An open reading frame, yabJ, immediately downstream from purR was found to have a role in the repression of purA by adenine. Repression by adenine was perturbed in a purR+ yabJ mutant, although guanosine regulation was retained. Mutations in the PurR PRPP binding motif abolished guanosine regulation in the yabJ mutant. Thus, PRPP appears to be required for upregulation by guanosine. The amino acid sequence of YabJ is homologous to the YER057c/YjgF protein family of unknown function.
There is an 11-step pathway for the de novo synthesis of IMP in Bacillus subtilis (22) and Escherichia coli (23). IMP is a branch point for the synthesis of AMP in two steps and the synthesis of GMP in two steps. A 12-gene pur operon encodes the enzymes required for the synthesis of IMP in B. subtilis (1). Two genes, purA and purB, are required to convert IMP into AMP. The purA gene is unlinked to the pur operon, while purB is in the operon. The purA gene encodes adenylosuccinate synthetase, and adenylosuccinate lyase is the product of purB. Purine biosynthesis is feedback regulated by end products of the pathway, and production of adenine and guanine nucleotides is balanced by regulation of the AMP and GMP branches. Expression of the pur operon is subject to dual regulation of transcription initiation and termination (1). The addition of adenine to cells results in the repression of transcription initiation, and the addition of guanosine promotes premature transcription termination in an mRNA leader region preceding the first structural gene. A purine repressor (PurR) mediates the regulation of transcription initiation. purR was cloned and overexpressed (20), and the protein was purified (19). The present studies were undertaken to assess the regulation of the branch from IMP to AMP. Earlier it was reported that adenine decreased the adenylosuccinate synthetase activity in B. subtilis and that guanosine increased the enzyme activity (17). Here we present evidence that adenine mediates a PurR-dependent repression of purA transcription and that guanosine leads to an upregulation of transcription. The repression by PurR is dependent upon a second protein, YabJ, which is homologous to a group of proteins of unknown function. On the other hand, the activation by guanosine is dependent upon an interaction of PRPP with PurR but not upon YabJ.
MATERIALS AND METHODS
Bacterial strains and vectors.
The bacterial strains and vectors used in this study are listed in Table 1. Since the purA promoter is lethal to E. coli in high-copy-number plasmids (12), strain KE94 was used for the propagation of plasmids pPAL1 and pPAL3. Plasmid pPAL4 contained an inactivated purA promoter and could therefore be amplified in DH5α F′. Strain XL2 Blue was used to propagate vectors containing the purR gene. The B. subtilis transformants were selected on either Penassay broth agar (Difco) or Luria-Bertani agar plates containing either chloroamphenicol or neomycin at 5 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| DE1 | Prototrophic revertant of W168 trpC2 | 1 |
| PAL1 | purA′-lucGR integrant | This study |
| PAL3 | purA′-lucGR integrant with a cis-control-site (−174 to −55) deletion | This study |
| PAL4 | purA′-lucGR integrant with a −10 promoter mutation | This study |
| NMW | purA′-lucGR integrant with purR::neo disruption | This study |
| N6H | purR6H Nmr ′yabJ integrant of PAL1 | This study |
| N3A | purR6H/D203A Nmr ′yabJ integrant of PAL1 | This study |
| N4A | purR6H/D204A Nmr ′yabJ integrant of PAL1 | This study |
| NPR | purR+ Nmr ′yabJ integrant of PAL1 | This study |
| E. coli | ||
| DH5α F′ | F′ φ80 dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdr17(rK− mK+) supE44 λ− thi-1 gyrA96 relA1 | 5 |
| XL2 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| KE94 | F′ hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK12 rpsL20 (str) xyl-5 mtl-1 supE44 λ− pcnB80 zad::Tn10; used to maintain low plasmid copy number | 11 |
| Plasmids | ||
| pPAL1 | Integration vector with transcriptional purA′-lucGR fusion | This study |
| pPAL3 | pPAL1 derivative with −174 to −55 cis control site replaced by Nmr gene | This study |
| pMW11 | pT7-7 derivative containing purR::neo disruption | 20 |
| pPAL4 | pPAL1 derivative with −10 promoter mutation | This study |
| pR6H | purR6H derivative of pT7-7 | 19 |
| pR6H3A | purR D203A mutation in pR6H | This study |
| pR6H4A | purR D204A mutation in pR6H | This study |
| pN6H2 | ′purR6H Nmr ′yabJ′ derivative of pGEM-7Zf(+) | This study |
| pN3A2 | ′purR6H/D203A Nmr ′yabJ′ derivative of pGEM-7Zf(+) | This study |
| pN4A2 | ′purR6H/D204A Nmr ′yabJ′ derivative of pGEM-7Zf(+) | This study |
| pNPR1 | ′purR Nmr ′yabJ′ derivative of pGEM-7Zf(+) | This study |
Construction and integration of a purA′-lucGR fusion.
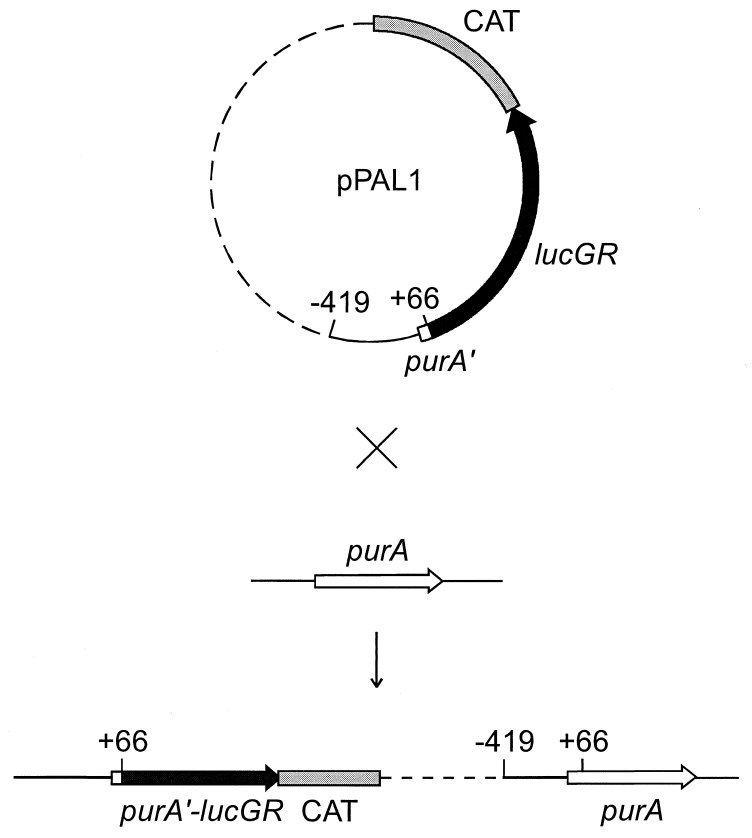
A purA′-lucGR fusion in plasmid pPAL1 was constructed in two steps. First, lacZ in pCATZ1 (2) was excised and replaced with the click beetle lucGR gene (21) from plasmid pCSS962 (9). Second, a fragment containing the 5′ end of purA from nucleotides −419 to +66 (relative to the start of transcription at +1) (12) was amplified by PCR from B. subtilis DE1 chromosomal DNA and inserted into the polylinker sequence immediately upstream of the lucGR gene. In this construction, codon 7 of purA is followed by 15 nucleotides of plasmid polylinker. Translation of purA terminates at a TGA in the polylinker. Translation of lucGR is expected to start at the initiator ATG which overlaps the purA TGA stop as follows: ATGA. The nucleotide sequence of the purA′-lucGR junction was verified. The resulting plasmid with the purA′-lucGR transcriptional fusion was named pPAL1. Plasmid pPAL1 was integrated into the chromosome of B. subtilis DE1 by homologous recombination as shown in Fig. 1.
FIG. 1.
Integration of pPAL1 into the B. subtilis chromosome. Vector sequences are shown by a dashed line, and purA upstream sequence is shown by a solid line.
Construction and integration of mutations in the purA control site and in the purR locus.
The purA gene contains an EcoRV site at nucleotide −174 (12). An MluI site at −55 was introduced into the purA control region in pPAL1 by site-directed mutagenesis. The resulting plasmid was digested with EcoRV and MluI, the 5′ cohesive end was filled by Klenow fragment, and a SmaI Nmr cartridge from pBEST 501 (6) was added to produce pPAL3. Plasmid pPAL3 thus contains a −174 to −55 cis-control-site deletion upstream of the purA′-lucGR fusion. pPAL3 was integrated into the chromosome of B. subtilis PAL1 by homologous recombination.
A mutation at the −10 site of purA was introduced into pPAL1 by a PCR method (10), resulting in a change of the −10 promoter element from TAAACT to TGCACT. The resulting plasmid, pPAL4, was integrated into the chromosome of B. subtilis DE1 in the same manner as pPAL1.
Disruption of purR was done by integration of plasmid pMW11, which contains a purR::neo disruption (20), into the chromosome of PAL1.
Mutations were constructed in the PurR PRPP binding site. Plasmid pR6H is a purR+ derivative in which six histidine codons are fused onto the 3′ end of purR (19). PurR mutations D203A and D204A were constructed by site-directed mutagenesis by using the method of Kunkel et al. (8). The His-tagged purR6H gene was excised from pR6H and transferred into M13mp18 for the mutagenesis. The mutations were verified by DNA sequencing. After mutagenesis, the mutant genes were returned to the pT7-7-derived vector to yield pR6H3A(D203A) and pR6H4A(D204A).
A series of integration vectors for the purR locus were constructed. First, the Nmr gene from pBEST 501 was inserted into the SmaI site of pGEM-7Zf(+) (Promega). Next, an HpaI-HindIII fragment of purR6H, purR6H(D203A), or purR6H(D204A), was inserted into the Ecl136II and HindIII sites. Finally, a fragment from codon 39 to codon 122 of yabJ downstream from purR was amplified by PCR and inserted into XhoI-SphI sites downstream from the Nmr gene to obtain plasmids pN6H2, pN3A2, and pN4A2, respectively. pNPR1 was prepared in the same way as pN6H2, pN3A2, and pN4A2, except that a wild-type HpaI-HindIII fragment from pMW10 (20) was used instead of His-tagged purR6H. pN6H2, pN3A2, pN4A2, and pNPR1 were integrated into the chromosome of PAL1 by a double crossover type of homologous recombination. These strains contain disrupted yabJ. The gene replacement was verified by Southern analysis and PCR.
PRPP inhibition of PurR binding to DNA.
Plasmid pR6H was used for overexpression and hyperproduction of PurR containing a C-terminal His tag (19). The two PurR PRPP binding site mutants, D203A and D204A, were hyperproduced from plasmids pR6H3A and pR6H4A, respectively, by the same procedure as for the wild type. The proteins were purified by using an Ni2+ affinity resin as described previously (19). Binding of the wild type and the two mutants to a DNA fragment containing the purA control region (−163 to +47) was determined as described previously (19). The 20-μl binding mixture contained 10 mM HEPES (pH 7.6), 100 mM KCl, 1 mM EDTA, 5 mM MgCl2, 10 fmol of 32P-labelled DNA fragment, 5 ng of purified protein, and varied concentrations of PRPP. Free DNA and PurR-DNA complexes were separated by electrophoresis on agarose gels and quantitated by counting radioactivity with a Packard instant imager.
Primer extension mapping.
The B. subtilis DE1 cells were grown in 200 ml of minimal medium (1) supplemented with 1 mM guanosine to an optical density of 650 nm (OD650) of 0.7. The cells were poured onto ice chilled to −20°C and collected by centrifugation for 10 min at 3,000 × g at 2°C. The total RNA was isolated by the chromosomal DNA isolation method for gram-positive bacteria (15) with slight modifications and additions as necessary. The cells were lysed with lysozyme (0.5 mg/ml) in the presence of 30 U of RNasin/ml (Promega) for 30 min, followed by proteinase K treatment in 1% sodium dodecyl sulfate at 55°C for 2 h. RNA was extracted once with phenol-chloroform and once with chloroform and precipitated with isopropanol. RNA pellet was dissolved in water containing 30 U of RNasin/ml. The RNA solution was treated with RNase-free DNase and extracted with phenol-chloroform. RNA was precipitated with isopropanol and dissolved in water containing 30 U of RNasin/ml. When possible, all solutions were treated with diethylpyrocarbonate. The yield of RNA was approximately 5 mg, with an A260/A280 ratio of 1.7.
The primer for primer extension was the same one that was used previously for mapping the transcription initiation site of B. subtilis purA (12). Approximately 450 ng of the fluorescent-labelled primer was annealed to 55 μg of total RNA in 15 μl of solution containing 10 mM Tris-HCl (pH 8.3), 150 mM KCl, and 1 mM EDTA, by maintaining the mixture at 65°C for 90 min and then letting it cool slowly to room temperature. Thirty microliters of solution containing 30 mM Tris-HCl (pH 8.3), 15 mM MgCl, 8 mM dithiothreitol, a mixture of the four deoxynucleoside triphosphates (1 mM each), 38 U of RNasin, and 18 U of avian myeloblastosis virus reverse transcriptase was added, and incubation was performed for 60 min at 42°C. The cDNA products were precipitated with ethanol and analyzed by the automatic ABI 377 sequencer by using GeneScan software (Perkin-Elmer). The length of the cDNA product was determined by comparing the retention time of the primer extension product with those of the products from the dideoxy sequencing reaction of pGEM-3Zf(+) (Promega) by using M13 reverse dye primer (Applied Biosystems) and dideoxy GTPs.
Luciferase assay.
Expression of purA was determined by measuring luciferase activity in strains with purA-lucGR integrated into the chromosomal purA gene. The cultures for luciferase assay were grown in minimal medium (1) supplemented either with 1 mM adenine, 1 mM guanosine, or both adenine and guanosine (1 mM each) or with no added purine compounds. The medium contained either chloramphenicol (strains PAL1 and PAL4) or neomycin (other integrants) at 5 μg/ml. The 5-ml overnight cultures were centrifuged, and the pellets were suspended in 50 ml of minimal medium containing the respective supplements. The cultures were grown to an OD650 of 0.5. From each culture, a 1-ml sample was taken, centrifuged, and resuspended in 5 ml of minimal medium containing the respective supplements. The 5-ml cultures were grown for 2 h. Samples of 1 ml were taken, and 100 μl of solution A (1 M K2HPO4 and 20 mM EDTA [pH 7.8]) was added to each sample. The final samples were frozen at −70°C. For the measurement of luciferase activity, the samples were thawed and centrifuged, and the supernatant was carefully removed. The cells were resuspended in 50 μl of the supernatant solution. One volume of cell lysis buffer 1 (Bio-Orbit) containing 2 mg of lysozyme/ml and 2 mg of bovine serum albumin (BSA)/ml was added, and the mixture was incubated at room temperature for 5 min. The lysate was centrifuged, and a 50-μl aliquot of the supernatant was sampled. One hundred microliters of Luciferin reagent and 100 μl of ATP reagent of GenGlow-100 kit (Bio-Orbit) were added to the extract, and the maximum light output was measured by using an LKB 1250 luminometer. The values were compared with a standard curve made by assaying known amounts of firefly luciferase standard in dilution buffer (1 mg of BSA/ml, 0.5× cell lysis buffer 1, 0.45× minimal medium, and 0.05× solution A). When necessary, the samples were diluted with dilution buffer. The results are expressed as attomoles of luciferase per 108 CFU.
RESULTS
Integration of a purA′-lucGR fusion and purR locus mutations into the B. subtilis chromosome.
A gene encoding luciferase was used as a reporter to monitor purA expression. To obtain a single-copy purA′-lucGR integrant, plasmid pPAL1 was recombined into the B. subtilis chromosome in strain DE1, as diagrammed in Fig. 1, to give strain PAL1. Strain PAL1 is purA+. Two purA′-lucGR derivatives, one with a promoter mutation (PAL4) and another with a cis-control-site deletion (PAL3), were constructed in a like manner. The integrations were verified by PCR amplification and DNA sequencing. Like strain PAL1, the two purA′-lucGR derivatives are purA+.
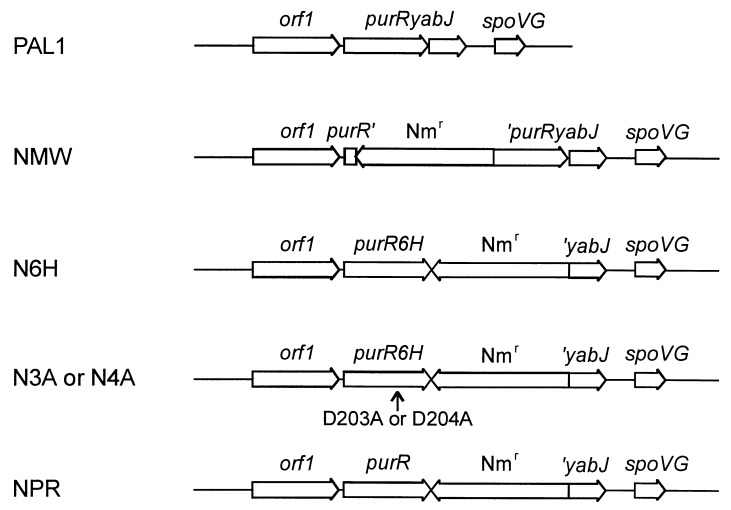
In order to evaluate regulation by purR, a series of mutations was constructed in the purR region. The purR locus in these strains, obtained by recombining purR plasmids into PAL1 (purA′-lucGR), is shown in Fig. 2. NMW contains a purR::neo disruption. Strain N6H (purR+) contains a functional repressor with a C-terminal His tag. In strain N6H (purR6H, ′yabJ), the downstream yabJ gene was disrupted. Mutations of Asp 203 or Asp 204 were incorporated into purR6H in N3A or N4A.
FIG. 2.
Gene arrangement of wild-type (PAL1) and various purR integrants. purR6H is a purR+ derivative with six His codons at the 3′ end, followed by two translation stop codons. Truncation of the yabJ or the purR yabJ operon is marked by an apostrophe. The site of mutations D203A and D204A is marked by an arrow.
Regulation of purA.
The level of purA expression was determined by using the lucGR gene as a reporter. In the purA′-lucGR transcriptional fusion, a DNA fragment containing nucleotides −419 to +66 of the purA operon was ligated immediately upstream from the lucGR gene. A basal luciferase activity of 498 amol per 108 CFU was obtained from strain PAL1 (purA′-lucGR) grown in medium without added purines (Table 2). For a luciferase control, a −10 promoter mutation was incorporated into purA′-lucGR in strain PAL4. The −10 promoter mutation abolished luciferase activity (data not shown), indicating that all of the activity was derived from purA′-lucGR expression. To examine regulation by PurR, purR and OpurA operator mutations were incorporated into the purA′-lucGR reporter strain. The levels of luciferase activity in these strains are given in Table 2. In the purR+O+purA wild type, there was 10-fold repression of purA expression by adenine and a 4.5-fold activation by guanosine. When adenine and guanosine were combined, repression by adenine overrode the activation by guanosine.
TABLE 2.
Effect of purR disruption or deletion of purA 5′-flanking region on transcription of purA
| B. subtilis strain | Operatorb | purR | Mean
luciferase activity ± SD for culture grown as
indicateda
|
|||
|---|---|---|---|---|---|---|
| No purines | Ade | Guo | Ade + Guo | |||
| PAL1 | + | + | 498 ± 153 | 48 ± 5.0 | 2,278 ± 428 | 34 ± 1.4 |
| NMW | + | − | 5,382 ± 817 | 7,514 ± 1,405 | 5,870 ± 2,114 | 7,611 ± 1,128 |
| PAL3 | Δ | + | 5,525 ± 915 | 8,852 ± 1,328 | 5,667 ± 548 | 8,275 ± 1,190 |
Values were obtained from three independent experiments. Ade, adenine; Guo, guanosine.
The cis control region is either wild type (+) or deleted (Δ). Bacteria were grown as described in Materials and Methods.
In the strains with a purR disruption or an operator deletion (nucleotides −174 to −55), regulation of purA was lost. In the regulatory mutants, basal expression was about 10-fold higher than in the purR+O+purA wild type, reflecting release from repression by the endogenous pool of adenine or adenine nucleotides. Basal expression was not repressed by the addition of adenine to cells or upregulated by the addition of guanosine in these mutants. These results support the view that both repression and upregulation are a consequence of the PurR interaction with the purA control site. Repression of PurA (adenylosuccinate synthetase) by adenine and upregulation by guanosine were reported previously, although the regulatory elements were not identified (17).
Primer extension mapping.
To rule out the possibility that activation by guanosine in the wild-type cells is due to a shift of the transcription initiation site, the 5′ end of the purA transcript in DE1 cells grown with excess guanosine was determined by primer extension mapping by using the same primer previously used for mapping the 5′ end of purA mRNA (12). The length of primer extension product was 117 nucleotides (data not shown), the same length as that previously determined with the cells grown in the absence of purines.
Role of yabJ in the regulation of purA.
To test the significance of the yabJ gene downstream from purR for regulation of purA, yabJ was disrupted to obtain strain N6H. The data in Table 3 show that regulation by purR6H in strain N6H was perturbed and is not similar to that by purR in PAL1 (Table 2). Repression of basal expression by adenine was abolished in N6H (purR6H ′yabJ), although the upregulation by guanosine was retained.
TABLE 3.
Regulation of purA in the PRPP binding mutants
| B. subtilis strain | purR | yabJ | Mean
luciferase activity ± SD for culture grown as
indicateda
|
|||
|---|---|---|---|---|---|---|
| No purines | Ade | Guo | Ade + Guo | |||
| NPR | purR | − | 1,186 ± 98 | 822 ± 134 | 3,415 ± 464 | 1,041 ± 196 |
| N6H | purR6H | − | 1,106 ± 221 | 1,357 ± 145 | 5,334 ± 628 | 2,173 ± 370 |
| N3A | purR6H/D203A | − | 726 ± 148 | 984 ± 211 | 932 ± 154 | 1,063 ± 461 |
| N4A | purR6H/D204A | − | 399 ± 74 | 799 ± 146 | 810 ± 59 | 820 ± 35 |
Values were obtained from three independent experiments. Ade, adenine; Guo, guanosine.
In order to verify that the loss of purA repression in N6H (purR6H ′yabJ purA′-lucGR) was due to the disruption of yabJ rather than to the addition of six His codons in purR, an integrant with a disrupted yabJ but an intact purR was constructed. This strain, NPR, is diagrammed in Fig. 2. Data in Table 3 show that regulation in strain NPR was similar to that in N6H, the purR6H ′yabJ integrant. Although repression by adenine was lost, high-level constitutive expression, as in strain NMW, was not seen, and upregulation by guanosine was retained. This result thus points to a role for yabJ in PurR repression by adenine.
PRPP inhibition of PurR binding to DNA.
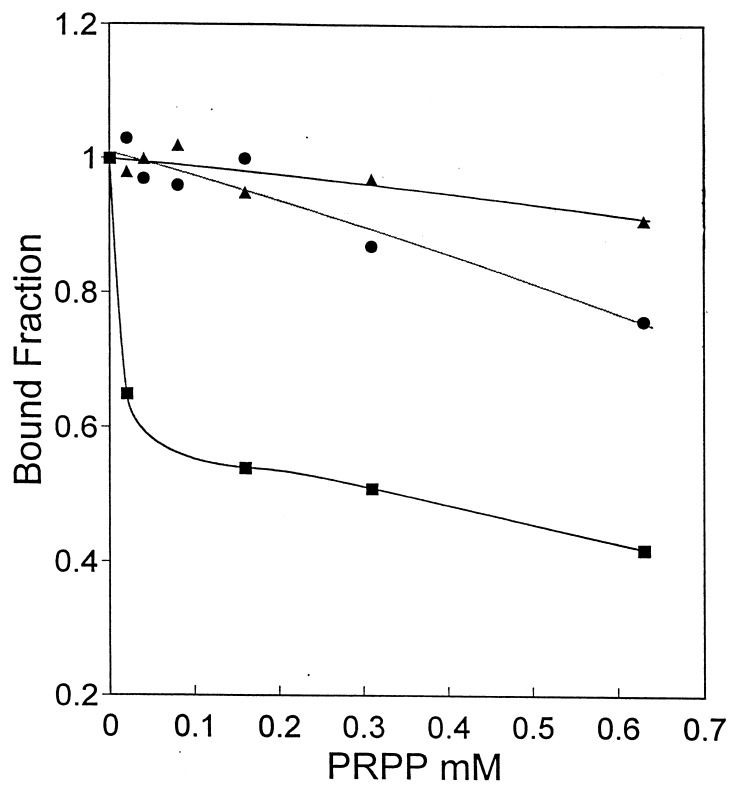
A Kd of 7.9 μM was determined previously for the interaction of PurR with purA-control-site DNA (19). Given that PRPP inhibits the binding of PurR to the pur operon control site (20), a similar inhibition by PRPP was expected for PurR binding to purA. The data in Fig. 3 show PRPP inhibition of the PurR-purA operator DNA interaction. For 50% inhibition of binding, approximately 30 μM PRPP was required, but complete inhibition was not attained even at 2.5 mM PRPP (data not shown).
FIG. 3.
PRPP inhibition of PurR binding to the purA control region DNA. The effect of PRPP on binding is shown for wild-type PurR (■) and two mutants, PurR D203A (●) and D204A (▴). The fraction of DNA bound to PurR in the absence of PRPP (0.82 for wild type, 0.81 for D203A, and 0.89 for D204A) was normalized to 1.0. The apparent Kd values for binding to operator DNA for the wild type and the mutants were similar.
PurR contains a PRPP binding sequence motif that is conserved in all type I phosphoribosyltransferases (20). Mutations were introduced into the two conserved aspartates of the PRPP binding motif to further evaluate PRPP’s role as an effector that modulates PurR binding to DNA. The corresponding aspartate side chains in glutamine PRPP amidotransferase have essential interactions with the C2 and C3 hydroxyl groups of PRPP (7). The two mutant proteins PurR D203A and PurR D204A each bound to purA operator DNA with an affinity similar to that of the wild type (data not shown). However, the binding of both PurR mutants was relatively insensitive to inhibition by PRPP (Fig. 3). Binding of the D204A repressor to DNA was more resistant to inhibition by PRPP than the D203A PurR. Loss of PRPP effector function likely results from defective binding to PurR, although PRPP binding was not determined directly.
Regulation of purA by PurR PRPP binding mutants.
The PurR D203A and D204A mutations were incorporated into the chromosome of strain PAL1 (purA′-lucGR) in order to determine the in vivo role of PRPP as a PurR effector. The two Asp mutants purR6H/D203A and purR6H/D204A, encoding repressors having a C-terminal His tag, were integrated into PAL1 to give yabJ mutant strains N3A (purR6H/D203A purA′-lucGR) and N4A (purR6H/D204A purA′-lucGR) (Fig. 2). The purR6H gene encodes a repressor with a His tag identical to that used for the in vitro PurR-purA operator DNA binding experiments (19) (Fig. 3). The results in Table 3 show that in the two PurR ′yabJ Asp mutants, strains N3A and N4A, repression by adenine was lost, as in the yabJ mutants with His-tagged or wild-type purR. In addition, the upregulation of guanosine was completely lost in both of the PurR PRPP binding mutants. It thus appears that the two regulatory events, repression by adenine and upregulation by guanosine, were separated in strain N6H. YabJ was required for repression by adenine but not for upregulation by guanosine. The PRPP effector site was necessary for upregulation by guanosine.
DISCUSSION
It is not surprising that regulation of purA expression should contribute to controlling de novo AMP synthesis and to balancing the production of AMP with GMP. The data in Table 2 establish that purA expression, monitored by a luciferase reporter gene, is repressed by the addition of adenine to cells and is upregulated by added guanosine. Mutant analysis indicates that both repression and upregulation are dependent upon the interaction of PurR with the purA control region. Direct in vitro evidence for this binding has been reported previously (19). The data are consistent with the view that transcriptional regulation of purA by PurR contributes to controlling the production of AMP and to maintaining the balanced synthesis of adenine and guanine nucleotides.
The data reported in Table 3 have brought to light an important new aspect of PurR function. Repression of purA by PurR depends upon yabJ, an overlapping downstream gene. This gene encodes a member of a protein family with unknown function. Data in Tables 2 and 3 show that yabJ+ is needed for PurR-mediated purA repression by adenine although not for upregulation by guanosine. The yabJ mutation thus appears to uncouple the repression and activation functions of PurR.
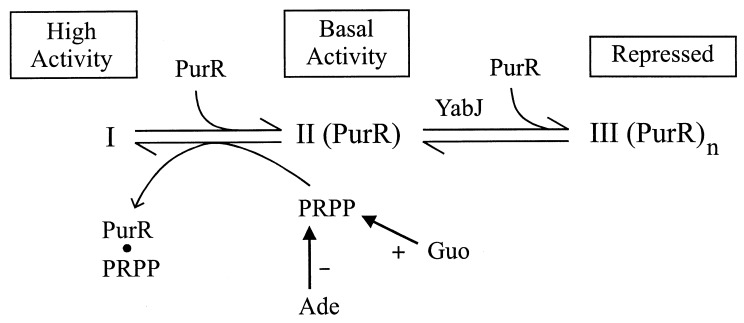
How can PurR mediate repression by adenine and upregulation by guanosine, and what role might yabJ have? A working model shown in Fig. 4 explains these results. First, we consider three states of the purA control region as follows: (i) little or no PurR bound, giving maximal purA expression; (ii) partial occupation by PurR, giving basal expression; and (iii) saturation by PurR, resulting in full repression. purA expression in strains NMW (purR::neo purA′-lucGR) and PAL3 (OpurAΔ1 purA′-lucGR) (Table 2) reflects the unoccupied state I control region that leads to high constitutive expression. In these mutants, there is no binding of PurR to the control region. State II, partial occupancy by PurR, giving basal expression, is seen in the wild-type grown without purines (Table 2). Partial occupancy of the purA control region results from the interplay between the endogenous purine compounds and the PRPP pool. PRPP, which inhibits PurR binding to the purA control region (Fig. 3), has been proposed to be the key regulatory molecule for de novo purine nucleotide synthesis in B. subtilis based on its exclusive ability to influence PurR-DNA binding in vitro (20). Adenine lowers the PRPP concentration in B. subtilis (17), thus allowing PurR to bind to the control region and shift the I⇆II equilibrium toward II. Guanosine, on the other hand, increases the cellular PRPP pool (17), promoting a shift toward state I. According to this model, YabJ is needed for the state II-to-state III conversion, in which additional PurR is bound and the control site is saturated with PurR. This YabJ-dependent step may correspond to the high PurR/control site binding stoichiometry detected in vitro at an elevated PurR concentration (19). Alternatively, YabJ could inhibit the ability of endogenous adenine to lower the intracellular PRPP level. In contrast to the state II-state III equilibrium, YabJ is not required for the state II-to-state I shift promoted by guanosine.
FIG. 4.
Hypothetical model explaining roles of YabJ and PRPP. PRPP is required to inhibit binding of PurR, leading to state I. The PRPP concentration in the cell is increased by Guo and decreased by Ade. Basal activity results from partial occupancy of the control region with PurR. Repression requires the binding of additional molecules of PurR (state III). YabJ is essential for this additional PurR binding, perhaps as a PurR-YabJ complex. The possible association of YabJ with PurR in state III is not addressed.
This model explains the loss of repression by adenine in the yabJ mutants N6H and NPR (Table 3). These strains exhibit basal expression when grown with excess adenine. Guanosine, however, increases the PRPP pool and shifts the equilibrium toward high state I activity. A mutation in the PurR PRPP site abolishes this PRPP-mediated shift in the yabJ mutant strains N3A and N4A (Table 3).
In an earlier report from one of our laboratories (20), Weng et al. stated that a yabJ disruption had no effect on the expression or regulation of purR or the pur operon. Unfortunately, this mutant was lost, and the result cannot be replicated. We assume that the earlier result was incorrect and that yabJ has a similar role in the regulation of purA, the pur operon, and purR.
What is YabJ and how does it work? The deduced amino acid sequence of YabJ (125 residues) is homologous to a group of 35 other proteins of unknown function from archaea, procaryotes, and eucaryotes. Some organisms (e.g., E. coli and yeast) encode several YabJ paralogs, whereas some have only one YabJ ortholog. In pairwise comparisons of these 35 sequences with YabJ, there is an identity of 21 to 53% over a span of 117 to 125 amino acids. A multiple alignment of all the sequences does not reveal any invariant residues, although approximately 10 conserved amino acids can be identified. YabJ belongs to the YER057c/YjgF protein family of unknown function (PROSITE accession no. PS01094). The family is defined by a conserved signature motif located at the C terminus of these proteins, consisting of the following amino acids: P-[AT]-R-[SA]-X-[LIVMY]-X2-[AK]-X-L-P-X4-[LIVM]-E. The consensus pattern is between amino acids 100 and 117 in YabJ. The YER057c/YjgF motif is conserved in all 36 YabJ homologs presently in sequence databases, although the degree of conservation varies to some extent.
The homologs from rat and human have 45 and 44% identity with YabJ, respectively. These proteins have been shown to inhibit cell-free protein synthesis at a high concentration (14, 18). Samuel et al. (16) found that the YabJ homolog Hrp12 from mouse had some similarity to heat shock proteins Hsp70 and Hsp90. They also noticed that the purified mouse Hrp12 could be phosphorylated in vitro with protein kinase C. Melloni et al. (13) reported that the bovine and goat YabJ homologs had the capacity to activate calpains. An aldR gene product from Lactococcus lactis (4), which is 52% identical to B. subtilis YabJ, has been suggested to interfere with branched-chain amino acid synthesis, although aldR does not encode an isoleucine biosynthetic enzyme. Mutations that allow thiamine synthesis in the absence of both PurF (glutamine phosphoribosylpyrophosphate amidotransferase) and the pentose phosphate pathway in Salmonella typhimurium have been localized in a gene encoding a YabJ homolog, YjgF. Moreover, in these mutants the isoleucine biosynthetic pathway seems to be affected as in L. lactis (3).
A common molecular function for YabJ and its 35 homologs is implied by their high degree of sequence identity and similar sizes. However, no certain biological function or common molecular function has emerged from studies of YabJ homologs. In some cases, as in L. lactis aldR and in the present study, the target seems to be related to the operon containing the gene for the YabJ homolog. Our work adds a new dimension to the question of the function of a YabJ homolog, which for the first time, can be located to a specific target. The effect of the disruption of YabJ on the regulation of transcription has been established by using a promoter-reporter system. Given the complex PurR-DNA interaction that has been studied in vitro (19), several possibilities exist for YabJ function, all of which require adenine dependence and guanosine independence in vivo. One possibility is that YabJ associates with PurR and promotes an interaction with DNA that is more stable than the interaction that is possible with PurR alone. A stoichiometry of two or six PurR dimers per pur operon was reported in studies with a DNA control-site fragment (19). Perhaps interaction of PurR and YabJ favors the higher binding stoichiometry and a more stable PurR-DNA interaction. Another possibility is that YabJ may stabilize PurR and increase its half-life. It has been speculated that some YabJ homologs may have a chaperone-like function (13). To test this hypothesis, it will be necessary to determine the intracellular level of PurR in yabJ+ and yabJ strains.
The results presented here provide important new information on the YER057c/YjgF protein family of unknown function. We have shown by using a promoter-reporter system that B. subtilis YabJ, a member of this family, affects the purine repressor-mediated regulation of purA. The established promoter-reporter system will be valuable for future in vivo studies of YabJ. In addition, a collaborative study of the three-dimensional structure of YabJ (19a) provides important clues about the function of YER057c/YjgF family members.
ACKNOWLEDGMENTS
We thank Janet Smith for critical reading of the manuscript.
This work was supported by a grant from the Finnish Ministry of Education, Academy of Finland (to P.M.) and by U.S. Public Health Service grant GM24658 (to H.Z.).
REFERENCES
- 1.Ebbole D J, Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilisencoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987;262:8274–8287. [PubMed] [Google Scholar]
- 2.Ebbole D J, Zalkin H. Bacillus subtilis puroperon expression and regulation. J Bacteriol. 1989;171:2136–2141. doi: 10.1128/jb.171.4.2136-2141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enos-Berlage J L, Langendorf M J, Downs D M. Complex metabolic phenotypes caused by mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J Bacteriol. 1998;180:6519–6528. doi: 10.1128/jb.180.24.6519-6528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goupil-Feuillerat N, Cocaign-Bousquet M, Godon J-J, Ehrlich S D, Renault P. Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol. 1997;179:6285–6293. doi: 10.1128/jb.179.20.6285-6293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 6.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilischromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krahn J M, Kim J H, Burns M R, Parry M R, Zalkin H, Smith J L. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry. 1997;36:11061–11068. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 9.Lampinen J, Koivisto L, Wahlsten M, Mäntsälä P, Karp M. Expression of luciferase genes from different origins in Bacillus subtilis. Mol Gen Genet. 1992;232:498–504. doi: 10.1007/BF00266255. [DOI] [PubMed] [Google Scholar]
- 10.Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 11.Lopilato J, Bortner S, Beckwith J. Mutation in a new chromosomal gene of Escherichia coli K-12, pcnB, reduces plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 12.Mäntsälä P, Zalkin H. Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP. J Bacteriol. 1992;174:1883–1890. doi: 10.1128/jb.174.6.1883-1890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melloni E, Michetti M, Salamino F, Pontremoli S. Molecular and functional properties of a calpain activator protein specific for μ-isoforms. J Biol Chem. 1998;273:12827–12831. doi: 10.1074/jbc.273.21.12827. [DOI] [PubMed] [Google Scholar]
- 14.Oka T, Tsuji H, Noda C, Sakai K, Hong Y, Suzuki I, Muñoz S, Natori Y. Isolation and characterization of a novel perchloric acid-soluble protein inhibiting cell-free protein synthesis. J Biol Chem. 1995;270:30060–30067. doi: 10.1074/jbc.270.50.30060. [DOI] [PubMed] [Google Scholar]
- 15.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 16.Samuel S J, Tzung S, Cohen S A. Hrp12, a novel heat-responsive, tissue-specific, phosphorylated protein isolated from mouse liver. Hepatology. 1997;25:1213–1222. doi: 10.1002/hep.510250525. [DOI] [PubMed] [Google Scholar]
- 17.Saxild H H, Nygaard P. Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools. J Gen Microbiol. 1991;137:2387–2394. doi: 10.1099/00221287-137-10-2387. [DOI] [PubMed] [Google Scholar]
- 18.Schmiedeknecht G, Kerkhoff C, Orsó E, Stohr J, Aslanidis C, Nagy G M, Knuechel R, Schmitz G. Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur J Biochem. 1996;242:339–351. doi: 10.1111/j.1432-1033.1996.0339r.x. [DOI] [PubMed] [Google Scholar]
- 19.Shin B S, Stein A, Zalkin H. Interaction of Bacillus subtilispurine repressor with DNA. J Bacteriol. 1997;179:7394–7402. doi: 10.1128/jb.179.23.7394-7402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Sinha, S., and J. L. Smith. Personal communication.
- 20.Weng M, Nagy P, Zalkin H. Identification of the Bacillus subtilis puroperon repressor. Proc Natl Acad Sci USA. 1995;92:7455–7459. doi: 10.1073/pnas.92.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood K V, Lam Y A, Seliger H H, McElroy W D. Complementary DNA coding beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- 22.Zalkin H. De novo purine nucleotide synthesis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 335–341. [Google Scholar]
- 23.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 561–579. [Google Scholar]