Abstract
Gene therapy would benefit from the effective editing of targeted cells with CRISPR-Cas9 tools. However, it is difficult to precisely assess the editing performance in vivo because the tissues contain many non-targeted cells, which is one of the major barriers to clinical translation. Here, in the Atoh1-GFP;Kcnq4+/G229D mice, recapitulating a novel mutation we identified in a hereditary hearing loss pedigree, the high-efficiency editing of CRISPR-Cas9 in hair cells (34.10% on average) was precisely detected by sorting out labeled cells compared with only 1.45% efficiency in the whole cochlear tissue. After injection of the developed AAV_SaCas9-KKH_sgRNA agents, the Kcnq4+/G229D mice showed significantly lower auditory brainstem response (ABR) and distortion product otoacoustic emission (DPOAE) thresholds, shorter ABR wave I latencies, higher ABR wave I amplitudes, increased number of surviving outer hair cells (OHCs), and more hyperpolarized resting membrane potentials of OHCs. These findings provide an innovative approach to accurately assess the underestimated editing efficiency of CRISPR-Cas9 in vivo and offer a promising strategy for the treatment of KCNQ4-related deafness.
Keywords: MT: RNA/DNA editing, gene therapy, CRISPR-Cas9, hearing loss, KCNQ4, DFNA2, AAV, editing efficiency, hair cells
Graphical abstract

This study precisely assessed the editing efficiency of CRISPR-Cas9 in hair cells in vivo and improved auditory function in a mouse model of autosomal dominant deafness.
Introduction
CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9) technology has been used in preclinical research for the treatments of many somatic cell diseases, such as hearing loss, Duchenne muscular dystrophy, and liver disease, etc.1, 2, 3, 4, 5, 6, 7 However, the tissues, including targeted and non-targeted cells, are generally used to extract cellular DNA for detection of editing efficiency, resulting in an inability to accurately assess the number of edited cells.1, 2, 3 In particular, the targeted hair cells in the cochlea accounted for approximately 1.5%, and the editing efficiency of cochlear tissue did not exceed 5% with the reported percentages of 0.60%, 0.92%, 2.20%, and 4.05%.4, 5, 6, 7 However, the hearing of treated mice was significantly improved, suggesting an inconsistency between editing efficiency and phenotypic restoration.4, 5, 6, 7 Therefore, there is still an urgent need to accurately evaluate the editing efficiency of CRISPR-Cas9 technology in the cells that are expected to be treated. The accurate measurement of editing efficiency is an important prerequisite for the evaluation of therapeutic efficacy and is one of the key factors affecting the translation of research findings into clinical practice.8
Hearing loss is the most common genetic sensory deficit in humans.9,10 More than 120 genes are known to be involved in hereditary deafness,11,12 but effective treatments for hereditary deafness are still limited in clinical practice. In situ gene therapy is the ideal intervention strategy for genetic disorders and is also beneficial to patients. Hearing loss is one of the most promising diseases to be cured by gene therapy owing to the small number of target cells in the cochlea and the availability of the therapeutic agent to be delivered locally to the inner ear. Several preclinical studies for the adeno-associated virus (AAV)-mediated gene replacement have been successfully performed in mouse models of recessive deafness to prevent hearing impairment.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 However, the gene replacement strategy is not suitable for the treatment of genetic mutations with dominant-negative effects.23 Fortunately, CRISPR-Cas9 technology provides promising approaches for the treatment of both hereditary hearing loss and acquired hearing loss.4, 5, 6, 7,24, 25, 26
KCNQ4 is a voltage-gated potassium channel protein, in which the pathogenic gene mutations could cause DFNA2 (deafness, autosomal dominant 2A; OMIM: 600101) displaying progressive hearing loss. It is reported that the pathogenic variants in KCNQ4 account for approximately 9.5% of autosomal dominant sensorineural hearing loss.27 Currently, over 30 KCNQ4 pathogenic point mutations have been implicated in DFNA2 patients.28, 29, 30 The degeneration of outer hair cells (OHCs) and hearing loss are attributed to the abolishment of the native KCNQ4-mediated K+ current by the dominant-negative effect of mutant subunit in heterozygotes with the missense mutation.30,31 In previous work, we identified a novel mutation KCNQ4: c.683G>A, p.G228D in the pedigree of DFNA2 patients and generated a novel mouse model to simulate progressive hearing loss in patients.32 Kharkovet et al. generated a knock-out mouse model with a deletion of Kcnq4 exons 6–8, and no hearing impairment was observed in Kcnq4+/− animals, which suggests that the disruption of KCNQ4 mutant allele is a potential strategy to treat hearing loss caused by KCNQ4 heterozygous pathogenic variants.31 The hair cells express GFP specifically in the Atoh1-GFP mice, where a portion of the Atoh1 enhancer sequence was randomly integrated into the mouse genome, directing the expression of the reporter gene GFP.33 It suggests the Atoh1-GFP mice could be utilized to precisely detect the editing efficiency of CRISPR-Cas9 technology in vivo.
In this study, we successfully established an innovative approach based on the Atoh1-GFP mice to accurately detect the editing efficiency of CRISPR-Cas9 technology in the hair cells. The hearing of Kcnq4+/G229D mice was also significantly improved after treatment with AAV_SaCas9-KKH_sgRNA. Our findings provide a direction for precise evaluation of editing efficiency of CRISPR-Cas9 in vivo and develop a potential therapeutic strategy for KCNQ4-related deafness, which will facilitate the application of CRISPR-Cas9 technology in the treatment of human diseases.
Results
Targeting the Kcnq4G229D allele with effective and specific SaCas9-KKH_sgRNAs
KCNQ4 (p.G228D) is a novel pathogenic mutation identified in a large Chinese family with DFNA2 by us, and we constructed a mouse model with the humanized homologous mutation (KCNQ4 p.G229D) to explore its pathogenic mechanism (Figure S1). The Kcnq4+/G229D mice exhibited mid-frequency and high-frequency hearing loss at 4 weeks and deteriorated to a severe-to-profound hearing loss at all frequencies at 12 weeks. The progressive degeneration of OHCs was observed from 4 to 12 weeks. The depolarized resting membrane potentials and reduced K+ currents in OHCs could induce the degeneration of hair cells, resulting in hearing loss in KCNQ4 p.G229D mutant mice.32
To investigate whether the hearing loss in Kcnq4+/G229D mice could be restored by CRISPR-Cas9 technology, efficient and specific sgRNAs against the Kcnq4G229D allele were screened. The SaCas9-KKH system was chosen because it could be packaged into a single AAV vector and could recognize a simple protospacer adjacent motif (PAM) sequence, 5′-NNNRRT-3′, allowing the design of more suitable sgRNAs.34
We designed four sgRNAs covering the pathogenic mutation (Figure 1A) and tested the editing efficiency of sgRNAs by transfecting plasmids expressing Cas9 and sgRNA into skin fibroblasts from Kcnq4+/+, Kcnq4+/G229D, and Kcnq4G229D/G229D neonatal mice (Figure 1B). The editing efficiencies were in the order of g3 (28.31% ± 3.76%), g1 (10.65% ± 2.79%), g2 (2.89% ± 1.33%) and g4 (0.70% ± 0.41%) in Kcnq4G229D/G229D fibroblasts, whereas no obvious editing efficiency was observed in Kcnq4+/+ fibroblasts (Figure 1C; Table S1). The editing efficiency of g3 was also the highest in Kcnq4+/G229D fibroblasts (Figures 1D and 1E). The result suggests that the terminal base “T” on the PAM sequence is critical for gRNA specificity, which is consistent with our previous study.5,6
Figure 1.
Targeting the Kcnq4G229D allele with the CRISPR-SaCas9-KKH system
(A) Design of SaCas9-KKH sgRNAs. The Kcnq4G229D mutation site is highlighted in red. The nucleotides of the PAM site are marked in green. (B) The schematic diagram for in vitro screening of sgRNAs in skin fibroblasts. (C) Indel frequencies of g1, g2, g3, and g4 in Kcnq4+/+ and Kcnq4G229D/G229D fibroblasts determined by next-generation sequencing. (D) Indel frequencies of g1, g2, g3, and g4 in Kcnq4+/G229D fibroblasts. (E) The ratio of indel frequency between mutant allele and wild-type allele calculated from (D). (F) Indel profiles from SaCas9-KKH_g3-transfected Kcnq4+/G229D fibroblasts. Negative numbers represent deletions, and positive numbers represent insertions. Sequences without indels (value = 0) are not included in the chart. (G) The percentage of indels causing in-frame or frameshift mutations in SaCas9-KKH_g3-transfected Kcnq4+/G229D fibroblasts. Dots represent individual biological replicates (n = 3) in (C)–(E). Data are shown as mean ± SD.
In Kcnq4G229D/G229D fibroblasts, the indel profiles showed that the majority of SaCas9-KKH_sgRNA-induced variants were deletions, and the ratios of frameshift mutation were 71.03%, 96.8%, 81.8%, and 72.2% for g1, g2, g3, and g4, respectively (Figures S2A and S2B). For g3, the deletion mutations were also the major indel events in Kcnq4+/G229D fibroblasts, where the percentage of frameshift mutations was 89.30% (Figures 1F and 1G). The most common mutations were −1 bp (2.44%, 12,323 reads), −5 bp (2.06%, 10,410 reads), −1 bp (1.79%, 9,015 reads), −1 bp (1.63%, 8,217 reads), and +1 bp (1.23%, 6,211 reads) frameshifts for g3 in Kcnq4G229D/G229D fibroblasts (Figure S2C). The results suggest that SaCas9-KKH_g3 performs best in vitro, which is used in subsequent experiments in vivo.
Precise and effective editing by the CRISPR-Cas9 system in hair cells
We made use of the KCNQ4 p.G229D murine model to accurately assess the editing efficiency of CRISPR-Cas9, because of the primary expression of KCNQ4 in hair cells.31 SaCas9-KKH_g3 was packaged into AAV-PHP.eB, which could transduce hair cells with high efficiency in our previous work.35 The AAV_SaCas9-KKH_g3 system was injected into the inner ears of Kcnq4+/G229D and Kcnq4G229D/G229D mice at postnatal days 1–2 (P1–P2), the basilar membranes were isolated at P5 and incubated ex vivo for 9 days (9 DIV [days in vitro]), followed by second-generation sequencing to analyze editing efficiencies (Figures 2A and 2B).
Figure 2.
Genome editing with the AAV_SaCas9-KKH_g3 system in mice
(A) The schematic diagram for the detection of precise editing efficiency in the hair cells. (B) Experimental overview for in vivo and ex vivo studies. (C) Comparison of the indel frequency between unsorted and sorted hair cells in Kcnq4+/G229D mice. (D) The percentage of indels causing in-frame or frameshift mutations after AAV_SaCas9-KKH_g3 injection into Kcnq4+/G229D mice. (E) Indel frequencies of the whole cochlear tissues in Kcnq4G229D/G229D mice at 2, 8, and 13 weeks after AAV_SaCas9-KKH_g3 injection, respectively. ∗p ≤ 0.05; ns, no significance; one-way ANOVA. (F) The presentation of most abundant reads in a Kcnq4G229D/G229D mouse after AAV_SaCas9-KKH_g3 injection at 13 weeks. Dashed lines, blue bases, green bases, and arrow indicate deletions, insertions, PAM sequence, and sgRNA sequence, respectively. Dots represent individual biological replicates (n = 3–6) in (C) and (E). Data are shown as mean ± SD.
Kcnq4G229D/G229D mice were mated with Atoh1-GFP mice to obtain Atoh1-GFP;Kcnq4+/G229D mice, in which hair cells were specifically labeled with GFP.33 AAV_SaCas9-KKH_g3 was injected into the inner ears of Atoh1-GFP;Kcnq4+/G229D mice, and subsequently the accurate editing efficiency was analyzed in the sorted GFP-positive hair cells (Figures 2A and S3). 34.10% ± 20.46% (up to 54.24%) indel frequency was observed for the Kcnq4G229D allele in GFP-sorted hair cells compared with only 1.45% ± 1.30% frequency in the whole cochlear tissue, and no obvious indels were observed in the uninjected group (Figure 2C; Table S1). For the wild-type allele in Kcnq4+/G229D mice, no obvious indels were observed in the sorted, unsorted, injected, and uninjected groups (Figure 2C; Table S1). These results indicate that a method is successfully established to accurately test the editing efficiency of the CRISPR-Cas9 system in hair cells and demonstrate that the hair cells could be edited by CRISPR-Cas9 with high efficiency in vivo.
The indel profile showed that the majority of AAV_SaCas9-KKH_g3-induced variants in the Kcnq4G229D allele were deletions, and the frameshift mutation was 70.62% for Kcnq4+/G229D (Figures 2D and S4). We noticed that there were a few types of indel mutations with −1 bp, −2 bp, and −3 bp mutations in the different samples, accounting for the major proportion of mutations (Figure S4), and the editing efficiencies displayed a scattered distribution between the different samples (from 14.44% to 54.24%; Figure 2C).
We observed the long-term editing efficiencies of g3 in the whole cochlear tissues at 2, 8, and 13 weeks, considering the difficulty to obtain sufficient hair cells from the adult mice. The Kcnq4G229D/G229D mice were chosen because the Kcnq4G229D allele in Kcnq4+/G229D mice was not pure and needed to be distinguished from the wild-type allele by the mutation site, and the editing efficiency would be underestimated if the point mutation was edited in Kcnq4+/G229D mice (e.g., the −40 bp deletion in Figure S2C). We observed a positive correlation between editing efficiencies and the age of mice, with 0.82% ± 0.41%, 2.82% ± 2.15%, and 7.06% ± 3.56% editing efficiencies at 2, 8, and 13 weeks, respectively (Figure 2E; Table S1). At week 13, the predominately edited indel types were still deletion mutations with the most common indels, for example −1 bp (1.47%), −2 bp (0.87%), and −1 bp (0.81%) frameshifts for g3 in Kcnq4G229D/G229D mice (Figure 2F). These results indicate that AAV_SaCas9-KKH_g3 can specifically edit the Kcnq4G229D alleles over a long period in vivo.
Improvement of auditory function in Kcnq4+/G229D mice
To investigate the therapeutic efficacy of the AAV_SaCas9-KKH_g3 system on auditory function in Kcnq4+/G229D mice, a total of 500 nL AAV_SaCas9-KKH_g3 at a titer of 1.16 × 1013 vg/ml was injected into the inner ears of P1–P2 mice via lateral wall of the cochlea. In AAV-injected mice at 8 and 12 weeks, we measured auditory brainstem response (ABR) representing the sound-evoked neural output of the cochlea and distortion product otoacoustic emission (DPOAE)5 representing the amplification provided by OHCs.
Since Kcnq4+/G229D mice exhibited progressive hearing loss with an ABR threshold of approximately 90 dB sound pressure level (dB SPL) at 12 weeks, the ABR thresholds were measured at frequencies ranging from 5.6 to 32 kHz at 8 and 12 weeks. Compared to uninjected ears, the ABR thresholds of treated ears were decreased significantly by 16.52, 16.30, 11.30, 11.09, 14.35, and 18.48 dB SPL at frequencies 5.6, 8, 11.3, 16, 22.6, and 32 kHz, respectively, revealing significant improvement of hearing at 8 weeks (Figures 3A and S5A). At 12 weeks, the ABR thresholds in the injected ears were ameliorated by 8–13 dB SPL across all frequencies (Figures 3B and S5B).
Figure 3.
Therapeutic efficacy of AAV_SaCas9-KKH_g3 system on auditory function in Kcnq4+/G229D mice
(A–D) Comparison of ABR (A–B) and DPOAE (C–D) thresholds between injected ears and uninjected ears in Kcnq4+/G229D mice at 8 and 12 weeks of age. The red, blue, and gray lines indicate the thresholds in the uninjected ears, injected ears, and wild-type mice. Asterisks indicate the statistical differences between uninjected ears and injected ears. Red and blue arrows indicate the mice in which the maximum stimulus sound intensity could not be elicited at the frequency, and these mice were included in the statistical analysis with the ABR threshold at 90 dB SPL and the DPOAE threshold at 80 dB SPL. (E–H) Comparison of amplitudes (E–F) and latencies (G–H) of ABR wave I between injected ears and uninjected ears at 5.6–32 kHz in 8-week-old and 12-week-old Kcnq4+/G229D mice. Amplitudes: 8-week, n = 10–19; 12-week, n = 2–17. Latencies: 8-week, n = 10–19; 12-week, n = 3–17. The ABR wave, which could not be elicited, was excluded from the statistical analysis in (E)–(H). Data are presented as mean ± SEM. In (A)–(D), ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, two-way ANOVA with Bonferroni correction. In (E)–(H), ∗p ≤ 0.05, ∗∗p ≤ 0.01, Mann Whitney test. Dots represent individual biological replicates in (E)–(H).
We observed lower DPOAE thresholds in the injected ears than in the uninjected ears at frequencies of 8, 11.3, 16, and 32 kHz at 8 and 12 weeks. At 8 weeks, the DPOAE thresholds of injected ears were decreased significantly by 14.17, 9.17, 11.39, and 10.83 dB SPL compared with the uninjected ears at frequencies of 8, 11.3, 16, and 32 kHz, respectively (Figures 3C and S5C). At 12 weeks, the DPOAE thresholds were reduced significantly by 7.67, 9.67, 10.67, and 14.67 dB SPL in treated ears at frequencies 8, 11.3, 16, and 32 kHz (Figures 3D and S5D).
To further characterize the hearing improvement, we also observed ABR wave I amplitudes indicating the number of neurons firing and ABR wave I latencies indicating the speed of transmission in injected mice at 8 weeks and 12 weeks.36 At 8 weeks, the ABR wave I amplitudes were higher in the injected ears at frequencies of 5.6, 8, and 11.3 kHz in comparison with uninjected ears (Figure 3E). The injected ears still showed higher ABR wave I amplitudes at frequencies of 5.6 and 16 kHz in 12-week-old mice (Figure 3F). Similarly, compared with uninjected ears, the shorter ABR wave I latencies suggest a reduced lag in cochlear nerve transmission (Figures 3G and 3H).19 The above results indicate that the hearing of Kcnq4+/G229D mice is ameliorated by the AAV_SaCas9-KKH_g3 system.
Evaluation of hair cell
To evaluate the preservation of hair cells by AAV_SaCas9-KKH_g3, we took the cochleae of adult mice and observed the number of surviving hair cells at 12 weeks when the thresholds of ABR and DPOAE exhibited obvious amelioration. The results showed that the survival of OHCs at middle and basal turns of injected ears were more abundant than that of uninjected ears (Figures 4A and 4B), consistent with the hearing improvement. Compared to 8-week-old Kcnq4+/+ mice, the morphology of hair cells stereocilia in Kcnq4+/G229D mice was normal and did not appear to show significant changes between the treated group and untreated group, such as inversion and fusion (Figure S6).
Figure 4.
Effects of AAV_SaCas9-KKH_g3 on survival and resting membrane potentials of OHCs in 12-week-old Kcnq4+/G229D mice
(A) Representative confocal microscopy images at the apex, mid, and base sections of the cochleae from the injected ear and uninjected ear. (B) Quantification of survival OHCs in injected ears compared with uninjected ears in the apical, middle, and basal turns. The number of animals: n = 4. (C) Resting membrane potentials of OHCs in injected ears compared to uninjected ears. The collected cells are from 7 to 12 animals, and the number of collected cells is 19–24. Data are presented as mean ± SEM. ns, no significance; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, t test. Dots represent individual biological replicates.
We observed that the resting membrane potentials (RMPs) were significantly depolarized in OHCs of Kcnq4+/G229D mice compared with Kcnq4+/+ mice (Figure 4C). To detect whether the AAV_SaCas9-KKH_g3 system can restore the resting potentials of OHCs in Kcnq4+/G229D mice, the RMPs of OHCs were measured by the patch-clamp technique. Compared with untreated mice, the OHCs of treated mice exhibited more hyperpolarized resting potentials, and there was no obvious difference between treated mice and wild-type mice (Figure 4C). The results suggest that the RMP of OHCs in Kcnq4+/G229D mice is rescued well by the AAV_SaCas9-KKH_g3 system.
Safety and efficacy of the AAV_SaCas9-KKH_g3 system
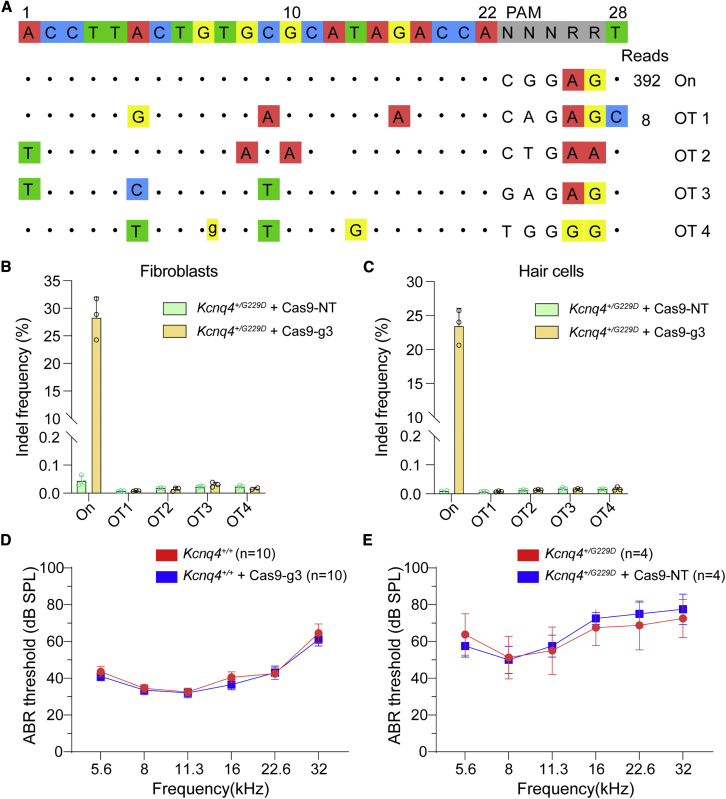
The off-target effect of CRISPR-Cas9 systems is an issue for gene therapy. To more comprehensively assess the specificity of g3, we employed the CIRCLE-seq (circularization for in vitro reporting of cleavage effects), an unbiased and highly sensitive off-target assay,37 to identify potential off-target editing sites associated with Cas9 and g3. We extracted genomic DNA from the tail tissue of Kcnq4G229D/G229D mice, circularized the sheared ∼400 bp DNA fragment, incubated with Cas9 protein and sgRNA3 complex, ligated the cleaved circular DNA to the adaptors, and identified the locations of DNA breaks by deep sequencing. The results showed that only one candidate off-target site associated with the g3 was identified (Figure 5A).
Figure 5.
Safety and efficacy of the AAV_SaCas9-KKH_g3 system
(A) Analysis of on-target and off-target effects. The off-target site 1 (OT1) was detected by CIRCLE-seq, and the other off-target sites (OT2–OT4) were predicted using CasOFFinder. (B) Detection of indel frequencies for the on-target and off-target sites in fibroblasts in vitro. (C) Detection of indel frequencies for the on-target and off-target sites in hair cells in vivo. (D) The ABR thresholds of wild-type mice at 4 weeks after AAV_SaCas9-KKH_g3 system injection. (E) The ABR thresholds of Kcnq4+/G229D mice at 8 weeks after AAV_SaCas9-KKH_NT injection. “NT” indicates sgRNA that does not target the mouse genome. In (B)–(C), dots represent individual biological replicates (n = 2–3); data are shown as mean ± SD. In (D) and (E), data are presented as mean ± SEM.
Then, four off-target sites, including off-target 1 (OT1) detected by CIRCLE-seq and OT2–OT4 predicted by CasOFFinder, were evaluated in fibroblasts and hair cells (Figure 5A; Table S2). No significant editing occurred at the four sites in treated mice compared with control (SaCas9-KKH + NT-gRNA, with a titer of 2 × 1013 vg/ml) (Figures 5B and 5C; Table S1). In summary, these results suggest that in vivo editing of the Kcnq4G229D allele mediated by AAV-delivered SaCas9-KKH_g3 is not accompanied by significant editing of candidate off-target sites.
AAV_SaCas9-KKH_g3 was injected into the inner ears of wild-type mice, and we observed that the hearing of wild-type mice was completely unaffected compared with the untreated group (Figure 5D), which was consistent with previous reports.6,25 We injected the AAV_SaCas9-KKH_NT-gRNA system into the inner ears of Kcnq4+/G229D mice and observed that the hearing of Kcnq4+/G229D mice was not improved compared with the untreated group (Figure 5E). The results suggest that the Kcnq4G229D allele is specifically disrupted by the AAV_SaCas9-KKH_g3, resulting in the improvement of hearing in Kcnq4+/G229D mice.
Discussion
KCNQ4 is one of the common causative genes of autosomal dominant sensorineural deafness, but there are currently no molecular therapeutics.27 The structural analysis of KCNQ4 protein may help to understand the role of KCNQ4 in the auditory system.38,39 In the present study, we established a method to accurately reflect the editing efficiency of hair cells and rescued the auditory function of Kcnq4+/G229D mice utilizing the CRISPR-Cas9 technology.
The editing efficiency of CRISPR-Cas9 in hair cells could not be precisely measured, hindering the accurate evaluation of therapeutic efficacy. Previously, we improved the editing efficiency by sorting out any GFP-positive cells including spiral ganglion and supporting cells transduced by the Anc80L65-GFP vector.25,40 Here, the indel frequency in the whole cochlear tissue was 1.45% ± 1.30% at 2 weeks, which is similar to the results of previous studies (0.60%–4.05%).4, 5, 6, 7 To precisely detect the editing efficiency of CRISPR-Cas9 in hair cells, we used the Atoh1-GFP mice in which cochlear hair cells were specifically labeled with GFP.33 Editing efficiency of up to 54.2% was observed in GFP-sorted hair cells (Figure 2C), indicating that the editing efficiency of CRISPR-Cas9 technology was successfully evaluated in hair cells in vivo, which suggests a novel approach for precise detection of editing efficiency in specific cell type. The results indicate that the high-efficiency editing could be achieved in non-dividing hair cells in vivo by the CRISPR-Cas9 system, which provides important support for the treatment of deafness mediated by CRISPR-Cas9 and offers a new option for investigating gene function by directly knocking out genes in vivo.
We found that the types of indels were simple, and the editing efficiencies were variable in treated mice (Figure S4; Table S1), probably resulting from a few hair cells obtained (Figure S3). Due to the limited number of hair cells (about 3,291) in the cochlear tissue of mouse,41 it is challenging to obtain more hair cells, and the digestion process of cochlear tissue may need to be optimized in future.42,43 Although the editing efficiency at the early stage could be detected by sorting GFP-positive hair cells, the editing efficiency of hair cells in the adult mice could not be accurately assessed because of the difficulty to obtain plenty of hair cells from the adult mice.44 An approach needs to be explored to collect enough adult hair cells in the field of hearing, which will promote the development of gene therapy for deafness and help researchers further understand the relationship between editing efficiency and hearing improvement.
Recently, Noh et al. improved the hearing of Kcnq4W276S/+ mice using CRISPR-Cas9 technology, which provides a potential strategy for the treatment of DFNA2 nonsyndromic hearing loss.7 However, there are several differences between our study and their study. Firstly, the therapeutic system used by Noh et al. was the delivery of split SpCas9 via dual-AAV vectors (Anc80L65); however, we utilized a single AAV (PHP.eB), which we confirmed had extremely high, efficient transduction of nearly 100% of inner hair cells and about 96.2%–98.6% of OHCs,6,35,45,46 to deliver SaCas9-KKH. We provided another potential therapeutic strategy for the treatment of KCNQ4-related deafness. György et al. demonstrated that SaCas9-KKH was superior to SpCas9 in specificity.5 Furthermore, remarkably, we provided an innovative approach to accurately assess the underestimated editing efficiency of CRISPR-Cas9 in vivo and observed approximately 34.10% ± 20.46% editing efficiency in hair cells. Last but not least, the hearing of Kcnq4W276S/+ mice was recovered to 7 weeks by Noh et al. However, the hearing of Kcnq4+/G229D mice was restored to at least 12 weeks by us.
The ABR and DPOAE results showed that the hearing of treated mice was improved significantly compared with the untreated group at 8 and 12 weeks (Figures 3 and S5). However, we also observed that the hearing of treated mice was not completely recovered in Kcnq4+/G229D mice. It is predicted that the underlying pathological progress is attributed to the death of non-functional hair cells, which affects function and survival of neighboring cells.47, 48, 49 Previous studies on the treatment of deafness have found that the rescued hair cells showed a delay in degeneration, and it is speculated that only rescue of partial hair cells is difficult to maintain hearing preservation for a long time.15,26,50 Noh et al. and Wu et al. found that the hearing improvement was positively correlated with the number of rescued cells.7,50 It is significant to study what percentage of the cells have to be rescued for a durable benefit in the future. Also, it is necessary to further improve the editing efficiency of targeted cells through optimization of gene-editing tools, including in vivo delivery approaches, and engineering of guide RNA or Cas9 protein.51,52 KCNQ4 is mainly expressed in OHCs, but also expressed in the inner hair cells as well as central auditory pathway in the cochlea.30,53, 54, 55 Perhaps due to haploinsufficiency, the truncated proteins resulting from KCNQ4 deletion mutations in patients were found to cause delayed, mild hearing loss, which needs to be taken into account for further application in humans.56 Therefore, another need is to further clarify the pathogenic mechanism of KCNQ4 in the future.
Taken together, we successfully established a novel methodology to accurately test the editing efficiency of the CRISPR-Cas9 system in hair cells; the auditory function of Kcnq4+/G229D mice was improved by the safe and effective AAV_SaCas9-KKH_g3 agents. However, there are still many aspects that need to be optimized before the genome-editing agents might be applied for clinical trials, including more efficient editing tools and additional safety assessments. Nevertheless, the present study will be beneficial in promoting the therapeutic application of gene-editing tools in treating hearing loss and human diseases.
Materials and methods
Animals and genotyping
The CRISPR-Cas9-mediated gene knock-in method was used to generate Kcnq4 gene mutant mice. In brief, the mixed solution containing the Cas9 mRNA, Kcnq4 gene-specific sgRNA, and Kcnq4 gene-specific single-stranded donor oligonucleotides was microinjected into fertilized zygotes. After culturing for about 24 h in vitro, the injected zygotes were transferred into the oviducts of pseudo-pregnant female mice. The Atoh1-GFP mouse strain33 was used in this study. All animals were bred in the Department of Laboratory Animal Science at Fudan University. All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of Fudan University and the Shanghai Medical Experimental Animal Administrative Committee, and we followed the NIH Guide for the Care and Use of Laboratory Animals. For genotyping of Kcnq4G229D mice, the tail was cut to extract genomic DNA with DirectPCR lysis reagents (Mouse Tail) (Viagen, #102-T). The Kcnq4 gene was amplified with forward and reverse primers in a 25 μL volume containing 2 μL lysate, 0.4 nM of each primer, and Taq PCR Master Mix (Biomed, #MT201). Amplification conditions were an initial 5 min denaturation at 94°C, 35 cycles of 30 s at 94°C, 30 s at 53°C, and 30 s at 72°C, and a final extension at 72°C for 5 min. PCR products were analyzed by Sanger sequencing.
Plasmid construction
The CRISPR-SaCas9-KKH system was selected for this study due to its capacity to be packaged and delivered via a single AAV vector and the presence of known PAM sequences (5′-NNNRRT-3′) that are suitable for a greater number of sgRNAs.34 We designed four sgRNAs covering the Kcnq4G229D mutation site and then ligated each sgRNA into the BsaI site of the pCMV_SaCas9-KKH_hU6 plasmid. The 60-bp sequence containing the Kcnq4G229D point mutation was constructed into the lentiviral vector using the ClonExpress II One Step Cloning Kit (Vazyme, #C112-01). The construction of each plasmid was mainly implemented by PCR and Gibson assembly.
Mouse primary fibroblast cell culture
Skin fibroblasts were derived from the Kcnq4G229D/G229D, Kcnq4+/G229D, and Kcnq4+/+ of P1 mouse pups. The pups were euthanized and cleaned with 75% ethanol, and skin fragments from the trunk and limbs were excised and submerged in cold 1×PBS (pH 7.4) (Gibco, #10010001). The skin fragments were minced into small pieces (∼1 mm2) with shears. The tissue pieces were treated at 37°C with cell culture-grade trypsin-EDTA (0.25%) (Thermofisher, #25200072) for 30 min with occasional pipetting up and down to break up any cell clumps. Warm culture medium (DMEM medium [Gibco, #11965092] with 10% fetal bovine serum [Gibco, #16140071], 100 U/ml penicillin and 100 mg/mL streptomycin [Millipore, #516106]) was added to stop the enzymatic digestion. The suspension was filtered through a 40-μm cell strainer (BD Falcon, #352350), and the filtered fluid was centrifuged at 200 × g for 5 min. The cell pellet was resuspended in a prepared culture medium and transferred to a six-well culture dish and incubated at 37°C with 5% CO2. Fibroblast cells were verified by Sanger sequencing.
Cochlear explant culture
The cochleae were dissected out of both the injected and uninjected ears of Kcnq4+/G229D or Atoh1-GFP;Kcnq4+/G229D P5 mice, and the organ of Corti was cultured in DMEM/F12 (Gibco, #11330032) with 1% N2 supplement (Gibco, #17502048), 2% B27 supplement (Gibco, #17504044), and 50 mg/mL ampicillin (Sigma, #A5354) at 37°C and 5% CO2 until P14.
Collection of GFP-positive hair cells
The organ of Corti from Atoh1-GFP;Kcnq4+/G229D mice was isolated 5 days after injection and cultured in vitro for 9 days; then the medium was discarded, and the basilar membrane was gently washed with 1xPBS. Approximately 100 μL of 0.25% trypsin-EDTA was added to the culture dish and gently puffed to keep the basilar membrane in suspension; then the suspension containing the basilar membrane was transferred to a 1.5-mL centrifuge tube. The basilar membrane was incubated at 37°C for 20–30 min, with occasional pipetting up and down to break up any cell clumps and gentle puffing every 10 min. When the basilar membrane became significantly smaller and finer, 100 μL of warm DMEM medium with 10% fetal bovine serum was added to stop the trypsin digestion. Approximately 200 μL cell suspension was filtered through a 40-μm cell strainer, and the filtered cell suspension was placed on ice. The GFP-positive cells were sorted out using a MoFlo XDP machine: SSC and FSC coordinate parameters were adjusted so that the main cell population was in the middle region, and then the cell population was selected as the R1 gate; in a new window, the GFP-positive cell population was selected as R2 gate with reference to the negative control using the coordinate parameters FITC-Log, and then it was sorted. The genomic DNA was extracted using QuickExtract DNA Extraction Solution (Epicentre, #QE09050).
Transfection and genomic DNA extraction
For sgRNA screening, a total of 1 μg of pCMV_SaCas9-KKH_hU6_sgRNA was transfected into the mouse primary fibroblasts with Lipofectamine 2000 (Life Technologies, #11668019) according to the manufacturer’s instructions, and genomic DNA was isolated from fibroblast cells for deep-sequencing analysis 5 days after transfection using the QuickExtract DNA Extraction Solution (Epicentre, #QE09050) following the manufacturer’s instructions. For the Atoh1-GFP;Kcnq4+/G229D mice, genomic DNA was extracted from the whole cochlear tissue of the inner ear that was injected with the therapeutic system at P1, dissected at P5, and cultured for another 9 days in vitro. Then the Corti tissue was treated at 37°C with cell culture-grade trypsin-EDTA (0.25%) for 30 min with occasional pipetting up and down to break up any cell clumps. A warm culture medium (DMEM medium with 10% fetal bovine serum) was added to stop the enzymatic digestion. GFP-positive cells were sorted by fluorescence-activated cell sorting. For the unsorted group, cells of the entire cochlear tissue were obtained from mice at 2, 8, and 13 weeks, respectively. The target sites were amplified by nested PCR and purified with a Gel Extraction Kit (QIAGEN, #28104) for deep sequencing. The primer sequences for amplifying the target DNA site were KCNQ4-On-F (5′-aacatctttgctacgtccgc-3′) and KCNQ4-On-R (5′-tactcgcccgccccaattt-3′) (Table S3), and the length of products was 224 bp. For Kcnq4G229D/G229D mice, the number of unedited sequences (cgcggtggcacctggaagctgttggACtccgtggtctatgcgcacagtaa) was counted (n1), the number of edited sequences (insertions or deletions) was counted (n2), and the editing efficiency was calculated as f = n2/(n1 + n2). For Kcnq4+/+ mice, the number of unedited sequences (cgcggtggcacctggaagctgttggGAtccgtggtctatgcgcacagtaa) was counted (n3), the number of edited sequences was counted (n4), and the editing efficiency was calculated as f = n4/(n3 + n4). In heterozygous mice, to analyze whether the edited sequence was derived from the wild-type or mutant allele, the “agctgttggA” sequence was used as the mutant allele and the “agctgttggG” sequence was used as the wild-type allele in the edited sequence. The original sequencing data were analyzed by fastp, seqkit, and csvtk.
Off-target analysis
We assessed the off-targeting of g3 on a genome-wide scale using CIRCLE-seq.57 Briefly, the genomic DNA of Kcnq4G229D/G229D mice was extracted, sheared, circularized, then cleaved in vitro with Cas9 protein and sgRNA, and finally, the off-target sites of gRNA were analyzed by next-generation sequencing. For the off-target sites identified by CIRCLE-seq, the target sites were amplified by PCR using the genomic DNA of the edited fibroblasts and the genomic DNA of the edited cells in mice as templates and then were analyzed by next-generation sequencing. In addition, we predicted potential off-target sites for g3 using Cas-OFFinder software,58 and specific primers were designed for these potential off-target sites. Off-target genomic sites were amplified from the edited genomic DNA, and the PCR primers are specified in Table S3.
AAV vector production
The titers of vectors were 1.16 × 1013 vg/ml and 2 × 1013 vg/ml for AAV_SaCas9-KKH_g3 and SaCas9-KKH + NT-gRNA, respectively. The plasmids SaCas9-KKH_g3 and SaCas9-KKH + NT-gRNA were sequenced before packaging into AAV-PHP.eB. The AAV vectors were produced by OBiO Technology (Shanghai) Corp., Ltd. (Shanghai, China).
Microinjection of the AAV CRISPR-Cas9 system into the inner ear
The method of injection was described in our previously published paper.6,25 Briefly, AAV carriers were injected into the inner ear of Kcnq4+/+, Kcnq4+/G229D, KcnqG229D/G229D, and Atoh1-GFP;Kcnq4+/G229D mouse pups. P1–P2 mice were anesthetized by placing them on ice, and the skin was incised behind the right ear to expose the greater auricular vesicle and stapedial artery. A total of 500 nL AAV_SaCas9-KKH_g3 or SaCas9-KKH + NT-gRNA was injected into the base cochlear turn via the lateral wall of the cochlea. After injection, the incision was closed with sutures. The pups were returned to their mothers after full recovery within 10 min. Standard postoperative care was used after the procedure.
Measurement of resting membrane potentials
As mentioned above, the cultured basilar membrane at P14 was bathed in extracellular solution59 containing 5.8 mM KCl, 144 mM NaCl, 0.9 mM MgCl2·6H2O, 1.3 mM CaCl2·2H2O, 0.7 mM NaH2PO4·H2O, 10 mM HEPES, and 5.6 mM D-glucose, pH adjusted to 7.4 with NaOH (∼300 mOsm/kg). The recording pipettes were filled with an internal solution composed of 142 mM KCl, 3.5 mM MgCl2·6H2O, 2.5 mM Mg-ATP, 0.1 mM CaCl2·2H2O, 5 mM HEPES, and 1 mM EGTA, pH adjusted to 7.3 with KOH (∼290 mOsm/kg). The hair cells in apical turns of cochleae were selected randomly, and the RMPs of hair cells were measured at zero current in the current-clamp mode of the whole-cell configuration using an EPC-10 USB amplifier with Patchmaster v2.90.5 software (HEKA Elektronik, Reutlingen, Germany). Igor Pro 6.22A and GraphPad Prism 8 were used to analyze data and generate graphs. A t test was used to evaluate the statistical significance in GraphPad Prism 8.0. Data are presented as means ± SEM.
Auditory testing
ABR and DPOAE measurements were recorded in a soundproof chamber using the RZ6 acoustic system (Tucker-Davis Technologies, Alachua, FL, USA) at 8 and 12 weeks after injection. According to our previously published methods,6,25 briefly, mice of either sex were anesthetized by intraperitoneal injection of xylazine (10 mg/kg) and ketamine (100 mg/kg), and three-needle electrodes were inserted separately into the subcutaneous tissue of the cranial vault between the two ears (reference electrode), the mastoid portion behind the left pinna (recording electrode), and the dorsal rump of the animal (grounding electrode). For ABR measurements, tone burst acoustic stimulus was raised from 20 to 90 dB SPL at frequencies of 5.6, 8, 11.3, 16, 22.6, and 32 kHz (in half-octave steps). ABR threshold was determined as the lowest sound pressure level at which any ABR wave peak could be visually detected and repeated compared to background noise. For the DPOAE measurements, primaries f1 and f2 (f2/f1 = 1.2) were presented with f2 from 5.6 to 32 kHz and L1 – L2 = 10 dB SPL. DPOAE thresholds were defined as the amplitude of the f2 level generating the DPOAE that was 3 dB SPL above the noise floor. The average noise floor level was below 0 dB SPL for all frequencies. After the animals recovered from anesthesia and regained voluntary movement, they were returned to the breeding center. In general, thresholds were defined by two independent observers.
Immunohistochemistry and confocal microscopy
After sacrificing the animals by cervical dislocation, the cochleae of 12-week-old adult mice were harvested.25 According to our previously published methods, briefly, the temporal bone was perforated, the round and oval windows were opened, and the cochlea was perfused, fixed with 4% paraformaldehyde, decalcified with 10% ethylene diamine tetraacetic acid (EDTA), permeabilized and infiltrated with 1% Triton X-100, and blocked with 10% bovine serum. The tissues were incubated with rabbit anti-Myosin VIIa (1:800 dilution, Proteus BioSciences, #25-6790), the appropriate Alexa-conjugated secondary antibodies (1:500 dilution, Donkey Anti-Rabbit IgG [H + L], Cy3, AlexJackson ImmunoResearch, 711-165-152), and DAPI for labeling the cell nuclei (1:1,000 dilution, Sigma, #D9542). Specimens were mounted on adherent microscope slides (Citotest, #188105). The fluorescent z stack images were collected using a Leica TCS SP8 laser scanning confocal microscope and a 40 × 1.30 oil objective.
Hair cell counting
For hair cell counting, the numbers of Myosin VIIa+ OHCs per 100 μm of the apical, middle, and basal turns of the cochlear were counted using Leica TCS SP8 software. Four independent samples were collected from wild-type, injected ear and contralateral uninjected ear of Kcnq4+/G229D mice. Results are expressed as the mean ± SEM.
Scanning electron microscopy
According to our previously published methods,25 briefly, the temporal bones of 8-week-old adult mice were harvested after cervical dislocation. The cochlea was fixed, decalcified, immersed, dissected, dehydrated, dried, and sprayed. Images were obtained with a high vacuum field emission scanning electron microscope (Hitachi SU-8010) at 2.2 kV (low magnification) or 10.0 kV (high magnification).
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2020YFA0908201, 2017YFA0103900), the National Natural Science Foundation of China (82171148, 81830029, 82171141, 82000992), the Science and Technology Commission of Shanghai Municipality (21S11905100, 21JC401000), Clinic Research Plan of SHDC (SHDC2020CR4083), “Shuguang Program” supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (20SG08).
Author contributions
Y.S. conceived and supervised the project. Y.S. H.L. and D.W. designed and developed the project. C.C. and D.W. jointly designed the experiments and analyzed the data for the project. C.C. and D.W. performed experiments related to the detection and analysis of editing efficiency in vitro and in vivo. C.C., B.H., J.L., and L.H. performed experiments related to the evaluation of hearing function in vivo. C.C., F.W., and Y.C. performed an electrophysiological assay. G.L. supervised the electrophysiological assay. C.C., D.W., and Y.C. wrote the manuscript. C.C., D.W., Y.C., Y.S., H.L., G.L., Z.C., L.Z., and D.L. reviewed and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors have no conflicts of interest to declare.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.07.016.
Supplemental information
Data availability
The data are available on request from the corresponding author.
References
- 1.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X., et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A., et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Haberle J., Matos J., Robinson M.D., et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 4.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y., et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.György B., Nist-Lund C., Pan B., Asai Y., Karavitaki K.D., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Solanes P., Spataro S., et al. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat. Med. 2019;25:1123–1130. doi: 10.1038/s41591-019-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y., Hu X., Wang D., Li D., Li Y., Wang F., Huang M., Gu X., Xu Z., Zhou J., et al. Gene editing in a Myo6 semi-dominant mouse model rescues auditory function. Mol. Ther. 2022;30:105–118. doi: 10.1016/j.ymthe.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noh B., Rim J.H., Gopalappa R., Lin H., Kim K.M., Kang M.J., Gee H.Y., Choi J.Y., Kim H.H., Jung J. In vivo outer hair cell gene editing ameliorates progressive hearing loss in dominant-negative Kcnq4 murine model. Theranostics. 2022;12:2465–2482. doi: 10.7150/thno.67781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morton C.C., Nance W.E. Newborn hearing screening--a silent revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 10.Korver A.M., Smith R.J., Van Camp G., Schleiss M.R., Bitner-Glindzicz M.A., Lustig L.R., Usami S.I., Boudewyns A.N. Congenital hearing loss. Nat. Rev. Dis. Primers. 2017;3:16094. doi: 10.1038/nrdp.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding N., Lee S., Lieber-Kotz M., Yang J., Gao X. Advances in genome editing for genetic hearing loss. Adv. Drug Deliv. Rev. 2021;168:118–133. doi: 10.1016/j.addr.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilgert N., Smith R.J., Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr. Mol. Med. 2009;9:546–564. doi: 10.2174/156652409788488775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akil O., Seal R.P., Burke K., Wang C., Alemi A., During M., Edwards R.H., Lustig L.R. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75:283–293. doi: 10.1016/j.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Q., Wang J., Li Q., Kim Y., Zhou B., Wang Y., Li H., Lin X. Virally mediated Kcnq1 gene replacement therapy in the immature scala media restores hearing in a mouse model of human Jervell and Lange-Nielsen deafness syndrome. EMBO Mol. Med. 2015;7:1077–1086. doi: 10.15252/emmm.201404929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien W.W., Isgrig K., Roy S., Belyantseva I.A., Drummond M.C., May L.A., Fitzgerald T.S., Friedman T.B., Cunningham L.L. Gene therapy restores hair cell stereocilia morphology in inner ears of deaf whirler mice. Mol. Ther. 2016;24:17–25. doi: 10.1038/mt.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulon D., Papal S., Patni P., Cortese M., Vincent P.F., Tertrais M., Emptoz A., Tlili A., Bouleau Y., Michel V., et al. Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J. Clin. Invest. 2018;128:3382–3401. doi: 10.1172/JCI94351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emptoz A., Michel V., Lelli A., Akil O., Boutet de Monvel J., Lahlou G., Meyer A., Dupont T., Nouaille S., Ey E., et al. Local gene therapy durably restores vestibular function in a mouse model of Usher syndrome type 1G. Proc. Natl. Acad. Sci. USA. 2017;114:9695–9700. doi: 10.1073/pnas.1708894114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isgrig K., Shteamer J.W., Belyantseva I.A., Drummond M.C., Fitzgerald T.S., Vijayakumar S., Jones S.M., Griffith A.J., Friedman T.B., Cunningham L.L., et al. Gene therapy restores balance and auditory functions in a mouse model of usher syndrome. Mol. Ther. 2017;25:780–791. doi: 10.1016/j.ymthe.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan B., Askew C., Galvin A., Heman-Ackah S., Asai Y., Indzhykulian A.A., Jodelka F.M., Hastings M.L., Lentz J.J., Vandenberghe L.H., et al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat. Biotechnol. 2017;35:264–272. doi: 10.1038/nbt.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taiber S., Cohen R., Yizhar-Barnea O., Sprinzak D., Holt J.R., Avraham K.B. Neonatal AAV gene therapy rescues hearing in a mouse model of SYNE4 deafness. EMBO Mol. Med. 2020;5:e13259. doi: 10.15252/emmm.202013259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Moyed H., Cepeda A.P., Jung S., Moser T., Kugler S., Reisinger E. A dual-AAV approach restores fast exocytosis and partially rescues auditory function in deaf otoferlin knock-out mice. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201809396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akil O., Dyka F., Calvet C., Emptoz A., Lahlou G., Nouaille S., Boutet de Monvel J., Hardelin J.P., Hauswirth W.W., Avan P., et al. Dual AAV-mediated gene therapy restores hearing in a DFNB9 mouse model. Proc. Natl. Acad. Sci. USA. 2019;116:4496–4501. doi: 10.1073/pnas.1817537116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minoda R., Miwa T., Ise M., Takeda H. Potential treatments for genetic hearing loss in humans: current conundrums. Gene Ther. 2015;22:603–609. doi: 10.1038/gt.2015.27. [DOI] [PubMed] [Google Scholar]
- 24.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu X., Wang D., Xu Z., Wang J., Guo L., Chai R., Li G., Shu Y., Li H. Prevention of acquired sensorineural hearing loss in mice by in vivo Htra2 gene editing. Genome Biol. 2021;22:86. doi: 10.1186/s13059-021-02311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh W.H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aay9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sloan-Heggen C.M., Bierer A.O., Shearer A.E., Kolbe D.L., Nishimura C.J., Frees K.L., Ephraim S.S., Shibata S.B., Booth K.T., Campbell C.A., et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia X., Zhang Q., Jia Y., Shu Y., Yang J., Yang H., Yan Z. Molecular basis and restoration of function deficiencies of Kv7.4 variants associated with inherited hearing loss. Hear. Res. 2020;388:107884. doi: 10.1016/j.heares.2020.107884. [DOI] [PubMed] [Google Scholar]
- 29.Namba K., Mutai H., Kaneko H., Hashimoto S., Matsunaga T. In silico modeling of the pore region of a KCNQ4 missense mutant from a patient with hearing loss. BMC Res. Notes. 2012;5:145. doi: 10.1186/1756-0500-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubisch C., Schroeder B.C., Friedrich T., Lutjohann B., El-Amraoui A., Marlin S., Petit C., Jentsch T.J. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 31.Kharkovets T., Dedek K., Maier H., Schweizer M., Khimich D., Nouvian R., Vardanyan V., Leuwer R., Moser T., Jentsch T.J. Mice with altered KCNQ4 K+ channels implicate sensory outer hair cells in human progressive deafness. EMBO J. 2006;25:642–652. doi: 10.1038/sj.emboj.7600951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui C., Zhang L., Qian F., Chen Y., Huang B., Wang F., Wang D., Lv J., Wang X., Yan Z., et al. A humanized murine model, demonstrating dominant progressive hearing loss caused by a novel KCNQ4 mutation (p.G228D) from a large Chinese family. Clin. Genet. 2022;102:149–154. doi: 10.1111/cge.14164. [DOI] [PubMed] [Google Scholar]
- 33.Lumpkin E.A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., Doetzlhofer A., White P., Groves A., Segil N., et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 34.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X., Wang J., Yao X., Xiao Q., Xue Y., Wang S., Shi L., Shu Y., Li H., Yang H. Screened AAV variants permit efficient transduction access to supporting cells and hair cells. Cell Discov. 2019;5:49. doi: 10.1038/s41421-019-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abadi S.P., Khanbabaee G.M., Sheibani K.M. Auditory brainstem response wave amplitude characteristics as a diagnostic tool in children with speech delay with unknown causes. Iran. J. Med. Sci. 2016;41:415–421. [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Med. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T., Wu K., Yue Z., Wang Y., Zhang F., Shen H. Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell. 2021;81:25–37.e24. doi: 10.1016/j.molcel.2020.10.037. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y., Liu H., Chen Y., Dong S., Wang F., Wang S., Li G.L., Shu Y., Xu F. Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel. Neuron. 2021;110:237–247.e4. doi: 10.1016/j.neuron.2021.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R., et al. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017;35:280–284. doi: 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehret G., Frankenreiter M. Quantitative analysis of cochlear structures in the house mouse in relation to mechanisms of acoustical information processing. J. Comp. Physiol. 1977;122:65–85. [Google Scholar]
- 42.Yamashita T., Zheng F., Finkelstein D., Kellard Z., Carter R., Rosencrance C.D., Sugino K., Easton J., Gawad C., Zuo J. High-resolution transcriptional dissection of in vivo Atoh1-mediated hair cell conversion in mature cochleae identifies Isl1 as a co-reprogramming factor. PLoS Genet. 2018;14:e1007552. doi: 10.1371/journal.pgen.1007552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi M.H., Qiu Y., Tian K.Y., Liang K., Chang H.M., Wang R.F., Chen E.F., Wang W.L., Zha D.J., Qiu J.H. Outer hair cells isolation from postnatal Sprague-Dawley rats. World J. Otorhinolaryngol. Head Neck Surg. 2019;5:14–18. doi: 10.1016/j.wjorl.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojanova Z.P., Kwan T., Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development. 2015;142:3529–3536. doi: 10.1242/dev.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu X., Hu X., Wang D., Xu Z., Wang F., Li D., Li G.L., Yang H., Li H., Zuo E., et al. Treatment of autosomal recessive hearing loss via in vivo CRISPR/Cas9-mediated optimized homology-directed repair in mice. Cell Res. 2022;32:699–702. doi: 10.1038/s41422-022-00624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Z., Li G., Cui C., Wang F., Wang X., Xu Z., Guo H., Chen Y., Tang H., Wang D., et al. Preventing autosomal-dominant hearing loss in Bth mice with CRISPR/CasRx-based RNA editing. Signal Transduct. Target. Ther. 2022;7:79. doi: 10.1038/s41392-022-00893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nayak G., Lee S.I., Yousaf R., Edelmann S.E., Trincot C., Van Itallie C.M., Sinha G.P., Rafeeq M., Jones S.M., Belyantseva I.A., et al. Tricellulin deficiency affects tight junction architecture and cochlear hair cells. J. Clin. Invest. 2013;123:4036–4049. doi: 10.1172/JCI69031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcotti W., Erven A., Johnson S.L., Steel K.P., Kros C.J. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J. Physiol. 2006;574:677–698. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazmierczak M., Harris S.L., Kazmierczak P., Shah P., Starovoytov V., Ohlemiller K.K., Schwander M. Progressive hearing loss in mice carrying a mutation in Usp53. J. Neurosci. 2015;35:15582–15598. doi: 10.1523/JNEUROSCI.1965-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J., Solanes P., Nist-Lund C., Spataro S., Shubina-Oleinik O., Marcovich I., Goldberg H., Schneider B.L., Holt J.R. Single and dual vector gene therapy with AAV9-PHP.B rescues hearing in Tmc1 mutant mice. Mol. Ther. 2021;29:973–988. doi: 10.1016/j.ymthe.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D., Zhang F., Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B., Niu Y., Ji W., Dong Y. Strategies for the CRISPR-based therapeutics. Trends Pharmacol. Sci. 2020;41:55–65. doi: 10.1016/j.tips.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beisel K.W., Nelson N.C., Delimont D.C., Fritzsch B. Longitudinal gradients of KCNQ4 expression in spiral ganglion and cochlear hair cells correlate with progressive hearing loss in DFNA2. Brain Res. Mol. Brain Res. 2000;82:137–149. doi: 10.1016/s0169-328x(00)00204-7. [DOI] [PubMed] [Google Scholar]
- 54.Beisel K.W., Rocha-Sanchez S.M., Morris K.A., Nie L., Feng F., Kachar B., Yamoah E.N., Fritzsch B. Differential expression of KCNQ4 in inner hair cells and sensory neurons is the basis of progressive high-frequency hearing loss. J. Neurosci. 2005;25:9285–9293. doi: 10.1523/JNEUROSCI.2110-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kharkovets T., Hardelin J.P., Safieddine S., Schweizer M., El-Amraoui A., Petit C., Jentsch T.J. KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc. Natl. Acad. Sci. USA. 2000;97:4333–4338. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamada F., Kure S., Kudo T., Suzuki Y., Oshima T., Ichinohe A., Kojima K., Niihori T., Kanno J., Narumi Y., et al. A novel KCNQ4 one-base deletion in a large pedigree with hearing loss: implication for the genotype-phenotype correlation. J. Hum. Genet. 2006;51:455–460. doi: 10.1007/s10038-006-0384-7. [DOI] [PubMed] [Google Scholar]
- 57.Lazzarotto C.R., Nguyen N.T., Tang X., Malagon-Lopez J., Guo J.A., Aryee M.J., Joung J.K., Tsai S.Q. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc. 2018;13:2615–2642. doi: 10.1038/s41596-018-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Flores M.C., Lee J.H., Park S., Zhang X.D., Sihn C.R., Ledford H.A., Wang W., Kim H.J., Timofeyev V., Yarov-Yarovoy V., et al. Cooperativity of K(v)7.4 channels confers ultrafast electromechanical sensitivity and emergent properties in cochlear outer hair cells. Sci. Adv. 2020;6:eaba1104. doi: 10.1126/sciadv.aba1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available on request from the corresponding author.