Abstract
The rhi genes of Rhizobium leguminosarum biovar viciae are expressed in the rhizosphere and play a role in the interaction with legumes, such as the pea. Previously (K. M. Gray, J. P. Pearson, J. A. Downie, B. E. A. Boboye, and E. P. Greenberg, J. Bacteriol. 178:372–376, 1996) the rhiABC operon had been shown to be regulated by RhiR and to be induced by added N-(3-hydroxy-7-cis-tetradecenoyl)-l-homoserine lactone (3OH,C14:1-HSL). Mutagenesis of a cosmid carrying the rhiABC and rhiR gene region identified a gene (rhiI) that affects the level of rhiA expression. Mutation of rhiI slightly increased the number of nodules formed on the pea. The rhiI gene is (like rhiA) regulated by rhiR in a cell density-dependent manner. RhiI is similar to LuxI and other proteins involved in the synthesis of N-acyl-homoserine lactones (AHLs). Chemical analyses of spent culture supernatants demonstrated that RhiI produces N-(hexanoyl)-l-homoserine lactone (C6-HSL) and N-(octanoyl)-l-homoserine lactone (C8-HSL). Both of these AHLs induced rhiA-lacZ and rhiI-lacZ expression on plasmids introduced into an Agrobacterium strain that produces no AHLs, showing that rhiI is positively regulated by autoinduction. However, in this system no induction of rhiA or rhiI with 3OH,C14:1-HSL was observed. Analysis of the spent culture supernatant of the wild-type R. leguminosarum bv. viciae revealed that at least seven different AHLs are made. Mutation of rhiI decreased the amounts of C6-HSL and C8-HSL but did not block their formation, and in this background the rhiI mutation did not significantly affect the expression levels of the rhiI gene or rhiABC genes or the accumulation of RhiA protein. These observations suggest that there are additional loci involved in AHL production in R. leguminosarum bv. viciae and that they affect rhiI and rhiABC expression. We postulate that the previously observed induction of rhiA by 3OH,C14:1-HSL may be due to an indirect effect caused by induction of other AHL production loci.
One of the most abundant proteins made by strains of Rhizobium leguminosarum bv. viciae is the rhiA gene product, which was first observed as a heavily stained band following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of stationary-phase cultures (7). rhiA is in a three-gene operon (rhiABC) that is under the regulatory control of the rhiR gene, which encodes a LuxR-type regulator. The rhiABC-rhiR gene cluster is located between genes involved in nitrogen fixation (nifHDK) and nodulation (nod) on the symbiotic plasmid pRL1JI (5). DNA hybridizations have shown that the rhi genes are adjacent to nodulation genes in other strains of R. leguminosarum bv. viciae (9a, 18). Analysis of several strains of R. leguminosarum (biovars viciae, trifolii, and phaseoli), Rhizobium meliloti, and Rhizobium sp. strain NGR234, using antibody to RhiA, showed that RhiA seems to be specific to R. leguminosarum bv. viciae (6, 7), indicating that it may play some host-specific role in the interaction between this biovar and its symbiotic partners (pea, vetch, lentil, and Lathyrus spp.).
Initially, no effect on nodulation or symbiotic nitrogen fixation was observed for bacteria containing transposon insertions in rhiA (7). Subsequently, it was found that mutation of rhiA could affect nodulation in some mutant strains that were already impaired for nodulation due to deletion of some of the nodulation genes (5). In particular, in the absence of the nodFEL genes, mutations of rhiA could be seen to decrease the low level of nodulation even further.
Analysis of rhiA-lacZ and rhiC-phoA gene fusions revealed that the rhiABC genes are strongly induced during the transition from late exponential to early stationary growth phase (15). The N-acyl-homoserine lactone (AHL) termed N-(3-hydroxy-7-cis-tetradecenoyl)-l-homoserine lactone (referred to hereafter as 3OH,C14:1-HSL) was identified as both an inducer of the rhiABC genes and a potent inhibitor of the growth of some strains of R. leguminosarum bv. viciae (15). Indeed the compound previously known as “small bacteriocin” (17, 35) had been purified and shown to be 3OH,C14:1-HSL (30). It was suggested that this AHL induces stationary phase in R. leguminosarum bv. viciae since it is thought to induce gene expression that inhibits growth but does not kill the cells (15).
Many strains of the R. leguminosarum biovars viciae, trifolii, and phaseoli (15, 17, 35, 37) make small bacteriocin (and thus probably make 3OH,C14:1-HSL) as does Rhizobium etli (26), whereas R. meliloti and the closely related Agrobacterium tumefaciens do not (17, 37). In addition, R. etli makes at least six other compounds that are probably AHLs (26). Mutation of one gene, raiI, in R. etli abolished the production of some autoinducers by R. etli, but the production of 3OH,C14:1-HSL was unaffected (26).
Rhizobium strains often contain multiple plasmids, and it is possible that different plasmids encode the production of different AHLs. In this regard it is not yet known if the small bacteriocin locus is located on the chromosome or on a plasmid in those strains which make small bacteriocin. The locus encoding production of small bacteriocin had been shown not to be located on the symbiotic plasmid pRL1JI (17, 37); nevertheless, 3OH,C14:1-HSL induces rhiABC gene expression. The regulation of the rhiABC genes is also affected by genes other than rhiR present on the symbiotic plasmid pRL1JI. It was observed (5, 10) that flavonoid inducers of nod gene expression decreased (by about 50%) the level of expression of the rhiABC operon and that this required the nod gene regulator NodD (5, 10). Furthermore, using a plasmid carrying a rhiA-lacZ fusion, Gray et al. (15) observed that an ethyl acetate extract containing 3OH,C14:1-HSL had different effects on rhiA gene expression in strains containing or lacking pRL1JI.
We wish to understand the physiological role of the rhi gene region. Analysis of RhiA protein with antiserum revealed that rhiA is expressed by bacteria in the rhizosphere but not by nitrogen-fixing bacteria in nodules (7). Database searches revealed no protein sequences with strong similarity to RhiA, RhiB, or RhiC, although it has been shown that RhiC has an N-terminal signal sequence that targets it to the periplasm (5). In this work we have further characterized the rhi gene region, identifying an AHL production locus that is involved in the induction of rhiABC expression.
MATERIALS AND METHODS
Microbiological techniques.
Rhizobium strains were grown in TY medium (2). Antibiotics were added as appropriate to maintain selection for plasmids. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600) by using an MSE Spectroplus spectrophotometer. β-Galactosidase activities were measured as described previously (27) by using a Titertek Multiscan Plus spectrophotometer (EFLAB). For measurements of β-galactosidase throughout growth, bacteria from 24-h cultures were diluted 1 in 200 to a starting OD600 of about 0.002. When added, AHLs were added at the start of growth to a final concentration of 0.1 μM. The AHLs N-hexanoyl-l-homoserine lactone (C6-HSL), N-(3-oxohexanoyl)-l-homoserine lactone (3O,C6-HSL), N-octanoyl-l-homoserine lactone (C8-HSL), N-(3-oxooctanoyl)-l-homoserine lactone (3O,C8-HSL), N-(3-oxobutanoyl)-l-homoserine lactone (3O,C4-HSL), and N-(3-hydroxy-tetradecenoyl)-l-homoserine lactone (3OH,C14:1-HSL) were synthesized (4) and kindly provided by Ram Chhabra, School of Pharmaceutical Sciences, University of Nottingham. Nodulation tests were done by using variety Frisson peas (Pisum sativum L.) as described previously (8, 23), with a minimum of 12 matched plants per test; two separate tests were carried out with similar results.
Bacterial strains and plasmids.
R. leguminosarum sp. strain 8401 lacks a symbiotic plasmid, and all Rhizobium strains used are based on 8401 (Table 1). A34 is a derivative of 8401 carrying the symbiotic plasmid pRL1JI. Strains A160 and A161, which are isogenic with A34, were made by conjugating derivatives of pRL1JI carrying rhiR1::Tn5 and rhiA4::Tn5 (7) into 8401. The rhiI15::Tn5 allele from pIJ7790 (see below) was recombined onto pRL1JI by marker exchange by using pPH1JI to select for recombinants (28). The derivative of pRL1JI carrying rhiI15::Tn5 was conjugated into 8401 to form A721, which was confirmed to lack pPH1JI (which transfers at a relatively low frequency).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| Strain | ||
| R. leguminosarum | ||
| 8401 | Strain lacking a symbiotic plasmid | 8 |
| A34 | Derivative of 8401 carrying pRL1JI (previously called 8401 pRL1JI) | 9 |
| A160 | Derivative of A34 carrying rhiR1::Tn5 | 5 |
| A161 | Derivative of A34 carrying rhiA4::Tn5 | This work |
| A721 | Derivative of A34 carrying rhiI15::Tn5 | This work |
| A. tumefaciens | ||
| C58.00 | Strain lacking the AT and Ti plasmids; does not make detectable AHLs | 36 |
| Plasmids | ||
| pRL1JI | R. leguminosarum bv. viciae native symbiotic plasmid | 19 |
| pIJ1089 | Cosmid carrying 30 kb of pRL1JI, including the nifHD, rhiABCR, and nodOTNMLEFDABCIJ genes | 9 |
| pIJ1242 | Derivative of pIJ1089 carrying rhiR1::Tn5 | 7 |
| pIJ1243 | Derivative of pIJ1089 carrying rhiA4::Tn5 | 7 |
| pIJ1642 | Derivative of pIJ1089 carrying rhiA5::Tn3HoHo1 (rhiA-lacZ) | 10 |
| pIJ1696 | Derivative of pIJ1089 carrying rhiI7::Tn3HoHo1 (rhiI-lacZ) | This work |
| pIJ1769 | rhiA-lacZ fusion | 5 |
| pIJ1891 | Cloning vector derived from pLAFR3 | 21 |
| pIJ7790 | Derivative of pIJ1089 carrying rhiI15::Tn5 | This work |
| pIJ7794 | 2.5-kb HindIII-SmaI fragment carrying rhiI in pIJ1891 | This work |
| pIJ7982 | rhiI-lacZ fusion in pMP220 | This work |
| pMP220 | lacZ promoter probe vector | 32 |
Mutagenesis of pIJ1089 with Tn5 and Tn3HoHo1 was done as described previously (9, 10). Derivatives of pIJ1089 were transferred to strain 8401 and screened for reduced RhiA production by SDS-PAGE analysis of proteins released from cells, following solubilization of proteins in SDS gel loading buffer, as described previously (10). Mutated plasmids were transferred to the Escherichia coli strain DH5α by transformation with DNA isolated from the Rhizobium strains. The location of Tn3HoHo1 within rhiI in pIJ1696 was determined by restriction enzyme mapping. The location of Tn5 within rhiI in pIJ7790 was determined by subcloning part of the Tn5 plus flanking DNA as a 4.9-kb EcoRI-BamHI fragment in pUC19 and sequencing the DNA by using a Tn5-specific primer.
Plasmid pIJ7794 was made by cloning the rhiI gene on a 2.5-kb HindIII-SmaI fragment subcloned from pIJ1089; the fragment was subcloned in pIJ1891, which had been first cut with BamHI, filled in with Klenow fragment of DNA polymerase, and finally digested with HindIII. The rhiI-lacZ gene fusion on pIJ7982 was made by cloning a 0.7-kb fragment carrying part of rhiI plus the 0.3-kb upstream region into pMP220. The 0.7-kb fragment was subcloned from one of the plasmids, generated by ExoIII nuclease deletion, for DNA sequencing.
Molecular biology techniques.
DNA cloning, ligations, transformation, restriction enzyme mapping, and DNA hybridization were done by standard methods (29). The DNA sequence of rhiI was determined on both strands by using an ordered series of ExoIII-generated deleted derivatives of the rhiI gene region that had been cloned as a blunt-ended HindIII-SmaI fragment in both orientations in the HincII site of pUC19. The sequencing reactions were carried out by using the Amersham Thermosequenase kit and an Applied Biosystems automated sequencer (ABI 377). Database searches of the protein sequence were done by using the BLAST and TFASTA (1) programs to find related sequences in the EMBL and SwissProt protein sequence databases.
Analysis of proteins.
Rhizobium strains were grown to an OD600 of 1.2 in 100 ml of TY medium, and the cells were harvested by centrifugation. The washed cells from 10 ml of culture were resuspended in 1 ml of 0.1 M Tris HCl (pH 8.0) and lysed by sonication (30-s sonication with an MSE Soniprep at full power, done six times). The protein extract was solubilized and separated by SDS-PAGE, and the gels were stained with Coomassie blue R250 or transferred to nitrocellulose and probed with RhiA antiserum as described previously (3, 7).
Assay of AHLs.
Cultures were grown for 48 h in TY medium to an OD600 of 1.0. The cells were removed by centrifugation, and the AHLs were extracted from culture supernatants as described previously (38). AHLs were analyzed by thin-layer chromatography (TLC) as described by Shaw et al. (31) but with use of Chromobacterium violaceum CV026 as the AHL indicator organism (22). CV026 can be used as a biosensor for exogenous AHLs because it produces the purple pigment violacein in response to added AHLs. Culture supernatants and synthetic AHL standards (as 1-mg ml−1 solutions in acetonitrile) were spotted (2 to 10 μl) onto aluminum-backed RP18 reverse-phase TLC plates (Merck) and dried in a stream of air. Samples were separated with 60% (vol/vol) methanol in water as the mobile phase. Once the solvent front had migrated to within 2 cm of the top of the chromatogram, the plate was removed from the chromatography tank, dried in air, and overlaid with a thin film of Luria-Bertani soft agar (0.7%, wt/vol) seeded with C. violaceum CV026. After overnight incubation at 30°C, AHLs were located by detection of purple spots against a white background.
Isolation, purification, and chemical characterization of AHLs.
Spent supernatant (4 liters) from stationary-phase cultures was extracted with dichloromethane (supernatant/dichloromethane ratio, 7:3). Dichloromethane was removed by rotary evaporation, and the residue was redissolved in 1.0 ml of acetonitrile and applied to a C8 reverse-phase semipreparative high-performance liquid chromatography (HPLC) column (Kromasil KR100-5C8 [250 by 8 mm] column; Hichrom, Reading, United Kingdom). Fractions were eluted with a linear gradient of acetonitrile in water (20 to 95%, vol/vol) over a 30-min period at a flow rate of 2 ml/min and monitored at 210 nm. Six fractions (F1 to F6), covering 5-min intervals, were collected and assayed for activity by using a variety of AHL reporter assays, including the C. violaceum CV026 detection system (22, 24) and the E. coli(pSB401) luxR plus luxI′::luxCDABE, and E. coli(pSB1075) lasR plus lasI′::luxCDABE luminescence detection systems (34, 39). No peaks of activity other than those corresponding with the four spots visualized with the TLC CV026 overlay assay described above were identified. Active fractions were rechromatographed by using an appropriate isocratic mobile phase of acetonitrile in water, and the fractions were assayed for AHL activity. Active subfractions were also reanalyzed on an analytical HPLC apparatus attached to a photodiode array system by using the same isocratic mobile phase (Waters 996 PDA system operating with a Millennium 2010 Chromatography manager; Watford, Hertfordshire, United Kingdom), and both retention time and spectral profiles were compared with those of a series of synthetic AHL standards. Following preparative HPLC, the final active subfractions were analyzed by HPLC-mass spectrometry (HPLC-MS) (Micromass Instruments, Manchester, United Kingdom) using an appropriate isocratic mobile phase. This technique couples the resolving power of C8 reverse-phase HPLC directly with MS such that the mass of the molecular ion (M + H) and its major component fragments can be determined for a compound with a given retention time. Samples eluting from the HPLC column were ionized by positive-ion atmospheric-pressure chemical-ionization MS and were analyzed at two different cone voltages (18 and 28 eV). The spectra obtained were compared with those of the synthetic AHL standard subjected to the same HPLC-MS conditions.
RESULTS
Identification of a novel locus producing AHLs.
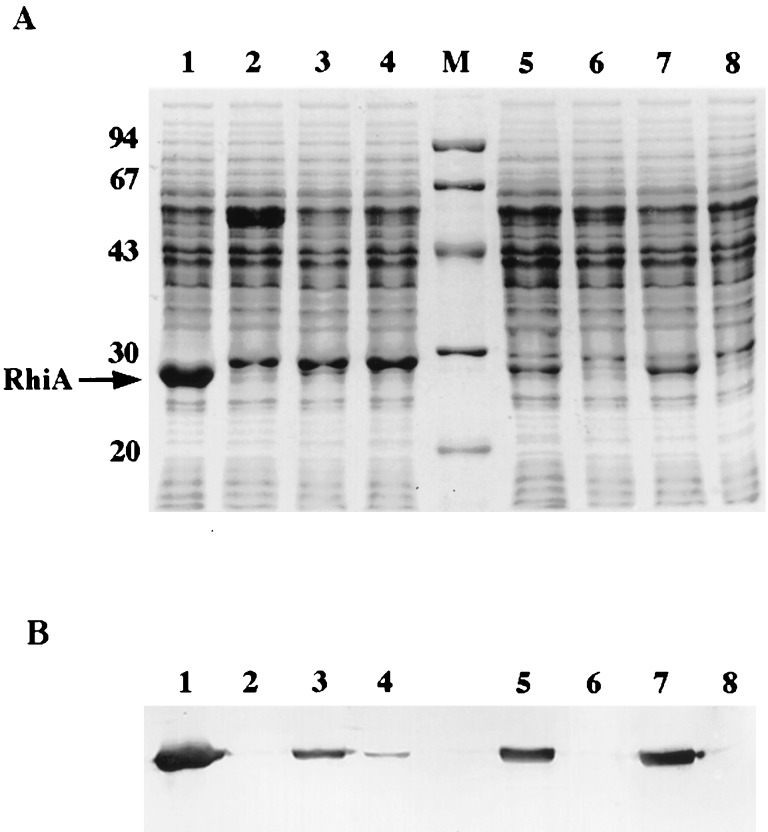
Plasmid pIJ1089 carries about 30 kb of DNA from the symbiotic plasmid pRL1JI. Identified genes on pIJ1089 include those involved with nitrogen fixation (nifHD) and nodulation (nodO, nodMNT, nodFEL, nodD, and nodABCIJ) and the rhizosphere-expressed genes rhiABC and rhiR. This plasmid directs the production of high levels of the RhiA protein with an Mr of 24,000 in strain 8401, which lacks a symbiotic plasmid (Fig. 1). pIJ1089 was mutagenized with Tn3HoHo1 or Tn5, the mutated derivatives were conjugated into strain 8401, and protein extracts of the transconjugants were analyzed by SDS-PAGE. Several mutants affected in production of the RhiA protein were identified, and DNA mapping or sequencing from the ends of the transposon confirmed that most of the mutations were in the structural and regulatory genes, rhiA and rhiR, respectively. However, with two of the mutated derivatives, pIJ1696 and pIJ7790 (carrying Tn3HoHo1 and Tn5, respectively), the sites of transposon insertions mapped to a new locus about 2 kb upstream of rhiA. As shown in Fig. 1A, strain 8401/pIJ1089 produces a prominent 24-kDa protein that is absent if rhiA is mutated and is very greatly reduced in intensity when rhiR is mutated (Fig. 1A, lanes 1, 2, and 4). With 8401/pIJ7790, the level of RhiA protein is significantly decreased (Fig. 1A, lane 3). Immunostaining performed by using antiserum to RhiA (Fig. 1B) confirmed that mutation of the new locus in pIJ7790 significantly reduces the level of rhiA expression. Similar observations (data not shown) were made with pIJ1696 (carrying Tn3HoHo1 in the region about 2 kb upstream of rhiA). We considered that the new locus, mutated in pIJ1696 and pIJ7790, may be involved in production of an AHL that influences expression of rhiA.
FIG. 1.
Effects on RhiA production of mutating rhiI. Cell extracts were separated by SDS-PAGE and either stained with Coomassie blue R250 (A) or transferred to nitrocellulose and immunostained with RhiA antiserum followed by a secondary antibody (anti-rabbit immunoglobulin G) coupled to alkaline phosphatase (B). The order of lanes in panel A is the same as that in panel B; 20 μg (panel A) or 10 μg (panel B) of protein was loaded in each lane. Extracts were from the following: (lane 1) A31/pIJ1089 (wild type); (lane 2) A31/pIJ1243 (rhiA4::Tn5); (lane 3) A31/pIJ7790 (rhiI15::Tn5); (lane 4) A31/pIJ1242 (rhiR1::Tn5); (lane 5) A34 (wild type); (lane 6) A161 (rhiA4::Tn5); (lane 7) A721 (rhiI15::Tn5); and (lane 8) A160 (rhiR1::Tn5). The markers (lane M) have Mr of 94,000, 67,000, 45,000, 30,000, and 20,000, as indicated.
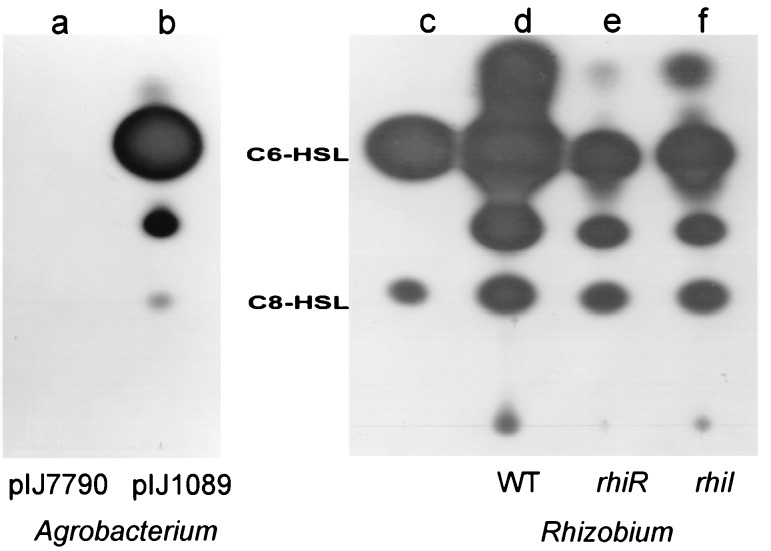
A. tumefaciens C58.00 makes no AHLs detectable by C. violaceum CV026 (Fig. 2) and so is a useful strain in testing for AHL production mediated by pIJ1089 and its derivatives (E. coli is not a good host for expression of R. leguminosarum genes). The culture supernatant of C58.00 carrying pIJ1089 was extracted with dichloromethane and separated by TLC. The chromatogram was overlaid with agar inoculated with the AHL-sensor strain C. violaceum CV026 to detect AHLs. As shown here (Fig. 2, lane b), introducing pIJ1089 into C58.00 results in the production of four components that activate pigment production by CV026. It should be noted that the intensity of staining does not reflect the relative amounts of individual AHLs made, since the sensor strain has different sensitivities for different AHLs (21). Two of the spots determined by pIJ1089 comigrate with the synthetic standards C6-HSL and C8-HSL. The crude extract was fractionated by HPLC by using a linear gradient of acetonitrile in water. When tested by the TLC CV026 overlay assay, fractions 3 and 4 were found to activate violacein production by CV026. Fractions 3 and 4 were further fractionated by using an isocratic acetonitrile-in-water mobile phase (percentages of acetonitrile in water, 25 and 35% [vol/vol], respectively). When active subfractions of fraction 3 were reanalyzed by analytical HPLC, the active compound was found to elute with the same retention time (17 min) and photodiode array absorption spectrum as synthetic C6-HSL (data not shown). To confirm the identities of the AHLs, active fractions were subjected to HPLC-MS on an isocratic mobile phase (percentages of acetonitrile in water were 25 and 50% [vol/vol] for subfractions from fractions 3 and 4, respectively). Positive-ion atmospheric-pressure chemical-ionization MS for fraction 3 revealed the presence of a molecular ion [M + H] of 200, corresponding to the C6-HSL, together with the characteristic fragmentation products at 102 and 99, which correspond to the homoserine lactone moiety and the C6 acyl side chain [CH3(CH2)4C O+], respectively. Similar analysis of the active subfraction of fraction 4 confirmed the presence of a molecular ion [M + H] of 228, corresponding to the C8-HSL, and breakdown products at 102 and 127, which correspond to the homoserine lactone moiety and the C8 acyl side chain [CH3(CH2)6C O+], respectively. The two additional putative AHLs detected by the TLC CV026 overlay assay (Fig. 2) did not migrate with any of the known AHLs. Characterization of these compounds is under way.
FIG. 2.
Identification of AHLs produced by RhiI. AHLs extracted from spent medium were separated by TLC and detected by using an overlay of agar containing C. violaceum CV026. Lanes a and b contain extracts of spent culture supernatants from A. tumefaciens C58.00 carrying either pIJ7790 (carrying the rhiI15::Tn5 mutation) (lane a) or pIJ1089 (lane b). Results similar to those seen in lane a (no AHLs were detected) were found with C58.00 carrying no introduced plasmid. Lane c contains the synthetic standards C6-HSL and C8-HSL. Lanes d, e, and f contain extracts of spent culture supernatants from the R. leguminosarum bv. viciae strains A34 (wild type), A160 (rhiR1::Tn5), and A721 (rhiI15::Tn5), respectively.
The plasmid (pIJ7790) defective for RhiA production (Fig. 1) was also defective in the production of all four spots detected by C. violaceum CV026 following TLC (Fig. 2, lane a). A similar observation was made with pIJ1696 (data not shown). These results demonstrate that pIJ1089 carries a locus that directs the production of C6-HSL and C8-HSL (and possibly other AHLs) and that transposon insertions in a region upstream of rhiA abolish this phenotype.
Characterization of rhiI.
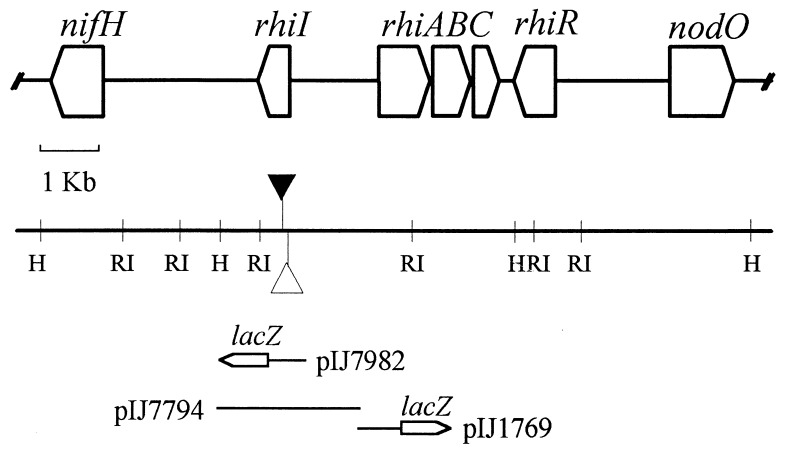
A 2.5-kb fragment of DNA corresponding to the region mutated by Tn5 and Tn3HoHo1 was subcloned to make pIJ7794, which was confirmed to be involved in AHL production by using C. violaceum CV026 as an AHL sensor (data not shown). The DNA sequence of the region was determined, and the sites of the transposon insertions were determined by restriction enzyme mapping and DNA sequencing. Both insertions are located in a short open reading frame (Fig. 3), which encodes a 185-amino-acid protein with similarity to several other proteins (including LuxI) that are involved in AHL production. We called the gene rhiI since it is involved in AHL production and mutations of this gene in pIJ1089 decrease rhiA expression as judged by the levels of RhiA protein detected by SDS-PAGE (Fig. 1; compare lanes 1 and 3).
FIG. 3.
Map of the rhi gene region. The open reading frames corresponding to the rhiI, rhiA, rhiB, rhiC, and rhiR genes are shown as arrows, and the locations of the rhiI7::Tn3HoHo1 and rhiI15::Tn5 insertions are indicated as open and closed triangles, respectively. The DNA cloned to form rhiI plasmid pIJ7794 and the rhiI-lacZ (pIJ7982) and rhiA-lacZ (pIJ1769) fusions is shown.
Figure 4 shows an alignment of the predicted sequence of RhiI with the most closely related proteins identified in database searches. Although RhiI shows clear similarities with the LuxI family of proteins, it is evident that RhiI has only about 15 to 20% identity with LuxI (Vibrio fischeri), LasI (Pseudomonas aeruginosa), RaiI (R. etli), RhlI (VsmI) (P. aeruginosa), and PhzI (Pseudomonas aureofaciens). Several other homologs were identified, but these had fewer similarities than those whose alignments are shown in Fig. 4. RhiI contains the highly conserved residues (12, 16, 25) found in multiple LuxI homologs (Arg-24, Phe-27, Trp-33, Glu-43, Asp-45, Asp-48, Arg-68, Glu-99, and Arg-102; numbered with reference to RhiI and marked in Fig. 4). In other RhiI homologs a Phe residue is normally found at position 82 (12), but in RhiI the conservatively substituted residue Tyr is found (Fig. 4).
FIG. 4.
Alignment of RhiI with related proteins. The predicted protein sequence of RhiI (rhii) was aligned with the sequences of related proteins by using the Genetics Computer Group programs Pileup and Prettybox. The aligned sequences (and their SwissProt database accession numbers) are LuxI (luxi) (P12747), LasI (lasi) (P33883), RaiI (raii) (U92712), RhlI (rhli) (P54291), and PhzI (phzi) (Q51522). The dots mark residues conserved in multiple LuxI homologs, and the open circle marks a Tyr residue that is usually found to be Phe in other homologs.
rhiI is regulated by RhiR in a cell density-dependent manner.
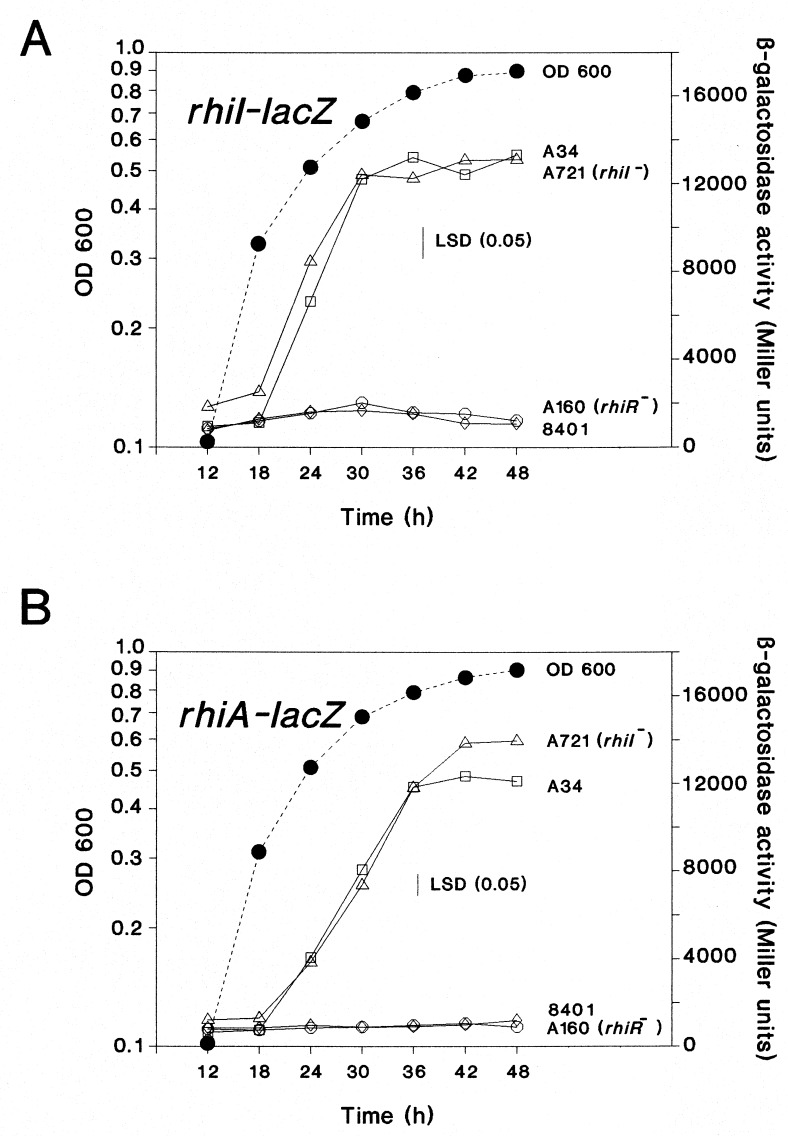
A rhiI-lacZ reporter plasmid (pIJ7982) was made by cloning a 0.7-kb fragment (containing 0.3 kb of DNA upstream of the predicted rhiI translation start site) into the lacZ fusion vector pMP220. The expression level of β-galactosidase was measured throughout growth in strain A34, in a derivative of it mutated in rhiR (A160), or in a strain lacking the symbiotic plasmid pRL1JI (8401). Parallel experiments were done with the rhiA-lacZ plasmid pIJ1769. In A34 there are very low levels of rhiI and rhiA expression early in growth, but the levels increase markedly in the late exponential phase and reach a plateau in stationary phase (Fig. 5). The expression of rhiI is rhiR dependent since very low levels of activity are seen with A160 (rhiR1::Tn5) or 8401 (which lacks pSym carrying rhiR). Therefore, rhiI (like rhiA) is regulated by RhiR in a cell density-dependent manner. We could identify no sequences upstream of rhiI or rhiA that showed strong similarity to the 20-bp region of dyad symmetry (lux box) found upstream of several genes regulated by LuxR-type proteins (13, 33). However, 45 bp upstream of the proposed translation start site of rhiI is a 22-bp region of dyad symmetry (Fig. 6). A similar sequence was found upstream of rhiA (Fig. 6), although the center of symmetry was slightly different. It remains to be demonstrated if these regions are involved in the binding of RhiR.
FIG. 5.
Expression of rhiI-lacZ and rhiA-lacZ. The expression of rhiI-lacZ (pIJ7794) (A) or rhiA-lacZ (pIJ1769) (B) was analyzed by measuring β-galactosidase throughout growth of strains A34 (wild type), 8401 (strain A34 lacking pRL1JI), A160 (rhiR1::Tn5), and A721 (rhiI15::Tn5). The growth curves (OD600) shown as broken lines correspond to those obtained with A34/pIJ7794 or A34/pIJ1769; the growth of the other strains was very similar. LSD, least significant difference.
FIG. 6.

Conserved DNA sequences found upstream of rhiI and rhiA. The sequences shown are found 45 and 146 bp upstream of the predicted translation start sites of rhiI and rhiA, respectively. Conserved residues (·) are marked, and the dyad symmetry is indicated with arrows.
rhiI and rhiA are induced by C6-HSL and C8-HSL.
The results presented above indicate that rhiI and rhiA are likely to be induced by AHLs. To analyze those AHLs that induce rhiI and rhiA, we made use of derivatives of pIJ1089 carrying lacZ transposon insertions. Mapping of Tn3HoHo1 in pIJ1696 revealed that the lacZ gene of Tn3HoHo1 was in the same orientation as rhiI and under the control of the rhiI promoter. Plasmid pIJ1642, a derivative of pIJ1089 carrying lacZ under the control of the rhiA promoter (rhiA5::Tn3HoHo1), was described previously (10). pIJ1696 and pIJ1642 were transferred into the AHL-nonproducer A. tumefaciens C58.00. This approach has the advantage of introducing the rhiR regulator gene on the same plasmid. (In preliminary experiments we found that there was no production of AHLs by E. coli DH5α carrying pIJ1089 and thus concluded that E. coli was not a good host for analysis of rhiI expression.) Derivatives of C58.00, carrying pIJ1696 or pIJ1642, were grown to early stationary phase, and the levels of rhiI and rhiA expression were determined by measuring the levels of β-galactosidase activity in cells. As shown in Table 2, in the absence of added AHLs there was a relatively low level of expression of rhiI-lacZ. Addition of C6-HSL gave the strongest induction of those AHLs tested, although induction was observed for several other AHLs (Table 2). However, no significant increase in activity was seen with 3OH,C14:1-HSL. Similar observations were made with C58.00 carrying pIJ1642 (rhiA5::Tn3HoHo1), in that C6-HSL was the strongest inducer of rhiA, lower levels of induction with C8-HSL, 3OH,C6-HSL, and 3OH,C8-HSL were seen, and no induction with 3OH,C14:1-HSL was observed.
TABLE 2.
Effects of added AHLs on rhiI-lacZ and rhiA-lacZ expressiona
| AHL tested | Expression (Miller units) of:
|
|
|---|---|---|
| rhiI-lacZ | rhiA-lacZ | |
| None (negative control) | 369.78 | 198.98 |
| C6-HSL | 2,493.28b | 1,079.54b |
| C8-HSL | 1,585.57b | 546.43b |
| 3O,C6-HSL | 896.01b | 196.35 |
| 3O,C8-HSL | 1,191.00b | 332.16b |
| 3OH,C4-HSL | 544.89b | 154.39 |
| 3OH,C14-HSL | 484.12 | 151.94 |
| LSD (P < 0.05) | 170.66 | 82.45 |
AHLs were tested in Agrobacterium sp. strain C58.00 carrying pIJ1642 or pIJ1696, using a final concentration of 0.1 μM each AHL.
Values are significantly different from that for the negative control (Student’s t test, P < 0.05).
LSD, least significant difference.
These data demonstrate that two of the AHLs produced by RhiI (C6-HSL and C8-HSL) act as inducers for rhiI and rhiA expression. However, no induction was observed for 3OH,C14:1-HSL, which was previously (15) shown to induce rhiA-lacZ expression. This suggests that the previously observed induction by 3OH,C14:1-HSL might be an indirect effect caused by regulation of other genes that influence rhiA and rhiI expression. It should be noted that the levels of expression of rhiA-lacZ or rhiI-lacZ in Agrobacterium strain C58.00 (Table 2) are considerably lower than those seen in R. leguminosarum bv. viciae A34 (Fig. 5). This is consistent with the hypothesis that other AHLs made by strain A34 but not made by C58.00 (Fig. 2) contribute to the enhanced expression of rhiI and rhiA in the Rhizobium background.
Genes on pRL1JI compensate for the absence of rhiI.
To analyze the phenotype of an rhiI mutant strain, the rhiI mutation on pIJ7790 was recombined onto pRL1JI and strain A721 (a derivative of A34 carrying rhiI15::Tn5) was constructed. The level of RhiA protein made by stationary-phase cells of A721 was analyzed: whereas mutation of rhiI on pIJ1089 significantly reduced the levels of RhiA formation (Fig. 1, lane 3), mutation of rhiI on pRL1JI had little or no observed effect on RhiA formation (Fig. 1, lane 7). The high level of RhiA in A721 (rhiI mutation on pRL1JI) compared with the level in strain 8401/pIJ7790 (rhiI mutation on pIJ1089) indicates that there may be a locus on pRL1JI that compensates for the absence of rhiI and that this locus is not contained in the 30-kb region of pRL1JI cloned in pIJ1089.
Measurements of the expression of rhiA-lacZ (pIJ1769) or rhiI-lacZ (pIJ7790) in A721 confirmed that the rhiI mutation in A721 does not reduce rhiA or rhiI expression (Fig. 5) compared with that for the control strain (A34). Therefore, although those AHLs produced by RhiI can induce rhiI and rhiA gene expression in strain C58.00 (Table 2), mutation of rhiI has little effect on rhiI and rhiA expression in the A34 background. These observations are consistent with a model in which rhiA and rhiI are regulated by RhiR not only in response to RhiI-made AHLs (such as C6-HSL and C8-HSL) but also in response to other AHLs that could be made by a product of another gene located elsewhere in the genome of A34. We used C. violaceum CV026 to analyze AHLs made by strain A34 and the derivatives of it carrying mutations in rhiI (A721) or rhiR (A160). As shown (Fig. 2, lane d), strain A34 makes many different compounds that are detected by this system; we estimate that (in addition to 3OH,C14:1-HSL that is not detected) there are at least six components that activate pigment production by C. violaceum CV026. Other components might be present but are not detected by this reporter system (22). Mutation of rhiI (Fig. 2, lane f) reduces the amount of C6-HSL, but an active component with the same mobility as C6-HSL is clearly made by the rhiI mutant (A721). This is consistent with the observations on RhiA production and rhiI-lacZ or rhiA-lacZ expression, which indicate that another locus in A721 may be involved in formation of C6-HSL. Mutation of rhiR (A160) has a slightly stronger effect on AHL production (Fig. 2, lane e) than mutation of rhiI. The difference between the rhiR and rhiI mutants could be explained if RhiR influences the expression of a gene present at another locus and involved in AHL production.
C. violaceum CV026 does not detect 3OH,C14:1-HSL, and so we measured the effect of mutating rhiI on production of this AHL using A34 as a sensor strain in a bacteriocin-like assay (35). Strain A721 did not induce a zone of growth inhibition, indicating that repression of 3OH,C14:1-HSL occurred normally and therefore mutation of rhiI did not affect the ability of pRL1JI to repress production of this AHL (data not shown). Strain A721 was also used as a sensor (lawn) in a similar assay, and its growth was as sensitive as that of the control strain (A34) to growth inhibition by 3OH,C14:1-HSL produced by strain 8401, showing that mutation of rhiI does not affect the growth sensitivity of A34 to 3OH,C14:1-HSL.
Influence of rhiI on nodulation.
Previous work with rhiA-lacZ fusions demonstrated that flavonoid inducers of nod gene expression decreased rhiA expression by about 50% and that this decrease was nodD dependent (5, 10). We measured expression of rhiI-lacZ using pIJ7982 in the presence and absence of the nod gene inducer hesperetin, under conditions similar to those shown in Fig. 5A. After 42 h of growth, the level of β-galactosidase activity in the cells grown with hesperetin (6,800 ± 610 U) was about half that seen when hesperetin was not added (11,300 ± 840 U). Therefore, like that of rhiA, rhiI expression is decreased by inducers of nod gene expression. No significant effect of hesperetin on expression of the rhiI-lacZ fusion in the nodD mutant A57 was observed (data not shown), confirming that the hesperetin-induced reduction of rhiI-lacZ expression is nodD dependent.
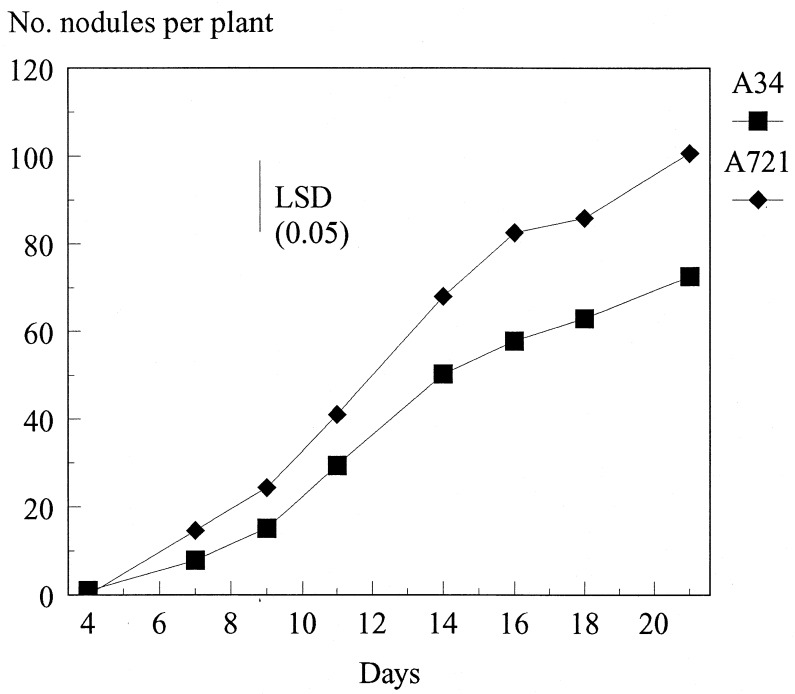
We tested nodulation ability of the rhiI mutant (A721) relative to that of the control strain A34. The mutant formed normal nitrogen-fixing nodules on peas; the final number of nodules formed was slightly (but significantly) higher than that for the control, although the rate of nodule formation was similar to that for the control (Fig. 7).
FIG. 7.
Effect of an rhiI mutation on nodulation. The average numbers of nodules formed by strains A34 (wild type) and A721 (rhiI15::Tn5) on Frisson peas are shown. The data shown are averages from one data set obtained with 16 plants. Similar results were found with two separate data sets. The difference in the final numbers of nodules formed is statistically significant (P < 0.05). LSD, least significant difference.
DISCUSSION
The rhiABC operon is conserved in all R. leguminosarum bv. viciae strains tested and has not been found in other rhizobia. This, taken together with the location of the rhi gene cluster (between the nod and nif genes), indicates that these genes may play some kind of role in the interaction between R. leguminosarum bv. viciae and at least some of its host legumes.
It is now evident that the rhiABC genes are regulated by RhiR in response to AHLs made by RhiI. The rhiI gene is regulated in the same way, thereby forming a positive autoregulatory loop that results in high levels of expression from the rhiI and rhiABC promoters. This explains the very high levels of RhiA protein detected in late-stationary-phase cells, in which it is certainly one of the most abundant proteins (7). However, rhiA expression is not totally dependent on rhiI since there appears to be another AHL production locus which can form AHLs that stimulate the expression of both the rhiABC operon and rhiI. Since rhiR mutants show very little expression of rhiI or rhiABC, it is evident that although there are other loci for AHL production, any regulatory genes that might be associated with those loci are not able to induce rhiABC or rhiI expression. However, there could be an indirect effect on rhiABC expression since AHLs, made by products of genes other than rhiI, can stimulate RhiR to induce rhiI and rhiABC expression. Thus, there is likely to be a degree of cross talk between different AHL production loci.
Previously the rhiA promoter was observed to be induced when 3OH,C14:1-HSL was added to wild-type cells during early-exponential-phase growth (15). However, RhiR activates rhiA in response to C6-HSL and C8-HSL but not 3OH,C14:1-HSL. Therefore, the most-probable explanation for the previous results is that 3OH,C14:1-HSL induces the expression of other AHLs, which in turn activate RhiR-mediated rhiABC and rhiI expression. The fact that rhiI and rhiR mutants still produce many short-chain AHLs is good evidence that there is at least one other AHL production locus in R. leguminosarum bv. viciae; indeed, in other (unpublished) work, we have cloned four AHL production loci from strain A34. Two other luxI-like genes in rhizobia have been described. In R. etli the raiI gene was sequenced and shown to be involved in the formation of several (chemically uncharacterized) AHLs (26). In Rhizobium sp. strain NGR234, a traI gene was identified in a symbiotic plasmid genome-sequencing project (11). Although RhiI described here is in the same family as these two proteins, it is not much more similar to RaiI or TraI proteins from rhizobia than to related proteins from several other bacteria (Fig. 4). R. leguminosarum bv. viciae strain A34 may have, in addition to rhiI, other AHL production genes homologous to traI and/or raiI. Perhaps the diversity of AHL production systems in Rhizobium may be related to the fact that many strains harbor multiple large plasmids. Different plasmids may have different AHL production systems, and strain 8401 (lacking a symbiosis plasmid) contains two plasmids thought to be greater than 300 and 500 kb in size (20). It remains to be determined if these plasmids harbor genes involved in AHL production.
It is not clear why R. leguminosarum bv. viciae should have the rhiI-rhiR regulatory system to induce expression of the rhiABC operon, although several lines of evidence relate this to some aspect of the interaction with leguminous plants. The observation that flavonoids which induce nod gene expression reduce rhiI expression suggests that the plant has the potential to influence the level of AHL production. However, the effect of flavonoids on rhi gene expression depends on the R. leguminosarum bv. viciae nod gene regulator NodD (10), indicating that the bacteria influence this decrease in rhiI and rhiABC expression. This is somewhat different from the observed inhibition of quorum-sensing regulated genes by halogenated furanones, which are thought to act as competitive inhibitors of AHL binding to LuxR-type regulators (14).
We do not yet know the biochemical role of the rhiABC gene products or why they should be regulated in a cell density-dependent manner. It is evident that in some way they influence the interaction with the plant since mutation of rhiA or rhiR significantly reduced nodulation in a strain lacking the nodFEL genes (5). Paradoxically, mutation of rhiI increased the final number of nodules formed. In R. etli, mutation of the raiI gene, which is also involved in AHL production, resulted in increased levels of bean nodulation (26). Thus, for two separate Rhizobium-legume interactions there is independent evidence that production of (at least) some of the AHLs inhibits nodulation under the growth conditions tested.
In the absence of significant protein sequence similarities to any other proteins of known function, it is difficult to predict the role of RhiA, RhiB, and RhiC. The RhiC protein appears to be located in the periplasm, possibly suggesting a role for the uptake of some metabolite, but our tests of growth of rhiABC mutants on various carbon sources have not identified any clear differences from the growth of the isogenic control strain (9a). It is evident that in R. leguminosarum bv. viciae quorum-sensing-based regulation is complex and may share similarities with the cascade of quorum-sensing-regulated genes in P. aeruginosa and V. fischeri. The reason for such complexity of regulation and for the apparent degree of redundancy of AHL production is not clear, and its elucidation will require characterization of the other AHL production loci in R. leguminosarum bv. viciae.
ACKNOWLEDGMENTS
We thank S. R. Chhabra for the generous gift of chemically synthesized AHLs, Y. Dessaux for suggesting the use of and providing Agrobacterium strain C58.00, and A. Davies for skilled technical assistance.
This work was supported in part by the BBSRC, a fellowship from the Universidad de Granada (to B.R.), and a contract (B104-CT96-0181) from the EU (DGXII-SSMI).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D J, Wood E A, Larkins A P, Galfrè G, Butcher G W, Brewin N J. Isolation of monoclonal antibodies reacting with peribacteroid membranes and other components of pea root nodules containing Rhizobium leguminosarum. Planta. 1988;173:149–160. doi: 10.1007/BF00403006. [DOI] [PubMed] [Google Scholar]
- 4.Chhabra S R, Stead P, Bainton N J, Salmond G P C, Stewart G S A B, Williams P, Bycroft B W. Autoregulation of carbapenem biosynthesis in Erwinia carotovora ATCC 39048 by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J Antibiot. 1993;46:441–454. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- 5.Cubo T, Economou A, Murphy G, Johnston A W B, Downie J A. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarumbiovar viciae. J Bacteriol. 1992;174:4026–4035. doi: 10.1128/jb.174.12.4026-4035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dibb N J. Plasmid-determined proteins in Rhizobium leguminosarum. Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1983. [Google Scholar]
- 7.Dibb N J, Downie J A, Brewin N J. Identification of a rhizosphere protein encoded by the symbiotic plasmid of Rhizobium leguminosarum. J Bacteriol. 1984;158:621–627. doi: 10.1128/jb.158.2.621-627.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downie J A, Hombrecher G, Ma Q-S, Knight C D, Wells B, Johnston A W B. Cloned nodulation genes of Rhizobium leguminosarumdetermine host-range specificity. Mol Gen Genet. 1983;190:359–365. [Google Scholar]
- 9.Downie J A, Knight C D, Johnston A W B. Identification of genes and gene products involved in nodulation of peas by Rhizobium leguminosarum. Mol Gen Genet. 1985;198:255–262. [Google Scholar]
- 9a.Downie, J. A. Unpublished observations.
- 10.Economou A, Hawkins F K L, Downie J A, Johnston A W B. Transcription of rhiA, a gene on a Rhizobium leguminosarum bv. viciae Sym plasmid, requires rhiR and is repressed by flavonoids that induce nodgenes. Mol Microbiol. 1989;3:87–93. doi: 10.1111/j.1365-2958.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 11.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobiumand legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanzelka B L, Stevens A M, Parsek M R, Crone T J, Greenberg E P. Mutational analysis of the Vibrio fischeriLuxI polypeptide: critical regions of an autoinducer synthase. J Bacteriol. 1997;179:4882–4887. doi: 10.1128/jb.179.15.4882-4887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch P R. Plasmid-determined bacteriocin production by Rhizobium leguminosarum. J Gen Microbiol. 1979;113:219–228. [Google Scholar]
- 18.Hombrecher G, Götz R, Dibb N J, Downie J A, Johnston A W B, Brewin N J. Cloning and mutagenesis of nodulation genes from Rhizobium leguminosarumTOM, a strain with extended host range. Mol Gen Genet. 1984;184:293–298. [Google Scholar]
- 19.Johnston A W B, Beynon J L, Buchanan-Wollaston A V, Setchell S M, Hirsh P, Beringer J E. High frequency transfer of nodulating ability between strains and species of Rhizobium. Nature. 1978;276:634–636. [Google Scholar]
- 20.Lamb J W, Hombrecher G, Johnston A W B. Plasmid-determined nodulation and nitrogen-fixation abilities in Rhizobium phaseoli. Mol Gen Genet. 1982;186:449–452. [Google Scholar]
- 21.Mavridou A, Barny M A, Poole P, Plaskitt K, Davies A E, Johnston A W B, Downie J A. Rhizobium leguminosarum nodulation gene (nod) expression is lowered by an allele-specific mutation in the dicarboxylate transport gene dctB. Microbiology. 1995;141:103–111. doi: 10.1099/00221287-141-1-103. [DOI] [PubMed] [Google Scholar]
- 22.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 23.Miller J A. Experiments in molecular genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1976. pp. 352–355. [Google Scholar]
- 24.Milton D L, Hardman A, Camara M, Chhabra S R, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsek M R, Schaefer A L, Greenberg E P. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosagene encoding an acylhomoserine lactone synthase. Mol Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 26.Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J Bacteriol. 1998;180:815–821. doi: 10.1128/jb.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossen L, Shearman C A, Johnston A W B, Downie J A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodABCgenes. EMBO J. 1985;4:3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schripsema J, de Rudder K E E, van Vliet T B, Lankhorst P P, de Vroom E, Kijne J W, van Brussel A A N. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw P D, Ping G, Daly S, Cronan J E, Jr, Rinehart K, Farrand S K. Detecting and characterizing acyl-homoserine lactone signal molecules by thin layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarumSym plasmid pRLIJI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 33.Stevens A M, Greenberg E P. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Throup P J, Camera M, Briggs G S, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 35.van Brussel A A N, Zaat S A J, Wijffelman C A, Pees E, Lugtenberg B J J. Bacteriocin smallof fast-growing rhizobia is chloroform soluble and is not required for effective nodulation. J Bacteriol. 1985;162:1079–1082. doi: 10.1128/jb.162.3.1079-1082.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaudequin-Dransart V, Petit A, Poncet C, Ponsonnet C, Nesme X, Jones J B, Bouzar H, Scott Chilton W, Dessaux Y. Novel Ti plasmids in Agrobacteriumstrains isolated from fig tree and chrysanthemum tumors and their opinelike molecules. Phytopathology. 1995;8:311–321. doi: 10.1094/mpmi-8-0311. [DOI] [PubMed] [Google Scholar]
- 37.Wijffelman C A, Pees E, Van Brussel A A N, Hooykaas P J J. Repression of small bacteriocin excretion in Rhizobium leguminosarum and Rhizobium trifoliiby transmissible plasmids. Mol Gen Genet. 1983;192:171–176. [Google Scholar]
- 38.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Chapon V, Bycroft B W, Salmond G P C, Lazdunski A, Stewart G S A B, Williams P. Multiple quorum sensing modulons interactively regulate virulence and secondary metabolism in Pseudomonas aeruginosa: identification of the signal molecules N-butanoyl-l-homoserine lactone and N-hexanoyl-l-homoserine lactone. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winson M K, Swift S, Hill P J, Sims C M, Griesmayr G, Bycroft B W, Williams P, Stewart G S A B. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5constructs. FEMS Microbiol Lett. 1998;16:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]